Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.102619
Revised: January 7, 2025
Accepted: February 25, 2025
Published online: April 15, 2025
Processing time: 153 Days and 7.8 Hours
Centromere protein A (CENPA) exhibits an increased expression level in primary human rectal cancer tissues, but its role has not been investigated.
To clarify the specific role and mechanism of CENPA in rectal cancer progression.
CENPA protein expression in rectal cancer tissues and cell lines were detected. CENPA was overexpressed and knocked down in SW837 and SW480 cells, and proliferation, invasion, apoptosis and epithelial-mesenchymal transition (EMT) marker protein levels were examined. O6-methylguanine DNA methyltransferase (MGMT) promoter methylation was assessed with methylation-specific poly
CENPA was upregulated in rectal cancer tissues and cell lines. CENPA overexpression promoted proliferation, invasion and EMT, and inhibited apoptosis in rectal cancer cells. Whereas CENPA knockdown showed the opposite results. Moreover, CENPA inhibited MGMT expression by promoting DNA methyltransferase 1-mediated MGMT promoter methylation. MGMT knockdown abolished the CENPA knockdown-mediated inhibition of rectal cancer cell progression. MGMT increased PTPN4 protein stability by inhibiting PTPN4 ubiquitination degradation via competing with ubiquitin-conjugating enzyme E2O for interacting with PTPN4. PTPN4 knockdown abolished the inhibitory effects of MGMT overexpression on rectal cancer cell progression. Moreover, CENPA knockdown inhibited xenograft tumor growth in vivo.
CENPA knockdown inhibited rectal cancer cell growth and attenuated xenograft tumor growth through regulating the MGMT/PTPN4 axis.
Core Tip: This study suggested that centromere protein A (CENPA) was upregulated in rectal cancer tissues and cell lines. CENPA overexpression promoted proliferation, invasion and epithelial-mesenchymal transition, and inhibited apoptosis in rectal cancer cells. Whereas CENPA knockdown showed the opposite results. Additionally, CENPA knockdown inhibited xenograft tumor growth in vivo. Mechanistically, CENPA inhibited O6-methylguanine DNA methyltransferase (MGMT) expression by promoting DNA methyltransferase 1-mediated MGMT promoter methylation. MGMT interacted with protein tyrosine phosphatase nonreceptor type 4 (PTPN4) and increased PTPN4 protein stability. CENPA knockdown inhibited rectal cancer progression through regulating the MGMT/PTPN4 axis.
- Citation: Xin MJ, Yuan Y. Centromere protein A knockdown inhibits rectal cancer through O6-methylguanine DNA methyltransferase/protein tyrosine phosphatase nonreceptor type 4 axis. World J Gastrointest Oncol 2025; 17(4): 102619
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/102619.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.102619
Rectal cancer represents a leading global malignancy with a poor prognosis; it ranks fourth in cancer mortality[1]. The majority of individuals with rectal cancer are diagnosed at an advanced stage due to its elusive nature[2]. Its incidence has increased, with an alarming trend of diagnosis in younger populations[3]. For those suffering from rectal cancer with visceral metastasis, the 5-year survival rate remains distressingly short at the 10.6% mark[4]. The standard clinical approaches to treatment encompass surgical removal, radiation therapy, chemotherapy, and immunotherapy[5]. As significant advancements in the management of rectal cancer, which results in a modest improvement in patient prognosis, the outcomes of rectal cancer patients remain seriously unsatisfactory due to the high incidence of metastasis and recurrence[6]. Therefore, studies should shed light on the mechanism of rectal cancer to search for more potent clinical therapies.
Centromere protein A (CENPA), which is present in every active centromeric region, is acknowledged as a distinctive indicator of centromeres[7]. CENPA is highly expressed in human cancer genome[8]. An increase in CENPA levels in numerous types of cancer is coupled to the advancement of various cancers, which suggests its potential as a prognostic and predictive biomarker for malignancies[9]. CENPA overexpression promoted clear-cell renal-cell carcinoma proliferation and metastasis by accelerating the cell cycle[10]. CENPA was remarkably overexpressed in gastric cancer tissues, which implies that CENPA may be a standalone prognostic factor for poor outcomes in gastric cancer patients[11]. Moreover, the overexpressed CENPA enhanced prostate cancer cell growth[12]. Notably, CENPA mRNA and protein were upregulated in primary human colorectal cancer tissues, which indicates that CENPA may participate in colorectal cancer progression[13]. Nevertheless, the role of CENPA in rectal cancer progression has rarely been investigated.
O6-Methylguanine DNA methyltransferase (MGMT) functions as a crucial DNA repair enzyme; it safeguards cells against the mutagenic and cytotoxic effects of alkylating agents[14]. Aberrant MGMT promoter methylation becomes the main cause of reduced MGMT expression in multiple cancers, including oesophageal[15], ovarian[16], rectal[17] and gastric cancers[18]. The silencing of the MGMT gene is related to cancer development and increased sensitivity to therapeutic methylating agents. Consequently, MGMT promoter methylation is recognised as a potential biomarker for early cancer detection. More importantly, current evidence implicates that MGMT hypermethylation in blood and tumor tissues of rectal cancer patients is linked to a decreased MGMT mRNA level, and MGMT methylation in blood is a viable biomarker capable of distinguishing between benign and malignant rectal tumors[19]. Thus, the role of MGMT promoter methylation in rectal cancer must be explored.
We explored the role of CENPA in rectal cancer progression and further explored the mechanisms of CENPA/MGMT axis in rectal cancer progression to provide a theoretical direction for rectal cancer treatment.
Rectal cancer tissues and corresponding adjacent tissues were collected from rectal cancer patients (n = 28; age 40-65 years) who underwent surgery at the Medical College of Henan Vocational University of Science and Technology. The patient group excluded those who had chemotherapy, radiation therapy, immunotherapy, or other cancer interventions. This study was reviewed and approved by the Ethic Committee of Medical College of Henan Vocational University of Science and Technology (Approval No. HVUYL414101416920231017001), and all participants signed a written informed consent.
Rectal cancer cells (SW480, HR8348, SW837, and SW1463) and normal colonic mucosa cell line FHC were obtained from ATCC and cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% foetal bovine serum (FBS) and penicillin-streptomycin solution at 37 °C in 5% CO2.
Overexpression plasmids of CENPA (pcDNA-CENPA) and MGMT (pcDNA-MGMT) were used for gene overexpression. Short hairpin RNAs targeting CENPA (sh-CENPA), MGMT (sh-MGMT) and protein tyrosine phosphatase nonreceptor type 4 (PTPN4) (sh-PTPN4) were used for gene knockdown. In addition, empty vector and sh-NC served as negative controls. These vectors and oligonucleotides were obtained from RiboBio (Guangzhou, China). The shRNA sequences were shown as follows: CENPA (5′-AGG AGA TCC GAA AGC TTC A-3′), MGMT (5′-GGA CAA GGA TTG TGA AAT GAA ACG CAC CA-3′), PTPN4 (5′-CGT CAT CAA CAC AAG CTA ATA-3′) and negative control (5′-CAT TGC TAT AGA GGC AGA T-3′). They were transfected into rectal cancer cells using Lipofectamine™ 3000 (Invitrogen, United States). Lipofectamine 3000 reagent (10 μL) and pcDNA/shRNA (10 μL) were each combined with Opti-MEM (250 μL) and incubated for 5 minutes at room temperature. Afterward, the two solutions were combined and allowed to incubate for an additional 20 minutes, followed by being added to cells had achieved 70% confluence (1 × 106 cells/well). The transfection process was carried out for a total of 48 hours. For lentiviral transfection, Once SW837 cells reached a confluence of 70%, 4 μL of lentiviral vectors at a titer of 1 x 108 TU/mL was introduced into each well (1 × 106 cells). The culture medium was changed every 24 hours. After a 72-hour post-infection period, the infection efficiency was evaluated by observing green fluorescence under a fluorescence microscope. Subsequently, the cells were subjected to selection with 4 μg/mL puromycin (Sigma, St Louis, MO, United States) for 2 weeks to establish a stable cell line.
Total proteins were extracted using radioimmunoprecipitation assay lysis buffer and separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. After being transferred to a polyvinylidene difluoride membrane, the membranes were blocked with 5% nonfat milk and incubated with primary antibodies at 4 °C overnight. The next step involved a 2 hours incubation with secondary antibodies. Bands were detected using an enhanced chemiluminescence system (Beyotime, China) and quantified with ImageJ software. The specific primary antibodies used are listed as follows: CENPA (1:1000, ab45694, Abcam), MGMT (1:1000, Abcam, ab108630), PTPN4 (1:1000, Novus, NBP1-80867), N-cadherin (1:5000, Abcam, ab76011), vimentin (1:1000, Abcam, ab92547), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:2500, Abcam, ab9485).
Cells were cultivated in 96-well plates and incubated for 0, 24, 48, and 72 hours. At each specified time, cell counting kit-8 (Dojindo, Japan) solution (10 μL/well) was added for another 2 hours incubation. Finally, the absorbance (450 nm) was measured using a microplate reader.
The Transwell chamber with 8 μm pores (Corning, NY, United States) were precoated with Matrigel matrix. A total of 5 × 104 transfected cells suspended in serum-free medium were seeded onto the upper chamber, facing (DMEM) with 10% FBS in the lower chamber. After a 24 hours incubation, cells invading the lower chamber were stained with crystal violet and counted under a microscope (Olympus, Tokyo, Japan).
Rectal cells (1 × 106 cells) were stained using the Annexin V-FITC and propidium iodide solution (Biolegend, San Diego, CA, United States) for 15 minutes in the dark. Finally, the cell apoptotic rate was detected with using flow cytometry (BD Bioscience, San Jose, CA, United States).
Tumour tissues were fixed with 4% paraformaldehyde, embedded in paraffin, and then cut into 4-μm-thick sections. They were then microwave heated in sodium citrate. Then, sections were blocked with goat serum and incubated with primary antibodies against CENPA, MGMT or PTPN4 overnight at 4 °C and with secondary antibodies at 37 °C for 1 hour. Next, the sections were stained using a DAB kit (Sangon Biotech, China) and assessed under a light microscope.
Intracellular RNAs were extracted using Trizol reagent and used for cDNA synthesis with PrimeScript II RT reagent kit (Takara, Madison, WI, United States). PCR amplification was performed using SYBR-green Premix Ex-Tag II (Takara, Dalian, China). The relative expression level of the MGMT mRNA were normalised to GAPDH and calculated through the 2-ΔΔCT method. The primer sequences are listed as follows: MGMT (forward: 5’-GCT GAA TGC CTA TTT CCA CCA-3’, reverse: 5’-CAC AAC CTT CAG CAG CTT CCA-3’), PTPN4 (forward: 5’-ATC TCC ACC GGG AAC TCC TA-3’, reverse: 5’-CGC TTG GGG AAG TAT GAA CCA-3’, and GAPDH (forward: 5’-CCA CTC CTC CAC CTT TGA C-3’, reverse: 5’-ACC CTG TTG CTG TAG CCA-3’).
SW837 cell supernatant was incubated with the anti-CENPA or anti-MGMT antibody at 4 °C overnight and then incubated with 100 μL protein A/G agarose beads overnight at 4 °C. Thereafter, the IP proteins were then collected and analysed via Western blot.
Methylation-specific PCR (MSP) was performed to MSP methylation in SW837 cells was examined using. In brief, DNA was obtained using a mammalian genomic DNA extraction kit (Beyotime, China) and then subjected to bisulfite modification using the EpiTect Bisulfite kit (Qiagen, Germany). PCR reaction was conducted using the AmpliTaq-Gold DNA Polymerase (Applied Biosystems, CA, United States). The primer sequences are as follows: Methylated MGMT (sense: 5’-GGA CGT TAA GGG TTT AGA GC-3’, antisense: 5’-CAA TAC ACG ACC TCG TCA C-3’) and unmethylated MGMT (sense: 5’-GGA TGT TAA GGG TTT AGA GT-3’, antisense, 5’-CAA TAC ACA ACC TCA TCA C-3’).
Chromatin immunoprecipitation (ChIP) assay was performed by using EZ-Magna ChIP TMA kit (Millipore, Billerica, MA, United States). SW837 cells were fixed with 1% formaldehyde for 10 minutes, followed by stopped with 0.125 M glycine treatment. Then, the cell supernatant was sonicated with several pulses to generate chromatin fragments (200-1000 bp). These fragments were incubated with anti-DNA methyltransferase 1 (DNMT1) (ab13537, Abcam) for immunoprecipitation at 4 °C overnight, followed by incubation with the ChIP-grade protein A/G magnetic beads for 2 hours. The precipitated products were analysed via quantitative reverse transcription PCR.
The tagged proteins were transfected into cells. Then, cells were treated with 20 μM proteasome inhibitor MG132 for 6 hours, followed by being lysed in RIPA buffer. Cell lysate was collected and incubated with anti-PTPN4 antibody and protein A/G magnetic beads overnight at 4 °C. PTPN4 ubiquitination was examined by immunoblotting with an anti-ubiquitin antibody.
BALB/c nude mice aged 4-6 weeks (16-18 g) were obtained from the animal experimental centre of the Medical College of Henan Vocational University of Science and Technology. The mice were maintained in sterile cages with 40%-55% humidity and 12 hours light/dark cycle at 22 ± 3 °C. All animal experiments were reviewed and approved by the Animal Ethics Committee of the Medical College of Henan Vocational University of Science and Technology (Approval No. HVUYL414101416920240603001). SW837 cells (2 × 106) transfected with lentivirus-carried sh-NC or sh-CENPA were subcutaneously injected into the left flanks of mice (n = 8 per group), and tumor volume was determined every 7 days. The nude mice were euthanised on day 28 after injection, and tumor tissues were excised for further experiments.
Data from triplicate experiments were presented as mean ± SE and analysed using SPSS 22.0 software. Data significance from two groups was analysed by Student’s t-test, and data from different groups were analysed through analysis of variance. P < 0.05 indicated statistically significant difference.
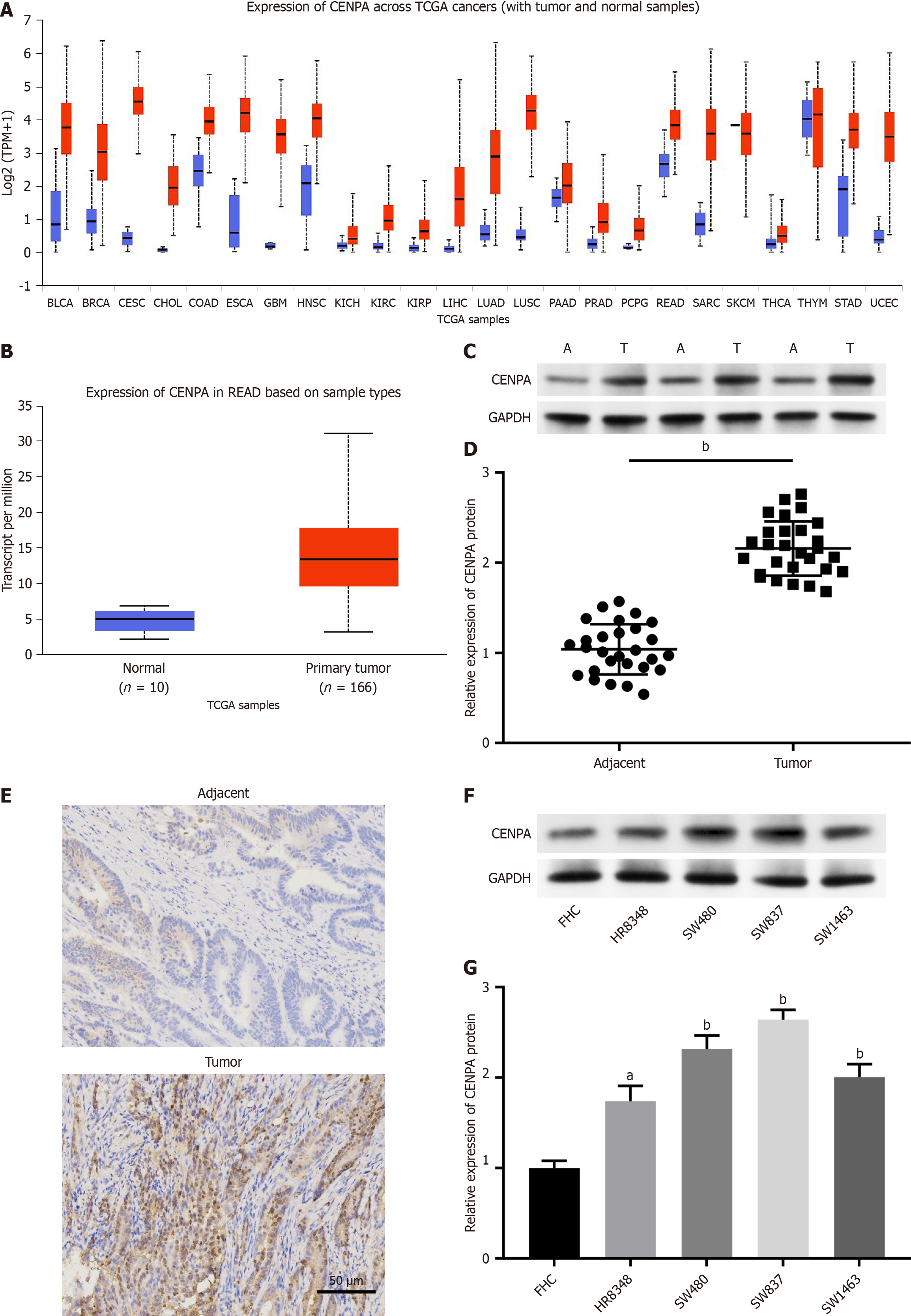
The UALCAN database showed the upregulation of CENPA in multiple cancers (Figure 1A), including rectal cancer (Figure 1B). Meanwhile, Western blot (Figure 1C and D) and immunohistochemistry (Figure 1E) results suggested that CENPA protein was upregulated in rectal cancer tissues compared with matched adjacent tissues. In addition, CENPA was upregulated in rectal cancer cell lines (SW480, HR8348, SW837, and SW1463) compared with that in human normal colonic mucosa cell line FHC (Figure 1F and G). Therefore, we speculated that CENPA might be involved in the progression of rectal cancer.
We then transfected pcDNA-CENPA or sh-CENPA into SW837 cells to investigate the role of CENPA in rectal cancer progression. We suggested that CENPA plasmids upregulated the protein expression of CENPA, and sh-CENPA transfection downregulated CENPA protein expression in SW837 and SW480 cells (Figure 2A and B). CENPA overexpression enhanced cell proliferation (Figure 2C and D) and invasion (Figure 2E-G) in SW837 and SW480 cells, and CENPA knockdown inhibited proliferation and invasion. CENPA overexpression inhibited apoptosis, and CENPA knockdown promoted apoptosis in SW837 and SW480 cells (Figure 2H-K). Moreover, the levels of epithelial-mesenchymal transition (EMT) marker proteins were measured. As shown in Figure 2L-O, CENPA overexpression decreased E-cadherin level those of and increased N-cadherin and Vimentin levels, whereas CENPA knockdown showed the opposite results.
We next explored CENPA-related mechanisms in rectal cancer progression. We first searched the proteins that had potential interaction relationship with CENPA through the BioGRID database (http://thebiogrid.org/). Among these proteins, we then screen 37 proteins that are aberrantly expressed in rectal cancer and have correlation with CENPA in rectal cancer (r > 0.5) through the Gene Expression Profiling Interactive Analysis database (http://gepia.cancer-pku.cn/). Next, among these 37 proteins, we screened 7 proteins that have been previously reported to be associated with the progression of colorectal and rectal cancers. We provided a schematic diagram in Figure 3A. Figure 3B showed the correlation between DNMT1 and CENPA in rectal cancer. Then, Co-immunoprecipitation (Co-IP) assay indicated that CENPA interacted with DNMT1 in SW837 cells (Figure 3C). MGMT promoter methylation was higher in tissue and blood of rectal cancer patients[19]. CpG islands inhabited the MGMT promoter region via the MethPrimer 2 website (Figure 3D). We observed that CENPA promoted DNMT1 enrichment in the MGMT promoter (Figure 3E). Moreover, CENPA overexpression enhanced the methylated MGMT level, whereas CENPA knockdown reduced that in SW837 cells (Figure 3F). Moreover, our results illustrate that CENPA overexpression reduced MGMT mRNA and protein levels in SW837 cells, and CENPA knockdown unveiled the opposite results (Figure 3G and H). Our results imply that CENPA facilitated DNMT1-mediated MGMT methylation and inhibited MGMT expression.
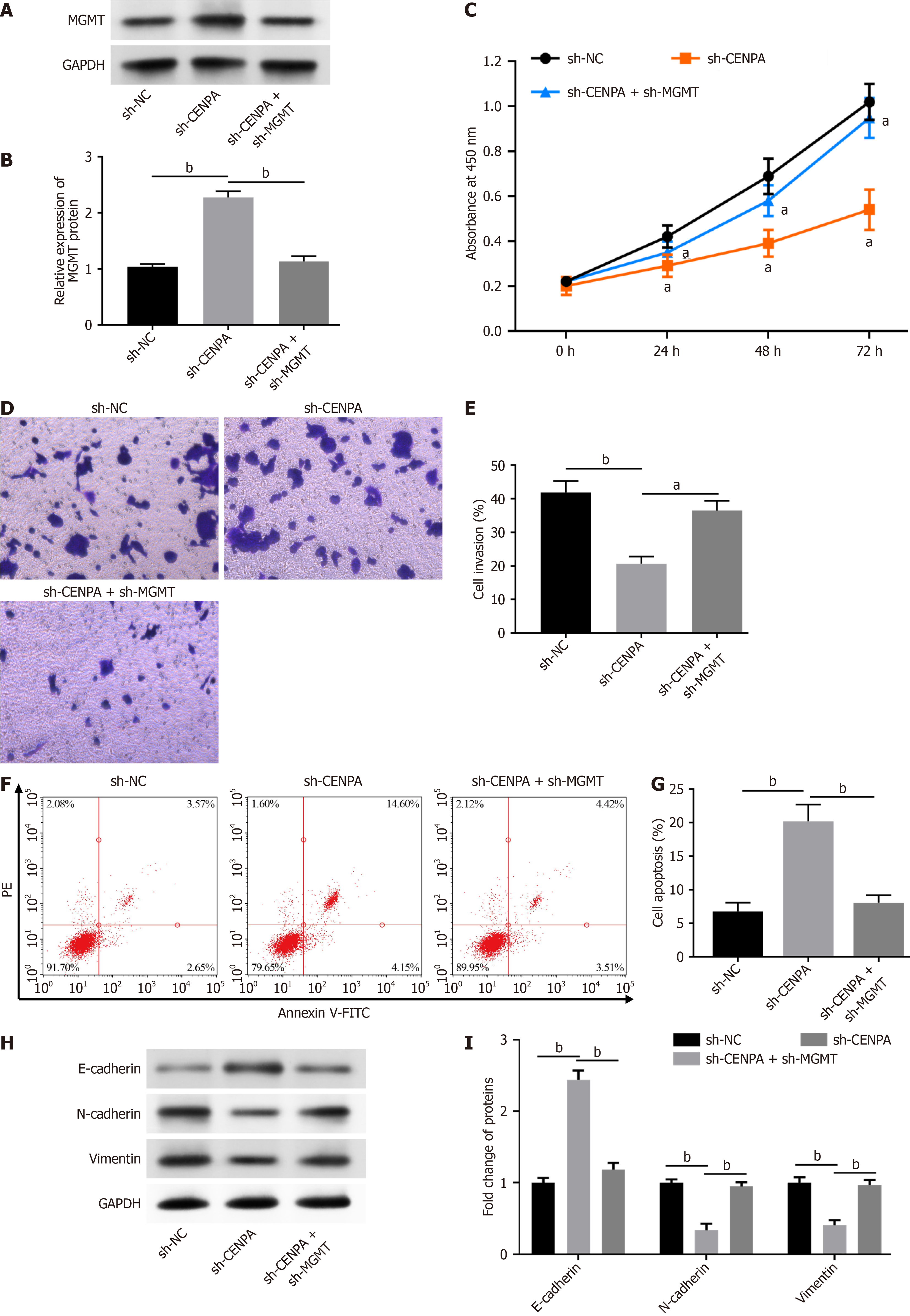
To explore whether CENPA plays regulatory roles in rectal cancer progression by regulating MGMT expression, we transfected SW837 cells with sh-CENPA and sh-MGMT. Transfection of sh-CENPA markedly enhanced MGMT expression, and transfection of sh-MGMT inhibited MGMT expression (Figure 4A and B). CENPA knockdown inhibited SW837 cell proliferation (Figure 4C) and invasion (Figure 4D and E), and MGMT knockdown abolished these effects. Moreover, CENPA knockdown induced SW837 cell apoptosis, which was abrogated by MGMT knockdown (Figure 4F and G). In addition, CENPA knockdown reduced N-cadherin and vimentin levels and increased that of E-cadherin, which was reversed by MGMT knockdown (Figure 4H and I). Collectively, CENPA knockdown inhibited rectal cancer development by promoting MGMT expression.
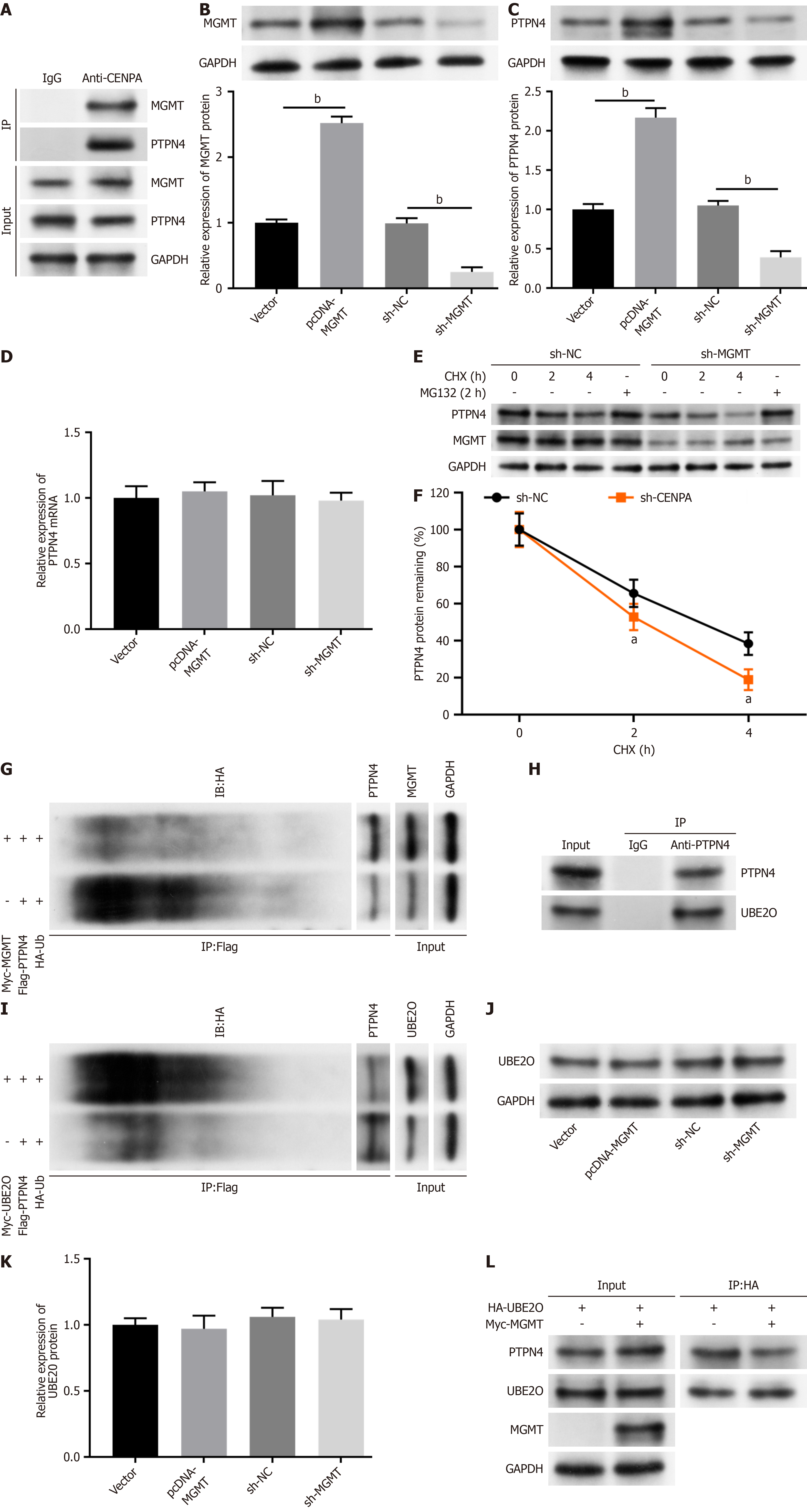
BioGRID tool revealed that possible interaction of MGMT with PTPN4 protein. Co-IP assay verified that that MGMT can interact with PTPN4 in SW837 cells (Figure 5A). Moreover, the transfection of pcDNA-MGMT dramatically increased MGMT and PTPN4 protein levels in SW837 cells, whereas sh-MGMT transfection reduced MGMT and PTPN4 protein levels (Figure 5B and C). We then investigated the mechanism of MGMT in regulating PTPN4 expression. The results showed that MGMT had no effect on PTPN4 mRNA expression (Figure 5D), indicating that MGMT regulates PTPN4 at a post-transcriptional level. We treated cells with protein synthesis inhibitor cycloheximide, and found that MGMT knockdown decreased PTPN4 protein stability (Figure 5E and F). However, proteasome inhibitor MG132 reversed MGMT knockdown-mediated PTPN4 protein inhibition (Figure 5E). Additionally, MGMT decreased PTPN4 ubiquitination (Figure 5G). We speculated that MGMT might decrease PTPN4 ubiquitination through regulating ubiquitination-related enzyme expression or preventing the interaction of PTPN4 to certain ubiquitination-related enzyme. We then searched proteins that had potential interaction with PTPN4 through the BioGRID database, and found a ubiquitin ligase, ubiquitin-conjugating enzyme E2O (UBE2O). Subsequent experiments suggested that UBE2O interacted with PTPN4 (Figure 5H) and promoted PTPN4 ubiquitination (Figure 5I). Furthermore, MGMT had no effect on UBE2O expression in SW837 cells (Figure 5J and K). However, the interaction between UBE2O and PTPN4 was reduced by MGMT overexpression (Figure 5L). These results suggested that MGMT might increase PTPN4 protein stability by inhibiting PTPN4 ubiquitination degradation via competing with UBE2O for interacting with PTPN4.
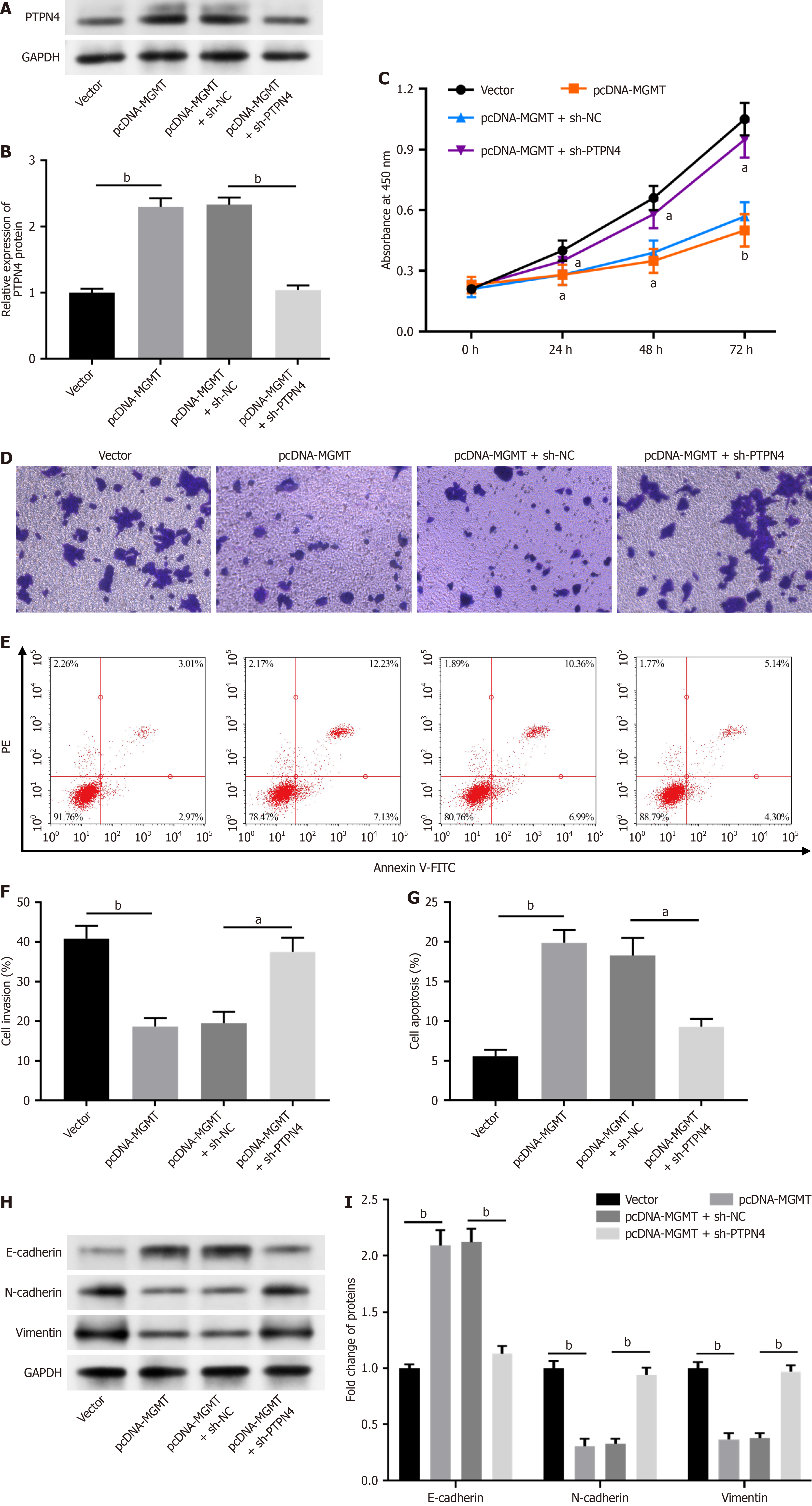
To further investigate whether MGMT modulates rectal cancer progression by regulating PTPN4, we transfected SW837 cells with pcDNA-MGMT and sh-PTPN4. The overexpression of MGMT evidently increased the PTPN4 level, and sh-PTPN4 decreased PTPN4 Level in SW837 cells (Figure 6A and B). MGMT overexpression inhibited SW837 cell proliferation (Figure 6C) and invasion (Figure 6D and E), and PTPN4 knockdown reversed these effects. Moreover, MGMT overexpression induced cell apoptosis (Figure 6F and G) in SW837 cells, which was abolished by PTPN4 knockdown. Moreover, MGMT overexpression reduced N-cadherin and vimentin levels and increased that of E-cadherin, and PTPN4 knockdown abolish these effects (Figure 6H and I). These results demonstrate that MGMT inhibited rectal cancer progression by reducing PTPN4 expression.
We finally detected the in vivo effect of CENPA on rectal cancer. CEPNA knockdown remarkably decreased tumor growth (Figure 7A-C). Moreover, immunohistochemistry assay demonstrated that sh-CENPA injection decreased the CENPA level and increased those of MGMT and PTPN4 in tumor tissues of xenograft mice (Figure 7D and E). Moreover, the proliferation marker Ki67 was substantially reduced in the sh-CENPA group (Figure 7F and G).
CENPA is upregulated in tumor tissues of various cancers, and its overexpression affects the behaviour of cancer cells, including proliferation, migration, invasion and apoptosis[10-12]. Current evidence has showed that CENPA exhibits excessive expression in primary colorectal cancer tissues[13], indicating its pivotal effect on colorectal cancer deve
Previous studies have revealed that CENPA play critical roles in the progression of various cancer types through directly or indirectly regulating gene transcription. CENPA promoted glutamine metabolism and endometrial cancer progression by directly regulating the transcriptional activity of solute carrier family 38 member 1[20]. CENPA promoted hepatocellular carcinoma progression by interacting with YY1 and cooperating as co-transcriptional complex to promote oncogene transcription[21]. CENPA recruited histone acetyltransferase GCN5 to the promoter region of karyopherin subunit alpha 2 to induce transcription activation, thus enhancing colon cancer cell growth and glycolysis[22]. Moreover, CENPA promoted clear cell renal cell carcinoma progression and metastasis by activating the Wnt/β-catenin signaling pathway[10]. Similarly, our results suggested that CENPA recruited DNMT1 to the MGMT promoter and indirectly regulating MGMT transcription in rectal cancer cells. Different from the previous literature[22], DNMT1 recruited by CENPA induced MGMT methylation and inhibits MGMT transcription.
MGMT promoter hypermethylation and its consequent epigenetic silencing are frequently observed as initial events in cancer development. Studies have pointed towards a significant downregulation of MGMT across multiple cancer types, with this reduction closely linked to the methylation of the MGMT promoter. The absence of MGMT protein is notably common in oesophageal cancer, with a strong correlation between MGMT methylation and its reduced expression[15]. MGMT promoter methylation is linked to the onset of ovarian cancer and thus the tumor’s histological type[16]. MGMT promoter methylation can be related to the prognosis of gastric cancer[18]. Genetic variations within the MGMT gene are related to the risk of colorectal cancer[23]. Furthermore, the current study illustrated the considerably higher frequency of MGMT methylation was in blood and tumor tissues of rectal cancer patients, which correlates with a decrease in MGMT mRNA levels[19]. Similarly, our findings demonstrated that CENPA overexpression reduced MGMT expression through promoting the recruitment of DNMT1 to the MGMT promoter and enhancing MGMT methylation level in rectal cancer cells. Moreover, MGMT knockdown counteracted the effects of CENPA knockdown on proliferation, invasion, EMT and apoptosis in rectal cancer cells, which indicates that CENPA promoted rectal cancer cell progression via epigenetic modification of MGMT.
PTPN4 is a nonreceptor protein tyrosine phosphatase participating in the regulation of cell behaviours. Upregulated PTPN4 suppresses cell proliferation and motility in HEK293T cells[24] and inhibits cell proliferation of kidney cells[25]. Moreover, PTPN4 regulates various cancer cells. MiRNA-183 enhanced lung adenocarcinoma cell metastasis via the inhibition of PTPN4 expression[26]. More notably, a low level of PTPN4 expression in rectal cancer has been significantly correlated with a poor prognosis. In rectal cancer cells, PTPN4 overexpression suppresses cell proliferation and colony formation, whereas its depletion accelerates cell growth and the cell cycle. Moreover, the deletion of PTPN4 has been linked to increased tumor formation in vivo[27]. However, the precise mechanism of PTPN4 in rectal cancer are not fully understood. Our study further revealed that PTPN4 can interact with MGMT and was positively regulated by MGMT in rectal cancer cells. Mechanistically, MGMT might increase PTPN4 protein stability by inhibiting PTPN4 ubiquitination degradation via competing with UBE2O for interacting with PTPN4. Moreover, PTPN4 knockdown counteracted the effects of MGMT overexpression on proliferation, invasion, EMT and apoptosis in rectal cancer cells. This finding indicates that MGMT inhibited rectal cancer cell progression by stabilizing PTPN4 protein expression.
This study may have certain inherent limitations. The small number of rectal cancer patients and the inadequate sample sizes for animal experiments could potentially impact the broader applicability and precision of our findings. Moreover, our study did not explore the association between CENPA and the prognosis of rectal cancer. patients. In subsequent research endeavors, we plan to expand the patient cohort and delve into the clinical expression and prognosis implications of CENPA in rectal cancer. Additionally, we aim to explore other potential molecular and signaling pathways through which SERPINB5 may exert its influence in rectal cancer, thereby laying a more robust experimental foundation for the clinical application of CENPA inhibition strategy.
To summarise, our research revealed the upregulation of CEPNA is in rectal cancer. CEPNA knockdown notably suppressed proliferation, invasion and EMT and induced apoptosis in rectal cancer cells. Such condition also resulted in a decreased growth of tumors in animal models. From a mechanistic perspective, CEPNA inhibited MGMT expression through promoting DNMT1-mediated promoter methylation. MGMT interacted with PTPN4 and increased PTPN4 protein stability. Our findings can potentially expand the therapeutic options for rectal cancer treatment.
| 1. | Kang L, Chen YG, Zhang H, Zhang HY, Lin GL, Yang YC, Chen WH, Luo SL, Chen N, Tong WD, Shen ZL, Xiong DH, Xiao Y, Zhang ZT, Wang JP. Transanal total mesorectal excision for rectal cancer: a multicentric cohort study. Gastroenterol Rep (Oxf). 2020;8:36-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 2. | Mu JF, Wang XD, Sun PD. Expression of miR-31 in rectal cancer patients and its effect on proliferation ability of rectal cancer cells SW837. Eur Rev Med Pharmacol Sci. 2018;22:8675-8681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 3. | Suresh RS, Garcia LE, Gearhart SL. Young-Onset Rectal Cancer: Is It for Real? Adv Surg. 2024;58:275-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 4. | Wan P, Bai X, Yang C, He T, Luo L, Wang Y, Fan M, Wang Z, Lu L, Yin Y, Li S, Guo Q, Song Z. miR-129-5p inhibits proliferation, migration, and invasion in rectal adenocarcinoma cells through targeting E2F7. J Cell Physiol. 2020;235:5689-5701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | Lee GC, Bordeianou LG, Francone TD, Blaszkowsky LS, Goldstone RN, Ricciardi R, Kunitake H, Qadan M. Superior pathologic and clinical outcomes after minimally invasive rectal cancer resection, compared to open resection. Surg Endosc. 2020;34:3435-3448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 6. | Junginger T, Goenner U, Hitzler M, Trinh TT, Heintz A, Wollschläger D. Local excision followed by early radical surgery in rectal cancer: long-term outcome. World J Surg Oncol. 2019;17:168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Mitra S, Srinivasan B, Jansen LET. Stable inheritance of CENP-A chromatin: Inner strength versus dynamic control. J Cell Biol. 2020;219:e202005099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 8. | Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, Ried T, Sung MH, Dalal Y. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin. 2015;8:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 113] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 9. | Sun X, Clermont PL, Jiao W, Helgason CD, Gout PW, Wang Y, Qu S. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int J Cancer. 2016;139:899-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 10. | Wang Q, Xu J, Xiong Z, Xu T, Liu J, Liu Y, Chen J, Shi J, Shou Y, Yue C, Liu D, Liang H, Yang H, Yang X, Zhang X. CENPA promotes clear cell renal cell carcinoma progression and metastasis via Wnt/β-catenin signaling pathway. J Transl Med. 2021;19:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 11. | Xu Y, Liang C, Cai X, Zhang M, Yu W, Shao Q. High Centromere Protein-A (CENP-A) Expression Correlates with Progression and Prognosis in Gastric Cancer. Onco Targets Ther. 2020;13:13237-13246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Saha AK, Contreras-Galindo R, Niknafs YS, Iyer M, Qin T, Padmanabhan K, Siddiqui J, Palande M, Wang C, Qian B, Ward E, Tang T, Tomlins SA, Gitlin SD, Sartor MA, Omenn GS, Chinnaiyan AM, Markovitz DM. The role of the histone H3 variant CENPA in prostate cancer. J Biol Chem. 2020;295:8537-8549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 13. | Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511-3516. [PubMed] |
| 14. | Kaina B, Margison GP, Christmann M. Targeting O⁶-methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy. Cell Mol Life Sci. 2010;67:3663-3681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 15. | Rehman AU, Saikia S, Iqbal MA, Ahmad I, Sadaf, Anees A, Aravinda PS, Mishra PK, Hedau S, Saluja SS, Medhi S, Husain SA. Decreased expression of MGMT in correlation with aberrant DNA methylation in esophageal cancer patients from North India. Tumour Biol. 2017;39:1010428317705770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Qiao B, Zhang Z, Li Y. Association of MGMT promoter methylation with tumorigenesis features in patients with ovarian cancer: A systematic meta-analysis. Mol Genet Genomic Med. 2018;6:69-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | An N, Shi Y, Ye P, Pan Z, Long X. Association Between MGMT Promoter Methylation and Breast Cancer: a Meta-Analysis. Cell Physiol Biochem. 2017;42:2430-2440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Ding Y, Yang Q, Wang B, Ye G, Tong X. The Correlation of MGMT Promoter Methylation and Clinicopathological Features in Gastric Cancer: A Systematic Review and Meta-Analysis. PLoS One. 2016;11:e0165509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Shalaby SM, El-Shal AS, Abdelaziz LA, Abd-Elbary E, Khairy MM. Promoter methylation and expression of DNA repair genes MGMT and ERCC1 in tissue and blood of rectal cancer patients. Gene. 2018;644:66-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Li S, Zhang Z, Li Z, Yang L, Liu J, Liu Y, Liu Y, Hou Y, Mei M, Huang Y. CENPA promotes glutamine metabolism and tumor progression by up-regulating SLC38A1 in endometrial cancer. Cell Signal. 2024;117:111110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 21. | Liao J, Chen Z, Chang R, Yuan T, Li G, Zhu C, Wen J, Wei Y, Huang Z, Ding Z, Chu L, Liang J, Zhang B. CENPA functions as a transcriptional regulator to promote hepatocellular carcinoma progression via cooperating with YY1. Int J Biol Sci. 2023;19:5218-5232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 22. | Liang YC, Su Q, Liu YJ, Xiao H, Yin HZ. Centromere Protein A (CENPA) Regulates Metabolic Reprogramming in the Colon Cancer Cells by Transcriptionally Activating Karyopherin Subunit Alpha 2 (KPNA2). Am J Pathol. 2021;191:2117-2132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Zhong Y, Huang Y, Huang Y, Zhang T, Ma C, Zhang S, Fan W, Chen H, Qian J, Lu D. Effects of O6-methylguanine-DNA methyltransferase (MGMT) polymorphisms on cancer: a meta-analysis. Mutagenesis. 2010;25:83-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Zhou J, Wan B, Shan J, Shi H, Li Y, Huo K. PTPN4 negatively regulates CrkI in human cell lines. Cell Mol Biol Lett. 2013;18:297-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 25. | Gu M, Meng K, Majerus PW. The effect of overexpression of the protein tyrosine phosphatase PTPMEG on cell growth and on colony formation in soft agar in COS-7 cells. Proc Natl Acad Sci U S A. 1996;93:12980-12985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Zhu C, Deng X, Wu J, Zhang J, Yang H, Fu S, Zhang Y, Han Y, Zou Y, Chen Z, Lin S. MicroRNA-183 promotes migration and invasion of CD133(+)/CD326(+) lung adenocarcinoma initiating cells via PTPN4 inhibition. Tumour Biol. 2016;37:11289-11297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | Zhang BD, Li YR, Ding LD, Wang YY, Liu HY, Jia BQ. Loss of PTPN4 activates STAT3 to promote the tumor growth in rectal cancer. Cancer Sci. 2019;110:2258-2272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |