Copyright
©The Author(s) 2023.
World J Clin Cases. Jul 6, 2023; 11(19): 4553-4566
Published online Jul 6, 2023. doi: 10.12998/wjcc.v11.i19.4553
Published online Jul 6, 2023. doi: 10.12998/wjcc.v11.i19.4553
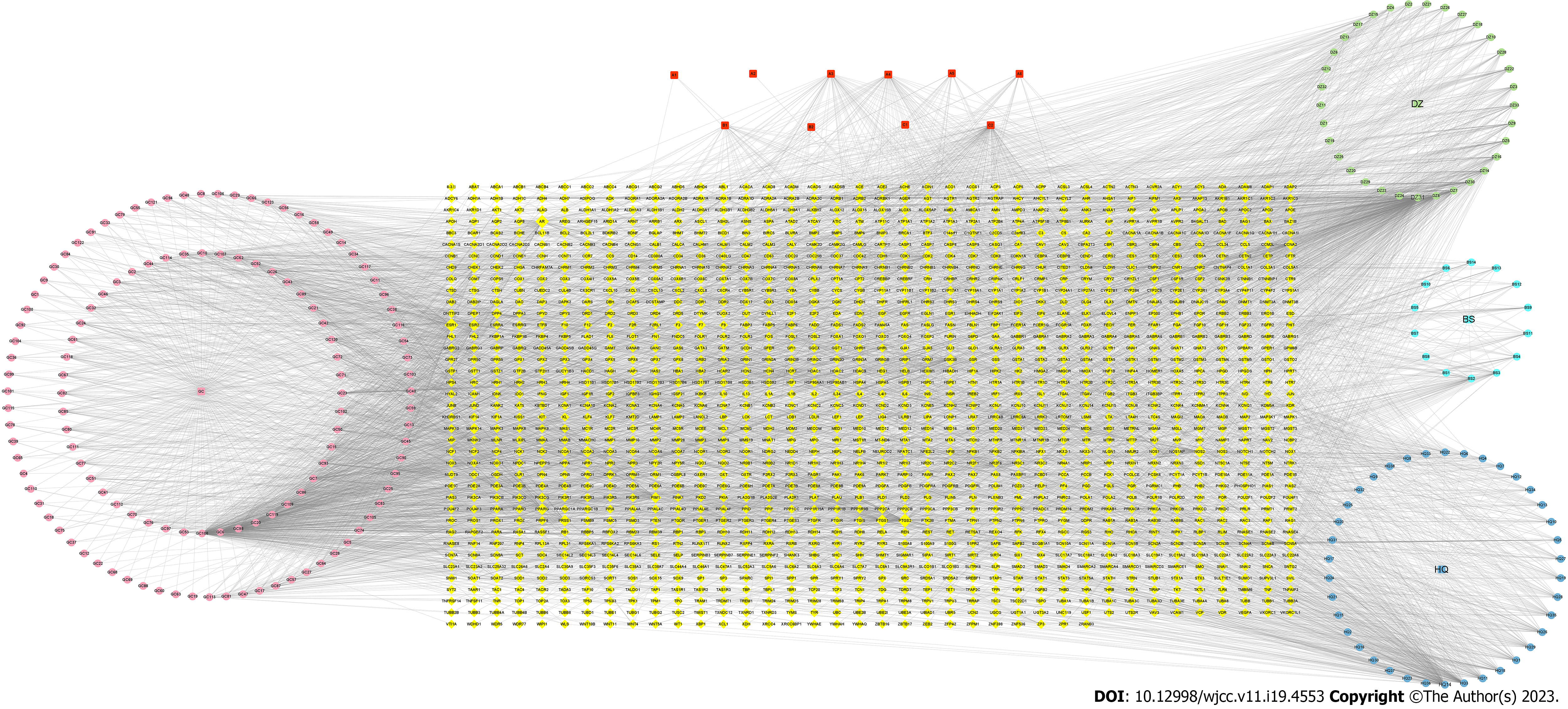
Figure 1 Drug-compound-target network.
The yellow ovals represent the potential genes. The pink circles represent the active ingredients of glycyrrhiza. The light blue circles represent the active ingredients of Paeonia lactiflora Pall. The dark blue circles represent the active ingredients of Scutellariae Radix. The green circles represent the active ingredients of Ziziphus jujuba Mill. The red squares represent the active ingredients of all herbs.
