Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103048
Revised: December 11, 2024
Accepted: January 16, 2025
Published online: April 15, 2025
Processing time: 139 Days and 10 Hours
Lotus plumule and its active components have demonstrated inhibitory effects on gastric cancer (GC). However, the molecular mechanism of lotus plumule against GC remains unclear and requires further investigation.
To identify the key hub genes associated with the anti-GC effects of lotus plumule.
This study investigated the potential targets of traditional Chinese medicine for inhibiting GC using weighted gene co-expression network analysis and bio
This study identified 26 genes closely associated with GC. Machine learning analysis and external validation narrowed the list to four genes: Aldo-keto reductase family 1 member B10, fructose-bisphosphatase 1, protein arginine methyltransferase 1, and carbonic anhydrase 9. These genes indicated a strong correlation with anti-GC activity.
Lotus plumule exhibits anti-GC effects. This study identified four hub genes with potential as novel targets for diagnosing and treating GC, providing innovative perspectives for its clinical management.
Core Tip: Lotus plumule exhibits promising efficacy in the prevention and treatment of gastric cancer. This extensive study has identified four pivotal hub genes associated with the anti-gastric cancer effects of lotus plumule and elucidated the upstream and downstream targets of one of these genes, which possess significant potential as novel therapeutic and diagnostic targets for gastric cancer. These findings offer groundbreaking and innovative insights into the clinical management of gastric cancer, potentially enhancing its treatment and facilitating early detection.
- Citation: Meng FD, Jia SM, Ma YB, Du YH, Liu WJ, Yang Y, Yuan L, Nan Y. Identification of key hub genes associated with anti-gastric cancer effects of lotus plumule based on machine learning algorithms. World J Gastrointest Oncol 2025; 17(4): 103048
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/103048.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.103048
Gastric cancer (GC) is a common malignancy of the digestive tract, ranking as the fifth most prevalent cancer and the fourth leading cause of cancer-related deaths worldwide, posing a significant clinical challenge[1,2]. The development of GC is closely associated with several factors, with Helicobacter pylori infection being the most prominent risk factor. Other contributing factors including age, high salt intake, and a diet low in fruits and vegetables are closely linked to the incidence of GC[3]. Most patients are diagnosed at an advanced stage due to subtle early symptoms and low rates of regular screening. Systemic treatment for GC includes chemotherapy, targeted therapy, and immunotherapy[4]. Perioperative chemotherapy is the standard treatment for resectable GC[5-8]. However, in patients with advanced metastatic GC, the disease often extensively metastasizes or invades vital organs and structures. Surgery may not completely remove all tumor lesions, and achieving a radical cure is often challenging. Additionally, chemotherapeutic drugs typically cause serious side effects and are prone to drug resistance. The potential value of traditional Chinese medicine (TCM) in treating GC is receiving increasing attention. TCM is widely used in cancer treatment as an important complementary and alternative healthcare system in Asian countries. TCM has built a rich experience and theoretical foundation for treating various diseases, including GC[9-11]. Recent studies have indicated that various forms of Chinese medicine, such as prescriptions, proprietary medicines, and single compounds, exhibit significant antitumor effects[12]. Clinically, TCM has been found to extend patients’ survival and improve their quality of life, demonstrating unique potential in cancer treatment[13].
Lotus plumule is derived from the plant Nelumbo nucifera Gaertn., a member of the water lily family, and refers to the young leaves and radicles inside mature seeds. The earliest record from the Five Dynasties and Southern Tang Dynasty, as noted by Li Shi-Zhen, describes lotus plumule as the green heart in the lotus seed, which can be used to treat heart-related conditions. According to the 2020 edition of the Chinese Pharmacopoeia, lotus plumule is characterized as bitter and cold, targeting the heart and kidney meridian. It has several therapeutic effects, including clearing the mind, communicating the heart and kidney, and stopping bleeding. Lotus plumule and its active components have been demonstrated to inhibit various tumors, such as gallbladder, gastric, liver, pancreatic, and colorectal cancers[14,15]. Lotus plumule contains alkaloids as one of its primary chemical constituents, with approximately 20 alkaloids identified. Studies have demonstrated that the main components are liensinine, neferine, and isoliensinine, which are bisbenzylisoquinoline alkaloids. Additionally, research has indicated that neferine has a specific therapeutic effect on GC, liver cancer, esophageal cancer, colorectal cancer, and various other tumors[16,17]. Moreover, it can exhibit antitumor properties and enhance the efficacy of cisplatin, doxorubicin, and paclitaxel. In addition, it increases cancer cell sensitivity to chemotherapy, leading to apoptosis, autophagy, and G1 arrest. Liensinine plays a tumor-suppressive role in various malignant tumors, and its inhibitory effect is achieved through tumor cell cycle arrest, promoting tumor cell apoptosis, and inhibiting tumor invasion and metastasis[18-20]. However, the molecular mechanism of lotus plumule against GC remains unclear and needs further investigation.
Network pharmacology is an approach used for multi-target verification in big data analysis. The multi-target, multi-level, and multi-component characteristics of TCM compounds align well with network pharmacology. This study predicted the targets and pathways of lotus plumule in treating GC using network pharmacology[21]. Weighted gene co-expression network analysis (WGCNA) is a systematic bioinformatics algorithm[22]. WGCNA is applied to experimental data to investigate the relationship between gene networks and traits of interest at the transcriptome level. WGCNA identifies co-expressed gene modules and explores their association with significant phenotypes. This method is valuable for identifying susceptibility genes associated with complex diseases and aiding in developing new drugs in biomedical research[23].
This study used WGCNA to identify GC-related modules and signature genes associated with GC. Key active components and core genes were identified for GC treatment using WGCNA and various databases. The enrichment analysis of gene ontology (GO) pathways and Kyoto encyclopedia of genes and genomes (KEGG) biological functions further revealed the mechanisms by which these components and genes may exert their effects. Next, a machine learning algorithm, primarily support vector machine-recursive feature elimination (SVM-RFE), random forest, and LASSO regression, was used to screen for characteristic genes[24]. These algorithms helped identify the characteristic genes associated with lotus plumule for GC treatment. The relationship between core genes and clinical relevance, patient prognosis, gene mutations, and immune infiltration was examined to clarify how lotus plumule exerts its therapeutic effects against GC.
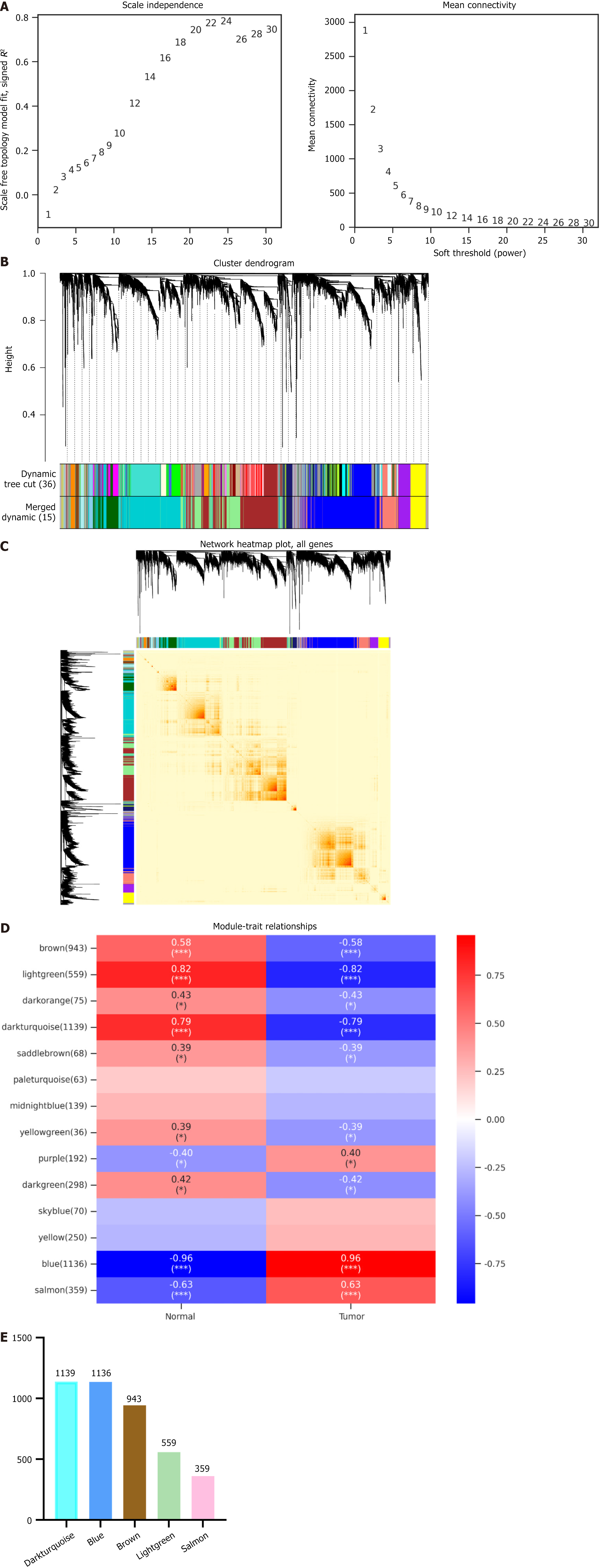
Targets of GC: The GSE118916 dataset was selected for analysis from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) using the keyword ‘gastric cancer’. Subsequently, WGCNA was performed to identify hub genes that exhibit a significant association with cancer-associated fibroblast scores. The GSE118916 expression profile was the first input, and genes with low expression fluctuation (SD ≤ 0.5) were filtered from 5588 genes across 30 samples. The power value was set, and the correlation coefficient and average network connectivity were computed. A weighted co-expression network model was constructed using the selected power values, which categorized the final 5588 genes into 15 distinct modules. Based on the WGCNA module division results, the module gene expression data was presented using cluster plots and bar charts. The Pearson correlation algorithm was applied to calculate the correlation coefficient and P value between module eigengenes and traits. Modules related to each trait were identified using an absolute correlation coefficient threshold of ≥ 0.3 and a P value < 0.05. Differentially expressed genes with log2 fold change (FC) > 1 and log2 FC < -1 were identified from GSE118916 datasets using GEO2R analysis. “Gastric cancer” was used as the keyword to retrieve GC-related targets from the GeneCards database (https://www.genecards.org/) and the Disgenet database (https://disgenet.com/home/). The final potential targets were obtained by combining the genes from the top five modules identified using WGCNA.
Targets of the drug: The chemical composition of lotus plumule was analyzed using the Traditional Chinese Medicine Systems Pharmacology database (https://old.tcmsp-e.com/tcmsp.php), with screening criteria set at 30% oral bioavailability and a drug-likeness threshold of 0.18. The simplified molecular input line entry system notation for the primary component was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and Novopro (https://www.novopro.cn/tools/mol2smiles.html) databases. The simplified molecular input line entry system notation of the primary component was entered into the SwissADME database (http://www.swissadme.ch/), and the preferred active component was identified when the gastrointestinal absorption results were classified as high and at least two affirmative responses were observed in the drug-likeness assessment. The Traditional Chinese Medicine Systems Pharmacology, SwissTargetPrediction (http://www.swisstargetprediction.ch/), and Similarity EnsemblAapproach (https://sea.bkslab.org/) databases were employed to predict the preferred active ingredients. An intersection map illustrating the number of components in lotus plumule was created using WeiShengXin website (https://www.bioinformatics.com.
Disease-drug intersection: The number of drugs and disease targets was entered in the GraphPad Prism software (version 10) to generate a bar graph representing both. The targets differentially expressed in GC from the GEO database were analyzed. The drug and GC targets were analyzed to identify 26 potential intersection targets using Jvenn (http://www.bioinformatics.com.cn/static/others/jvenn/example.html). A total of 26 targets were sorted based on their log2 FC value, and the top 10 downregulated genes were selected and graphed using WeiShengXin. The GEO data corresponding to the intersection of 26 targets were imported into WeiShengXin to create heatmaps. Finally, the 26 intersection targets were analyzed using WeiShengXin’s principal co-ordinates analysis tool to generate a principal component analysis (PCA) plot.
Protein-protein interaction enrichment analysis: A total of 26 target proteins were input into the STRING database (https://cn.string-db.org/), with “Homo sapiens” selected as the organism. The resulting data were downloaded and imported into Cytoscape to construct the topological network map. Data were calculated using betweenness, closeness, and degree in the CytoNCA plug-in and screened by Degree 10 to identify 10 potential core targets. The degree values of the 10 potential core targets were entered into GraphPad Prism software to plot the bars.
Correlation of core targets: The ten potential core targets were sequentially input into the HOME for Researchers (https://www.home-for-researchers.com/#/) to obtain the correlation between the two targets. The heatmap was generated to illustrate the correlation within the group.
GO analysis: A total of 26 intersection targets were uploaded to the Metascape database (https://metascape.org/gp/index.html). The top five entries of biological processes (BP), molecular functions (MF), and cellular components (CC) results in GO were imported into Sangerbox (http://sangerbox.com/home.html). A lollipop chart was generated, with red, blue, and green colors representing BP, CC, and MF, respectively.
KEGG analysis: Twenty-six intersection targets were input into the Metascape database using the previously specified parameters. The results were curated and imported into Sangerbox to create a KEGG enrichment analysis circle plot and a Sankey plot of the pathway using WeiShengXin.
Application of machine learning in hub gene screening: The LASSO regression was conducted using the ‘glmnet’ software package. SVM-RFE was implemented with the “e1071” and “caret” software packages. Random forest was conducted using the “randomForest” software package. Finally, an intersection analysis was performed to identify the core targets.
Expression of genes: The identified key targets were analyzed for clinical relevance. Initially, the Sagnerbox online analysis tool was used with screening conditions set to the data source: The Cancer Genome Atlas (TCGA) + Genotype-Tissue Expression Proje. The data were transformed using the following formula: Log2 (x + 0.001). A non-parametric test method was used to determine the expression levels of the target mRNA. Next, the targets were entered into the GEPIA2.0 website (http://gepia.cancer-pku.cn/). In the expression analysis section, the ‘Profile’ option under ‘Expression DIY’ was selected to visualize the copy number of the targets, providing an alternative perspective on their expression. Subsequently, the target was entered into the University of Alabama at Birmingham cancer data analysis (UALCAN) website (https://ualcan.path.uab.edu/index.Inhtml), where the “CPTAC” module was selected, and the tumor type was set to “GC”. A protein expression bin plot for the target proteins under total protein conditions was generated. Next, the target was input into the Gene Set Cancer Analysis (GSCA) online website (https://guolab.wchscu.cn/GSCA/#/), where “Expression and Subtype” under the “Expression” section was selected to generate box plots of GC subtypes for analyzing the target expression. Finally, the protein was input into the GEPIA2.0 website, where the “Stage Plot” under “Expression DIY” in the expression analysis section was selected to generate a violin plot depicting the gastric cancer staging.
Pathological information and prognosis analysis of targets: The Human Protein Atlas database (https: //www.proteinatlas.org/) was accessed by selecting “TISSUE” as “STOMACH”, and navigating to the "PATHOLOGY" section, and specifying “CANCER” as “stomach cancer” to obtain the immunohistochemical expression profile of the target in normal gastric tissue and GC tissue. Immunofluorescence maps of the targets were obtained from the “SUBCELL” module of the Human Protein Atlas database to determine their localization within the cells. The protein was entered into the Kaplan-Meier plotter, with GC selected to generate a prognostic survival map for the target.
Correlation between hub gene processes and GC: The single nucleotide variation (SNV) of the hub genes in GC was investigated. The seven deleterious mutations were missense, nonsense, frame shift insertion, splice site, frame shift deletion, in-frame deletion, and in-frame insertion. Non-deleterious mutations include silent, intron, intergenic region, 3’ untranalated region (UTR), 5’ UTR, 3’ flanking region, and 5’ flanking region.
Subsequently, the core target was input into the GSCA website, with “STAD” selected as the tumor type and “SNV” chosen within the “Mutation” module to identify the SNV mutation sites and types. The heatmap percentage summarized the frequency of deleterious SNV mutations in GC, and the copy number variation (CNV) summary was selected under CNV to obtain information on the heterozygous and homozygous mutations of the targets. The mutation association map of the core genes was entered under “Mutational Landscape” on the CAMOIP website. Boxplots exhibiting microsatellite instability (MSI) expression levels of the core genes were obtained under the “MANTIS score” in the "Immunogenicity” section of the CAMOIP website.
Methylation of the targets: The core targets were imported into the TCGA dataset on the UALCAN site, with “Stomach adenocarcinoma” and “Methylation” selected to map gene methylation levels. Methylation and prognostic analysis of the targets were performed using the Tumor Immune Dysfunction and Exclusion (TIDE) website (http://tide.dfci.harvard.edu/). On the TIDE website, core genes were imported, and under the "query gene" section, “Methylation” was selected under results, and “Stomach” was chosen for “Cancer”. The “cytotoxic T lymphocyte Cor” data were downloaded to obtain the correlation between gene methylation levels and cytotoxic T lymphocyte (CTL) markers. The “Risk” results were also downloaded to generate survival curves for gene hypermethylated and hypomethylated subpopulations.
The core target was imported into the Sangerbox platform, where the immune infiltration analysis in the pan-cancer analysis was selected. “TCGA-STAD” was chosen under “CODE” to generate a scatter plot illustrating the correlation between StromalScore, ImmuneScore, and ESTIMATEScore for the core target. The immune checkpoint gene analysis was performed for pan-cancer, specifically selecting “STAD” under “CODE” to create a heatmap depicting the correlation between the core target and the immune checkpoint. The Cell-omics Data Coordinate Platform website (https://db.cngb.org/cell) provides a comprehensive resource for accessing single-cell expression maps. By navigating to the Cell-omics Data Coordinate Platform section (https://db.cngb.org/cdcp/), users can select specific datasets related to stomach adenocarcinoma cancer type and specify the species as “Human”.
The core genes were entered into the “Drug” module on the GSCA website to assess drug sensitivity and mRNA expression for GDSC and Cancer Therapeutics Response Portal (CTRP) systems. The core targets were imported into the TISIDB website (http://cis.hku.hk/TISIDB/simple_search_result.Php), where the “Drug” section was selected to generate a network map illustrating the relationship between the core gene and immunotherapy.
A map of methylation levels of potential core genes in normal and GC subtype groups was generated using TCGA data from the UALCAN database. Additionally, the TIDE database (http://tide.dfci.harvard.edu) offers extensive data for analysis. Researchers can use the TIDE platform to query genes and examine the correlation between methylation levels and CTL markers in GC subtypes. Furthermore, the database enables the generation of survival curves for subgroups characterized by hypermethylation and hypomethylation.
Upstream transcription factors were obtained from Gene Transcription Regulation Database, KnockTF, and FIMO_JASPAR, and intersection analysis was performed. Similarly, downstream targets were retrieved from GeneMANIA, STRING, and HitPredict, followed by intersection analysis.
The protein entry number was obtained from UniProt, and the corresponding protein PDB file was input into AlphaFold. The file was imported into GRAMM to obtain the docking results, which were further analyzed using the PDBePISA tool to calculate the docking binding energy. The preferred active ingredient’s SDF structure was retrieved from PubChem. This structure and the core target files were imported into the CB-Dock 2 website (https://cadd.labshare.cn/cb-dock2/php/blinddock) for analysis. The analysis was conducted, the Vina score was recorded, and PDB files were downloaded. These files were imported into PyMOL software to visualize some of the docking results.
Targets of GC: For the WGCNA analysis, a power value of 24 was selected, which resulted in a high correlation coefficient and mean network connectivity (Figure 1A). A weighted co-expression network model was constructed using the selected power values, dividing 5588 genes into 15 modules. The grey module contained genes that could not be assigned to any specific module and lacked reference significance. Subsequently, dynamic modules were identified in different cohorts, with each module containing at least 50 genes (Figure 1B and C). The correlation coefficient and P value of module eigengene and traits were calculated using the Pearson correlation algorithm (Figure 1D). The top five modules in the correlation coefficient and P value were selected, and the number of genes in each module is presented in a bar chart (Figure 1E). The top five modules containing 1139, 1136, 943, 559, and 359 genes were identified as potentially relevant to GC via WGCNA analysis.
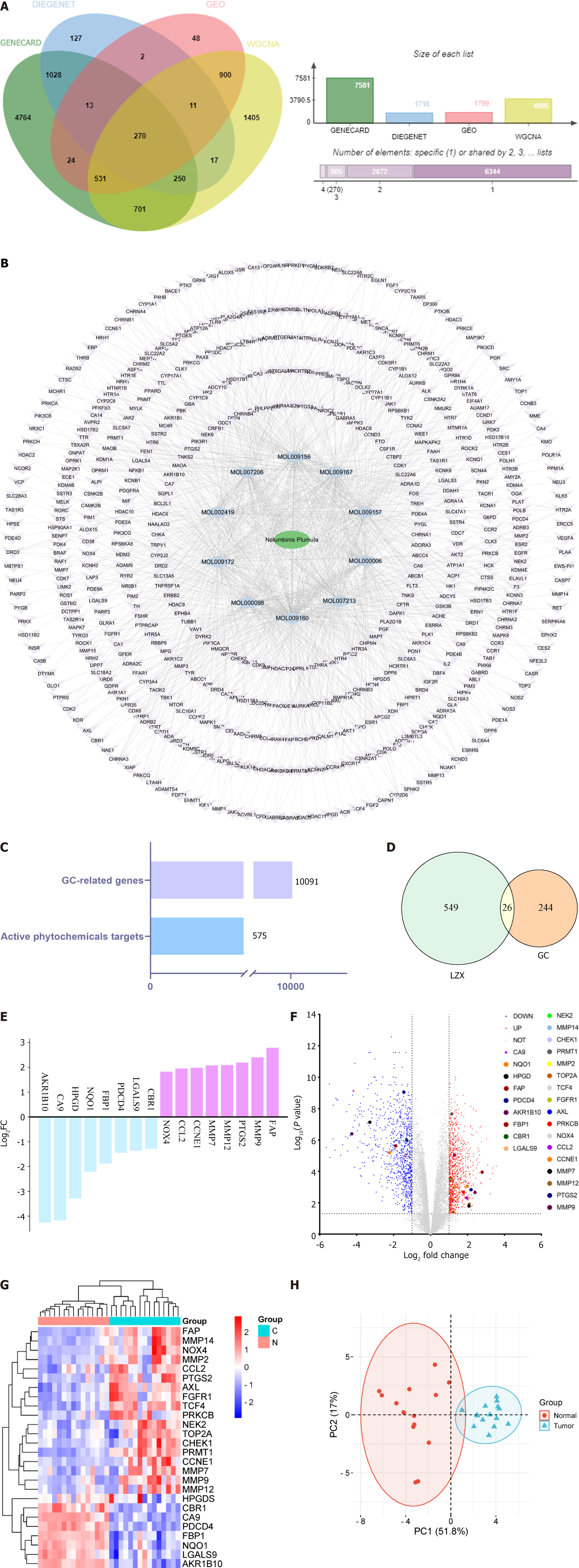
The GSE118916 dataset from the GEO database, comprising 30 samples (15 normal and 15 GC), was selected for further analysis. The GSE118916 dataset revealed a total of 1799 differentially expressed genes. Simultaneously, 7581 potential GC targets were identified using the GeneCards database, and the Disgenet database yielded 1718 potential GC targets. By integrating these potential GC-related genes with those obtained from WGCNA analysis, 10091 GC targets were identified (Figure 2). The intersection of these data yielded 270 intersection genes (Figure 2A), which were used for further analysis.
Drug targets: In the comprehensive analysis of the pharmacological characteristics of lotus plumule, the network pharmacology method was used to identify 10 preferred active ingredients. Cytoscape software was employed to construct the active component-target network map (Figure 2B) to explore the relationship between these components and drug action. In this network, purple nodes represent drug targets, blue nodes denote active drug ingredients, and green nodes indicate the drug itself, resulting in 575 pharmacological targets (Figure 2C).
A total of 26 intersection targets related to GC and lotus plumule were identified (Figure 2D). The log2 FC values of these targets were sorted to emphasize the genes with the most significant expression differences (Figure 2E). These 26 differential genes are also illustrated in a volcano map (Figure 2F) and heatmap (Figure 2G). Besides, PCA was performed on the intersection targets to represent the 26 samples (Figure 2H). In the PCA chart, the red dots represent normal, while the blue dots denote tumor tissue. The PC scores for PC1 and PC2 were 51.8% and 17%, with minimal overlap and obvious separation. The PC1 axis exhibited a negative distribution for the normal group and a more positive distribution for the tumor group, demonstrating the reliability of the GEO data.
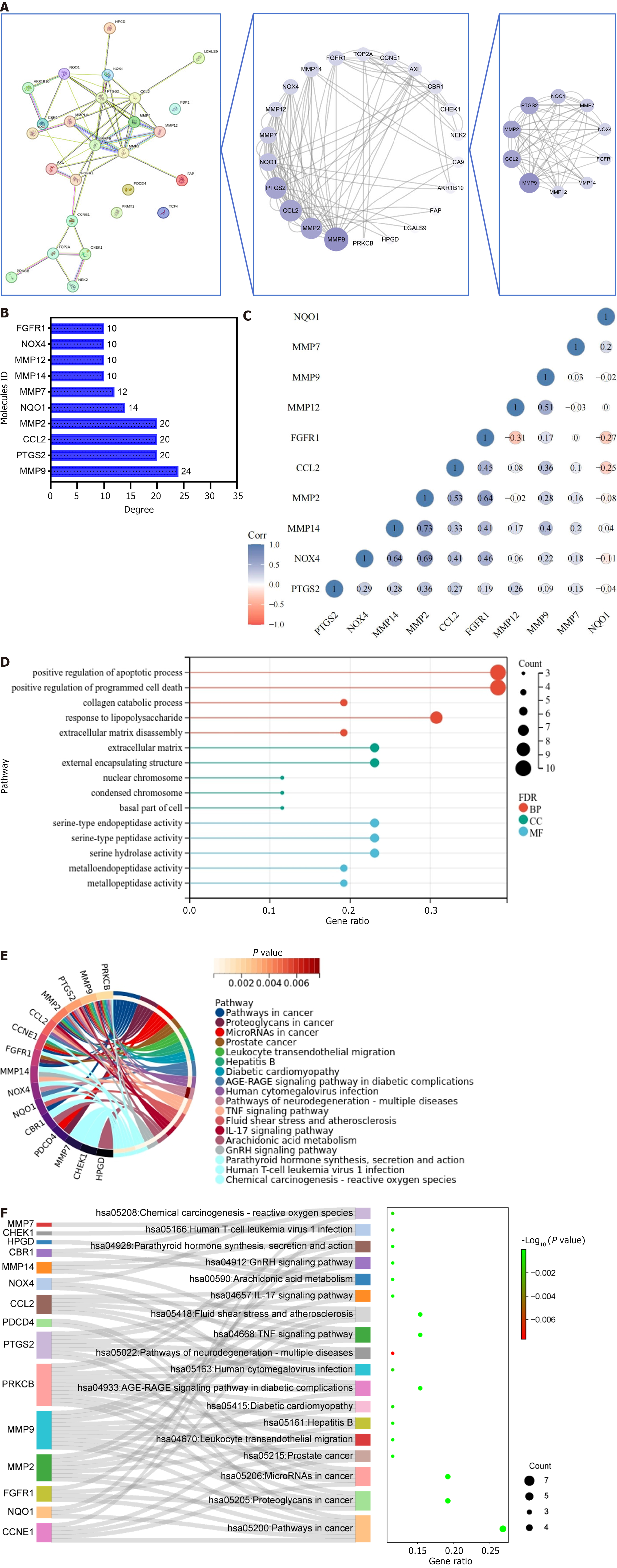
Cytoscape software was used to identify 26 key targets of GC. Figure 3A illustrates the 10 targets that were potentially central to GC treatment. Based on the degree value, Figure 3B depicts the contribution of each target in the topological map, highlighting targets such as matrix metallopeptidase 9 (MMP9), prostaglandin-endoperoxide synthase 2, C-C motif chemokine ligand 2, MMP2, and others. Correlation analysis presented in Figure 3C revealed a positive correlation between most targets, while NAD(P)H quinone dehydrogenase 1, MMP12, and fibroblast growth factor receptor 1 exhibited a negative correlation trend. These results demonstrated that these targets exhibited a close relationship with strong interactions.
GO analysis of 26 targets was conducted to investigate their roles in influencing and regulating tumor genesis and progression via biological behavior. In the GO analysis, red, blue, and green colors represent BP, CC, and MF biological categories, respectively. Larger circles indicate more targets enriched in the entry, as illustrated in Figure 3D. The analysis identified 222 BP entries co-enriched with the 26 targets, primarily associated with processes such as cell apoptosis, programmed cell death, protein metabolism, and other related biological activities. Ten CC entries were identified, indicating that the intersection targets were predominantly located within the extracellular matrix, external encapsulation structures, nuclear chromosomes, condensed chromosomes, and cell bases. Moreover, the analysis identified 19 MF entries, suggesting that the intersection targets are mainly linked to biological functions, including regulating protein serine/threonine kinase, oxidoreductase, and metalloenzyme activity. KEGG enrichment analysis was performed to investigate how lotus plumule exerts anti-GC effects by regulating signaling pathways. A total of 26 targets were enriched in 18 signaling pathways, as illustrated in Figure 3E. These pathways mainly included cancer pathogenesis pathway, proteoglycans in cancer, and tiny RNA in cancer.
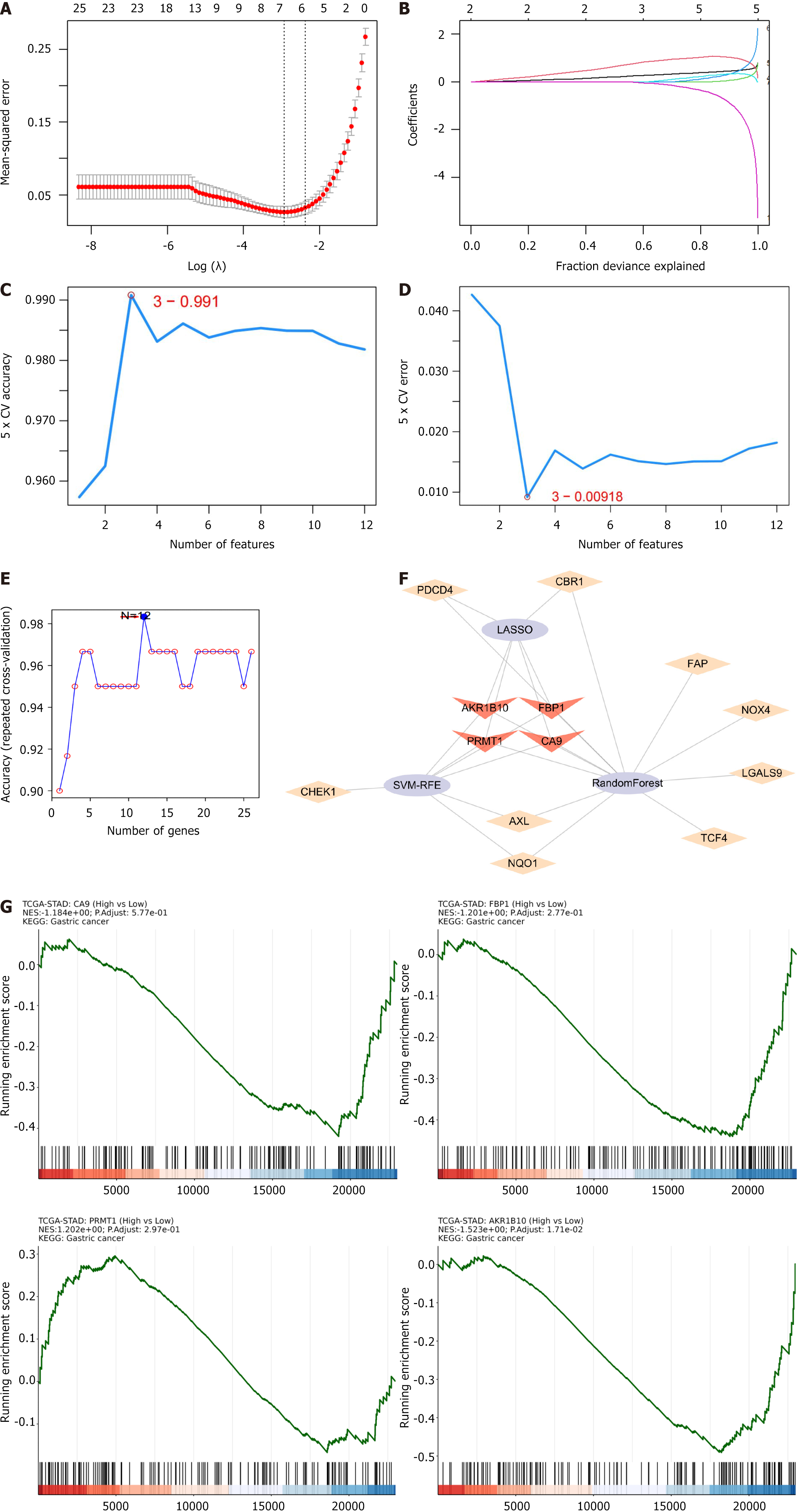
Machine learning was applied to screen for core genes among the 26 target genes. Initially, six genes were identified using the LASSO regression algorithm (Figures 4A and B). Seven genes were subsequently extracted using the SVM-RFE algorithm (Figures 4C and D). Ten genes were selected using the random forest algorithm (Figure 4E). Finally, four hub genes were identified by intersecting these gene subpopulations: Aldo-keto reductase family 1 member B10 (AKR1B10), fructose-bisphosphatase 1 (FBP1), protein arginine methyltransferase 1 (PRMT1), and carbonic anhydrase 9 (CA9) (Figure 4F).
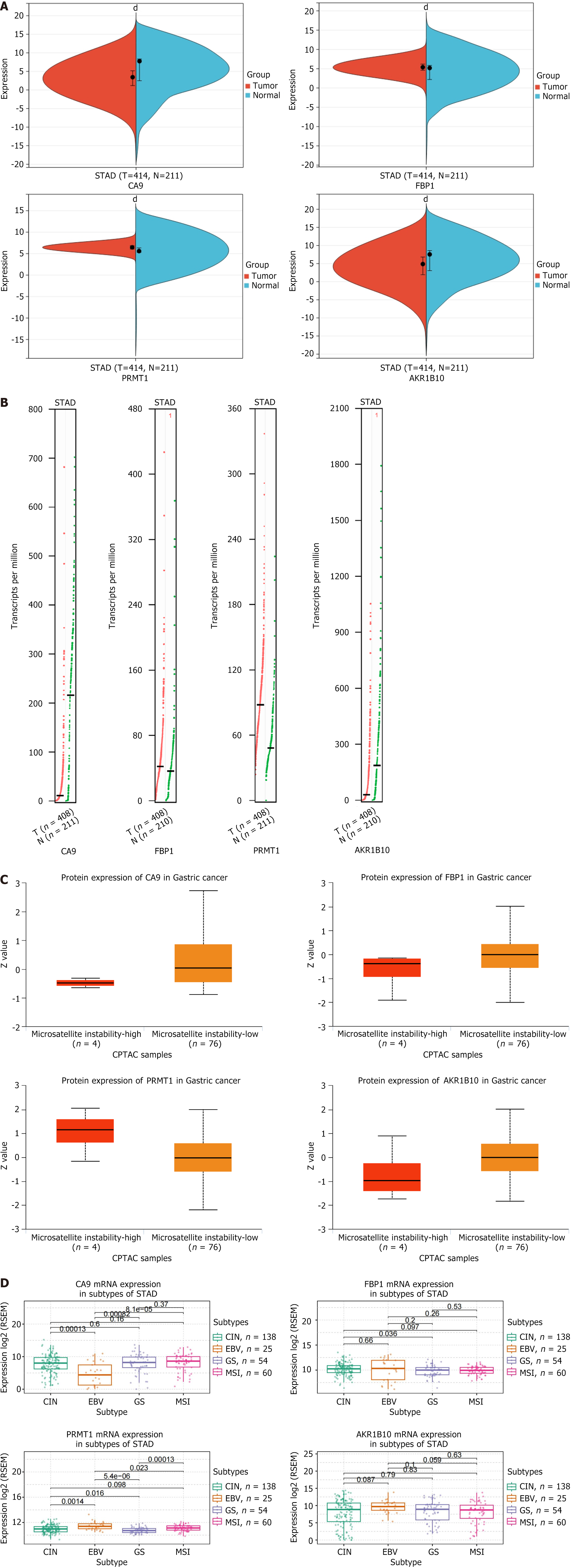
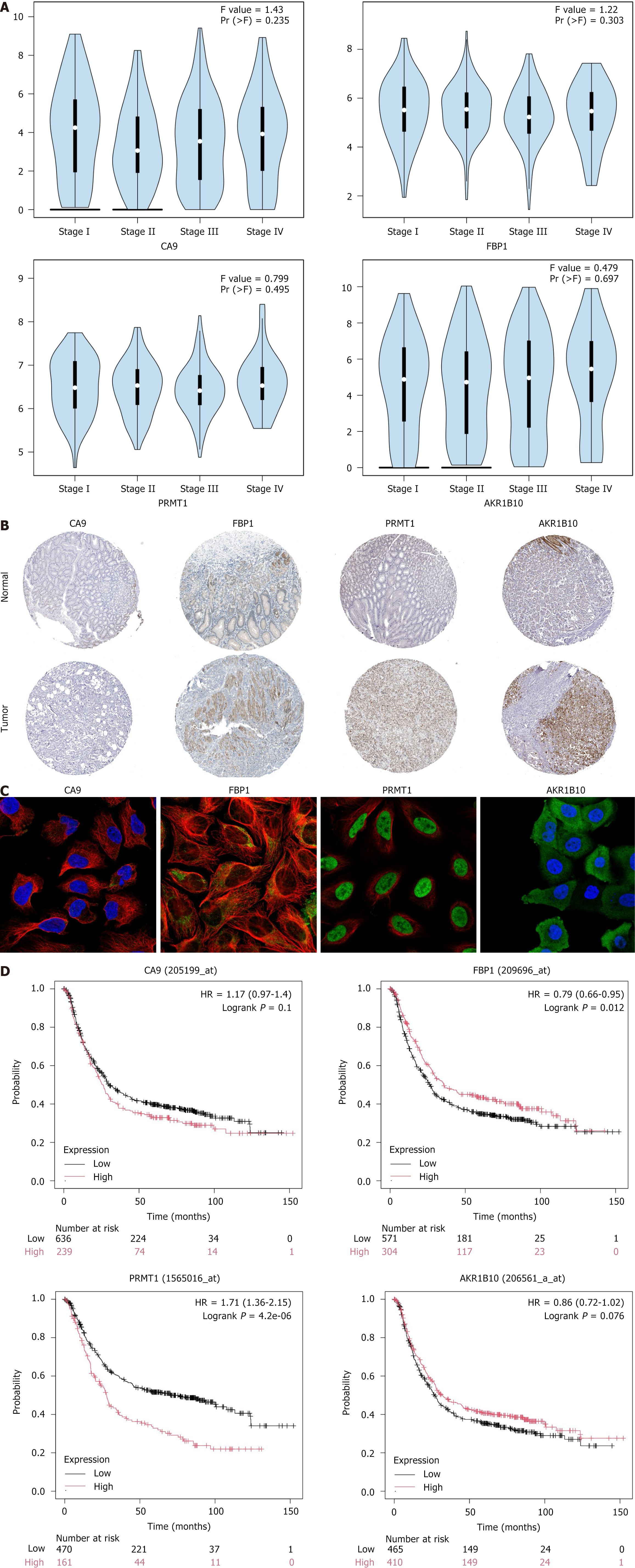
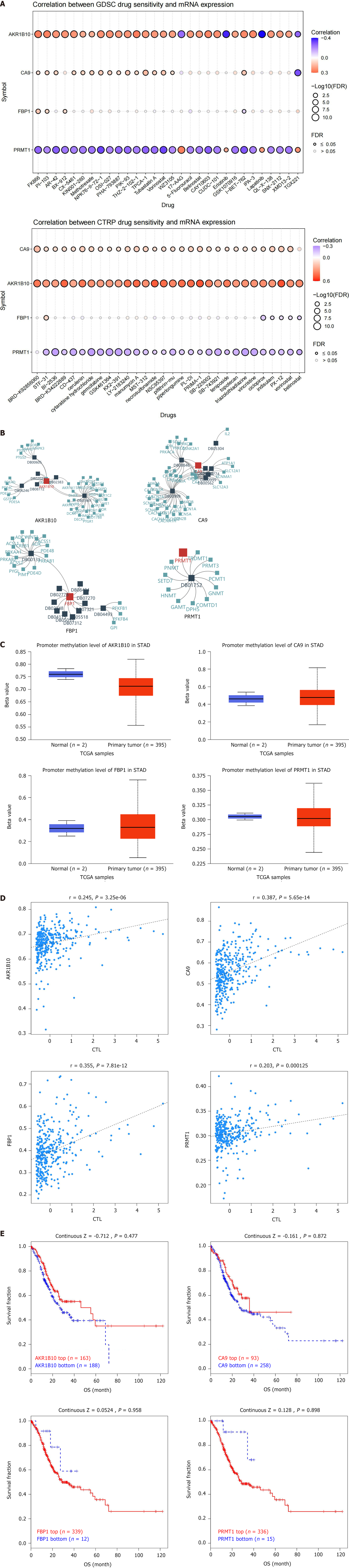
Clinical correlation analysis of AKR1B10, FBP1, PRMT1, and CA9 revealed statistically significant results (P < 0.001) (Figure 4G). The hub gene expression was analyzed at the transcriptional and translation levels to explore their association with GC. As depicted in Figure 5A-C, the analysis revealed that at the mRNA level, the hub genes CA9 and AKR1B10 were highly expressed in GC tissues, and FBP1 and PRMT1 were highly expressed in normal tissues. CA9 and AKR1B10 were highly expressed at the copy number level in the GC group. At the protein level, FBP1, CA9, and AKR1B10 were highly expressed in the GC microsatellite stable group, whereas PRMT1 was not. GC is classified into four molecular subtypes: EB virus (EBV) positive, MSI, genome stability (GS), and chromosomal instability (CIN). The various molecular subtypes exhibited a strong correlation with patient prognosis and survival outcomes. Specifically, the EBV group was associated with the most favorable prognosis, followed by the MSI and CIN groups, whereas the GS group was linked to the poorest prognosis. Figure 5D illustrates significant differences in CA9 mRNA levels between CIN and EBV (P = 0.00013), between GS and EBV (P = 0.0082), and EBV and MSI (P = 8.1e-05). For FBP1 mRNA, a significant difference was observed between CIN and GS (P = 0.036). PRMT1 mRNA exhibited significant differences between CIN and EBV (P = 0.0014), between CIN and GS (P = 0.016), between EBV and GS (P = 5.4e-06), between EBV and MSI (P = 0.023), and between GS and MSI (P = 0.00013). Continuing the analysis of the hub genes and clinical stage of GC (Figure 6A), AKR1B10, FBP1, PRMT1, and CA9, revealed no significant difference among different GC stages. As depicted in Figure 6B, immunohistochemistry results revealed a significant increase in AKR1B10, FBP1, and PRMT1 expression in GC tissues. Their localization images across various tumor tissues were acquired to elucidate the localization of hub genes in human tumor tissues and facilitate subsequent research. As illustrated in Figure 6C, immunofluorescence results indicated that PRMT1 was predominantly expressed in the nucleus, while AKR1B10 was primarily localized in the cytoplasm. As presented in Figure 6D, the prognostic significance of hub genes in patients with GC revealed that except for CA9, AKR1B10, FBP1, and PRMT1 were associated with overall survival. Higher expression levels of FBP1 and AKR1B10 correlated positively with patient prognosis, indicating longer survival, whereas PRMT1 expression was negatively correlated with overall survival.
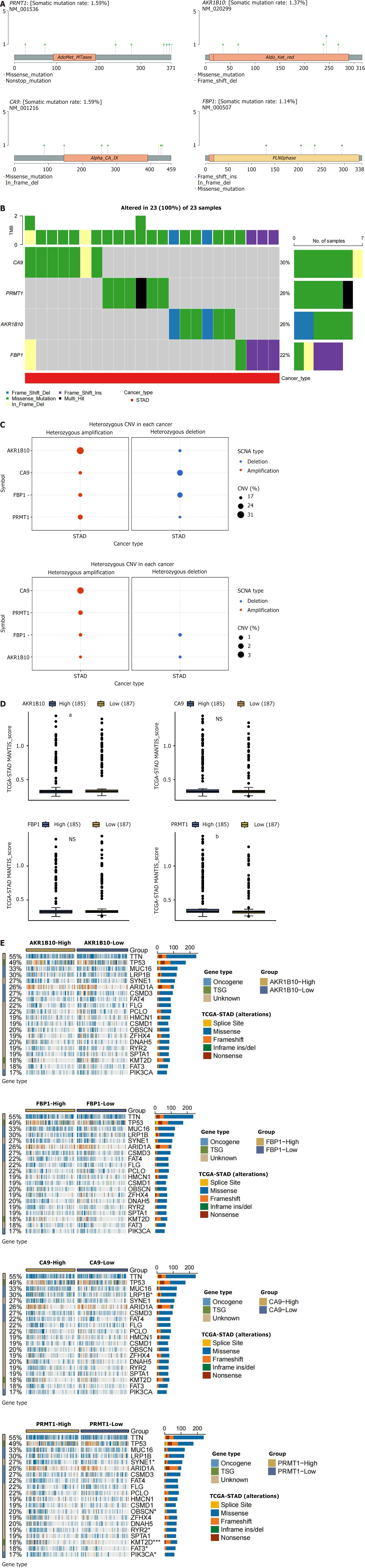
Given that tumorigenesis involves genomic alterations, the four hub genes were examined for SNV and CNV mutations. Figure 7A illustrates the SNV mutation sites and types. The somatic mutation rates for CA9, AKR1B10, FBP1, and PRMT1 were 1.59%, 1.37%, 1.14%, and 1.59%, respectively. As presented in Figure 7B, analysis of the frequency of harmful hub mutations revealed that CA9 exhibited the highest mutation frequency (30%), followed by PRMT1 and AKR1B10 (26% each). FBP1 exhibited the lowest frequency (22%). CNV included heterozygous and homozygous mutations, with homozygous mutations causing more severe disease outcomes. Figure 7C displays amplifications (red) and deletions (blue), with bubble size indicating mutation frequency. The analysis identified predominant amplifications in four hub genes, with CA9 mutations being the most frequent. High expression levels of AKR1B10 and PRMT1 promoted pronucleus expression. MSI occurs when the DNA mismatch repair mechanism fails during the process of DNA replication, leading to a change in microsatellite length due to the inability of the body to repair repetitive fragments. As illustrated in Figure 7D, MSI expression levels of the hub genes AKR1B10 and PRMT1 significantly differ between high and low expression groups of the two genes. Figure 7E illustrates variations in driver genes associated with CA9, such as low-density lipoprotein receptor related protein 1B (P < 0.05). Additionally, PRMT1 was associated with significant differential expression in spectrin repeat-containing nuclear envelope protein 1 (P < 0.05), obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF (P < 0.05), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (P < 0.05), and lysine methyltransferase 2D (P < 0. 001).
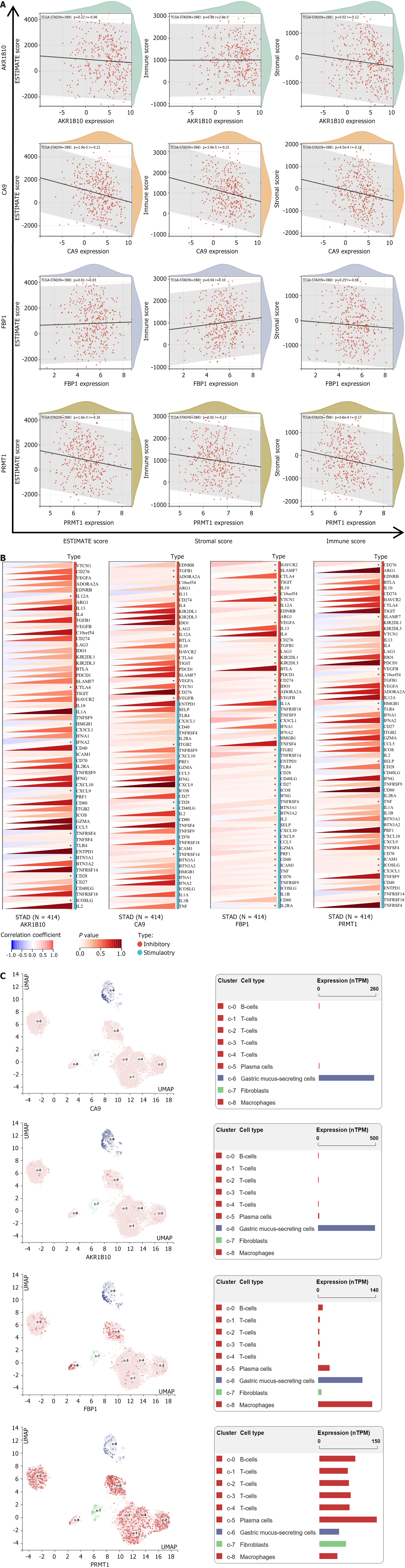
FBP1 exhibited a positive correlation with all three GC scores to evaluate the immunological relevance of GC targets (Figure 8A). Figure 8B indicates a positive correlation between the four targets and immune checkpoint molecules, suggesting their potential role in immune checkpoint regulation in GC. Single-cell sequencing was employed to annotate hub genes within cell subsets. As illustrated in Figure 8C, CA9 and AKR1B10 were predominantly expressed in gastric mucus-secreting cells. FBP1 was expressed in macrophages and gastric mucus-secreting cells, whereas PRMT1 exhibited widespread expression across various cell types.
The GDSC and CTRP databases were used to identify antitumor drugs (Figure 9A). CA9, AKR1B10, and FBP1 demonstrated positive associations with most drugs in GDSC and CTRP analyses, whereas PRMT1 exhibited limited associations. The TISIDB database was used to investigate the relationship between the hub gene and their targeted drugs. In Figure 9B, the red box denotes the hub gene, the black box indicates the targeted drug, and the green box represents other targeted drug targets. Notably, the core targets of the drug components did not include active components of lotus plumule, providing the foundation for future experimental validation of anti-GC therapies and the development of new clinical adjuvant or surrogate treatments.
Four hub genes were analyzed for their correlation with methylation. Figure 9C illustrates that no significant differences were observed for AKR1B10, FBP1, PRMT1, and CA9 in GC. Figure 9D displays a positive association between hub genes and CTL markers following methylation. Figure 8E presents a survival analysis comparing patients with high methylation to those with low methylation levels, which revealed no statistically significant difference for AKR1B10, FBP1, PRMT1, and CA9.
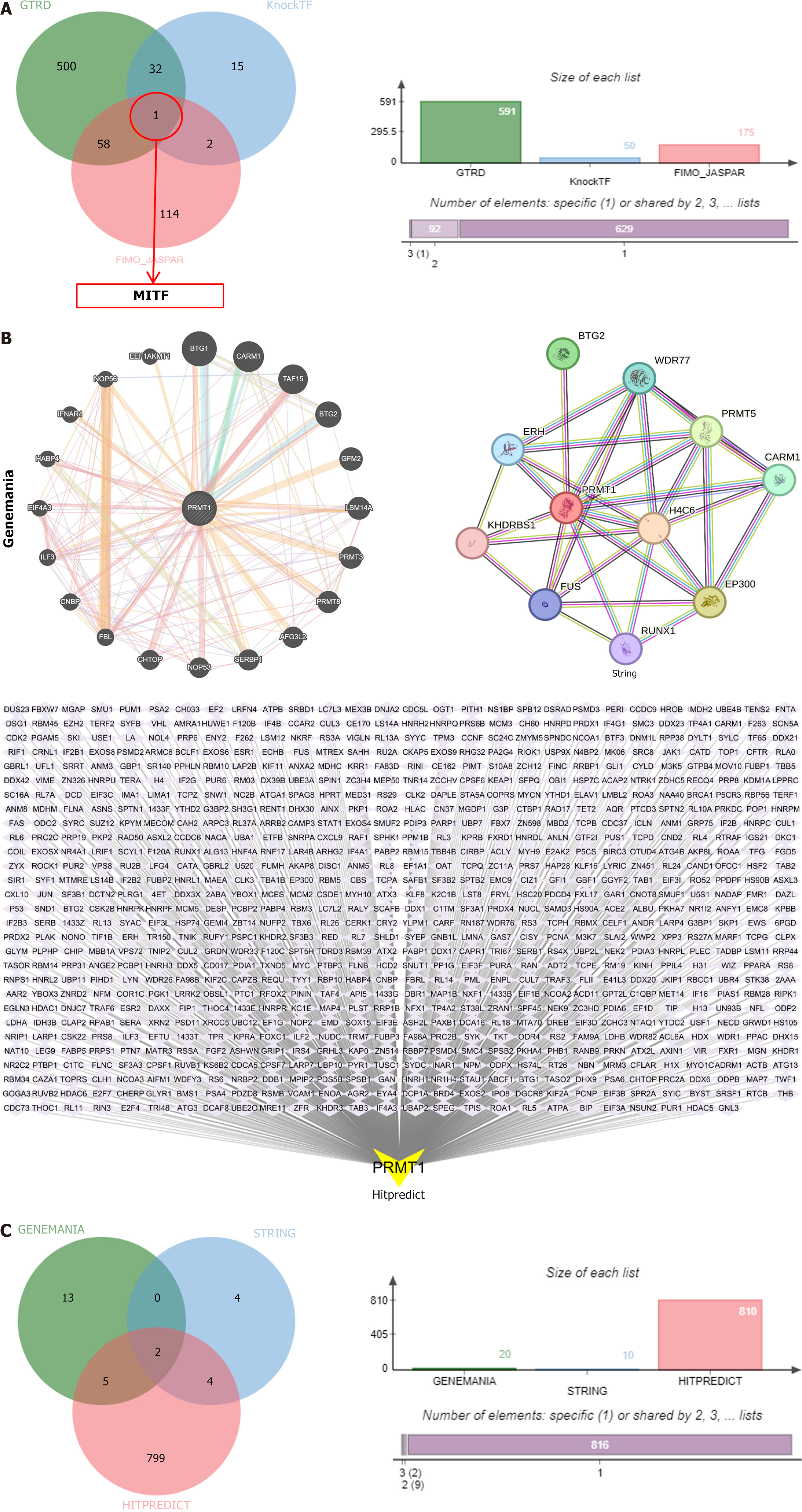
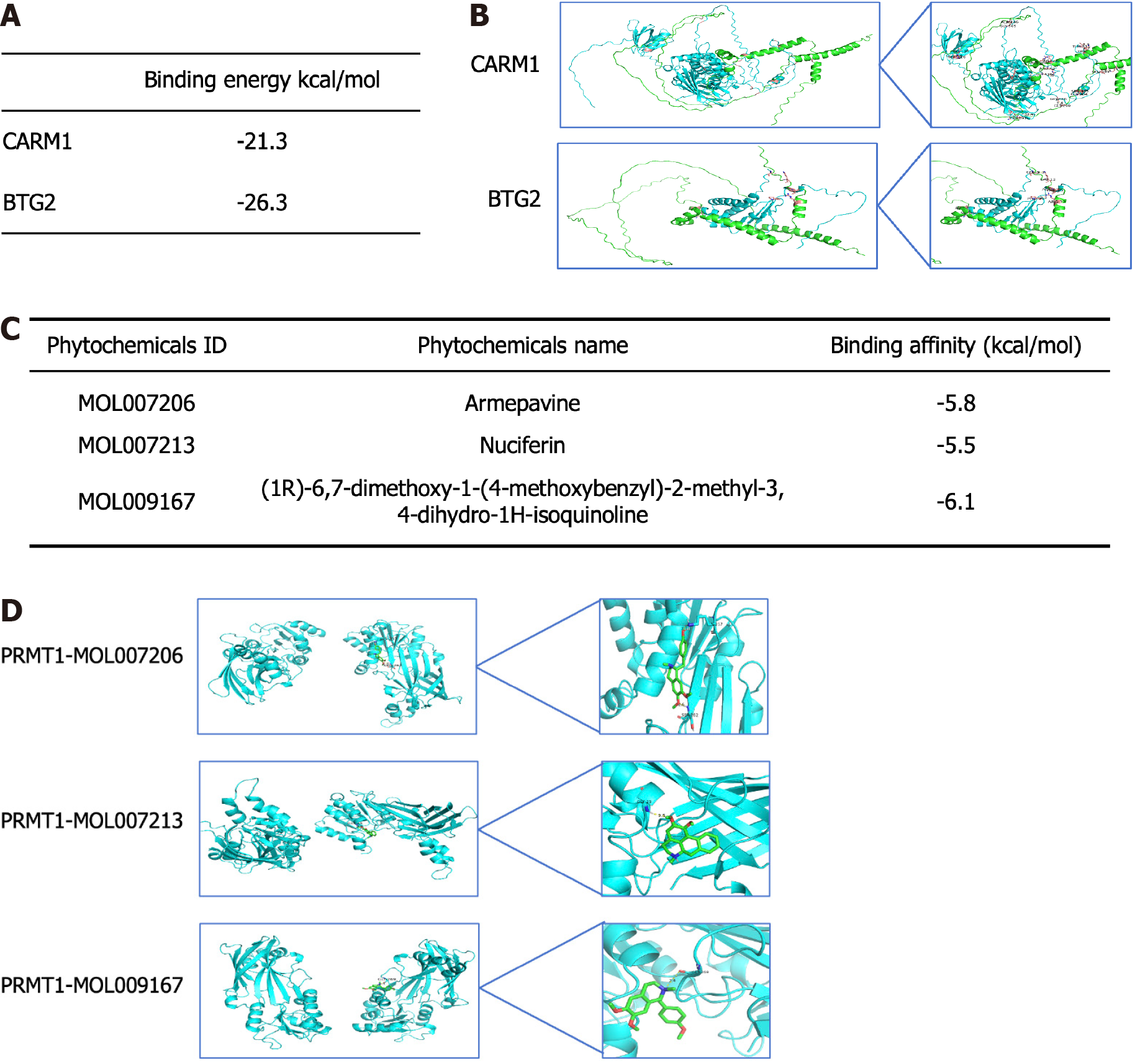
Based on the above analysis, PRMT1 was selected for further investigation. The Gene Transcription Regulation Database, knockTF, and FIMO-JASPAR databases identified melanocyte inducing transcription factor (MITF) as the transcription factor of PRMT1 (Figure 10A). The downstream targets of PRMT1 were identified in GeneMANIA, STRING, and HitPredict databases, revealing two downstream target proteins (Figure 10B and C). PRMT1 and the downstream targets coactivator associated arginine methyltransferase 1 and B-cell translocation gene 2 (BTG2) were subjected to protein-protein docking to determine the binding energy (Figure 11A) and visualized (Figure 11B). The results revealed that PRMT1 binds to coactivator associated arginine methyltransferase 1 with a binding energy of -21.3 kcal/mol, and PRMT1 binds to BTG2 with a binding energy of -26.3 kcal/mol. BTG2 demonstrated a stronger bind affinity to PRMT1 and was selected as the final downstream target. Molecular docking experiments were conducted to predict the binding affinity of drug-active components to GC targets, simulating their drug absorption and metabolism in the human body. Subsequently, the active components containing the PRMT1 target were molecular docked with PRMT1, resulting in binding energies of -5.8, -5.6, and -6.1 kcal/mol, respectively (Figure 11C), and visualized (Figure 11D).
This study explored the molecular mechanisms underlying the therapeutic effect of lotus plumule against GC by identifying the potential targets of this TCM using WGCNA analysis and bioinformatics. Functional and pathway enrichment analyses revealed that the intersecting targets are closely linked to tumor cell proliferation, apoptosis, migration, and regulation of immune-inflammatory responses. Using LASSO and random forest analyses, four core genes, AKR1B10, FBP1, PRMT1, and CA9, were identified as key targets for antitumor effects. Analysis at the trans
AKR1B10, a member of the aldo-keto reductase superfamily, is involved in various BP across different organisms, including detoxification, osmoregulation, hormone metabolism, lipid synthesis, diabetic complications, and tumorigenesis. High expression of AKR1B10 has been observed in liver cancer[25], mammary cancer[26], and lung carcinoma[27,28], highlighting its significant role in tumor development. In GC, AKR1B10 inhibits tumor cell migration, invasion, and adhesion by regulating integrin subunit alpha 5[29]. Additionally, low expression of AKR1B10 promotes M2 macrophage polarization, the predominant immune infiltration pattern in GC, thereby facilitating malignant transformation[30]. AKR1B10 overexpression may independently predict poor survival in patients with GC undergoing gastrectomy with D2 lymph node dissection[31]. Knockdown of AKR1B10 significantly promoted tumor growth and increased levels of vimentin and E-cadherin. Accordingly, AKR1B10 is identified as a tumor suppressor in GC and represents a promising therapeutic target[32]. Moreover, AKR1B10 expression is associated with lymph node metastasis, poor prognosis, and reduced response to neoadjuvant chemotherapy, making it a potential predictor of treatment response in locally advanced GC[33].
FBP1 is a key enzyme in gluconeogenesis that regulates the rate of this process by catalyzing the irreversible hydrolysis of fructose 1,6-bisphosphate to fructose 6-phosphate and inorganic phosphate, influencing cellular glycolysis. Recent studies highlight its significant role in cancer, revealing abnormal FBP1 expression in various tumors, which is closely associated with tumor occurrence, development, and prognosis. These findings suggest promising prospects for developing molecular markers, targeted therapies, and prognostic tools for tumors. Gastric cancer mesenchymal stem cells can also suppress natural killer cell activity by upregulating FBP1 expression, providing a novel approach for GC immunotherapy targeting the natural killer cell matrix[34]. Snail positively regulates glucose metabolism by modulating FBP1 in GC. Targeting the Snail-FBP1 signaling axis could prevent primary tumor epithelial-mesenchymal transition and glycolysis[35]. Downregulation of FBP1 promotes GC metastasis by inducing epithelial-mesenchymal transition, suggesting that FBP1 may serve as a prognostic marker and therapeutic target[36].
CA9 plays a crucial role in maintaining acid-base balance and facilitating carbon dioxide and bicarbonate transport, thereby regulating intracellular and extracellular pH levels. This process contributes to conditions such as hypoxia and an acidic tumor microenvironment. The CA9 inhibitor SLC-0111 exhibits anticancer efficacy against GC cell lines and resensitizes resistant cells to 5-fluorouridine, taxane-derived, and platinum-based drugs[37]. CREB acts as a key negative regulator of CA9 in GC, with its overexpression significantly inhibiting CA9 expression, providing insight into the negative regulatory mechanism of CA9 in GC[38].
PRMT1, a member of the arginine methyltransferase family, is a marker of transcriptional activation and is involved in controlling gene transcription, mRNA splicing, protein stability, DNA damage signaling, and cell fate determination. PRMT1 interacts with various transcription factors and gene promoters, with its overexpression and aberrant splicing linked to tumorigenesis in cancers such as breast, lung, bladder, and leukemia. Metformin disrupts signal transducer and activator of transcription 1 phosphorylation by binding to it, inhibiting PRMT1 expression, and promoting GC progression[39]. PRMT1 also recruits MLX interacting protein to the β-catenin promoter, inducing β-catenin transcription and activating the β-catenin signaling pathway, which promotes GC cell migration and metastasis[40]. Some studies have demonstrated that the c-Fos protein undergoes autophagic degradation and PRMT1-mediated R287 methylation, protecting c-Fos from this process. This protective mechanism is associated with clinical pathological variables and the prognosis of gastric tumors. The findings of a previous study indicated that PRMT1-mediated stabilization of the c-Fos protein promotes gastric tumorigenesis[41]. PRMT1 overexpression enhances migration and invasion while inhibiting proliferation, whereas PRMT1 knockdown reverses these effects[42]. PRMT1 downregulation in GC cells via RNA interference reduced sensitivity to cisplatin and 5-fluoridine. Consequently, PRMT1 expression in GC predicts poor prognosis and recurrence following adjuvant chemotherapy[43].
In molecular biology, transcription factors are proteins that bind to specific nucleotide sequences upstream of a gene, regulating its transcription by influencing the binding of RNA polymerase to DNA templates. Downstream target proteins, located downstream in signaling pathways, are regulated by upstream signaling molecules and affect biochemical responses and cellular functions. Signals are transmitted from the extracellular environment to intracellular space via enzymatic reactions, ultimately affecting specific downstream target proteins. These proteins play diverse roles, including regulating gene expression, influencing the cell cycle, and promoting or inhibiting cell growth and differentiation.
The MITF/PRMT1/BTG2 signaling axis, involving one of the selected hub genes, might exhibit antitumor effects. MITF, characterized by its basic helical-loop-helix and leucine zipper structure, plays a pivotal role in melanocyte development, differentiation, survival, pigment production, and cell cycle regulation. Furthermore, MITF has been implicated in various cancers, including gastric, liver, pancreatic, lung, and papillary RCC. A previous study revealed that miR-585-5p inhibits GC proliferation and metastasis by mediating the interaction between CREB1, MAPK1, and MITF[44]. Additionally, CSE1L was found to reduce MITF and GPNMB expression, activating the PI3K/Akt/mTOR and MEK/ERK pathways, thereby inhibiting tumor growth and metastasis in GC[45].
The BTG2 gene, a BTG/TOB anti-proliferative family member, is associated with tumor cell proliferation, metastasis, and cell cycle regulation. Consequently, BTG2 could be a potential target for GC therapy. These findings suggest that the miR-27a-3p/BTG2 axis may serve as a valuable diagnostic biomarker and therapeutic target for patients with GC[46]. BTG2 significantly inhibited the growth and proliferation of GC cells, reducing their malignant phenotypes. Accordingly, BTG2 is a potential gene therapy target[47].
This study identified key targets of lotus plumule treatment for GC using WGCNA, LASSO, and other analyses, highlighting their clinical relevance. However, limited time and funding did not allow in-depth mechanistic validation. Future research should investigate the causal relationships between these core genes and immune cell infiltration, as well as their roles in GC progression using in vivo and in vitro models. Additionally, direct experimental evidence at the cellular level is lacking, particularly regarding protein interactions via limited proteolysis-coupled mass spectrometry. Verification of upstream and downstream protein interactions in signaling pathways using co-immunoprecipitation and glutathione-S-transferase pull-down, along with gene knockout, overexpression studies, and immunohistochemical staining, is necessary. These comprehensive experiments will help clarify the pathological roles of these core targets in GC and may contribute to developing new therapeutic strategies.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64362] [Article Influence: 16090.5] [Reference Citation Analysis (175)] |
| 2. | Yang WJ, Zhao HP, Yu Y, Wang JH, Guo L, Liu JY, Pu J, Lv J. Updates on global epidemiology, risk and prognostic factors of gastric cancer. World J Gastroenterol. 2023;29:2452-2468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 72] [Cited by in RCA: 185] [Article Influence: 92.5] [Reference Citation Analysis (13)] |
| 3. | Yang K, Lu L, Liu H, Wang X, Gao Y, Yang L, Li Y, Su M, Jin M, Khan S. A comprehensive update on early gastric cancer: defining terms, etiology, and alarming risk factors. Expert Rev Gastroenterol Hepatol. 2021;15:255-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 4. | Guan WL, He Y, Xu RH. Gastric cancer treatment: recent progress and future perspectives. J Hematol Oncol. 2023;16:57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 399] [Reference Citation Analysis (5)] |
| 5. | Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 1073] [Article Influence: 268.3] [Reference Citation Analysis (0)] |
| 6. | Ilson DH. Advances in the treatment of gastric cancer: 2019. Curr Opin Gastroenterol. 2019;35:551-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 7. | Wang FH, Zhang XT, Tang L, Wu Q, Cai MY, Li YF, Qu XJ, Qiu H, Zhang YJ, Ying JE, Zhang J, Sun LY, Lin RB, Wang C, Liu H, Qiu MZ, Guan WL, Rao SX, Ji JF, Xin Y, Sheng WQ, Xu HM, Zhou ZW, Zhou AP, Jin J, Yuan XL, Bi F, Liu TS, Liang H, Zhang YQ, Li GX, Liang J, Liu BR, Shen L, Li J, Xu RH. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2023. Cancer Commun (Lond). 2024;44:127-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 120] [Article Influence: 120.0] [Reference Citation Analysis (0)] |
| 8. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2827] [Article Influence: 565.4] [Reference Citation Analysis (5)] |
| 9. | Xu W, Li B, Xu M, Yang T, Hao X. Traditional Chinese medicine for precancerous lesions of gastric cancer: A review. Biomed Pharmacother. 2022;146:112542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 10. | Zhang Y, Lou Y, Wang J, Yu C, Shen W. Research Status and Molecular Mechanism of the Traditional Chinese Medicine and Antitumor Therapy Combined Strategy Based on Tumor Microenvironment. Front Immunol. 2020;11:609705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 11. | Lu Y, Liu H, Yang K, Mao Y, Meng L, Yang L, Ouyang G, Liu W. A comprehensive update: gastrointestinal microflora, gastric cancer and gastric premalignant condition, and intervention by traditional Chinese medicine. J Zhejiang Univ Sci B. 2022;23:1-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 12. | Yu X, Xia K, Wu S, Wang Q, Cheng W, Ji C, Yang W, Kang C, Yuan Z, Li Y. Simultaneous determination and pharmacokinetic study of six components in beagle dog plasma by UPLC-MS/MS after oral administration of Astragalus Membranaceus aqueous extract. Biomed Chromatogr. 2022;36:e5488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Wang K, Chen Q, Shao Y, Yin S, Liu C, Liu Y, Wang R, Wang T, Qiu Y, Yu H. Anticancer activities of TCM and their active components against tumor metastasis. Biomed Pharmacother. 2021;133:111044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 224] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 14. | Zheng B, Zheng Y, Zhang N, Zhang Y, Zheng B. Rhoifolin from Plumula Nelumbinis exhibits anti-cancer effects in pancreatic cancer via AKT/JNK signaling pathways. Sci Rep. 2022;12:5654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 15. | Chen S, Li X, Wu J, Li J, Xiao M, Yang Y, Liu Z, Cheng Y. Plumula Nelumbinis: A review of traditional uses, phytochemistry, pharmacology, pharmacokinetics and safety. J Ethnopharmacol. 2021;266:113429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 16. | Bharathi Priya L, Huang CY, Hu RM, Balasubramanian B, Baskaran R. An updated review on pharmacological properties of neferine-A bisbenzylisoquinoline alkaloid from Nelumbo nucifera. J Food Biochem. 2021;45:e13986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | Marthandam Asokan S, Mariappan R, Muthusamy S, Velmurugan BK. Pharmacological benefits of neferine - A comprehensive review. Life Sci. 2018;199:60-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 18. | Shen Y, Bian R, Li Y, Gao Y, Liu Y, Xu Y, Song X, Zhang Y. Liensinine induces gallbladder cancer apoptosis and G2/M arrest by inhibiting ZFX-induced PI3K/AKT pathway. Acta Biochim Biophys Sin (Shanghai). 2019;51:607-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Chang M, Ding S, Dong X, Shang X, Li Y, Xie L, Song X, Song X. Liensinine Inhibits Cell Growth and Blocks Autophagic Flux in Nonsmall-Cell Lung Cancer. J Oncol. 2022;2022:1533779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 20. | Yang JH, Yu K, Si XK, Li S, Cao YJ, Li W, Zhang JX. Liensinine inhibited gastric cancer cell growth through ROS generation and the PI3K/AKT pathway. J Cancer. 2019;10:6431-6438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | Zhao L, Yu X, Wu S, Xia K, Wang Y, Qin P, Huang Z, Kang C, Yuan Z, Li Y. Pharmacokinetic profiling and network pharmacology of honey-fried Licorice: An Integrative workflow to study traditional Chinese medicines (TCMs). J Chromatogr B Analyt Technol Biomed Life Sci. 2024;1248:124353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 22. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10254] [Cited by in RCA: 16333] [Article Influence: 960.8] [Reference Citation Analysis (0)] |
| 23. | Tian Z, He W, Tang J, Liao X, Yang Q, Wu Y, Wu G. Identification of Important Modules and Biomarkers in Breast Cancer Based on WGCNA. Onco Targets Ther. 2020;13:6805-6817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 24. | Lv JH, Hou AJ, Zhang SH, Dong JJ, Kuang HX, Yang L, Jiang H. WGCNA combined with machine learning to find potential biomarkers of liver cancer. Medicine (Baltimore). 2023;102:e36536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 25. | Kim SY, Shen Q, Son K, Kim HS, Yang HD, Na MJ, Shin E, Yu S, Kang K, You JS, Yu KR, Jeong SM, Lee EK, Ahn YM, Park WS, Nam SW. SMARCA4 oncogenic potential via IRAK1 enhancer to activate Gankyrin and AKR1B10 in liver cancer. Oncogene. 2021;40:4652-4662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Li J, Guo Y, Duan L, Hu X, Zhang X, Hu J, Huang L, He R, Hu Z, Luo W, Tan T, Huang R, Liao D, Zhu YS, Luo DX. AKR1B10 promotes breast cancer cell migration and invasion via activation of ERK signaling. Oncotarget. 2017;8:33694-33703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 27. | Duan W, Liu W, Xia S, Zhou Y, Tang M, Xu M, Lin M, Li X, Wang Q. Warburg effect enhanced by AKR1B10 promotes acquired resistance to pemetrexed in lung cancer-derived brain metastasis. J Transl Med. 2023;21:547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 44] [Reference Citation Analysis (0)] |
| 28. | Jang TH, Lin SC, Yang YY, Lay JD, Chang CL, Yao CJ, Huang JS, Chuang SE. The Role of AKR1B10 in Lung Cancer Malignancy Induced by Sublethal Doses of Chemotherapeutic Drugs. Cancers (Basel). 2024;16:2428. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 29. | Yao H, Hu J, Shao Y, Shah Q, Zheng S. Aldo-keto Reductase 1B10 Restrains Cell Migration, Invasion, and Adhesion of Gastric Cancer via Regulating Integrin Subunit Alpha 5. Turk J Gastroenterol. 2023;34:1197-1205. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 30. | Wu Y, Hao Y, Zhuang Q, Ma X, Shi C. AKR1B10 regulates M2 macrophage polarization to promote the malignant phenotype of gastric cancer. Biosci Rep. 2023;43:BSR20222007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Liu YY, Liu YW, Huang GK, Hung KC, Lin YH, Yeh CH, Yin SM, Tsai CH, Chen YH. Overexpression of AKR1B10 Predicts Poor Prognosis in Gastric Cancer Patients Undergoing Surgical Resection. Curr Oncol. 2022;30:85-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | Shao X, Wu J, Yu S, Zhou Y, Zhou C. AKR1B10 inhibits the proliferation and migration of gastric cancer via regulating epithelial-mesenchymal transition. Aging (Albany NY). 2021;13:22298-22314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Ahmed SMU, Jiang ZN, Zheng ZH, Li Y, Wang XJ, Tang X. AKR1B10 expression predicts response of gastric cancer to neoadjuvant chemotherapy. Oncol Lett. 2019;17:773-780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 34. | Han F, Guo S, Huang C, Cui L, Zhao Y, Ma J, Zhu M, Chen Z, Wang M, Shen B, Zhu W. Gastric cancer mesenchymal stem cells inhibit natural killer cell function by up-regulating FBP1. Cent Eur J Immunol. 2021;46:427-437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 35. | Yu J, Li J, Chen Y, Cao W, Lu Y, Yang J, Xing E. Snail Enhances Glycolysis in the Epithelial-Mesenchymal Transition Process by Targeting FBP1 in Gastric Cancer. Cell Physiol Biochem. 2017;43:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 36. | Li J, Wang Y, Li QG, Xue JJ, Wang Z, Yuan X, Tong JD, Xu LC. Downregulation of FBP1 Promotes Tumor Metastasis and Indicates Poor Prognosis in Gastric Cancer via Regulating Epithelial-Mesenchymal Transition. PLoS One. 2016;11:e0167857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 37. | Andreucci E, Biagioni A, Peri S, Versienti G, Cianchi F, Staderini F, Antonuzzo L, Supuran CT, Olivo E, Pasqualini E, Messerini L, Massi D, Lulli M, Ruzzolini J, Peppicelli S, Bianchini F, Schiavone N, Calorini L, Magnelli L, Papucci L. The CAIX inhibitor SLC-0111 exerts anti-cancer activity on gastric cancer cell lines and resensitizes resistant cells to 5-Fluorouracil, taxane-derived, and platinum-based drugs. Cancer Lett. 2023;571:216338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 38. | Wang G, Cheng Z, Liu F, Zhang H, Li J, Li F. CREB is a key negative regulator of carbonic anhydrase IX (CA9) in gastric cancer. Cell Signal. 2015;27:1369-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 39. | Wang K, Chen Y, Zhang M, Wang S, Yao S, Gong Z, Fei B, Huang Z. Metformin suppresses gastric cancer progression by disrupting the STAT1-PRMT1 axis. Biochem Pharmacol. 2024;226:116367. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 40. | Wang F, Chen S, Peng S, Zhou X, Tang H, Liang H, Zhong X, Yang H, Ke X, Lü M, Cui H. PRMT1 promotes the proliferation and metastasis of gastric cancer cells by recruiting MLXIP for the transcriptional activation of the β-catenin pathway. Genes Dis. 2023;10:2622-2638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 41. | Kim E, Rahmawati L, Aziz N, Kim HG, Kim JH, Kim KH, Yoo BC, Parameswaran N, Kang JS, Hur H, Manavalan B, Lee J, Cho JY. Protection of c-Fos from autophagic degradation by PRMT1-mediated methylation fosters gastric tumorigenesis. Int J Biol Sci. 2023;19:3640-3660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 42. | Zhang Y, Wang D, Zhang M, Wei H, Lu Y, Sun Y, Zhou M, Gu S, Feng W, Wang H, Zeng J, Gong A, Xu M. Protein arginine methyltransferase 1 coordinates the epithelial-mesenchymal transition/proliferation dichotomy in gastric cancer cells. Exp Cell Res. 2018;362:43-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Altan B, Yokobori T, Ide M, Mochiki E, Toyomasu Y, Kogure N, Kimura A, Hara K, Bai T, Bao P, Suzuki M, Ogata K, Asao T, Nishiyama M, Oyama T, Kuwano H. Nuclear PRMT1 expression is associated with poor prognosis and chemosensitivity in gastric cancer patients. Gastric Cancer. 2016;19:789-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Wang Y, Li M, Zeng J, Yang Y, Li Z, Hu S, Yang F, Wang N, Wang W, Tie J. MiR-585-5p impedes gastric cancer proliferation and metastasis by orchestrating the interactions among CREB1, MAPK1 and MITF. Front Immunol. 2022;13:1008195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | Li Y, Yuan S, Liu J, Wang Y, Zhang Y, Chen X, Si W. CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. J Cell Physiol. 2020;235:2071-2079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 46. | Zhou L, Liang X, Zhang L, Yang L, Nagao N, Wu H, Liu C, Lin S, Cai G, Liu J. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget. 2016;7:51943-51954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 47. | Zhang L, Huang H, Wu K, Wang M, Wu B. Impact of BTG2 expression on proliferation and invasion of gastric cancer cells in vitro. Mol Biol Rep. 2010;37:2579-2586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |