INTRODUCTION
Poly (ADP-ribose), or PAR, is a polymer structure in which ADP-ribose is covalently bound to various amino acid residues of target genes or linked to each other via ribose-ribose bonds[1]. The polymerization of ADP-ribose from the donor molecule, nicotinamide adenine dinucleotide (NAD)+, is catalyzed by PAR polymerase (PARP). This biochemical action yields a linear or branched conformation, depending on whether the ADP-ribose is attached to the 2’-OH terminus of a growing chain or an ADP-ribose residue adjacent to the PARylation target[2,3]. The formation of PAR enables it to interact non-covalently with other molecules and act as a regulator mediating physiological signals involving potential molecular biological and biochemical reactions[4]. Recent studies have shown that PAR could a significant role in the entire process of carcinogenesis, including initiation, promotion, progression, and metastasis[5]. In other words, PAR can be associated with a major network to understand malignant transformation as a multistep process of cell cycle and signaling regulation, including genomic instability, which represents cancer cell proliferation and differentiation from a functional perspective[6,7]. Despite the involvement of PAR in all major post-translational modification processes in carcinogenesis, the synthesis and hydrolysis of PAR are tightly controlled, with a half-life of only a few minutes[8]. In an environment where the hydrolysis of PAR is delayed, the excessive accumulation of intact PAR freely dissociated from the target can conversely induce cell death[9]. An approach to cancer treatment that leverages this unique entity, which can simultaneously control the survival and death of cancer cells, is considered a promising avenue to explore. In this respect, this editorial briefly introduces the general knowledge about PAR and intriguing methods to induce anticancer effects by regulating PAR.
THE DUAL ROLE OF PAR INVOLVED IN CANCER CELL SURVIVAL AND DEATH
The biosynthesis of PAR is part of various physiological processes that are intrinsically linked to the survival of cancer cells. Therefore, continuous efforts are being made to uncover the relationship between PAR and carcinogenesis. The numerous functions of PAR can be categorized into three main categories based on their roles: overcoming genomic instability in response to DNA damage, regulating proliferation and differentiation, and mitosis[10-17]. Firstly, PAR acts as an epigenetic regulator under conditions of genetic instability caused by external or internal stress[11]. PARylation on histone proteins is associated with an open chromatin structure, crucial for maintaining DNA integrity[10]. At sites of DNA damage, this function is vital for recruiting relevant factors for DNA damage repair, such as X-ray repair cross-complementing and proliferating cell nuclear antigen[16,17]. Secondly, PAR directly regulates the recruitment of transcription factors and cofactors[14]. PARP-1 triggers the synthesis of PAR, which in turn influences the upregulation of oncogenes or the downregulation of tumor suppressor genes. This activity plays a key role in the growth and differentiation of cancer cells[13,14]. Lastly, PAR plays a role in mitosis. PARylation is responsible for the regulation of chromosomal proteins involved in chromosome segregation and interaction with spindle checkpoint components[10,15]. It is also crucial for the formation and maintenance of spindle polarity through covalent modification with spindle pole proteins that cross-link microtubule ends at the spindle poles[15]. This process facilitates the relentless proliferation characteristic of malignancy, ensuring the continuous replication and distribution of oncogenic genetic material necessary for cancer progression[15]. In contrast, the beneficial role of PAR in cancer cell survival may shift significantly under certain stress-inducing conditions. PARylation by PARP-1 is initially linked to DNA repair; however, its overactivation can lead to cell death due to a metabolic catastrophe and represents a caspase-independent cell death mechanism, distinct from conventional apoptosis[9]. This occurs when excessive DNA damage causes PARP-1 to overconsume NAD+, an essential substrate for synthetizing PAR, resulting in an energy crisis[18]. The process involves a complex interplay of cellular mechanisms, including the reversal of mitochondrial ATP synthase activity and the depletion of NAD+ levels[18,19]. Parthanatos represents another form of cell death by PAR accumulation, which leads to PAR translocation from the nucleus to the cytoplasm and alters mitochondrial function[20,21]. It is characterized by the release of apoptosis-inducing factor (AIF) from mitochondria to the nucleus, resulting in chromatin condensation and large-scale DNA fragmentation. Since AIF is known to be a binding protein with a high affinity for PAR, morphologically branched PAR would act as a more potent inducer of AIF release[20,21]. In summary, the functions of PAR within cancer cells are under strict regulation, they simultaneously exhibit dual roles - promoting survival or inducing death, contingent upon the internal conditions of cancer cells. The unique characteristic that the conjugated form of a single molecule can dictate both events of cancer cell fate underscores the potential of PAR as a therapeutic target.
HOW TO CONQUER CANCER BY CONTROLLING THE DOUBLE-EDGED SWORD, PAR?
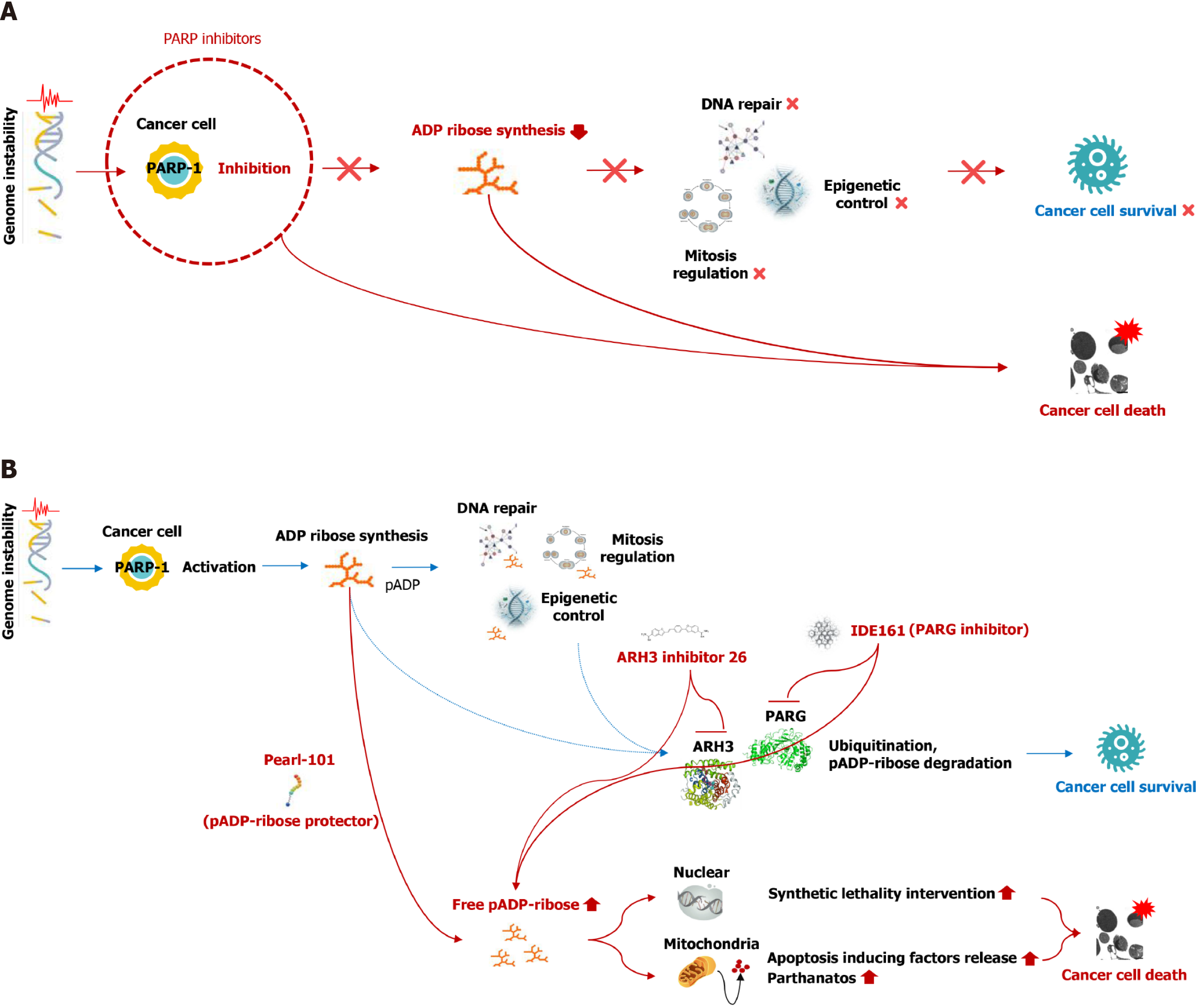
Given the substantial contribution of PAR in promoting cancer cell survival first, strategies that inhibit PAR synthesis could be prioritized in an anticancer area. It has been realized with the development of PARP inhibitors work by impeding the function of PARPs, the enzymes integral to DNA repair processes[22]. PARP inhibitors were designed to treat cancers with breast cancer susceptibility gene mutations, however, one of their key mechanisms of action is the regulation of PARylation[23]. By inhibiting PARylation, PARP inhibitors disrupt the survival of cancer cells, leading to their death (Figure 1A). This mechanism could be harnessed to extend their application in treating several types of cancer. Therefore, the recent research is also focused on the preclinical and clinical advancement of PARP inhibitors as standalone treatments and in combination with other therapies for gastrointestinal cancer[24]. Meanwhile, developing strategies to increase PAR levels could potentially enhance the anticancer effect by considering the mechanisms through which PAR induces cancer cell death. There are two primary strategies for preserving PAR levels within cancer cells: the first involves inhibiting ADP-ribosylhydrolases, such as PAR glycohydrolase (PARG) and (ADP-ribosyl)hydrolase 3 (ARH3), while the second strategy aims to protect PAR itself from these hydrolases (Figure 1B)[25,26]. The most advanced PARG inhibitor, IDE161, is currently under development by IDEAYA Biosciences. IDE161 operates by inhibiting the hydrolysis of PAR chains, leading to an accumulation of PAR and subsequently resulting in synthetic lethality (Figure 1B)[27]. IDE161 is under evaluation in a Phase 1 clinical trial, and this trial is strategically aimed at estrogen receptor-positive, human epidermal growth factor receptor 2-negative breast cancer, as well as other solid tumors with homologous recombination deficiency including colorectal cancer (NCT05787587)[28]. ARH3 inhibitor 26 (AI26) is another recognized agent for ADP-ribosylhydrolase inhibition[26]. This novel small molecule selectively targets ARH3, binding to its catalytic pocket and inhibiting its enzymatic activity[26]. When cells are pretreated with AI26, there is an accumulation of PAR, leading to defects in DNA damage repair and the induction of synthetic lethality (Figure 1B). This reveals the potential of AI26 as a chemotherapeutic strategy in cancer treatment[26]. However, it is crucial to note that research on AI26 is still in progress, and further studies are required to fully comprehend its therapeutic benefits. In addition to the conventional approach using inhibitory agents, there exists a unique method that promotes intracellular PAR accumulation by protecting the PAR chain. Pearl-101, an anticancer drug currently under non-clinical development by PearlsInMires, employs this unique method (http://eng.pearlsinmires.com/). It recombines an endogenous peptide to specifically bind to PAR, highlighting a novel approach in cancer therapeutics. Pearl-101 directly protects the PAR chain, thereby disrupting the activity of ADP-ribosylhydrolase. This disruption leads to an accumulation of PAR in cancer cells, which subsequently triggers cancer cell death by parthanatos and synthetic lethality intervention with DNA fragmentation (Figure 1B). This approach has the benefit of specifically targeting areas where PAR is excessively synthesized, thereby ensuring precise action within cancer cells while minimizing adverse effects on normal cells. Taken together, inhibitors of PARP and ADP-ribosylhydrolases, and a PAR protector are being developed for cancer treatment as distinct methods to either reduce or accumulate PAR. Considering the distinct targets of these drugs, a synergistic anticancer effect would also be achieved through a combined approach such as a fixed-dose combination in addition to use as a single agent.
Figure 1 Strategies for inducing cancer cell death through poly (ADP-ribose) regulation.
A: The anticancer effect is achieved by diminishing the synthesis of poly (ADP-ribose) (PAR), a process mediated by the inhibition of PAR polymerase-1; B: Promising strategies for the development of innovative anticancer treatments focused on PAR accumulation. The enzymes responsible for PAR degradation, such as PAR glycohydrolase or (ADP-ribosyl)hydrolase 3 (ARH3), are inhibited by IDE161 (IDEAYA Biosciences) or ARH3 inhibitor 26. Pearl-101, a molecule currently under development by PearlsInMires, plays a crucial role in maintaining the structural integrity of the PAR chain. The accumulation of PAR can lead to the death of cancer cells through a process known as synthetic lethality intervention and the release of apoptosis-inducing factors. PAR: Poly (ADP-ribose); PARG: PAR glycohydrolase; ARH3: (ADP-ribosyl)hydrolase 3; PARP-1: Poly (ADP-ribose) polymerase-1; pADP: Poly(ADP-ribose).
CONCLUSION
The dual nature of PAR, which can both support and challenge cancer cell survival, provides a wealth of opportunities for scientific and clinical exploration. As such, ongoing research into the effects of PAR on cancer cell fate is anticipated to be a promising area, and this suggests that PAR could emerge as a significant biomarker in clinical settings and may play a crucial role in anti-cancer strategies.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country of origin: South Korea
Peer-review report’s classification
Scientific Quality: Grade A
Novelty: Grade A
Creativity or Innovation: Grade A
Scientific Significance: Grade A
P-Reviewer: Cui M S-Editor: Li L L-Editor: A P-Editor: Zhao YQ