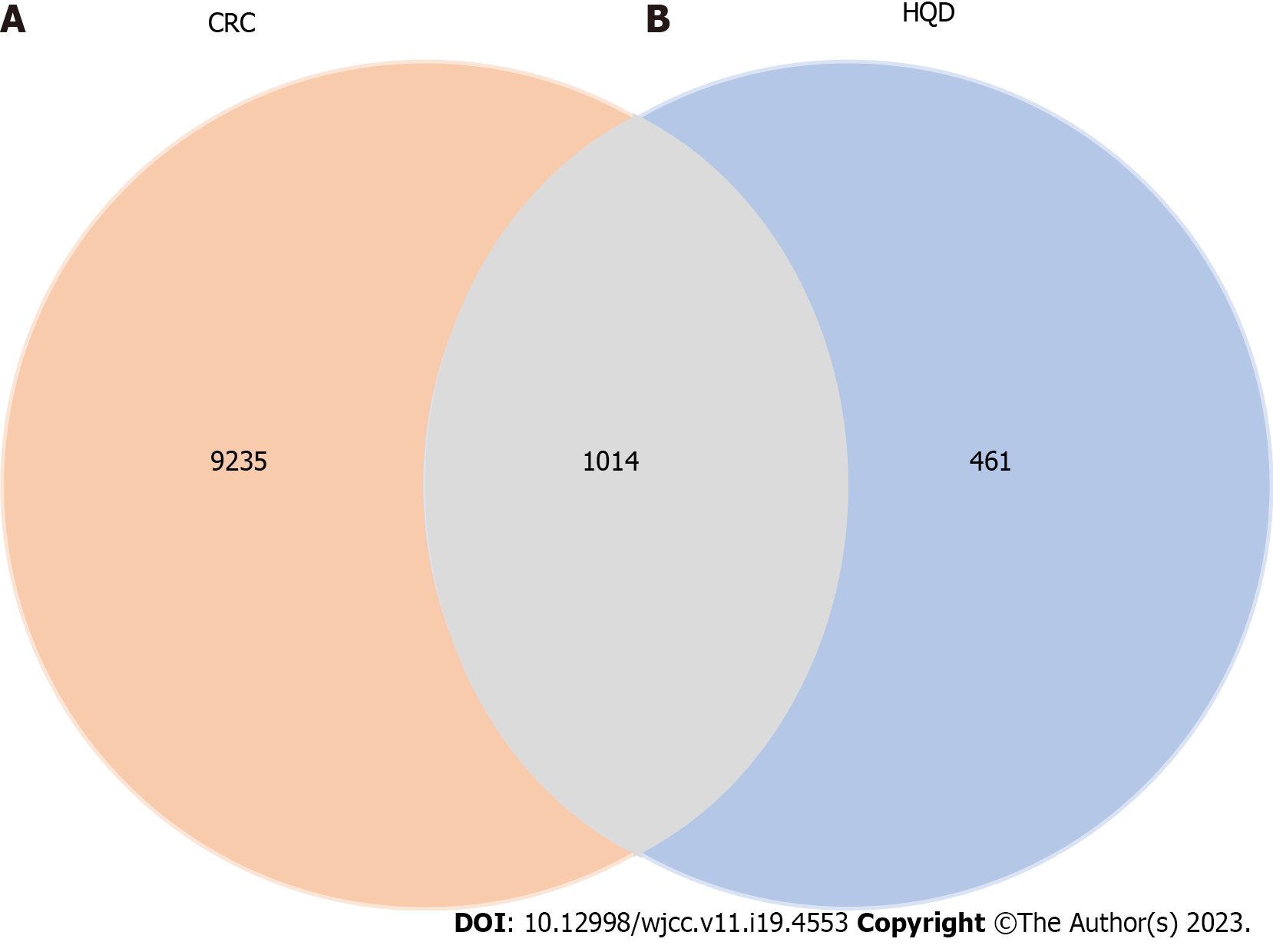
Figure 2 Huangqin decoction–colorectal cancer intersection target.
A: Targets of Huangqin decoction; B: Colorectal cancer-related targets. CRC: Colorectal cancer; HQD: Huangqin decoction.
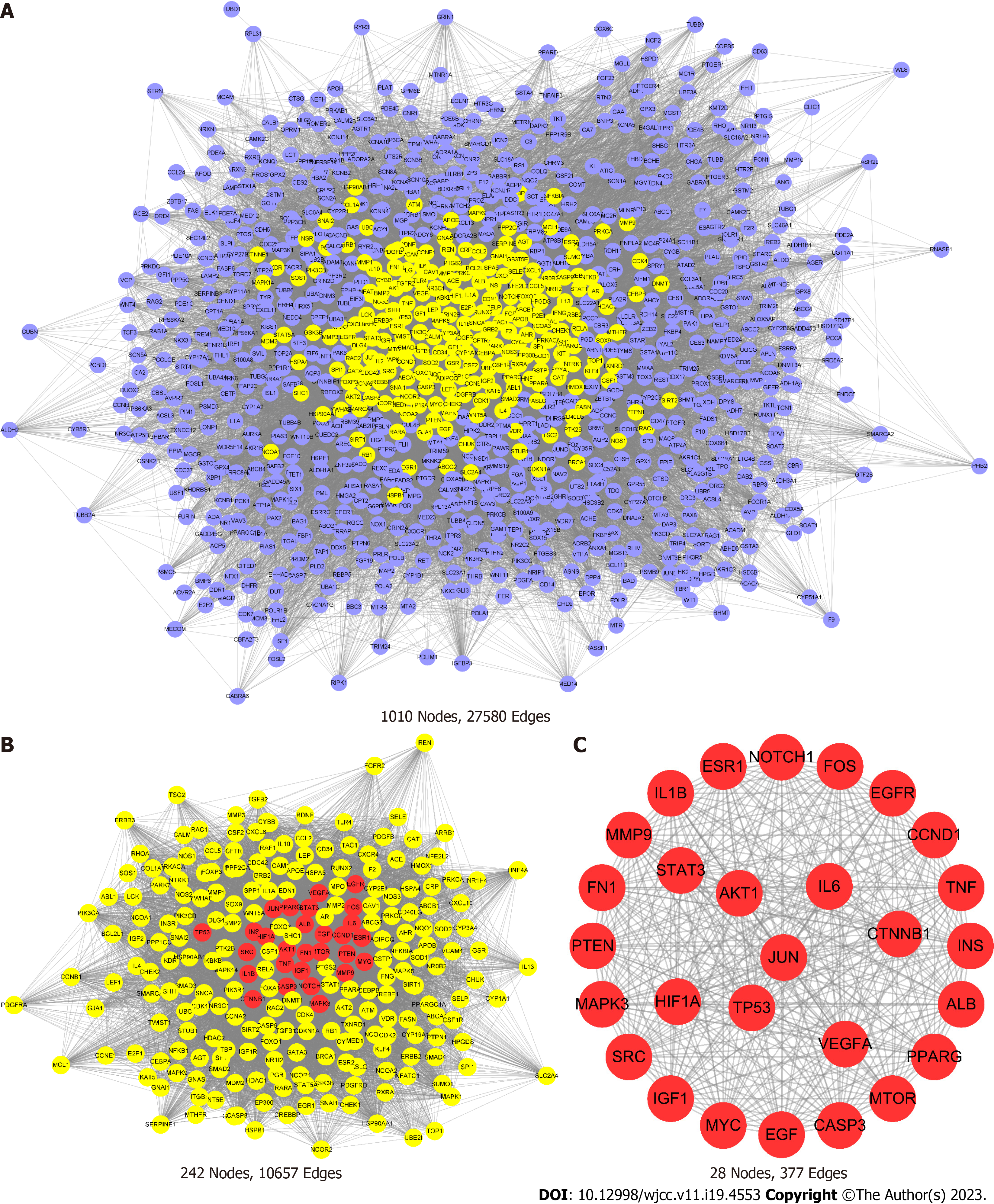
Figure 3 The core target screening process.
A: All potential targets of Huangqin decoction in the treatment of colorectal cancer (purple nodes); B: Core targets screened based on CytoNCA (yellow nodes); C: The top 28 targets screened by betweenness, closeness, degree, eigenvector, local average connectivity and network values of topology parameters. JUN: Jun proto-oncogene, AP-1 transcription factor subunit; STAT3: Signal transducer and activator of transcription 3; TP53: Tumor protein p53; VEGFA: Vascular endothelial growth factor; AKT1: AKT serine/threonine kinase 1 (AKT1); MAPK: Mitogen-activated protein kinase; EGFR: Epidermal growth factor receptor.
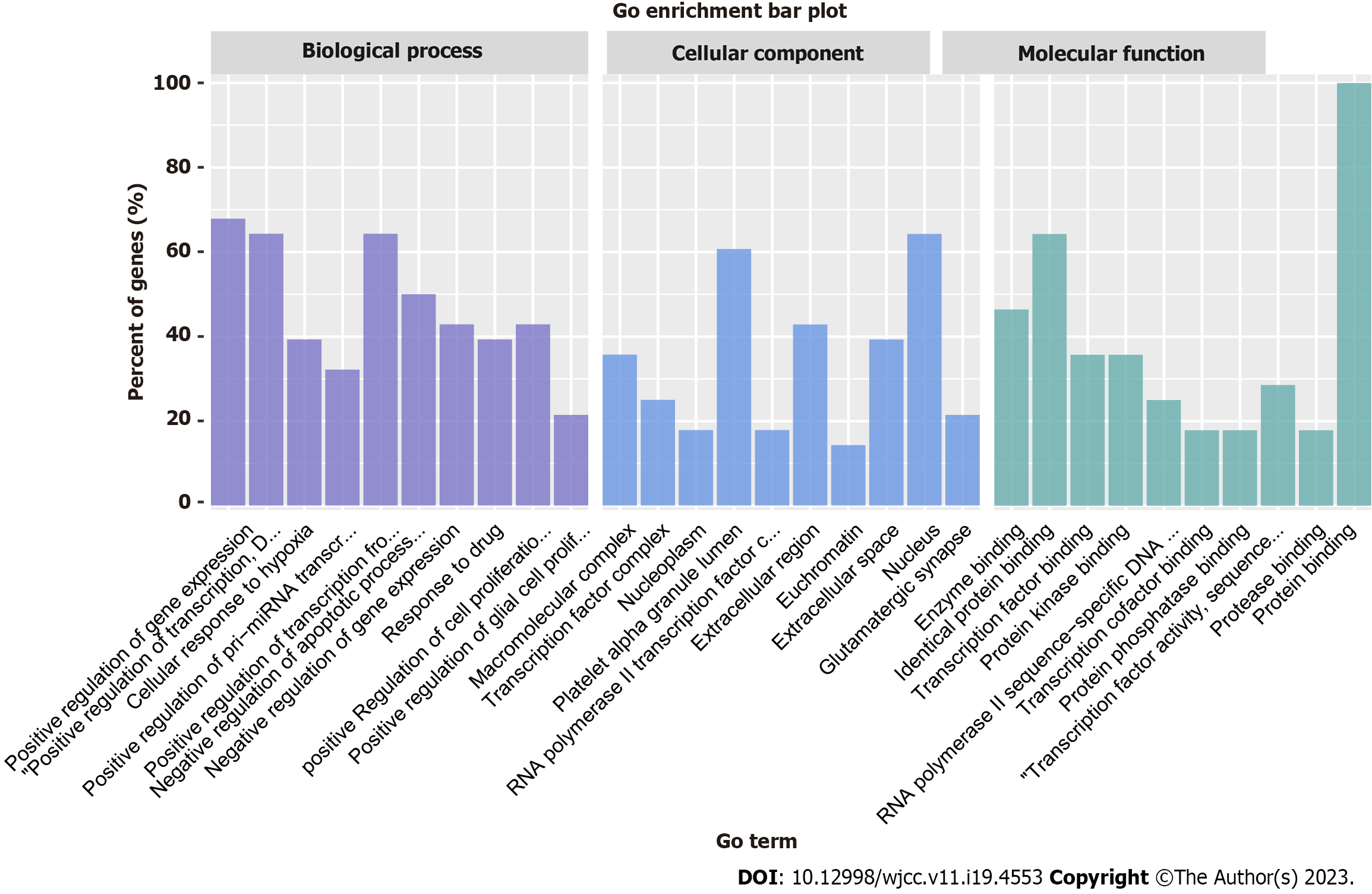
Figure 4 Gene ontology functional enrichment analysis.
The purple bars represent biological processes, the blue bars represent cellular components, and the green bars represent molecular functions. GO: Gene ontology.
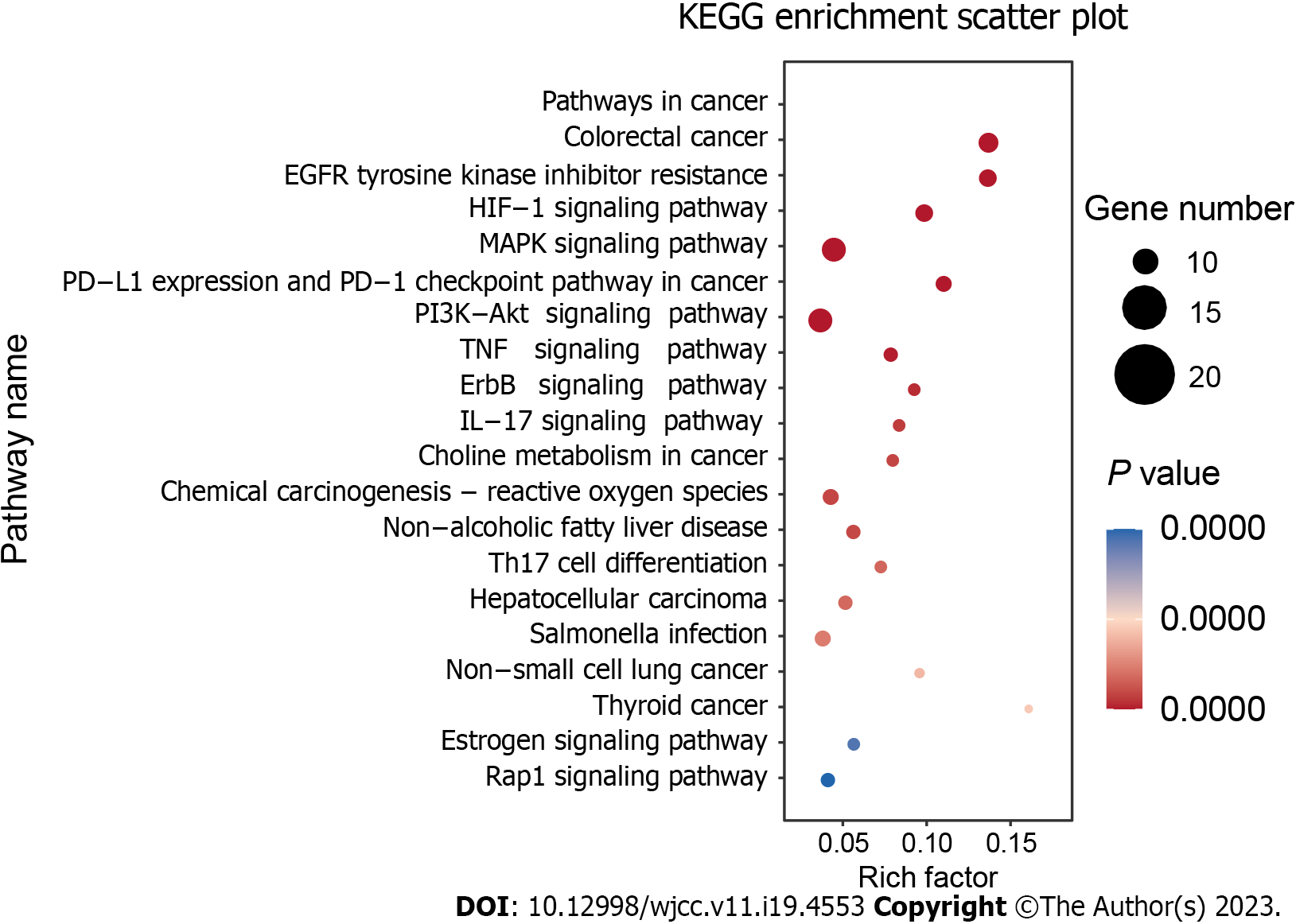
Figure 5 Kyoto Encyclopedia of genes and genomes pathway enrichment analysis.
The top 20 signaling pathways of Huangqin decoction are involved in the treatment of colorectal cancer. The size of nodes is proportional to the number of genes enriched. KEGG: Kyoto Encyclopedia of Genes and Genomes; EGFR: Epidermal growth factor receptor; MAPK: EGFR: Mitogen-activated protein kinase.
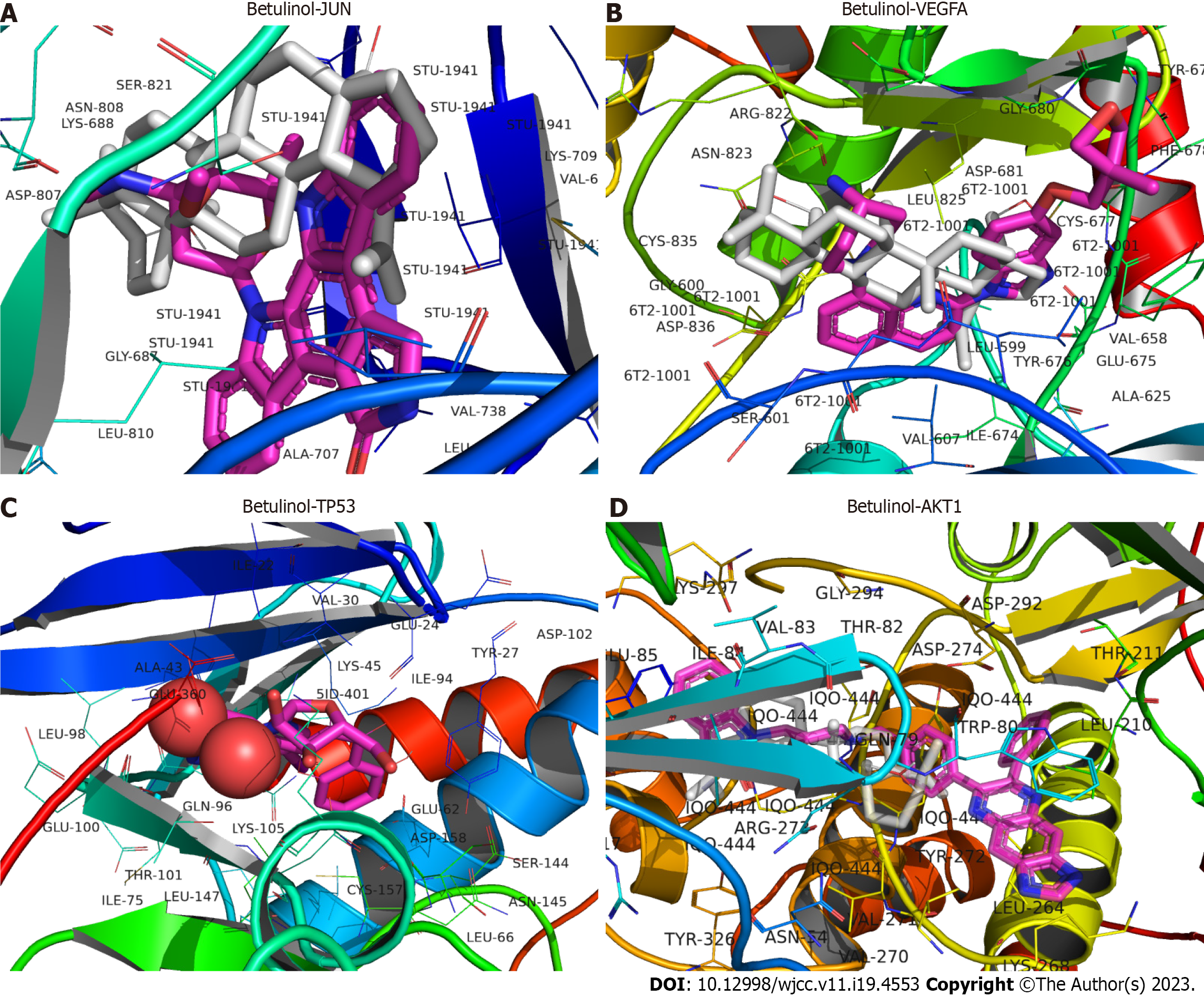
Figure 6 The docking model of the core active ingredient betulinol with the target proteins.
A: Docking of betulinol to Jun proto-oncogene, AP-1 transcription factor subunit; B: Docking of betulinol to vascular endothelial growth factor; C: Docking of betulinol to tumor protein p53; D: Docking of betulinol to AKT1 serine/threonine kinase 1. JUN: Jun proto-oncogene, AP-1 transcription factor subunit; VEGFA: Vascular endothelial growth factor; TP53: Tumor protein p53; AKT1: AKT serine/threonine kinase 1.
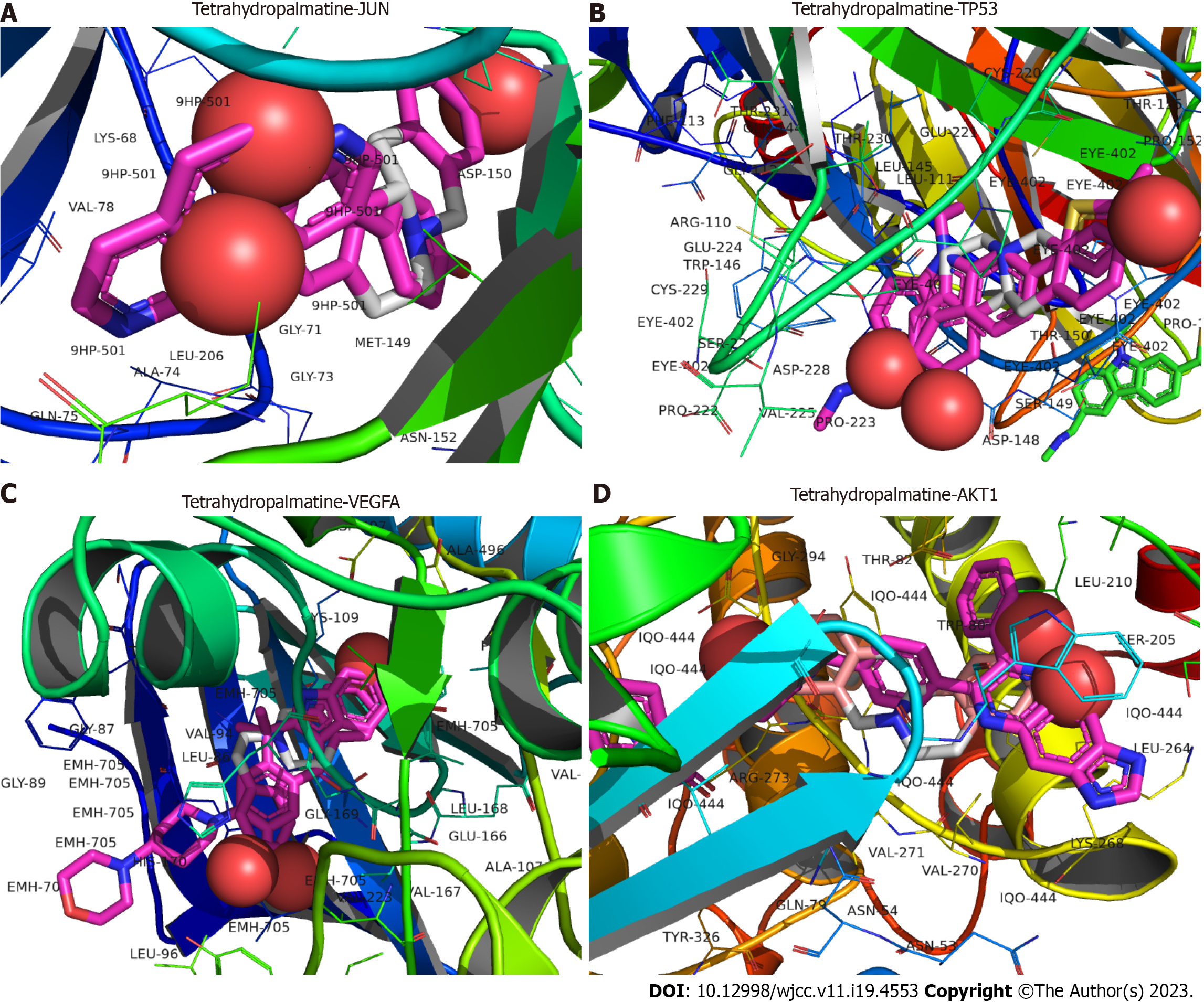
Figure 7 The docking model of the key active ingredient tetrahydropalmatine to the target proteins.
A: Docking of tetrahydropalmatine to Jun proto-oncogene, AP-1 transcription factor subunit; B: Docking of tetrahydropalmatine to tumor protein p53; C: Docking of tetrahydropalmatine to vascular endothelial growth factor; D: Docking of tetrahydropalmatine to AKT1 serine/threonine kinase 1. JUN: Jun proto-oncogene, AP-1 transcription factor subunit; TP53: Tumor protein p53; VEGFA: Vascular endothelial growth factor; AKT1: AKT serine/threonine kinase 1.
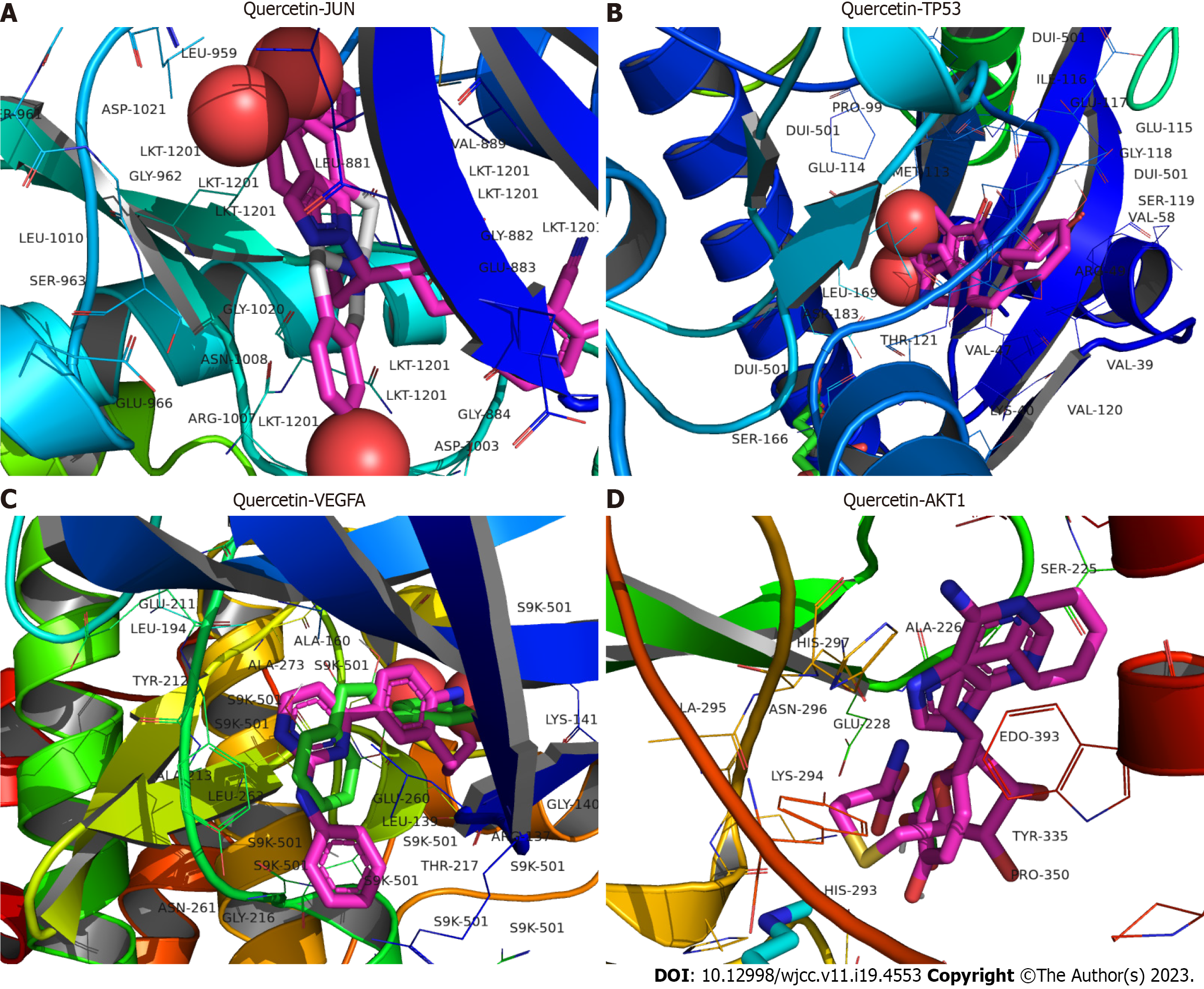
Figure 8 The docking model of the key active ingredient quercetin to the target proteins.
A: Docking of quercetin to Jun proto-oncogene, AP-1 transcription factor subunit; B: Docking of quercetin to tumor protein p53; C: Docking of tetrahydropalmatine to vascular endothelial growth factor; D: Docking of tetrahydropalmatine to AKT1 serine/threonine kinase 1. JUN: Jun proto-oncogene, AP-1 transcription factor subunit; TP53: Tumor protein p53; VEGFA: Vascular endothelial growth factor; AKT1: AKT serine/threonine kinase 1.
- Citation: Li YJ, Tang DX, Yan HT, Yang B, Yang Z, Long FX. Network pharmacology and molecular docking-based analyses to predict the potential mechanism of Huangqin decoction in treating colorectal cancer. World J Clin Cases 2023; 11(19): 4553-4566
- URL: https://www.wjgnet.com/2307-8960/full/v11/i19/4553.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i19.4553