Published online Jul 15, 2024. doi: 10.4251/wjgo.v16.i7.3069
Revised: May 5, 2024
Accepted: May 22, 2024
Published online: July 15, 2024
Processing time: 97 Days and 19.6 Hours
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide. As liver cancer often presents no noticeable symptoms in its early stages, most patients are diagnosed at an advanced stage, complicating treatment. Therefore, the identification of new biomarkers is crucial for the early detection and treatment of HCC. Research on exportin-5 (XPO5) could offer new avenues for early diagnosis and improve treatment strategies.
To explore the role of XPO5 in HCC progression and its potential as a prognostic biomarker.
This study assessed XPO5 mRNA expression in HCC using The Cancer Genome Atlas, TIMER, and International Cancer Genome Consortium databases, correlating it with clinical profiles and disease progression. We performed in vitro experiments to examine the effect of XPO5 on liver cell growth. Gene Set Enrichment Analysis, Kyoto Encyclopedia of Genes and Genomes, and Gene Ontology were used to elucidate the biological roles and signaling pathways. We also evaluated XPO5’s impact on immune cell infiltration and validated its prognostic potential using machine learning.
XPO5 was significantly upregulated in HCC tissues, correlating with tumor grade, T-stage, and overall survival, indicating poor prognosis. Enrichment analyses linked high XPO5 expression with tumor immunity, particularly CD4 T cell memory activation and macrophage M0 infiltration. Drug sensitivity tests identified potential therapeutic agents such as MG-132, paclitaxel, and WH-4-023. Overexpression of XPO5 in HCC cells, compared to normal liver cells, was confirmed by western blotting and quantitative real-time polymerase chain reaction. The lentiviral transduction-mediated knockdown of XPO5 significantly reduced cell proliferation and metastasis. Among the various machine learning algorithms, the C5.0 decision tree algorithm achieved accuracy rates of 95.5% in the training set and 92.0% in the validation set.
Our analysis shows that XPO5 expression is a reliable prognostic indicator for patients with HCC and is sig
Core Tip: This study reveals the pivotal role of exportin-5 (XPO5), a miRNA transport protein, in the progression and prognosis of hepatocellular carcinoma (HCC). Bioinformatic databases and in vitro assays demonstrated that XPO5 expression was upregulated in HCC tissues, correlating with aggressive tumor features and poor patient outcomes. Furthermore, XPO5 levels influenced immune cell behavior, particularly in T cells and macrophages, suggesting its potential as a target for immunotherapy. This study suggests that XPO5 could be used as a biomarker for the prognosis of HCC and as a novel therapeutic target.
- Citation: Li H, Li F, Wang BS, Zhu BL. Prognostic significance of exportin-5 in hepatocellular carcinoma. World J Gastrointest Oncol 2024; 16(7): 3069-3081
- URL: https://www.wjgnet.com/1948-5204/full/v16/i7/3069.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i7.3069
Liver cancer is the third most prevalent cause of cancer-related mortality worldwide, with hepatocellular carcinoma (HCC) representing the majority of all liver cancer cases[1,2]. Despite advancements in detection and treatment, the 5-year relative survival rate remains low (18.4%). These findings highlight the urgent need for novel diagnostic and prognostic markers[3,4].
Exportin-5 (XPO5) is a nuclear protein essential for the transport of miRNAs from the nucleus to the cytoplasm and plays a crucial role in cellular regulation and proliferation[5,6]. The functional site of XPO5 indicates a potential risk of carcinogenesis[7]. Moreover, its overexpression is associated with cell proliferation, migration, and invasion in HCC, indicating its oncogenic role[8,9].
To investigate XPO5 expression and function in HCC, we conducted a bioinformatics analysis using The Cancer Genome Atlas (TCGA), which revealed significant XPO5 overexpression. This finding was validated using the International Cancer Genome Consortium (ICGC) dataset. Machine learning algorithms, such as the support vector machine (SVM) and Naive Bayes, were used to evaluate the diagnostic and prognostic value of XPO5. This reflects the increasing use of machine learning in medical research because of its accuracy and sensitivity[10-12].
In addition, in vitro experiments were conducted to further investigate the role of XPO5 in HCC using five cell lines. WRL68, SK-HEP-1, Li-7, SNU-449, and SNU-398 cells were analyzed using quantitative real-time polymerase chain reaction and western blotting to confirm XPO5 overexpression, which was consistent with the bioinformatic findings. Further experiments were conducted using cell lines with significantly different expressions to validate the impact of XPO5 knockdown on HCC cell behavior.
In conclusion, we confirmed the significant overexpression of XPO5 in HCC and its potential as a diagnostic and prognostic marker using a comprehensive approach that combined bioinformatics analysis, machine learning, and in vitro experiments. This study highlights the pivotal role of XPO5 in HCC progression and its potential as a target for the
The training sets for patients with HCC were sourced from TCGA database along with the corresponding clinical information and RNA-sequencing data. An additional dataset of patients with HCC was acquired from the ICGC database for validation. Patients without survival-related data were excluded from subsequent analyses, and the selection of the dataset was based on the use of high-quality gene expression data and complete clinical information. Initially, 377 and 231 patients with HCC from TCGA and ICGC databases, respectively, were selected. Patients without basic survival data, which are essential for longitudinal outcome analysis, were excluded. Furthermore, patients were excluded if they lacked complete demographic information, were not diagnosed with HCC, or had undergone prior malignancies or treatments that could affect the assessment of genomic integrity and treatment outcomes.
We investigated the correlation between XPO5 expression and clinical outcomes in patients with HCC. Patients were categorized into high and low XPO5 expression groups based on the median expression levels. The Z-score (indicating the number of standard deviations from the mean) was calculated for each data point. Data points with Z-scores greater than 3 or less than -3 were considered outliers. We analyzed a comprehensive set of clinicopathological factors, including age, sex, tumor grade, TNM stage, and vital status. This approach ensures a broad representation of clinical conditions. The dataset was cleaned thoroughly by removing entries containing incomplete information and potential duplicates. Quality control was performed to eliminate outliers. Kaplan-Meier survival curves were primarily used to construct a prognostic classifier. This study aimed to determine the effect of XPO5 expression on HCC prognosis. This study aimed to evaluate the impact of different levels of XPO5 expression on patient survival and clinical outcomes to determine its potential as a prognostic marker for HCC. Kaplan-Meier analyses were conducted to generate survival curves adjusted for age and sex.
To elucidate the molecular mechanisms of action of XPO5 in HCC, we used a multifaceted approach. Initially, we conducted gene co-expression analysis using public databases, such as TCGA, to identify genes that were co-expressed with XPO5. This helped shed light on the potential regulatory networks and functional roles of XPO5 in HCC. We analyzed tumor-infiltrating immune cells in liver cancer using the TIMER database. The analyzed cell types included CD8+ T cells, CD4+ T cells, and macrophages. We aimed to elucidate the relationship between XPO5 expression and immune regulation. We utilized the CIBERSORT algorithm to conduct a differential analysis of the 22 types of tumor-infiltrating immune cells. This allowed us to observe the XPO5 expression patterns across immune cell types, thereby enhancing our understanding of their immune regulatory mechanisms.
We compared stromal, immune, and ESTIMATE scores between high and low XPO5 expression groups to determine the impact of XPO5 expression on the tumor microenvironment (TME) and to gain insights into its role in HCC progression. We evaluated the potential of CTLA4 and PD-1 receptor blockers in predicting immunotherapy responsiveness in patients with HCC having different XPO5 expression levels using immunophenotypic scores (IPS) sourced from the Tumor Immunohistochemical Atlas. This study focused on immunotherapy and aimed to advance personalized treatment approaches while uncovering XPO5 mechanisms in HCC immunotherapy.
In this study, we evaluated the drug susceptibility of HCC based on XPO5 expression levels using the Genomics of Drug Sensitivity in Cancer database (https://www.cancerrxgene.org/). For the drug sensitivity analysis, we selected 29 drugs commonly used for treating HCC. After statistical calculations, four drugs were selected for presentation (P < 0.05). The half-maximal inhibitory concentration (IC50) was calculated to quantify drug susceptibility. To facilitate comparison, normalization was applied to calculate the IC50 values. We used OncoPredict, a software package that is known for its accuracy in predicting drug response profiles. Statistical analyses were conducted to compare drug sensitivity between the groups with high and low XPO5 expression using rigorous methods to ensure statistical significance. Our process was designed to ensure reproducibility, allowing researchers to replicate their findings using the same database, IC50 calculation methods, and statistical analyses. This ensures the reliability and scholarly rigor of our approach to drug sensitivity assessment in HCC.
We developed a structured approach to assess the integration of machine learning algorithms for the prognosis of HCC. We selected four algorithms, namely Bayesian classifiers, neural networks, SVMs, and Decision Tree C5.0, based on their clinical applicability and accuracy potential. Data collection involved gathering extensive clinical and gene expression datasets from two cohorts of patients with HCC to ensure a rich foundation for analysis. During preprocessing, the data were cleaned, the numerical ranges were normalized, and key features were selected to streamline the dataset. The data were then divided into training and testing sets for model training. Cross-validation was employed to enhance the model’s robustness and mitigate overfitting. Performance metrics, including accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC), were used to evaluate the clinical prognostic efficacy of each algorithm. This systematic approach aims to effectively apply machine learning to HCC prognosis in clinical settings.
WRL68 cells were cultured in MEM medium supplemented with penicillin (final concentration: 100 U/mL), streptomycin (final concentration: 100 μg/mL), and 100 mL/L fetal bovine serum (FBS). When the cells reached 90% confluence, the spent medium was removed, and the cells were washed twice with 2.0 mL of PBS. The cells were then treated with 2.0 mL of 2.5 mL/L trypsin-0.2 mL/L EDTA solution and observed under a microscope. The digestion was terminated by adding 2 mL of complete medium once the cells were rounded. Cells were collected using gentle pipetting, followed by centrifugation at 800 rpm and 4 °C. The supernatant was discarded, and the cell pellet was resuspended in a complete medium for further culturing. The medium was refreshed every alternate day.
For real-time PCR analysis, cells were seeded in 6-well plates at a density of 5 × 105 cells/well. Upon reaching 90% confluence, the cells were subjected to the respective treatments. After the treatment, the cells were harvested, the supernatant was removed by centrifugation, and the cells were lysed using RNAiso Plus (0.5 mL). RNA was extracted, quantified, and reverse-transcribed. The relative expression levels of the target mRNA were calculated using the 2-ΔΔCt method, with GAPDH as the reference gene (Supplementary Table 1).
For Western blot experiments, cells were lysed, and protein concentrations were determined using the BCA method. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes for immunoblotting with specific antibodies.
SNU449 and SNU398 cells were seeded logarithmically into 24-well plates at a density of 5 × 104 cells/well 24 h before transfection. The cells were cultured in a complete medium supplemented with 100 mL/L FBS. Prior to lentiviral infection, the cells were pretreated with a serum-free medium for 2 h. Following the mapping of multiplicity of infection (MOI) values, the optimal MOI was determined to be 30. The lentivirus was then added based on the specified MOI values and the cells were incubated at 37 °C with 50 mL/L CO2 for 12 h. The infection efficiency was assessed 72 h post-infection.
To perform the wound healing assay, lines were drawn on the back of a 6-well plate using a UV-sterilized marker. Cells in the logarithmic growth phase were prepared as single-cell suspensions and seeded into 6-well plates. A scratch was created using a sterile 200 μL pipette tip, and the detached cells were removed by washing with PBS. The cells were incubated in serum-free medium at 37 °C with 50 mL/L CO2, and images were captured under a microscope at 100 × magnification 48 h after the scratch was inflicted.
To perform the Transwell invasion assay, the Matrigel was thawed overnight at 4 °C. The Matrigel was diluted in the pre-cooled medium at a 1:9 ratio. The upper chamber of the Transwell was coated with 40 μL of the diluted Matrigel and allowed to solidify at 37 °C for 2-4 h. Subsequently, the cells were seeded into the upper chamber, and the Transwell was placed into a 24-well plate containing 600 μL of 100 mL/L FBS medium. After incubating at 37 °C for 24 h, the invading cells were fixed, stained, and counted under a microscope at 200 × magnification.
TCGA database was used as the source of survival information for patients with HCC. Statistical analyses were performed using R software, with the SurvMiner package used as appropriate. Univariate and multivariate Cox regression analyses were used to identify the independent prognostic variables. Subsequently, a prognostic nomogram was constructed using the R software, which incorporated variables such as age, sex, tumor grade, TNM stage, and XPO5 expression. The predictive efficacy of the nomogram was validated using receiver operating characteristics and calibration curves.
To clarify the different biological processes between the high and low XPO5 expression groups, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. Additionally, we used Gene Set Enrichment Analysis (GSEA) to identify relevant signaling pathways. The database versions used in this study were GO version 2022-06-15, KEGG Release 103.1, and GSEA v4.3.0.
R software (version 4.0.5) was used to analyze all public databases. Pearson’s chi-squared test was used for categorical variables. The statistical significance of the difference in overall survival (OS) between the high- and low-expression groups was assessed using Kaplan-Meier analysis and the log-rank test. The prognostic factors were evaluated using univariate and multivariate Cox regression analyses. In this study, the Mann-Whitney U test was used to compare the expression levels of immune cells and to evaluate the activation scores of immune pathways using GSEA.
All in vitro experimental data were processed, analyzed, and graphically presented using GraphPad Prism 9 (version 9.4.0). The figures were created using Adobe Illustrator 2022 (Version 2022). Data are presented as mean ± SD. Statistical differences among groups were evaluated using one-way analysis of variance, followed by Tukey’s post-hoc test.
The Benjamini-Hochberg procedure was employed to control the false discovery rate and mitigate the risk of Type I errors due to multiple hypothesis testing.
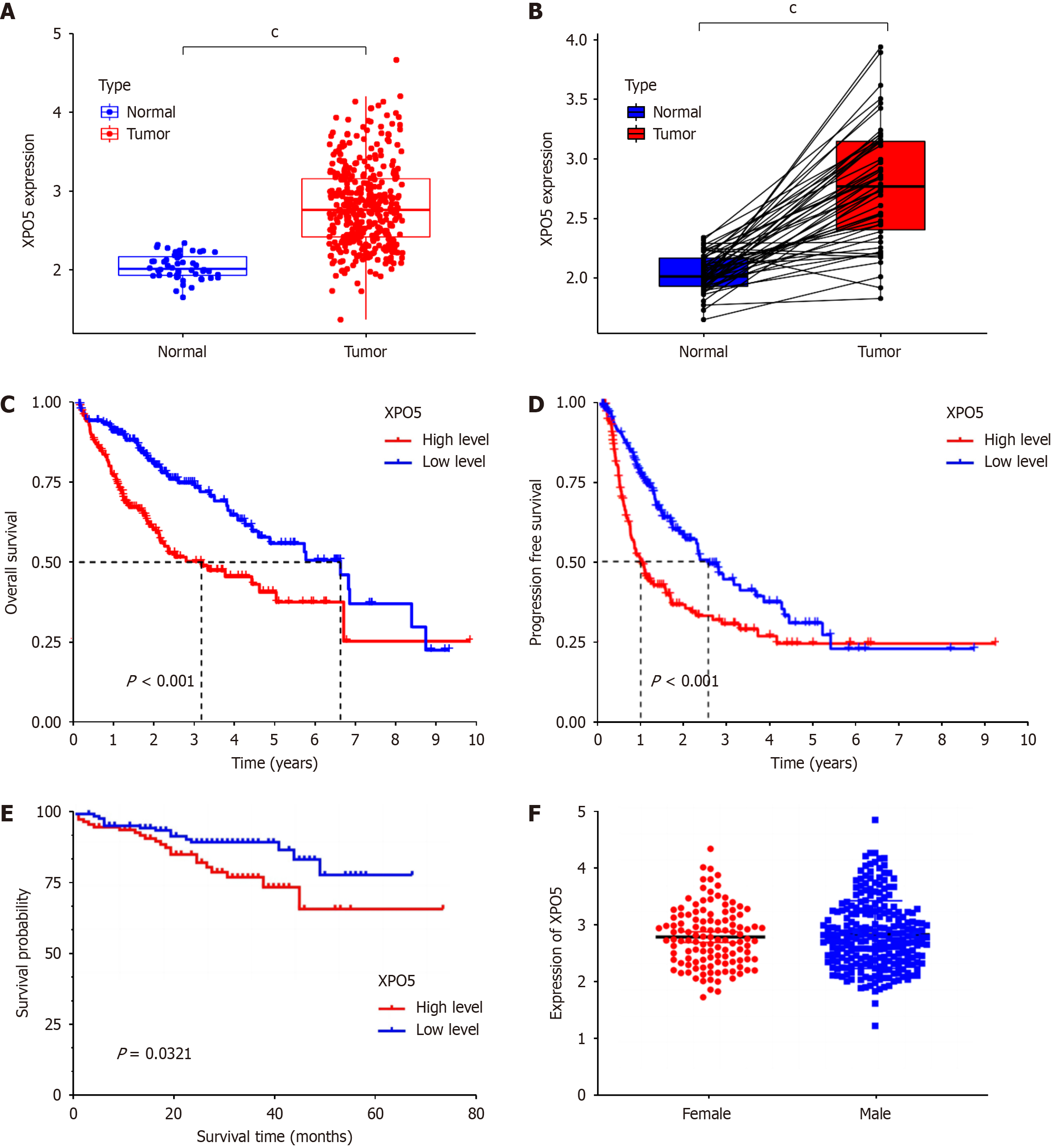
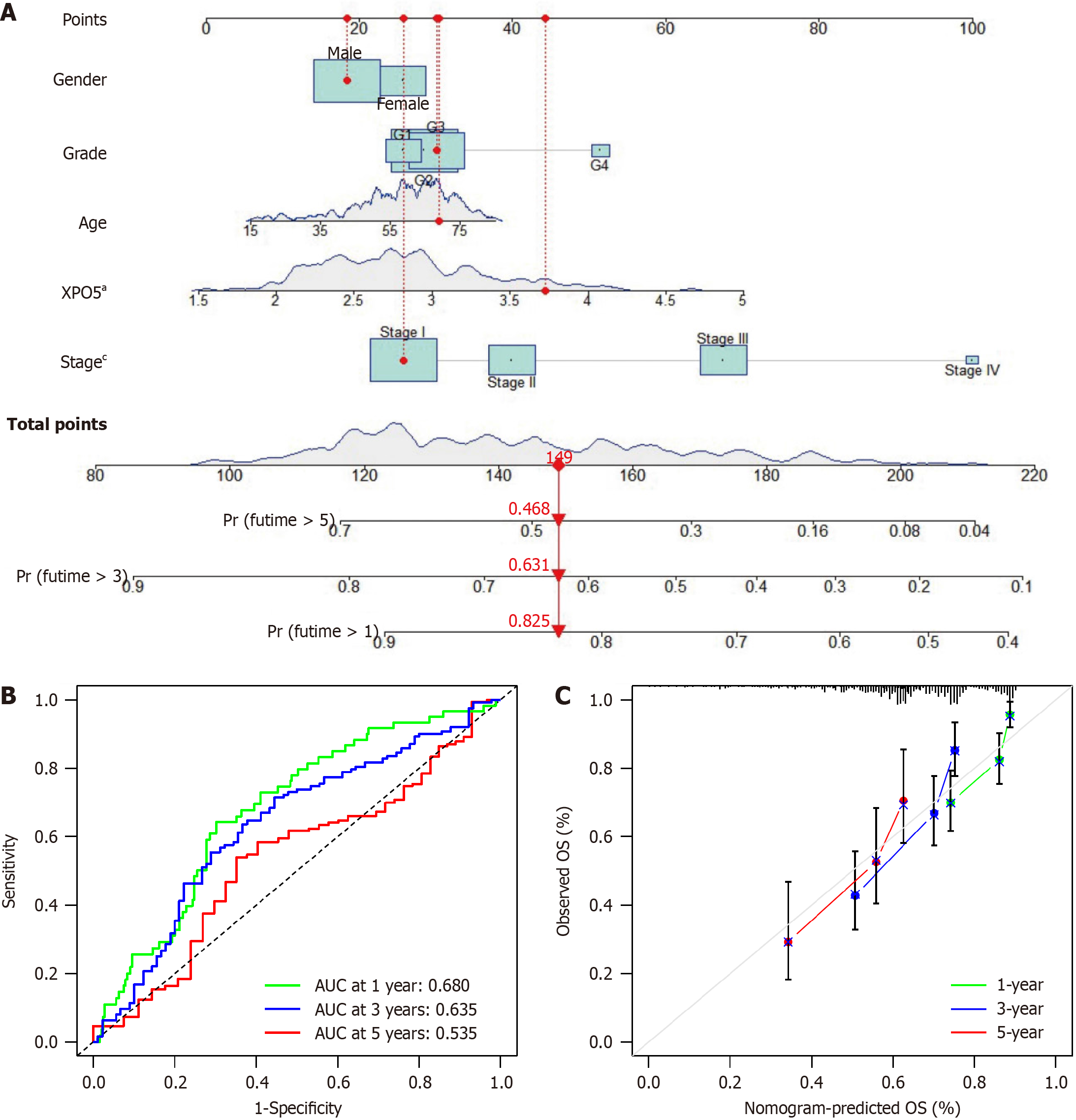
After analyzing the dataset from TCGA database regarding the XPO5 gene, we found that XPO5 is overexpressed in many cancers. The results of the t-test of XPO5 expression levels in HCC and normal liver tissues revealed significant upregulation of XPO5 expression in cancerous tissues (P < 0.001; Figure 1A and B). High XPO5 expression was associated with worse OS and progression-free survival outcomes (Figure 1C and D). This was confirmed by the ICGC LIRI-JP cohort study of 231 patients with HCC, which demonstrated reduced survival in the group with high XPO5 expression (P = 0.0321; Figure 1E). No significant sex differences were observed in XPO5 expression (P = 0.5021; Figure 1F). We developed a prognostic nomogram to predict the 1-, 3-, and 5-year survival probabilities of patients with HCC. The nomogram incorporates sex, grade, age, XPO5 expression, and tumor stage. This tool calculates the survival probabilities by summing the scores for each variable. The AUC values for the 1-, 3-, and 5-year outcomes were 0.680, 0.635, and 0.535, respectively, indicating good predictive accuracy. Calibration plots confirmed the validity of the nomogram (Figure 2).
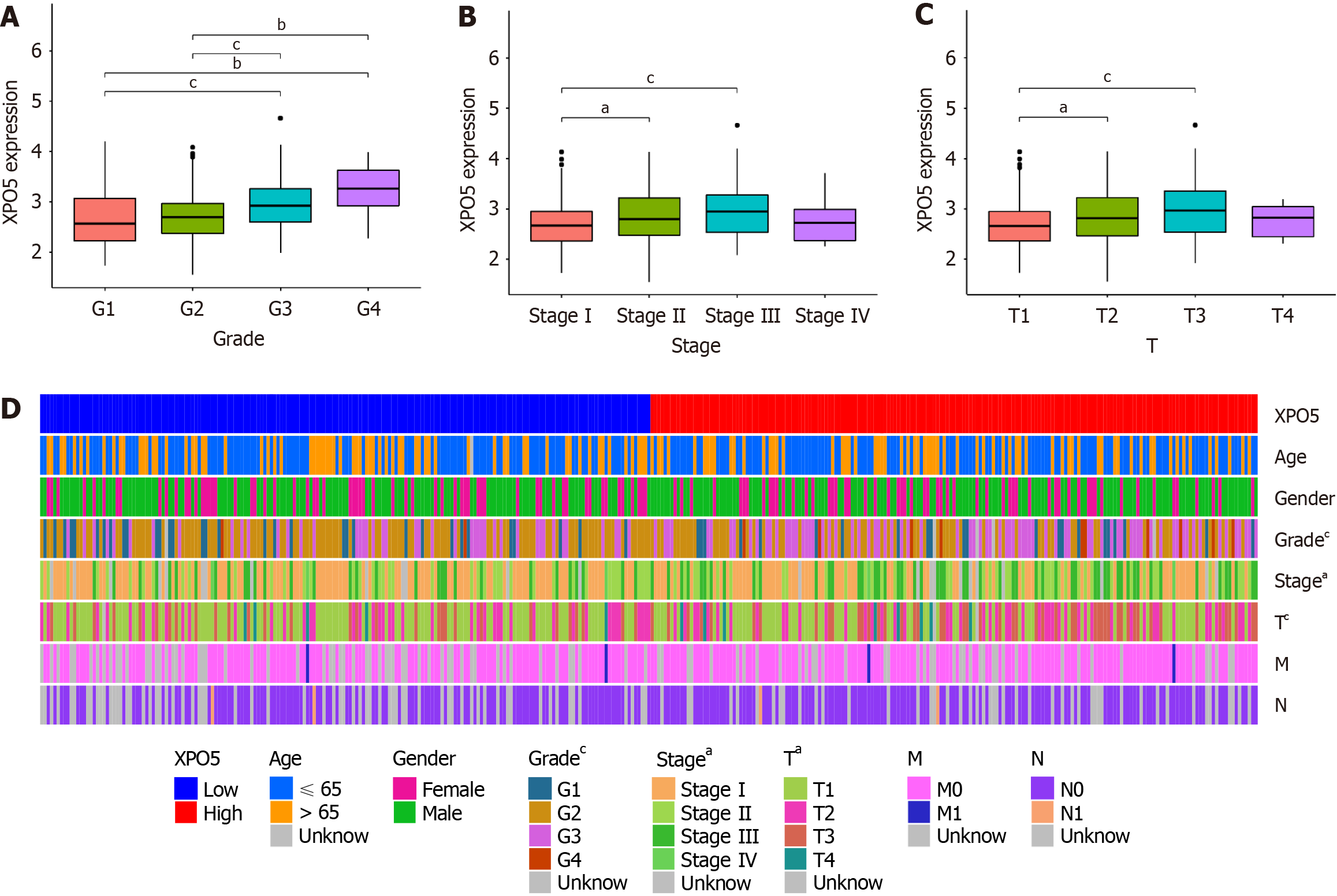
Further examination revealed strong correlations between high XPO5 expression and adverse clinical parameters, including grade, stage, and T stage (Figure 3A-C). These correlations are depicted in a heat map along with other clinicopathological features (Figure 3D). This comprehensive analysis emphasizes XPO5’s potential as a prognostic biomarker in HCC, affecting survival outcomes and correlating it with key clinical characteristics. Our findings indicate that XPO5 plays a significant role in HCC prognosis and patient management, providing opportunities for further research and clinical applications.
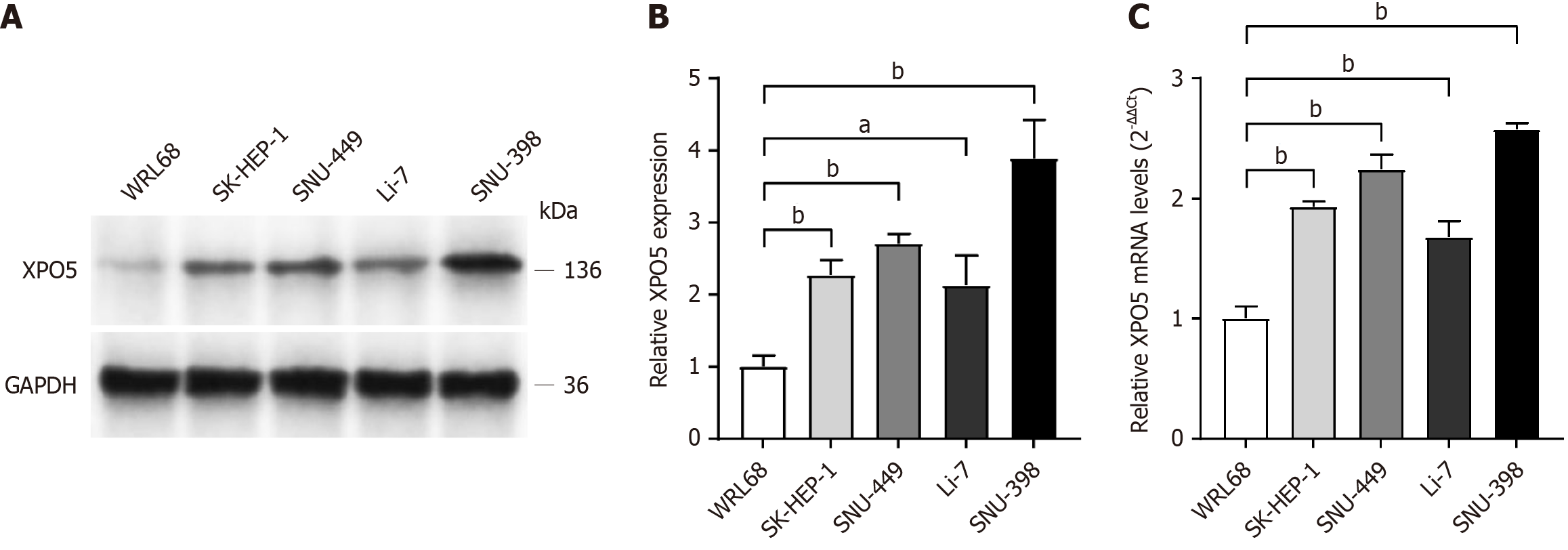
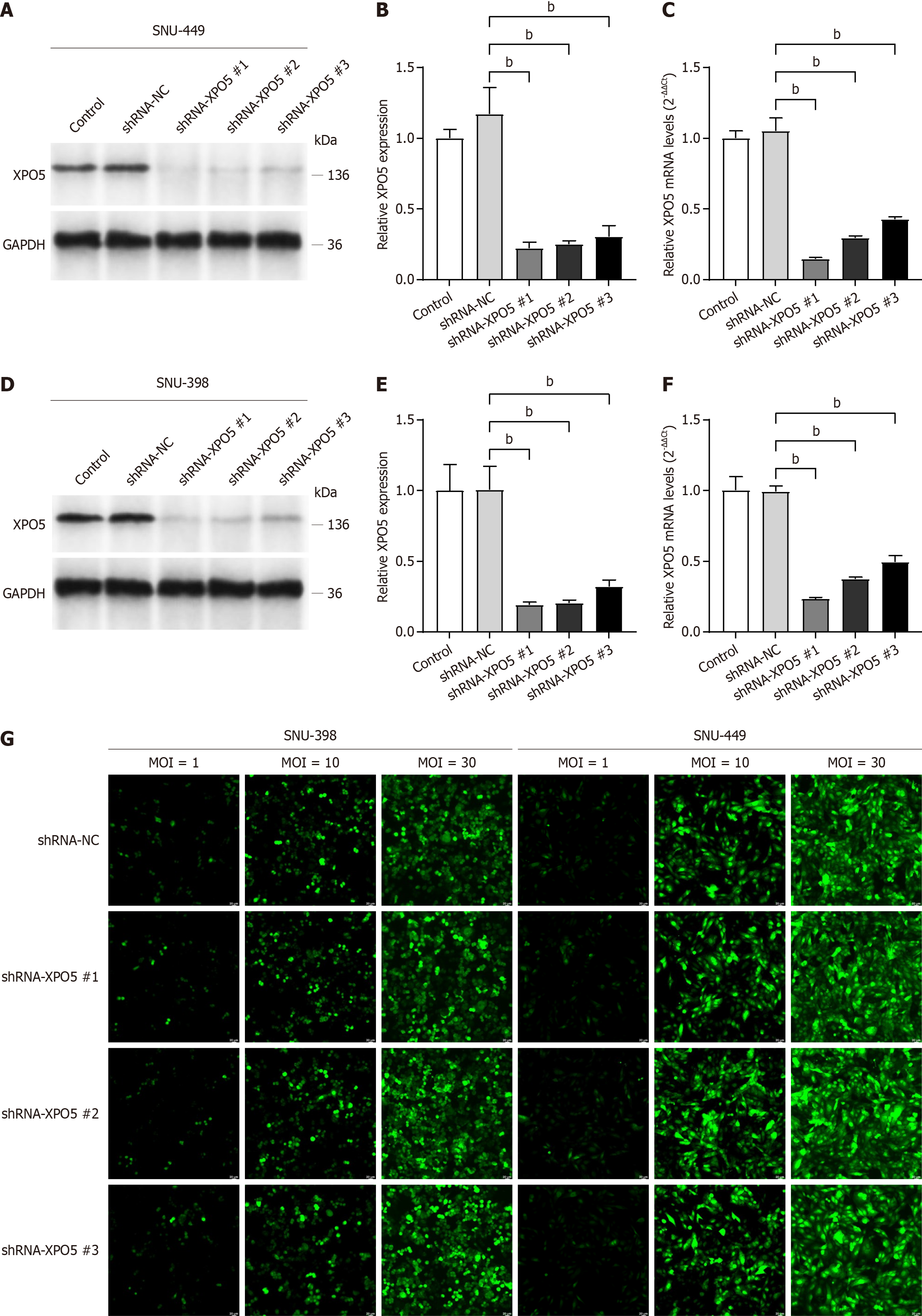
In the later stages of database exploration and data analysis, we conducted in vitro experiments to validate the initial findings. After reviewing the relevant literature and consulting the appropriate websites, we selected four HCC cell lines (SK-HEP-1, SNU-449, Li-7, and SNU-398) and one normal liver cell line (WRL68). Comparative analysis showed a significant increase in both XPO5 protein and mRNA expression levels in HCC cell lines compared to those in the normal liver cell line WRL68.
Based on these distinct expression patterns, we selected two cell lines, SNU-449 and SNU-398, which had markedly different XPO5 expression levels, for subsequent functional validation (Figure 4). Transfection efficiency was thoroughly verified at the end of the process, enabling us to select the plasmid with the optimal transfection effect for subsequent experiments (Figure 5). To ensure effective lentiviral transfection, we meticulously mapped the MOI values (Figure 5G).
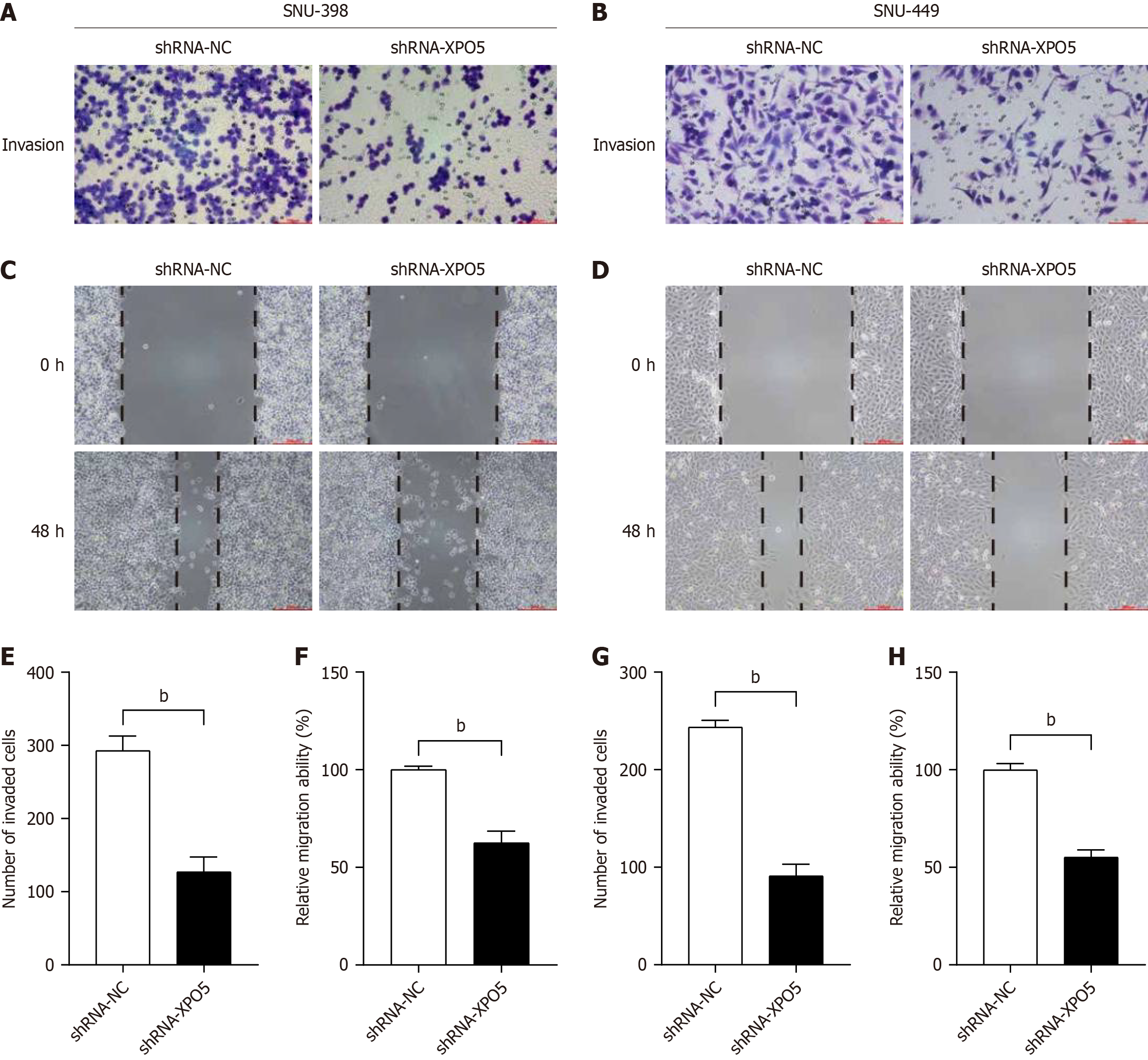
Our XPO5 knockdown experiments yielded compelling results, showing significant inhibition of HCC cell proliferation. A comparative analysis was conducted with the control group (shRNA-NC), which demonstrated that the XPO5-knockdown groups for SNU-398 and SNU-449 cells exhibited significant reductions in both invasive and migratory capacities (Figure 6). These findings suggest that the knockdown of XPO5 significantly suppresses the growth, proliferation, and metastasis of HCC cells.
In conclusion, our in vitro experiments provided evidence supporting the role of XPO5 in modulating cellular processes in HCC. These findings provide insight into the potential therapeutic implications of targeting XPO5 in HCC.
Univariate and multivariate Cox regression analyses were conducted to evaluate the potential of risk variables as individual prognostic indicators. In univariate analysis, elevated XPO5 expression [P = 0.001, hazard ratio (HR): 1.776, 95% confidence interval (CI): 1.253-2.518] and advanced tumor stage (P < 0.001, HR: 1.680, 95%CI: 1.369-2.062) were identified as robust indicators associated with poorer survival outcomes (Supplementary Figure 1A).
The prognostic relevance of XPO5 expression (P = 0.013, HR: 1.606, 95%CI: 1.103-2.339) and tumor stage (P < 0.001, HR: 1.639, 95%CI: 1.328-2.022) in relation to poorer OS was further confirmed using multivariate Cox regression analysis (Supplementary Figure 1B).
Upon examination of the Cox regression model, the P values of the likelihood ratio test, Wald test, and score (log-rank) test were all less than 0.001. This indicates that the model was verified by the residual analysis, as evidenced by the mean value of the residuals, which was -0.08794533. Additionally, the autocorrelation plot of the residuals demonstrated no apparent correlation between the data residuals (Supplementary Figure 1C). The lack of correlation between the variables indicated that the Cox model had a superior overall fit to the data.
Our study clarifies the intricate correlation between XPO5 expression and immune cell dynamics in HCC. We found significant associations with CD4 memory resting T cells, monocytes, and resting mast cells, suggesting the immunomodulatory roles of XPO5 (Supplementary Figure 2). Further analysis revealed positive correlations between Macrophages M0 and activated CD4 memory T cells, suggesting the involvement of XPO5 in immune activation. In contrast, monocytes and CD4 memory resting T cells displayed inverse correlations, indicating nuanced interactions within the HCC TME (Supplementary Figure 2B and C).
These correlations were then validated (Supplementary Figure 3A). Further analysis showed that XPO5 was associated with immune checkpoint molecules in various cancers (Supplementary Figure 3B) and positively correlated with tumor mutation burden (r = 0.27, P < 0.05) (Supplementary Figure 4A). TME analysis revealed that the low XPO5 expression group had higher stromal, immune, and ESTIMATE scores, indicating the impact of XPO5 on the TME (Supplementary Figure 4B). Stratification by PD1 and CTLA4 status showed significant differences in the IPS between the XPO5 expression groups (Supplementary Figure 5A), with a negative correlation between XPO5 expression and chemotherapeutic sensitivity (Supplementary Figure 5B).
This study revealed the role of XPO5 in immune interactions, prognosis, and treatment response in HCC. These findings offer insights for future research and the identification of potential therapeutic targets.
In this study on HCC, we evaluated the performance of four machine learning algorithms in predicting patient clinical outcomes: Bayesian classifier, neural network, SVM, and Decision Tree C5.0. The Decision Tree C5.0 algorithm was the most effective, achieving an accuracy rate of 95.5% in the training set and maintaining the highest accuracy of 92.0% in the validation set (Supplementary Figure 6A and B). The Decision Tree C5.0 algorithm had the highest accuracy rate at 93.2%, followed by SVM at 91.4%, Bayesian classifier at 86.5%, and neural network at 85.5%.
Additionally, sensitivity and specificity analysis, as represented in the AUC plots (Supplementary Figure 6C and D), confirmed the superior predictive capability of the Decision Tree C5.0 algorithm during the training and validation phases. Our findings highlight the robustness and reliability of the Decision Tree C5.0 algorithm in the prognostic prediction of HCC. This makes it an invaluable asset for clinical decision-making and enhances patient care through optimized treatment strategies.
Our investigation using TCGA dataset revealed varying levels of XPO5 expression across multiple cancer types compared to adjacent normal tissues. XPO5 expression significantly increased in cancers such as lung squamous cell carcinoma, HCC, and lung adenocarcinoma, highlighting its prominence in these tumors compared to their normal counterparts. Notably, KICH and KIRC cells exhibited decreased XPO5 expression (Supplementary Figure 7). Further analysis, supported by immunohistochemistry data from the Human Protein Atlas, showed that XPO5 was substantially upregulated in the nucleus and cytoplasm of tumor cells, in contrast to the para-cancer and HCC tissues (Supplementary Figure 8). These findings highlight the complexity and heterogeneity of XPO5 expression in the cancer landscape and emphasize its potential role in cancer biology. This study provides a foundation for understanding the effects of XPO5 and suggests potential avenues for future research to explore its mechanistic roles in cancer progression and therapeutic targeting.
HCC is a leading cause of cancer-related mortality worldwide[13]. The prognosis of HCC is challenging because of its complex etiology[14]. This study provides an in-depth examination of the molecular characteristics, oncogenic potential, prognostic significance, and influence of XPO5 on immune response, and drug sensitivity in HCC. The role of XPO5 as an oncogenic factor has been emphasized by its upregulation in various cancers in TCGA dataset analyses[15-17]. The utility of this biomarker in the diagnosis and prognosis of HCC can be confirmed using immunohistochemical assays[18,19]. Survival analysis using TCGA and ICGC databases showed that high XPO5 expression was associated with poor survival outcomes. This contradicts a previous study from 2016 but highlights the novelty of our findings[20].
Our study investigated the association between XPO5 and clinical variables, excluding age, sex, and gender, indicating its broad impact on outcomes in patients with HCC. Nomogram models are applicable in a wide range of contexts, allowing for the integration of multiple relevant clinical variables to predict disease outcomes[21-24]. We used a nomogram model to identify XPO5 and histopathological stage as independent risk factors, demonstrating the model’s predictive accuracy, especially for short-term prognostications.
Bioinformatics analyses, which included GO and KEGG pathways, revealed that XPO5 is involved in crucial cellular processes and may have adverse effects on outcomes in patients with HCC. Additionally, we explored the correlation between XPO5 and various immune cells to uncover its intricate connection with immune modulation in HCC. Our findings generally align with the literature, indicating a positive correlation between high TME scores and better HCC prognosis despite the mixed results in database studies regarding tumor mutation burden and TME scores[25-27]. We also investigated the impact of the expression of specific genes, including XPO5, on drug responsiveness in HCC. High levels of XPO5 expression decreased the effectiveness of immune checkpoint therapy but increased sensitivity to certain chemotherapeutic agents, such as MG-132 and paclitaxel. This finding suggests the complex role of HCC treatment strategies. To validate these findings, we conducted in vitro experiments using HCC cell lines and found that reducing XPO5 expression significantly impaired cell proliferation, migration, and invasion. This study highlights the potential of XPO5 as a therapeutic target for HCC. Machine learning is a commonly utilized tool for the diagnosis and prediction of various diseases and cancers[28-30], which confirmed the predictive value of XPO5 in distinguishing HCC from noncancerous tissues in our study. The Decision Tree C5.0 algorithm exhibited superior performance. These computational findings support our experimental and analytical results and provide a complete understanding of the significance of XPO5 in HCC research.
Our study, which relied on public databases and included potential sample variability, identified XPO5 as a crucial biomarker of HCC prognosis and treatment. This conclusion was supported by robust in vitro and computational validations. Future studies should expand on these findings to validate XPO5’s role in HCC therapeutics and diagnostics. This will contribute to a better understanding of the molecular underpinnings of HCC and pave the way for the development of novel therapeutic approaches.
The limitations of public databases stem from varied data collection standards, methodologies, and diverse sample origins (geographic locations, ethnicities, and socioeconomic backgrounds). These factors may influence the disease progression and treatment responses. In the future, we intend to conduct a pooled comprehensive evaluation and analysis of the data from various studies with the objective of enhancing the statistical power and generalizability of the results. Furthermore, in vivo experiments will be conducted to validate this study when feasible.
This study highlights the importance of XPO5 expression in the prognosis of HCC. Elevated XPO5 Level is an independent prognostic indicator, with higher expression correlating with poorer patient outcomes. Further analysis validated the prognostic significance of XPO5 in HCC, suggesting its potential as a diagnostic and therapeutic biomarker. In summary, our study highlights the potential of XPO5 as a focal point in oncological research for the diagnosis and treatment of HCC through the integration of data analytics and in vitro experiments.
We thank all the authors for their contributions in this study. I would like to express my sincere thanks to the staff of TCGA and ICGC databases and the patients who source the data. Their contributions to science are incalculable.
| 1. | Kinsey E, Lee HM. Management of Hepatocellular Carcinoma in 2024: The Multidisciplinary Paradigm in an Evolving Treatment Landscape. Cancers (Basel). 2024;16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 36] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64104] [Article Influence: 16026.0] [Reference Citation Analysis (174)] |
| 3. | Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10573-10583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 294] [Cited by in RCA: 381] [Article Influence: 38.1] [Reference Citation Analysis (6)] |
| 4. | Chaiteerakij R, Addissie BD, Roberts LR. Update on biomarkers of hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2015;13:237-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 5. | Zhang X, Liu F, Yang F, Meng Z, Zeng Y. Selectivity of Exportin 5 binding to human precursor microRNAs. RNA Biol. 2021;18:730-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 6. | Wu K, He J, Pu W, Peng Y. The Role of Exportin-5 in MicroRNA Biogenesis and Cancer. Genomics Proteomics Bioinformatics. 2018;16:120-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 7. | Shao Y, Shen Y, Zhao L, Guo X, Niu C, Liu F. Association of microRNA biosynthesis genes XPO5 and RAN polymorphisms with cancer susceptibility: Bayesian hierarchical meta-analysis. J Cancer. 2020;11:2181-2191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Shigeyasu K, Okugawa Y, Toden S, Boland CR, Goel A. Exportin-5 Functions as an Oncogene and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23:1312-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 9. | Melo SA, Esteller M. A precursor microRNA in a cancer cell nucleus: get me out of here! Cell Cycle. 2011;10:922-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 2019;19:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 614] [Cited by in RCA: 552] [Article Influence: 92.0] [Reference Citation Analysis (0)] |
| 11. | Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284:603-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 486] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 12. | Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019;19:281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 288] [Cited by in RCA: 539] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 13. | Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, Negri E. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 477] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 14. | Beumer BR, Buettner S, Galjart B, van Vugt JLA, de Man RA, IJzermans JNM, Koerkamp BG. Systematic review and meta-analysis of validated prognostic models for resected hepatocellular carcinoma patients. Eur J Surg Oncol. 2022;48:492-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Khan M, Khan Z, Uddin Y, Mustafa S, Shaukat I, Pan J, Höti N. Evaluating the Oncogenic and Tumor Suppressor Role of XPO5 in Different Tissue Tumor Types. Asian Pac J Cancer Prev. 2018;19:1119-1125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 16. | Wen J, Gao Q, Wang N, Zhang W, Cao K, Zhang Q, Chen S, Shi L. Association of microRNA-related gene XPO5 rs11077 polymorphism with susceptibility to thyroid cancer. Medicine (Baltimore). 2017;96:e6351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Leaderer D, Hoffman AE, Zheng T, Fu A, Weidhaas J, Paranjape T, Zhu Y. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int J Mol Epidemiol Genet. 2011;2:9-18. [PubMed] |
| 18. | Hu X, Bao M, Huang J, Zhou L, Zheng S. Identification and Validation of Novel Biomarkers for Diagnosis and Prognosis of Hepatocellular Carcinoma. Front Oncol. 2020;10:541479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Debes JD, Romagnoli PA, Prieto J, Arrese M, Mattos AZ, Boonstra A; On Behalf Of The Escalon Consortium. Serum Biomarkers for the Prediction of Hepatocellular Carcinoma. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 20. | Li Y, Wang X, He B, Cai H, Gao Y. Downregulation and tumor-suppressive role of XPO5 in hepatocellular carcinoma. Mol Cell Biochem. 2016;415:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Yang M, Du C, Zhang Q, Ma Q, Li R. Nomogram Model for Predicting Hematoma Expansion in Spontaneous Intracerebral Hemorrhage: Multicenter Retrospective Study. World Neurosurg. 2020;137:e470-e478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Zhang B, Liu Q, Zhang X, Liu S, Chen W, You J, Chen Q, Li M, Chen Z, Chen L, Dong Y, Zeng Q, Zhang S. Clinical Utility of a Nomogram for Predicting 30-Days Poor Outcome in Hospitalized Patients With COVID-19: Multicenter External Validation and Decision Curve Analysis. Front Med (Lausanne). 2020;7:590460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Xu J, Zuo Y, Sun J, Zhou M, Dong X, Chen B. Application of clinical nomograms to predicting overall survival and event-free survival in multiple myeloma patients: Visualization tools for prognostic stratification. Front Public Health. 2022;10:958325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 24. | Ren J, Sun P, Wang Y, Cao R, Zhang W. Construction and validation of a nomogram for patients with skin cancer. Medicine (Baltimore). 2021;100:e24489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 25. | Lin P, Wen DY, Chen G, Dang YW, He Y, Yang H. Predictive value of hypoxia, metabolism and immune factors for prognosis in hepatocellular carcinoma: a retrospective analysis and multicenter validation study. J Cancer. 2020;11:4145-4156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Huang S, Zhang J, Lai X, Zhuang L, Wu J. Identification of Novel Tumor Microenvironment-Related Long Noncoding RNAs to Determine the Prognosis and Response to Immunotherapy of Hepatocellular Carcinoma Patients. Front Mol Biosci. 2021;8:781307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 27. | Zhang FP, Huang YP, Luo WX, Deng WY, Liu CQ, Xu LB, Liu C. Construction of a risk score prognosis model based on hepatocellular carcinoma microenvironment. World J Gastroenterol. 2020;26:134-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 28. | Rabiei R, Ayyoubzadeh SM, Sohrabei S, Esmaeili M, Atashi A. Prediction of Breast Cancer using Machine Learning Approaches. J Biomed Phys Eng. 2022;12:297-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 29. | Gaur K, Jagtap MM. Role of Artificial Intelligence and Machine Learning in Prediction, Diagnosis, and Prognosis of Cancer. Cureus. 2022;14:e31008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |