Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2842
Revised: March 10, 2024
Accepted: April 15, 2024
Published online: June 15, 2024
Processing time: 154 Days and 11 Hours
Gastrointestinal neoplasm (GN) significantly impact the global cancer burden and mortality, necessitating early detection and treatment. Understanding the evolution and current state of research in this field is vital.
To conducts a comprehensive bibliometric analysis of publications from 1984 to 2022 to elucidate the trends and hotspots in the GN risk assessment research, focusing on key contributors, institutions, and thematic evolution.
This study conducted a bibliometric analysis of data from the Web of Science Core Collection database using the "bibliometrix" R package, VOSviewer, and CiteSpace. The analysis focused on the distribution of publications, contributions by institutions and countries, and trends in keywords. The methods included data synthesis, network analysis, and visualization of international collaboration networks.
This analysis of 1371 articles on GN risk assessment revealed a notable evolution in terms of research focus and collaboration. It highlights the United States' critical role in advancing this field, with significant contributions from institutions such as Brigham and Women's Hospital and the National Cancer Institute. The last five years, substantial advancements have been made, representing nearly 45% of the examined literature. Publication rates have dramatically increased, from 20 articles in 2002 to 112 in 2022, reflecting intensified research efforts. This study underscores a growing trend toward interdisciplinary and international collaboration, with the Journal of Clinical Oncology standing out as a key publication outlet. This shift toward more comprehensive and collaborative research methods marks a significant step in addressing GN risks.
This study underscores advancements in GN risk assessment through genetic analyses and machine learning and reveals significant geographical disparities in research emphasis. This calls for enhanced global collaboration and integration of artificial intelligence to improve cancer prevention and treatment accuracy, ultimately enhancing worldwide patient care.
Core Tip: This study offers a novel bibliometric analysis of global trends in gastrointestinal neoplasms risk assessment from 1984 to 2022. It reveals a pivotal shift towards genetic insights and machine learning in risk assessment, highlighting the leading roles of the United States and China in research contributions. The findings demonstrate significant geographical disparities in research focus and methodologies, underscoring the need for enhanced global collaboration. This analysis not only maps the current landscape but also sets a direction for future research, emphasizing the integration of advanced technologies and international cooperation in the field.
- Citation: Fu QQ, Ma L, Niu XM, Zhao HX, Ge XH, Jin H, Yu DH, Yang S. Trends and hotspots in gastrointestinal neoplasms risk assessment: A bibliometric analysis from 1984 to 2022. World J Gastrointest Oncol 2024; 16(6): 2842-2861
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2842.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2842
Gastrointestinal neoplasms (GNs) represent a heterogeneous cohort of malignancies that manifest through a spectrum of etiologies, pathophysiological pathways, and clinical outcomes, significantly exacerbating the global burden of cancer morbidity and mortality. According to the Global Cancer Statistics Report 2024, although the incidence of GNs has exhibited a downward trend since 2011/2012, there has been an annual increase of 1%-2% in incidence rates among individuals younger than 55 years. This trend positions colorectal cancer as the second predominant cause of cancer-related fatalities worldwide[1]. This alarming statistic accentuates the imperative health challenges posed by GNs, thereby underscoring the need for the development of comprehensive risk assessment methodologies. Such methodologies should adequately reflect the complex nature of these malignancies by incorporating considerations of genetic predispositions, environmental contributors, lifestyle factors, and their interactions.
Historically, the assessment of GN risk has predominantly leveraged epidemiological and clinical data[2-4]. Although these conventional approaches have yielded significant insights, they frequently do not fully account for the complex interplay of factors that influence GN risk. In contrast, recent advancements in the fields of biostatistics, bioinformatics, and systems biology have introduced a novel paradigm for risk assessment[5,6]. These cutting-edge methodologies enable the amalgamation of heterogeneous data types, ranging from genomic to environmental variables, thereby facilitating a more comprehensive and nuanced understanding of GN risk.
Furthermore, the advent of machine learning and artificial intelligence (AI) in the analysis of large-scale health datasets heralds unprecedented possibilities for the prediction and management of GN risks. These technological innovations have the capability to unearth patterns and correlations within the data that may remain obscure to human investigators. Such insights are instrumental in elucidating the multifactorial dimensions of GN risk, ultimately contributing to the advancement of novel preventative and therapeutic strategies[7,8].
Our study addresses the existing gap in synthesizing and analyzing the evolution of GN risk assessment methodologies. Through a comprehensive bibliometric analysis of literature spanning nearly four decades, we aim to trace the progression from traditional epidemiological approaches to contemporary computational models. Our findings highlight a significant shift toward multidisciplinary techniques, reflecting the growing acknowledgment of GNs as complex diseases necessitating comprehensive management strategies.
This bibliometric analysis represents a pioneering effort in the GN research domain by charting the historical development and current landscape of risk assessment methodologies and identifying key trends, emerging focus areas, and research gaps. We propose a novel hypothesis positing that the integration of interdisciplinary approaches-capitalizing on the latest developments in biostatistics, computational modeling, and genomics-will markedly improve the precision and relevance of GN risk assessments.
Our study's overarching goal is to lay the groundwork for future research in GN risk assessment, advocating for a paradigm shift toward more integrative and predictive models. By delineating the trends and emerging areas within the GN risk assessment research, we endeavor to forge new paths in prevention, early detection, and management strategies for GNs. By constructing more precise and predictive GN risk assessment models, we expect to provide substantial benefits to GN patients, including earlier detection, more accurate prognosis evaluation, and more effective prevention strategies, ultimately reducing the disease burden and improving patient outcomes.
Compared with other databases, such as Scopus and PubMed, the Web of Science (WOS) is one of the most frequently utilized academic database sources and is universally acknowledged for its unparalleled comprehensiveness and reliability for use in in bibliometric analyses[9]. For the purpose of this investigation, relevant literature was meticulously searched for and retrieved from the WOS Core Collection database (WOSCC) on October 25, 2022. The WOSCC, along with its encompassing editions-namely, the Science Citation Index Expanded, Social Sciences Citation Index, Conference Proceedings Citation Index-Science, Emerging Sources Citation Index, Conference Proceedings Citation Index–Social Science and Humanities, Arts and Humanities Citation Index, Book Citation Index–Science, Book Citation Index-Social Sciences and Humanities, Index Chemicus, and Current Chemical Reactions-were utilized. The search strategy used the following terms: (1) TS = ("colorectal cancer" OR "intestinal cancer" OR "CRC" OR "colon cancer" OR "colorectal carcinoma"); (2) TS = ("rectal cancer" OR "rectal carcinoma" OR "carcinoma of the rectum" OR "carcinoma of the colon"); (3): TS = ("colorectal neoplasm" OR "colorectal tumor*" OR "colon tumor*" OR "colon neoplasm" OR "sigmoid neoplasm"); and (4) TS = ("risk assessment" OR "risk evaluation" OR "risk appraisal" OR "exposure rate"). The final dataset was defined as (1 OR 2 OR 3) AND 4.
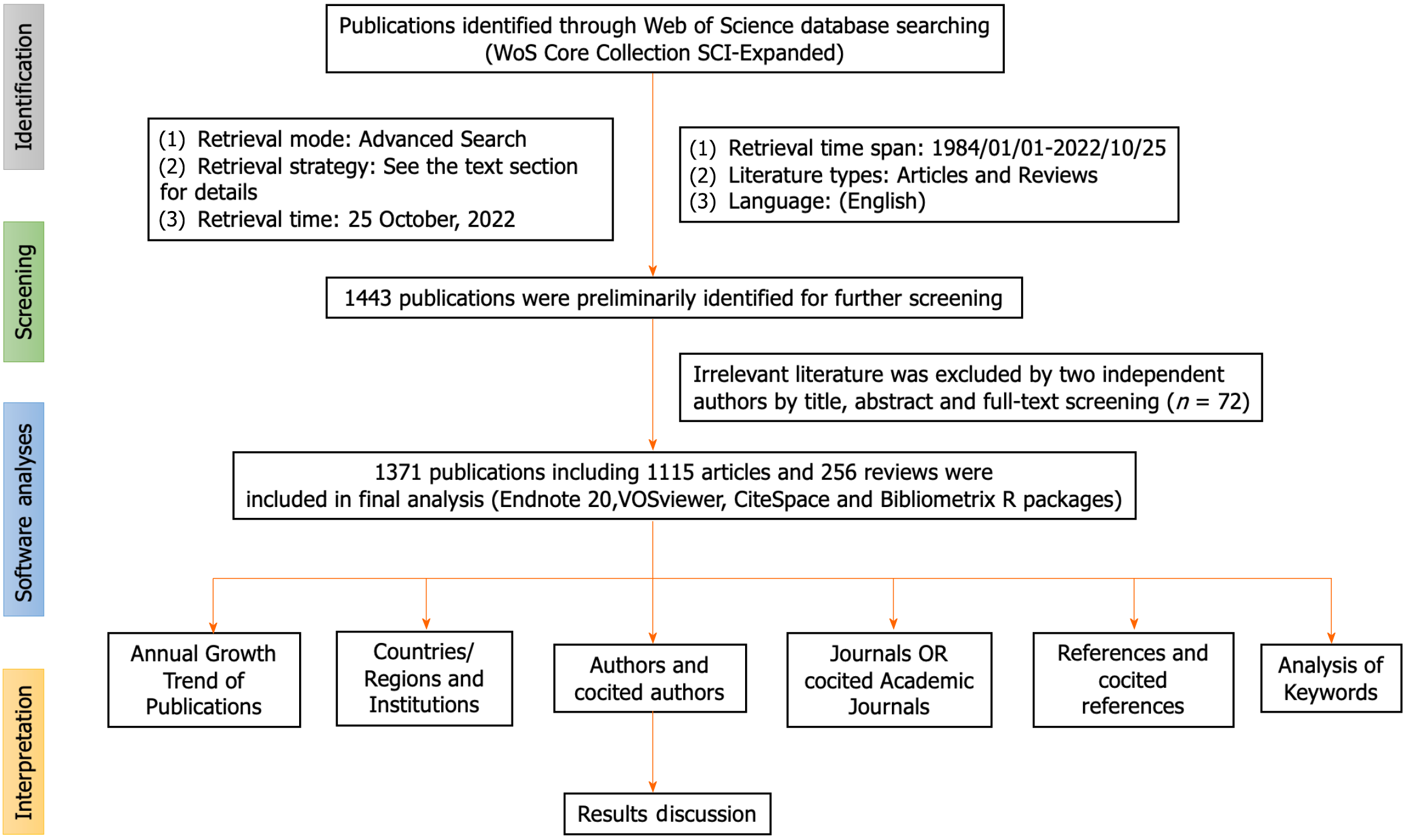
An initial examination of the literature yielded a preliminary pool of 1443 documents identified as potentially eligible for inclusion in this study. Following this, a meticulous screening process was undertaken by two investigators, Yang S and Fu QQ, who performed an exhaustive manual review of the titles, abstracts, and full texts of the literature. This scrupulous examination was aimed at excluding irrelevant materials based on disease typologies, research objectives, therapeutic modalities, outcome indicators, and other pertinent criteria. Following this stringent selection process, a total of 1371 articles and reviews were deemed appropriate for further data analysis. The strategic methodology employed for the systematic compilation and retrieval of relevant literature is visually summarized in Figure 1 and Table 1.
| Parameter | Description |
| Database | Web of Science Core Collection |
| Search period | 25-Oct-22 |
| Search strategy | Keywords related to colorectal cancer and risk assessment, combined as (1 OR 2 OR 3) AND 4 |
| Screening process | Two investigators manually screened titles, abstracts, and full texts, resulting in 1371 articles and reviews selected for analysis |
| Analysis software | VOSviewer (1.6.18), CiteSpace (V6.1.R4), Bibliometrix (4.1.2) |
| Analysis focus | Coauthorship, keyword cooccurrence, international collaboration, cocitation, clustering analysis, citation trends, and research topic evolution over time |
| Visualization Techniques | Node size reflects citation or occurrence frequency; line connections indicate relationships; color variations represent different clusters or average occurrence years |
For the execution of bibliometric and visualization analyses, this study employed three distinct bibliometric software packages: VOSviewer version 1.6.18 (Leiden University, The Netherlands), CiteSpace V6.1.R4 (Drexel University, United States), and the Bibliometrix R package version 4.1.2 (University of Naples Federico II, Italy). VOSviewer, developed by van Eck and Waltman[10], is a complimentary Java-based tool designed for bibliometric mapping and clustering analysis. This research focused on analyzing coauthorship and keyword co-occurrence among countries, institutions, and authors. Typically, the magnitude of the nodes within these visual representations corresponds to the quantity of citations or instances of specific elements, such as authors, countries, institutions, or keywords. The connections denoting coauthorship, cocitation, or co-occurrence are illustrated through lines interlinking the nodes, whereas different clusters or their mean years of occurrence are signified by the coloration of both nodes and lines[11,12].
Moreover, CiteSpace, devised by Professor Synnestvedt et al[13], serves as an additional instrumental software for the visualization and construction of bibliometric networks. Within the context of this analysis, CiteSpace facilitated the visualization of international collaboration among journals and authors, cocitation of references, and cluster analysis. It also aided in identifying the reference literature that garnered the highest citation count over a designated timeframe, which is indicative of a period of heightened popularity within the study's domain.
Furthermore, CiteSpace enabled the examination of citation bursts in the literature, delineating the emergence of new references. This feature allowed for the portrayal of the interrelationship between citing and cited journals through the superimposition of journal dual maps and the creation of a keyword network visualization diagram. This diagram was instrumental in exploring the temporal evolution of research topics. The configuration parameters set for CiteSpace included a time span from 1984 to 2022, with one-year intervals per slice, a selection criterion of the top 30 most cited references, and network pruning options (minimum spanning tree, pruning of sliced networks).
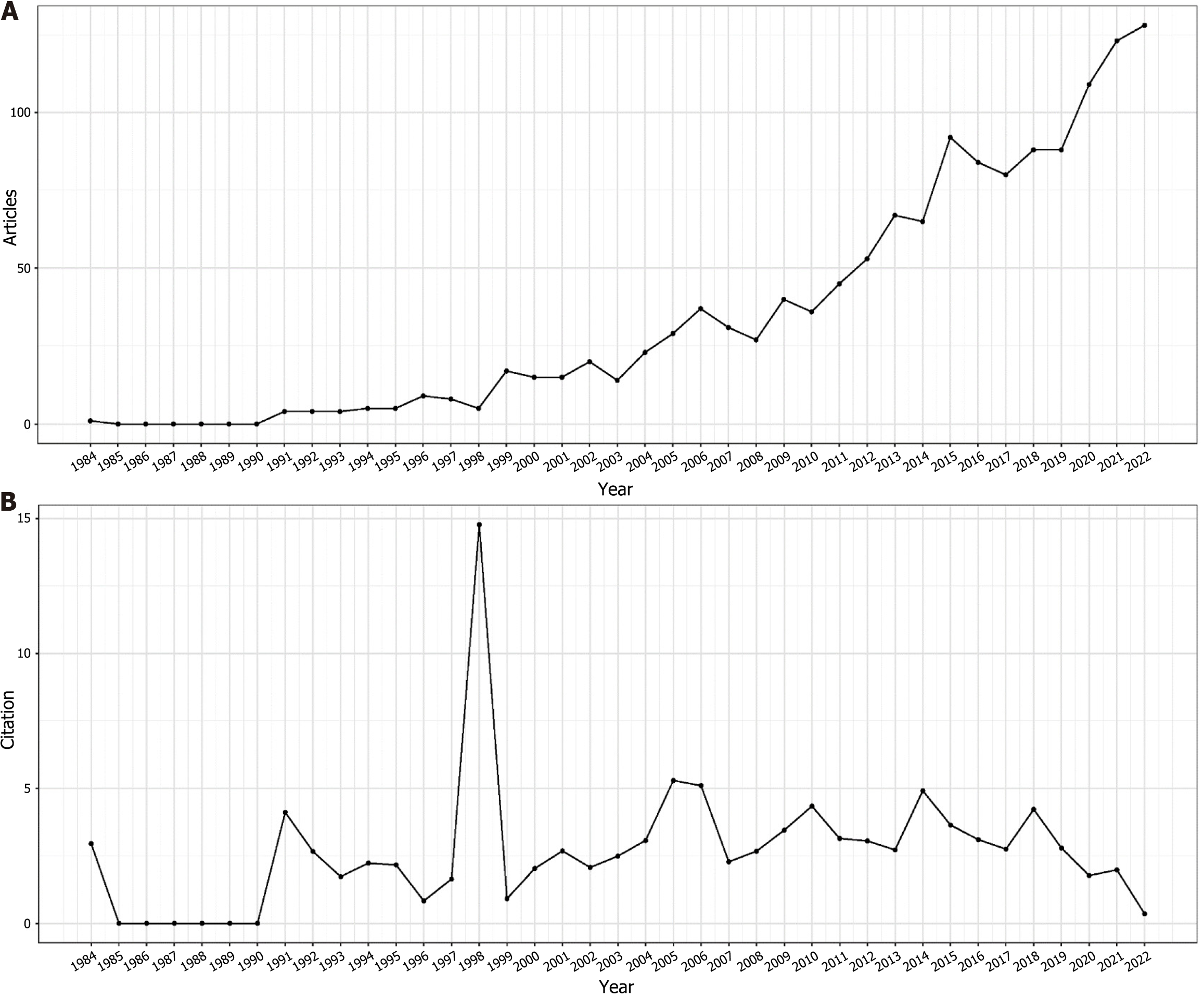
A rigorous analysis was conducted on a corpus of 1443 papers, employing the previously described search methodology. After meticulous review, 72 papers deemed redundant or irrelevant were excluded, resulting in a final sample of 1371 articles for bibliometric analysis. Most of these articles were identified as original research (n = 1115; 81.3%), with the remainder being literature reviews (n = 256; 18.7%). With a specific focus on the area of GN risk assessment, the WOS database contributed a total of 1371 relevant articles. The chronological distribution of these global publications, spanning from 1984 to 2022, is visually presented in Figure 2 highlights the mean annual citation frequencies associated with GN risk assessment research. Notably, significant advancements in this field have occurred in the last five years, accounting for approximately 45% of the total relevant literature. The annual output of global publications increased from 20 articles in 2002 to an impressive count of 112 in 2022. Prior to 2011, the annual publication rate consistently remained below 50, marking a significant increase in research interest after 2012. As of October 25, 2022, a cumulative total of 112 articles have been published within the year, indicating a steady upward trend. The average annual citation count reveals an arc-shaped upward trajectory, peaking in 1998 with an average of 16.62 citations per article, as detailed in Table 2.
| Year | Publications | Mean total citations per year | Mean total citations per article | Year | Publications | Mean total citations per year | Mean total citations per article |
| 2022 | 112 | 0.35 | 0.99 | 2002 | 20 | 2.38 | 47.65 |
| 2021 | 118 | 7.93 | 7.63 | 2001 | 15 | 3.06 | 64.33 |
| 2020 | 109 | 4.41 | 8.83 | 2000 | 15 | 2.3 | 50.67 |
| 2019 | 88 | 5.57 | 16.72 | 1999 | 17 | 1.03 | 23.65 |
| 2018 | 88 | 7.38 | 29.53 | 1998 | 5 | 16.62 | 398.8 |
| 2017 | 80 | 4.4 | 22 | 1997 | 8 | 1.84 | 46 |
| 2016 | 84 | 4.64 | 27.86 | 1996 | 9 | 0.92 | 24 |
| 2015 | 92 | 5.19 | 36.35 | 1995 | 5 | 2.4 | 64.8 |
| 2014 | 65 | 6.75 | 53.97 | 1994 | 5 | 2.46 | 69 |
| 2013 | 67 | 3.63 | 32.66 | 1993 | 4 | 1.91 | 55.5 |
| 2012 | 53 | 3.97 | 39.68 | 1992 | 4 | 2.92 | 87.75 |
| 2011 | 45 | 4 | 43.98 | 1991 | 4 | 4.51 | 139.75 |
| 2010 | 36 | 5.42 | 65.08 | 1990 | 0 | 0 | 121 |
| 2009 | 40 | 4.24 | 55.15 | 1989 | 0 | 0 | 47.65 |
| 2008 | 27 | 3.24 | 45.33 | 1988 | 0 | 0 | 64.33 |
| 2007 | 31 | 2.74 | 41.06 | 1987 | 0 | 0 | 50.67 |
| 2006 | 37 | 6.06 | 96.92 | 1986 | 0 | 0 | 23.65 |
| 2005 | 29 | 6.22 | 105.76 | 1985 | 0 | 0 | 398.8 |
| 2004 | 23 | 3.59 | 64.57 | 1984 | 1 | 3.18 | 46 |
| 2003 | 14 | 2.89 | 54.86 | NA | NA | NA | NA |
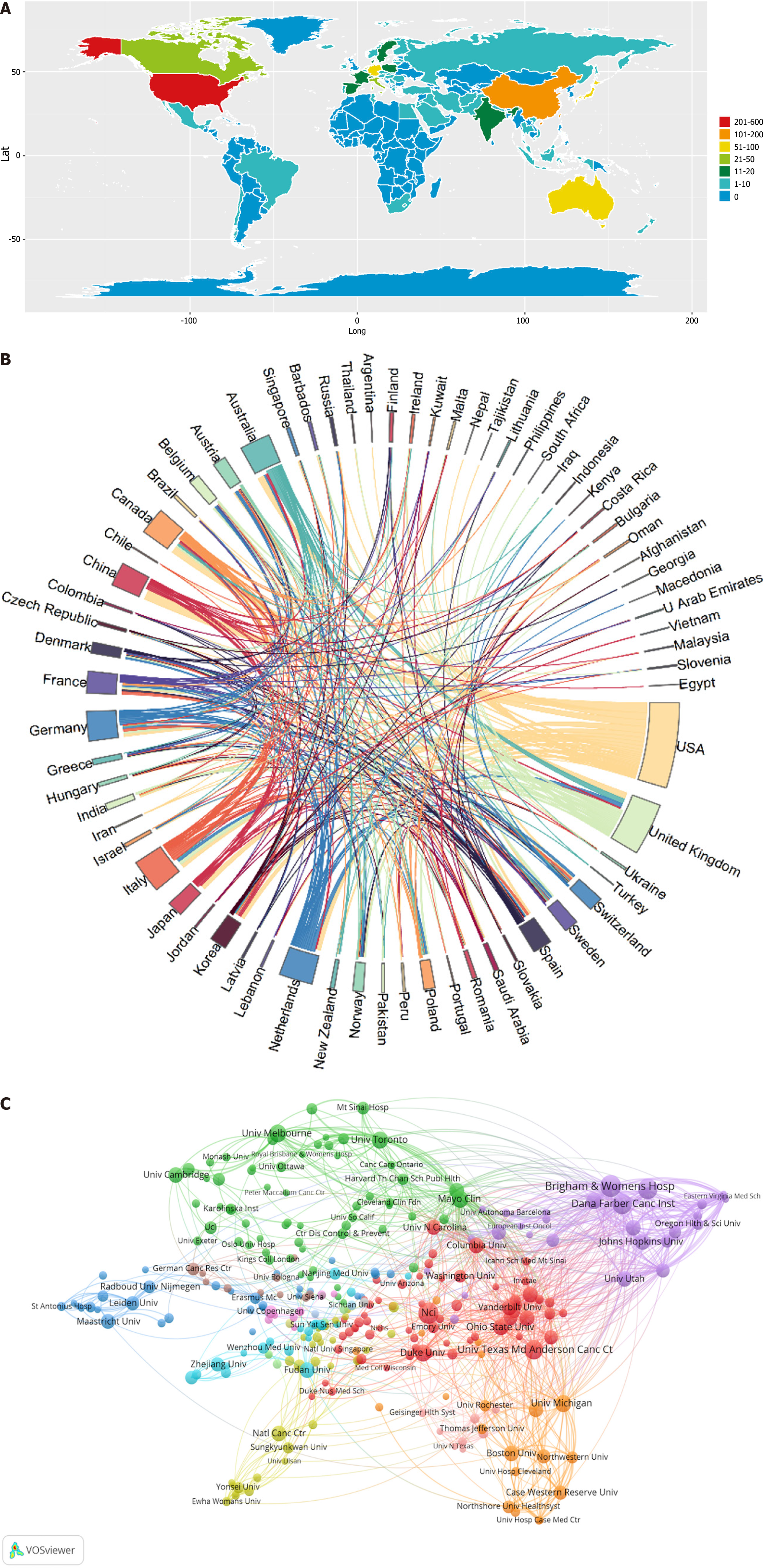
This field has garnered attention, resulting in publications from authors across 60 countries/regions. An examination of the geographical distribution of the global productivity map revealed the top twenty nations/regions contributing to GN risk assessment research (Table 3). When setting the threshold to a minimum of more than one publication, 68 countries were included in our analysis of global collaboration utilizing the Bibliometrix R package (Figure 3A). The United States emerged as a pivotal center of research, demonstrating close collaborative ties with China, Australia, Canada, and the United Kingdom. Notably, Australia and the United Kingdom have also engaged in consistent collaborative efforts in this research field (Figure 3B).
| Rank | Country | Articles | Freq | SCP | MCP | MCP_Ratio |
| 1 | United States | 516 | 0.38025 | 433 | 83 | 0.1609 |
| 2 | China | 199 | 0.14665 | 176 | 23 | 0.1156 |
| 3 | United Kingdom | 74 | 0.05453 | 49 | 25 | 0.3378 |
| 4 | Japan | 65 | 0.0479 | 57 | 8 | 0.1231 |
| 5 | Germany | 54 | 0.03979 | 37 | 17 | 0.3148 |
| 6 | Australia | 52 | 0.03832 | 26 | 26 | 0.5 |
| 7 | Italy | 47 | 0.03464 | 38 | 9 | 0.1915 |
| 8 | Netherlands | 47 | 0.03464 | 37 | 10 | 0.2128 |
| 9 | Canada | 34 | 0.02506 | 19 | 15 | 0.4412 |
| 10 | Korea | 32 | 0.02358 | 30 | 2 | 0.0625 |
| 11 | Spain | 18 | 0.01326 | 12 | 6 | 0.3333 |
| 12 | France | 17 | 0.01253 | 11 | 6 | 0.3529 |
| 13 | Denmark | 16 | 0.01179 | 12 | 4 | 0.25 |
| 14 | India | 15 | 0.01105 | 13 | 2 | 0.1333 |
| 15 | Sweden | 14 | 0.01032 | 13 | 1 | 0.0714 |
| 16 | Poland | 11 | 0.00811 | 6 | 5 | 0.4545 |
| 17 | Switzerland | 11 | 0.00811 | 8 | 3 | 0.2727 |
| 18 | Brazil | 10 | 0.00737 | 9 | 1 | 0.1 |
| 19 | Iran | 10 | 0.00737 | 10 | 0 | 0 |
| 20 | Norway | 10 | 0.00737 | 7 | 3 | 0.3 |
The exploration of risk assessment concerning GNs encompassed analyses from 2178 distinct institutions. Table 4 presents the leading twenty academic entities based on their publication output, emphasizing contributions primarily originating from the United States, a hub for a significant portion of these scientific investigations. Notably, institutions such as Brigham and Women's Hospital, the National Cancer Institute, Harvard University, the Dana-Farber Cancer Institute, and the MD Anderson Cancer Center of the University of Texas were identified as prolific contributors to this field of research.
| Rank | Institutions | Publications | Percentage of 1371 | Total link strength | Total citations | Average citation per paper |
| 1 | Brigham and Womens Hosp | 41 | 2.99 | 168 | 2855 | 69.63 |
| 2 | Nci | 39 | 2.84 | 42 | 2060 | 52.82 |
| 3 | Harvard Univ | 36 | 2.63 | 120 | 3721 | 103.36 |
| 4 | Dana Farber Canc Inst | 35 | 2.55 | 142 | 2635 | 75.29 |
| 5 | Univ Texas Md Anderson Canc Ctr | 30 | 2.19 | 79 | 1074 | 35.8 |
| 6 | Univ Melbourne | 24 | 1.75 | 81 | 759 | 31.63 |
| 7 | Univ Michigan | 23 | 1.68 | 92 | 1042 | 45.3 |
| 8 | Univ Toronto | 23 | 1.68 | 75 | 667 | 29 |
| 9 | Duke Univ | 22 | 1.6 | 75 | 994 | 45.18 |
| 10 | Mayo Clin | 22 | 1.6 | 45 | 1283 | 58.32 |
| 11 | Johns Hopkins Univ | 21 | 1.53 | 104 | 3262 | 155.33 |
| 12 | Univ Washington | 21 | 1.53 | 71 | 1372 | 65.33 |
| 13 | Ohio State Univ | 20 | 1.46 | 53 | 1030 | 51.5 |
| 14 | Columbia Univ | 19 | 1.39 | 102 | 661 | 34.79 |
| 15 | Mem Sloan Kettering Canc Ctr | 19 | 1.39 | 89 | 2076 | 109.26 |
| 16 | Natl Canc Ctr | 19 | 1.39 | 65 | 402 | 21.16 |
| 17 | Stanford Univ | 19 | 1.39 | 47 | 1889 | 99.42 |
| 18 | Univ Cambridge | 19 | 1.39 | 37 | 777 | 40.89 |
| 19 | Univ Utah | 19 | 1.39 | 28 | 2008 | 105.68 |
| 20 | Vanderbilt Univ | 19 | 1.39 | 26 | 971 | 51.11 |
Remarkably, Johns Hopkins University was distinguished as having the highest average citation count per publication, achieving an impressive 155.33 citations. Collectively, the top twenty institutions were responsible for the publication of 490 articles, which represented 35.76% of the total research outputs in this area. Furthermore, the utilization of the VOSviewer for network visualization offered an insightful analysis of institutional collaborations, revealing a graphical depiction that included at least three publications per institution.
As depicted in Figure 3C, the network map consisted of 308 nodes and 3396 links, with Brigham and Women's Hospital total link strength (TLS) = 168, the Dana-Farber Cancer Institute (TLS = 142), and Harvard University (TLS = 120) exhibiting the highest TLS among the foremost institutions. This analysis underscored that collaborative efforts were predominantly interinstitutional rather than international, with Brigham and Women's Hospital demonstrating extensive partnerships with numerous academic and research entities that extent beyond the United States to include European, Canadian, and various other international organizations.
The investigation included a comprehensive analysis of 402 authors and 7991 cocited authors, as delineated in Tables 5 and 6, which highlight the twenty most prolific contributors and cocited authors in the field of GN risk assessment research. The construction of an author network diagram has emerged as a pivotal tool for examining the contributions and connections within this academic domain, thereby enhancing the identification of potential collaborative networks and partners for future inquiries into GN risk assessment.
| Rank | Author | Publications | Percentage of 1371 | Total citations | Average citation per paper |
| 1 | Syngal S | 24 | 1.75 | 2273 | 94.71 |
| 2 | Wang Y | 18 | 1.31 | 178 | 9.89 |
| 3 | Liu Y | 15 | 1.09 | 195 | 13 |
| 4 | Zhang Y | 13 | 0.95 | 145 | 11.15 |
| 5 | Li Y | 12 | 0.88 | 82 | 6.83 |
| 6 | Xu J | 12 | 0.88 | 136 | 11.33 |
| 7 | Orlando LA | 11 | 0.8 | 137 | 12.45 |
| 8 | Walter FM | 10 | 0.73 | 228 | 22.8 |
| 9 | Wang J | 10 | 0.73 | 52 | 5.2 |
| 10 | Zhang L | 10 | 0.73 | 239 | 23.9 |
| 11 | Emery JD | 9 | 0.66 | 160 | 17.78 |
| 12 | Li J | 9 | 0.66 | 245 | 27.22 |
| 13 | Li S | 9 | 0.66 | 240 | 26.67 |
| 14 | Li X | 9 | 0.66 | 130 | 14.44 |
| 15 | Lindor NM | 9 | 0.66 | 874 | 97.11 |
| 16 | Stoffel EM | 9 | 0.66 | 601 | 66.78 |
| 17 | Verma M | 9 | 0.66 | 379 | 42.11 |
| 18 | Wang X | 9 | 0.66 | 430 | 47.78 |
| 19 | Zhang J | 9 | 0.66 | 43 | 4.78 |
| 20 | Burt RW | 8 | 0.58 | 1568 | 196 |
| Rank | Cocite author | Citations | Centrality |
| 1 | Lynch HT | 134 | 0.12 |
| 2 | Vasen HFA | 125 | 0.03 |
| 3 | Hampel H | 89 | 0.04 |
| 4 | Giardiello FM | 79 | 0.13 |
| 5 | Jarvinen HJ | 68 | 0.04 |
| 6 | Umar A | 65 | 0.05 |
| 7 | Siegel RL | 63 | 0.01 |
| 8 | Berg AO | 63 | 0.04 |
| 9 | Bray F | 56 | 0 |
| 10 | Jemal A | 55 | 0.03 |
| 11 | Ferlay J | 54 | 0.03 |
| 12 | Syngal S | 53 | 0.03 |
| 13 | Kastrinos F | 53 | 0.04 |
| 14 | Boland CR | 49 | 0.1 |
| 15 | Rex DK | 39 | 0.06 |
| 16 | Chen SN | 38 | 0.02 |
| 17 | Easton DF | 37 | 0.05 |
| 18 | Aaltonen LA | 37 | 0.17 |
| 19 | Lindor NM | 36 | 0.02 |
| 20 | Gail MH | 35 | 0.03 |
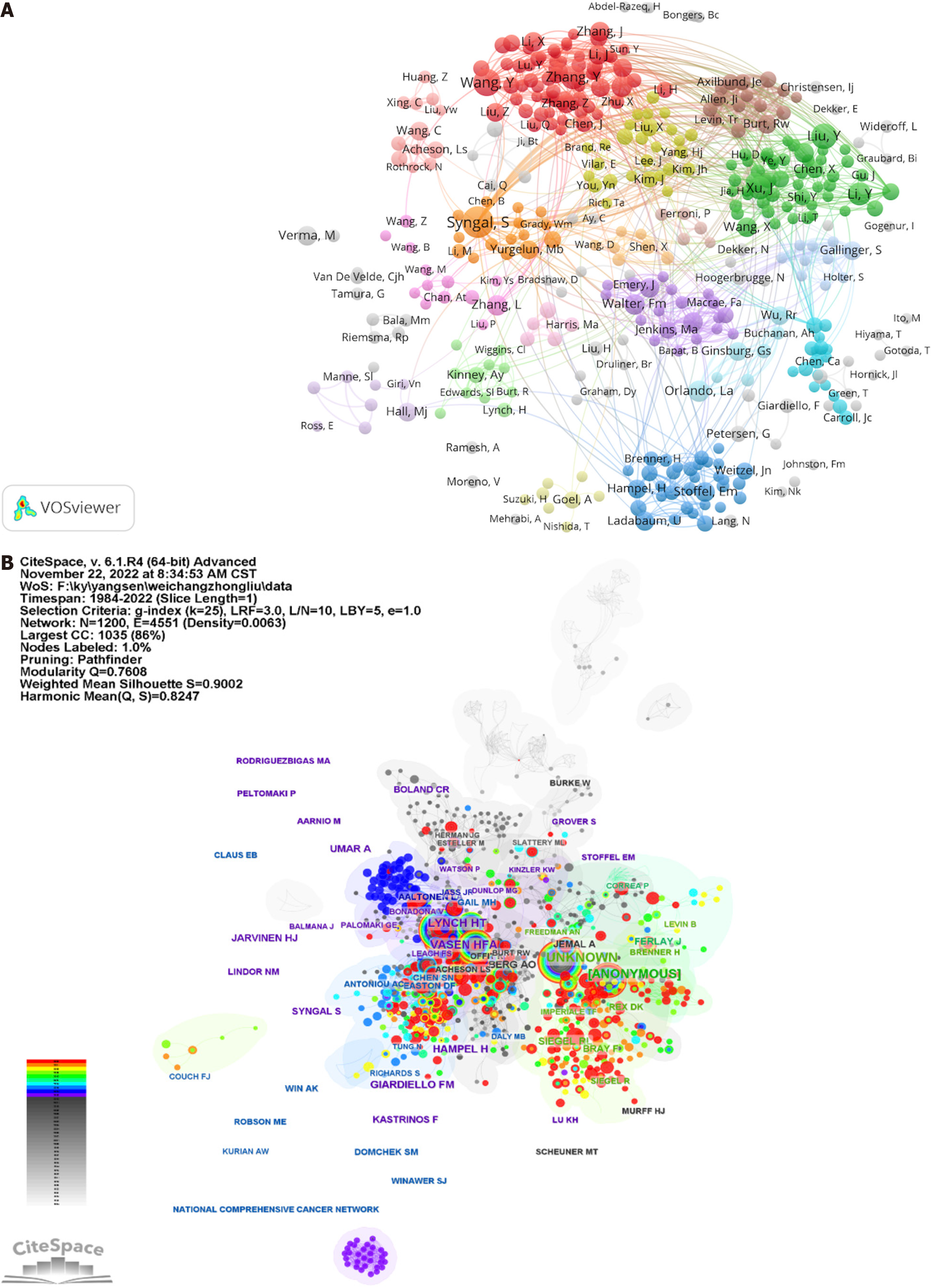
Moreover, domain maps, collaboratively crafted by the researchers, are visually depicted through the application of VOSviewer, as shown in Figure 4A. It is of particular interest that several author clusters demonstrate strong subgroup linkages, notably the clusters centered on Syngal S, Wang Y, and Liu Y, represented by the colors orange, red, and green, respectively. Nevertheless, collaboration predominantly occurs among authors with shared nationalities and/or institutional affiliations, thus highlighting the critical need for fostering interdisciplinary, interinstitutional, and international collaboration. Such collaborations are pivotal, as they facilitate mutual learning across diverse teams, thereby accelerating the innovative and varied advancement of research in the field of GN risk assessment.
The visualization of the author network in Figure 4B illustrates the extensive collaborative endeavors among scholars. Circles, symbolizing individual authors, are interconnected by lines that represent the degree of collaboration, where thicker lines indicate a closer collaborative relationship. The depiction utilizes various colors to denote different years of collaboration. The modular Q value is employed as an indicator of the clustering effect within the network, with higher values signifying more pronounced clustering. Additionally, the silhouette value is utilized to gauge the homogeneity of the network. Betweenness centrality (BC) is another metric that measures the significance of nodes within a network to determine their centrality. Notably, a node with a BC value of 0.1 is indicative of its critical role in linking multiple nodes.
According to Table 5, the three most distinguished authors are Syngal S (with 24 publications), Wang Y (with 18 publications), and Liu Y (with 15 publications). In Figure 4B, authors such as Aaltonen LA, Giardiello FM, and Lynch HT demonstrate significant BC values of 0.17, 0.13, and 0.12, respectively, surpassing the established threshold of 0.10. This evidence supports their influential contributions within this scholarly domain. Intriguingly, Syngal S, Wang Y, and Liu Y also rank among the authors with the highest BC values, further underscoring their centrality in the field. The top three cocited authors, Lynch HT, Vasen HFA, and Hampel H, with cocitation counts of 134, 125, and 89, respectively, as identified in Table 6, are associated with significant network nodes, highlighting their prominence in the GN risk assessment research.
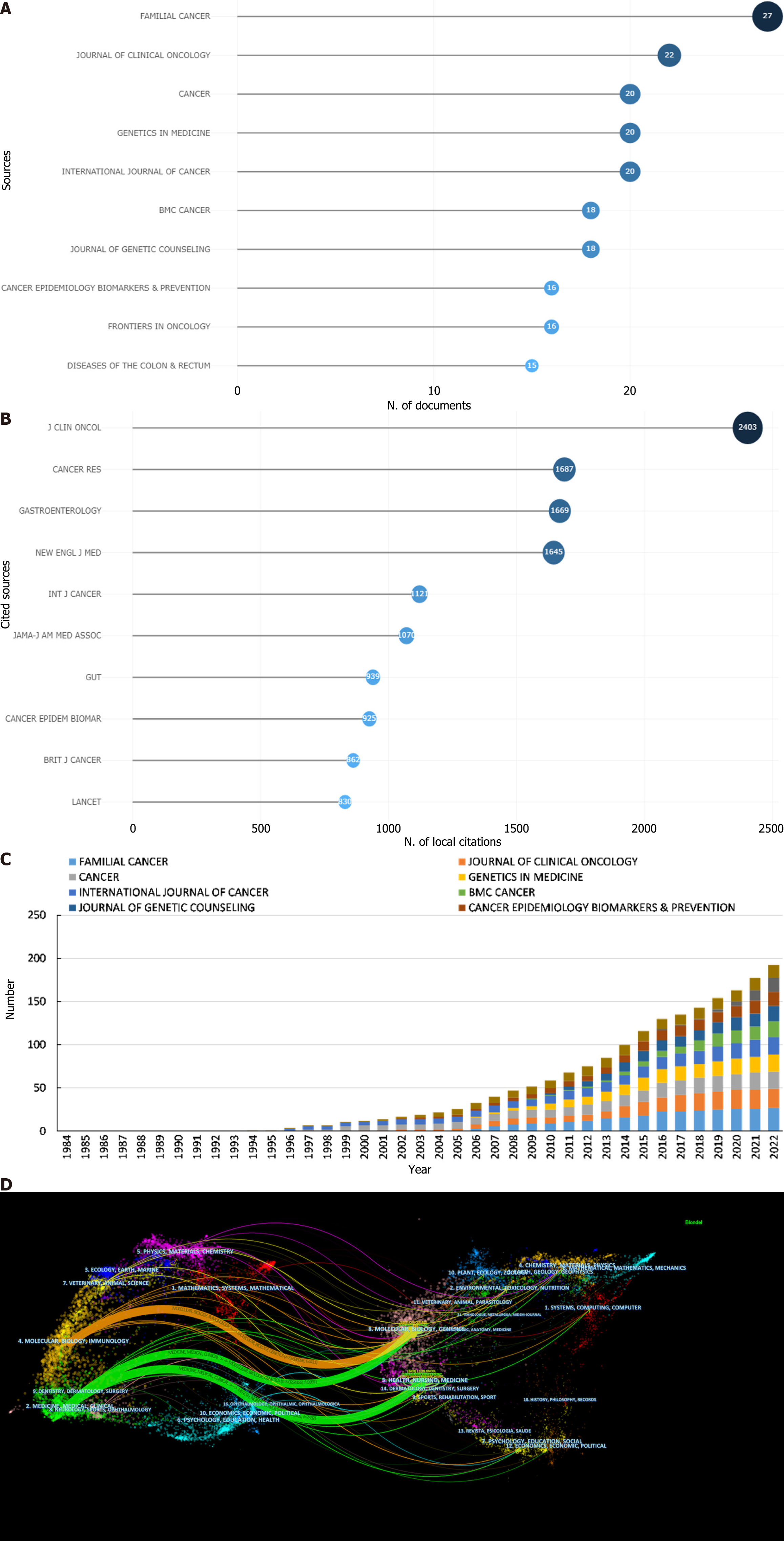
Among the 579 publications analyzed, the leading academic journals dedicated to GN risk assessment research were identified as Familial Cancer (with 27 articles), Journal of Clinical Oncology (22 articles), and Cancer (20 articles), as depicted in Figure 5A and Table 7. A geographic analysis of the top ten journals revealed that a majority, 60% (6 out of 10), are based in the United States, followed by contributions from Switzerland (20%; 2 out of 10), the Netherlands (10%; 1 out of 10), and England (10%; 1 out of 10). Among these, the Journal of Clinical Oncology stands out with an impressive impact factor (IF) of 50.717 for 2021, leading the field for journals publishing more than 15 articles on this topic. It is followed by Genetics in Medicine (IF 2021 = 8.86) and the International Journal of Cancer (IF 2021 = 7.32). Similarly, the Journal of Clinical Oncology achieved the highest H-index (H = 18), indicating significant academic influence, followed closely by Genetics in Medicine (H = 15) and the International Journal of Cancer (H = 13).
| Rank | Journal | Countries | Count | IF (2021) | JCR (2021) | H-index | Total citations |
| 1 | Familial Cancer | Netherlands | 27 | 2.45 | Q3 | 11 | 387 |
| 2 | Journal of Clinical Oncology | United States | 22 | 50.717 | Q1 | 18 | 2823 |
| 3 | Cancer | United States | 20 | 6.92 | Q1 | 10 | 1092 |
| 4 | Genetics in Medicine | United States | 20 | 8.86 | Q1 | 15 | 871 |
| 5 | International Journal of Cancer | Switzerland | 20 | 7.32 | Q1 | 13 | 668 |
| 6 | BMC Cancer | England | 18 | 4.64 | Q2 | 11 | 268 |
| 7 | Journal of Genetic Counseling | United States | 18 | 2.717 | Q3 | 10 | 492 |
| 8 | Cancer Epidemiology Biomarkers & Prevention | United States | 16 | 4.09 | Q2 | 11 | 363 |
| 9 | Frontiers in Oncology | Switzerland | 16 | 5.738 | Q2 | 11 | 611 |
| 10 | Diseases of The Colon & Rectum | United States | 15 | 4.41 | Q1 | 8 | 298 |
The analysis of the quartile distribution shows that 50% of the journals are classified within the first quartile (Q1), representing the top 25% of the IF distribution, with an additional three journals in the second quartile (Q2), which covers the 25% to 50% range. The Journal of Clinical Oncology has the highest overall citation frequency (cited 2823 times), IF (2021 = 50.717), and H-index (18), highlighting its critical role in the progression of research in this field. As shown in Figure 5B and Table 8, among the top ten cocited journals, 60% (6 out of 10) were from the United States, and 30% (3 out of 10) were from England.
| Rank | Cocited Journal | Countries | Total citations | IF (2021) | JCR (2021) |
| 1 | Journal of Clinical Oncology | United States | 2403 | 50.717 | Q1 |
| 2 | Cancer Research | United States | 1687 | 13.31 | Q1 |
| 3 | Gastroenterology | United States | 1669 | 33.88 | Q1 |
| 4 | New England Journal of Medicine | United States | 1645 | 176.079 | Q1 |
| 5 | International Journal of Cancer | Switzerland | 1121 | 7.32 | Q1 |
| 6 | Jama-Journal of The American Medical Association | United States | 1070 | 157.335 | Q1 |
| 7 | GUT | England | 939 | 31.79 | Q1 |
| 8 | Cancer Epidemiology Biomarkers & Prevention | United States | 925 | 4.09 | Q2 |
| 9 | British Journal of Cancer | England | 862 | 9.08 | Q1 |
| 10 | Lancet | England | 830 | 202.731 | Q1 |
Remarkably, 90% of the journals are categorized within the highest quartile (Q1), underscoring the quality and impact of the research in this discipline. The cumulative publication output of the top 10 most prolific journals from 1984 to 2022 is depicted in Figure 5C, showing a clear trend over time. Additionally, Figure 5D introduces a dual-map overlay that maps the distribution of journal topics, illustrating the interdisciplinary nature of the cited and citing journals across various fields. This representation highlights three primary citation pathways, emphasizing the interaction between molecular/biology/immunology and medicine/medical/clinical journals with molecular/biology/genetics and health/nursing/medicine journals, as indicated by distinct colored lines that signify the pathways of reference based on the z score-scaled citation frequency. This graphical depiction not only provides insight into the thematic interconnections but also underscores the dynamic interplay of research domains in advancing the understanding of GNs.
Table 9 delineates the ten most important references integral to the assessment of GN risk. Remarkably, Vasen HFA was identified as the predominant reference, accounting for 74 citations, followed by Umar A, with 67 citations. The journals with the greatest impact, serving as pivotal markers of academic influence, include the prestigious New England Journal of Medicine (IF 2021 = 176.079), the Journal of Clinical Oncology (IF 2021 = 50.717), and Gastroenterology (IF 2021 = 33.88).
| Title | First author | Journal | Year | IF (2021) | JCR (2021) | Citations |
| New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC | Vasen HFA | Gastroenterology | 1999 | 33.88 | Q1 | 74 |
| Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. | Umar A | Journal of The National Cancer Institute | 2004 | 11.816 | Q1 | 67 |
| Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer | Jarvinen HJ | Gastroenterology | 2000 | 33.88 | Q1 | 59 |
| The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) | Vasen HFA | Diseases of The Colon & Rectum | 1991 | 4.41 | Q1 | 49 |
| A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: Meeting Highlights and Bethesda Guidelines | Rodriguezbigas MA | Journal of The National Cancer Institute | 1997 | 11.816 | Q1 | 46 |
| A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer | Boland CR | Cancer Research | 1998 | 13.31 | Q1 | 42 |
| Hereditary Colorectal Cancer | Lynch HT | New England Journal of Medicine | 2003 | 176.079 | Q1 | 42 |
| Screening for the Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) | Hampel H | New England Journal of Medicine | 2005 | 176.079 | Q1 | 41 |
| Projecting Individualized Probabilities of Developing Breast Cancer for White Females Who Are Being Examined Annually | Gail MH | Journal of The National Cancer Institute | 1989 | 11.816 | Q1 | 40 |
| Feasibility of Screening for Lynch Syndrome Among Patients With Colorectal Cancer | Hampel H | Journal of Clinical Oncology | 2008 | 50.717 | Q1 | 34 |
In Supplementary Figure 1A, a graphical depiction elucidates the complex network of reference cocitations investigated in this study. The findings revealed a pronounced clustering effect within a cohesive network characterized by a modular Q of 0.9347 and an average silhouette width (S) of 0.8808. Conducted through the use of CiteSpace, the cocitation analysis encompasses the period from 1984 to 2022, incorporating the top 10% most cited references annually (Supplementary Figure 1B). This methodology not only highlights emergent research focal points but also provides a temporal view of the evolution of cocitation references.
The analysis of clusters revealed twenty distinct groups, with the notable clusters of "Lynch syndrome" (0) and "inherited cancer" (1) receiving significant focus. Initial studies were concentrated on "BRCA genes" (8) and "predictive preventive personalized medicine" (19). Currently, research emphasis has shifted toward risk assessment for GNs, as evidenced by the predominant research themes of "Lynch syndrome" (0), "risk assessment" (4), and "CPG-island methylation" (56).
Supplementary Figure 2 features the top 30 references distinguished by their significant citation impact scores. A marked increase in citations within this field since 1998 indicates that scholarly interest has grown. Numerous cocitation references that originated in the early stages of the research in this field continue to attract significant citations, highlighting the persistent relevance of GN risk assessment as a central theme of research.
The investigation of keyword co-occurrence offers valuable insights into prevailing areas of interest and suggests potential paths for future inquiry within the specified field. An examination of 2880 keywords extracted from abstracts and titles revealed that 307 terms satisfied the established criteria. These terms encapsulate the core themes prevalent across the analyzed body of literature. A detailed examination of these relevant keywords facilitates the identification of key concepts and aids in developing an integrated framework for research focused on risk assessment in the context of GNs.
In this study, the top deciles of keywords were identified using VOSviewer and were subsequently categorized, as presented in Table 10. This categorization highlights both established and nascent areas within the examined domain. The organization of these keywords into six distinct clusters is detailed in Supplementary Figure 3A, with each cluster represented by a specific color (red, yellow, green, purple, dark blue, or light blue). Prominent terms that exhibit a high frequency of occurrence include "genetic testing," "colorectal cancer," "risk assessment," "stomach cancer," and "Lynch syndrome."
| Rank | Keywords | Total link strength | Frequency |
| 1 | Colorectal cancer | 417 | 200 |
| 2 | Risk assessment | 411 | 169 |
| 3 | Gastric cancer | 168 | 77 |
| 4 | Cancer | 167 | 70 |
| 5 | Lynch syndrome | 162 | 63 |
| 6 | Genetic testing | 136 | 49 |
| 7 | Colon cancer | 102 | 45 |
| 8 | Genetic counseling | 131 | 44 |
| 9 | Prognosis | 108 | 44 |
| 10 | Family history | 117 | 38 |
Furthermore, Supplementary Figure 3B elaborates on the contextual relevance of the temporal trends associated with these keywords, acting as a marker for the studies’ leading areas of focus, thematic concentrations, and their evolution over time. Notably, consistent referencing of terms such as "machine learning" (2021-2022), "complications" (2020-2022), "health risk assessment" (2020-2022), "mass screening" (2011-2021), and "early detection of cancer" (2016-2021) during the last two years (2021 and 2022) indicates sustained scholarly engagement with specific research themes in recent periods.
Risk assessment holds critical importance in crafting effective cancer prevention and treatment strategies globally[14,15]. This paper explores the prevailing research directions in GN risk assessment over the past three decades by employing bibliometric analysis. By integrating various factors, such as lifestyle choices, genetic predispositions, and environmental influences, a comprehensive evaluation of the risks associated with GNs is facilitated[16,17].
Through bibliometric analysis, this study identified 1371 publications from 1984 to 2022, highlighting a growing body of research focused on GN risk assessment. The United States stands out as a leader in both the volume of publications and their citation impact scores, with mainland Chinese journals ranking second. Brigham and Women's Hospital is noted for producing the greatest number of publications, with Syngal S recognized as a leading researcher in this field. The core of these publications revolves around genetic factors, screening methods, and biomarkers for assessing GN risk. Cocitation analysis revealed thematic clusters focused on genetics, epigenetic alterations, and genetic testing, with Lynch syndrome emerging as a recent research focus, as indicated by keyword analysis.
Global overview of GN risk assessment research: The forefront of research into the risk assessment of GNs is led by the United States and China; their pivotal roles are showcased through an extensive analysis of global contributions by countries and academic entities. The United States stands out for its leading position in publication frequency and the significant impact of its research citations. England has emerged as the primary European contributor, notable for its scholarly output and citation impact within this field. The variation in the incidence and mortality rates of GNs worldwide may reflect differences in diet, lifestyle, environmental factors, and genetic predispositions, highlighting the differential risk levels across various regions[18-20]. This diversity underscores the existence of regions at higher risk and others at comparatively lower risk of developing these neoplasms[21,22]. Additionally, disparities in advancements in GN risk assessment research among countries and institutions are attributed to the uneven distribution of resources and priorities. With their substantial resources, developed nations and leading academic centers deliver groundbreaking findings[23], while resource-limited countries and smaller institutions struggle to make significant research contributions[24].
Further analysis revealed that China has produced the second-largest body of research on GN risk assessment in the last three decades. However, its citation impact score per paper lags behind those of more developed countries, a gap possibly linked to its economic development and growing public health consciousness[25,26]. Despite increasing global attention and international collaboration, research in China is still in its nascent stages, with evident gaps in study design and data analysis compared to developed countries[27]. Hence, there is a pressing need for enhanced collaboration with globally recognized research institutes to adopt advanced research methodologies and insights, aiming to improve the quality and impact of China's GN risk assessment research.
Predominance of genetic assessment in GN risk research: The vital role of genetic assessment in determining the risk associated with GN is increasingly recognized within the scientific community[28,29]. This recognition is underscored by the identification of numerous genetic mutations, especially within the BRCA1 and BRCA2 genes, which are significantly correlated with the initiation and progression of GN, thereby underscoring their role in colorectal cancer susceptibility[30-32]. Genetic testing plays a crucial role in identifying these mutations, enabling the implementation of preventive measures and the refinement of screening protocols to enhance early detection.
Our analysis also revealed that terms such as Lynch syndrome, hereditary nonpolyposis colorectal cancer (HNPCC), and epigenetic alterations frequently cluster together in cocitation analyses, highlighting the importance of genetic factors in GN risk. This is further supported by the identification of keywords closely linked to genetic aspects, emphasizing the pivotal role of genetic determinants in GN pathogenesis by affecting key physiological processes such as genomic stability and cellular proliferation[33]. The genetic underpinnings of many GNs indicate that mutations and genetic predispositions significantly contribute to cancer etiology[34]. Conditions such as familial adenomatous polyposis and HNPCC notably increase GN risk, with mutations in genes such as MLH1, MSH2, and APC being closely linked to these neoplasms[35-37]. This evidence strongly supports the need for regular screening in individuals with a significant family history to facilitate early intervention.
However, it is crucial to recognize that genetic assessments, while offering valuable insights into susceptibility, do not precisely predict the onset or progression of GN[38]. Given the multifaceted nature of GN development, a comprehensive approach that includes consistent monitoring and considers a broad range of risk factors remains indispensable for individuals at increased genetic risk.
Trends and advancements in GN risk assessment studies: The amalgamation of cluster analysis and outbreak detection analysis has succinctly synthesized the prevailing methodologies for assessing the GN risk. Contemporary investigations into GN risk assessment encompass multifaceted dimensions, including familial history, lifestyle factors, inflammatory status, biomarkers, genetic testing, imaging, and the burgeoning domain of machine learning.
An examination of the keyword trend graph reveals that, half a decade ago, scholarly attention was predominantly directed toward inquiries about genetic testing and family history. Notably, familial adenomatous polyposis emerged as a sustained focal point in the research spanning from 1999 to 2017. This prolonged focus can be attributed to the elucidation of its pathogenesis, affirming familial adenomatous polyposis as an autosomal dominant colorectal adenomatous syndrome stemming from germline mutations in the APC oncogene. Genetic testing, therefore, has emerged as a pivotal tool for individuals to comprehend their susceptibility to familial adenomatous polyposis and associated genetic variants[39]. Concurrently, family history studies illuminate transmission modes and incidence rates, significantly enhancing early diagnostic capabilities, implementing effective preventive measures, and optimizing treatment modalities for both patients and their families.
In tandem with the exponential growth of big data and AI technologies, their pervasive integration into GN risk assessment studies has become increasingly evident. Over the past half-decade, the scholarly focus has shifted toward lifestyle interventions and the application of machine learning. Notably, the utilization of machine learning in GN risk assessment models has emerged as a prominent research direction. Various risk assessment models, such as the Gail model, Tyrer-Cuzick model, colorectal cancer risk assessment tool model, BOADICEA model, and mismatch repair probability model, have been developed[40,41]. Leveraging expansive epidemiological databases and clinical datasets combined with machine learning algorithms enables the derivation of more nuanced and precise risk prediction models. This augments the accuracy and feasibility of GN risk assessment, facilitating a more comprehensive understanding of the complex interplay between risk factors and disease outcomes.
Although we rigorously adhered to bibliometric research methods, certain limitations warrant acknowledgment. Primarily, due to the study's exclusive focus on articles within the WoSCC database, a substantial number of relevant papers from alternate databases such as Medline, SCOPUS, and the Cochrane Library were regrettably excluded. The selection of the WoSCC database for in-depth scrutiny was necessitated by the utilization of CiteSpace, which is intricately linked to this particular repository. Second, the inherent limitations of bibliometric techniques become apparent, as they primarily consider publication quantity and citation metrics in gauging the significance and quality of scientific research while disregarding other pivotal dimensions. Additionally, bibliometric methodologies are limited to retrospective analyses of existing literature and lack the ability to promptly capture the latest advancements and trends in scientific research. Third, acknowledging the inherent bias toward higher citation rates in review articles, our amalgamation of publications and reviews into a singular analysis was motivated by the overarching aim of providing a comprehensive overview of the field of GN risk assessment without sacrificing any pertinent nodes. Consequently, we opted against segregating the incidence, recurrence, and metastasis risks from the mortality, and nutritional risks associated with GNs to present a holistic perspective.
This study meticulously charts the evolution of the GN risk assessment research, emphasizing the critical role of genetic factors and the emergent potential of machine learning in refining risk prediction. By employing bibliometric analysis of 1371 publications spanning from 1984 to 2022, we uncovered significant geographical disparities in research contributions, with the United States leading in research output and impact. Our findings highlight the paramount importance of genetic assessments, underscored by the identification of key mutations associated with increased risk, and the transformative impact of machine learning and big data analytics in enhancing predictive accuracy. Despite these advances, the need for global collaboration to equalize research capabilities across regions remains pressing, suggesting that future efforts should aim to democratize access to advanced technologies and methodologies in risk assessment.
We advocate for policy-makers and research institutions to prioritize support for genetic research and the integration of AI-driven analytics into risk assessment frameworks. Such initiatives promise not only to improve the precision of current models but also to pave the way for more personalized approaches to cancer prevention and treatment. Overcoming the identified geographical and methodological disparities requires a concerted international effort, fostering an environment where innovations can be shared and applied universally. As we navigate the complexities of GN risk, the emphasis on technological advancement and international collaboration will be pivotal in shaping the future of cancer research, with the ultimate goal of enhancing patient care and outcomes worldwide.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country of origin: China
Peer-review report’s classification
Scientific Quality: Grade A
Novelty: Grade A
Creativity or Innovation: Grade B
Scientific Significance: Grade A
P-Reviewer: Rahman MM, Bangladesh S-Editor: Qu XL L-Editor: A P-Editor: Zhang XD
| 1. | Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2279] [Cited by in RCA: 4734] [Article Influence: 4734.0] [Reference Citation Analysis (3)] |
| 2. | Grady WM, Yu M, Markowitz SD. Epigenetic Alterations in the Gastrointestinal Tract: Current and Emerging Use for Biomarkers of Cancer. Gastroenterology. 2021;160:690-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 3. | Kastrinos F, Kupfer SS, Gupta S. Colorectal Cancer Risk Assessment and Precision Approaches to Screening: Brave New World or Worlds Apart? Gastroenterology. 2023;164:812-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 52] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 4. | Na YS, Kim SG, Cho SJ. Risk assessment of metachronous gastric cancer development using OLGA and OLGIM systems after endoscopic submucosal dissection for early gastric cancer: a long-term follow-up study. Gastric Cancer. 2023;26:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 5. | Shulman RM, Deng M, Handorf EA, Meyer JE, Lynch SM, Arora S. Factors Associated With Racial and Ethnic Disparities in Locally Advanced Rectal Cancer Outcomes. JAMA Netw Open. 2024;7:e240044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 6. | Dermawan JK, Kelly C, Gao Z, Smith S, Jadeja B, Singer S, Tap WD, Chi P, Antonescu CR. Novel Genomic Risk Stratification Model for Primary Gastrointestinal Stromal Tumors (GIST) in the Adjuvant Therapy Era. Clin Cancer Res. 2023;29:3974-3985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 7. | Leśniewska M, Patryn R, Kopystecka A, Kozioł I, Budzyńska J. Third Eye? The Assistance of Artificial Intelligence (AI) in the Endoscopy of Gastrointestinal Neoplasms. J Clin Med. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Rompianesi G, Pegoraro F, Ceresa CD, Montalti R, Troisi RI. Artificial intelligence in the diagnosis and management of colorectal cancer liver metastases. World J Gastroenterol. 2022;28:108-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 72] [Article Influence: 24.0] [Reference Citation Analysis (3)] |
| 9. | García-Muñoz C, Cortés-Vega MD, Heredia-Rizo AM, Martín-Valero R, García-Bernal MI, Casuso-Holgado MJ. Effectiveness of Vestibular Training for Balance and Dizziness Rehabilitation in People with Multiple Sclerosis: A Systematic Review and Meta-Analysis. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010;84:523-538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4505] [Cited by in RCA: 5155] [Article Influence: 322.2] [Reference Citation Analysis (1)] |
| 11. | Wu H, Tong L, Wang Y, Yan H, Sun Z. Bibliometric Analysis of Global Research Trends on Ultrasound Microbubble: A Quickly Developing Field. Front Pharmacol. 2021;12:646626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 12. | Liu Y, Kotheeranurak V, Quillo-Olvera J, Facundo VI, Sharma S, Suvithayasiri S, Jitpakdee K, Lin GX, Mahatthanatrakul A, Jabri H, Khandge AV, Aher RB, Wu MH, Ho AWH, Wong NMR, Wing LS, Akbary K, Patel KK, Pakdeenit B, Chen KT, Lokanath YK, Jaiswal MS, Suen TK, Hasan GA, Sabal LA, Kim JS. A 30-Year Worldwide Research Productivity of Scientific Publication in Full-Endoscopic Decompression Spine Surgery: Quantitative and Qualitative Analysis. Neurospine. 2023;20:374-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 13. | Synnestvedt MB, Chen C, Holmes JH. CiteSpace II: visualization and knowledge discovery in bibliographic databases. AMIA Annu Symp Proc. 2005;2005:724-728. [PubMed] |
| 14. | Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag CJ, Laversanne M, Vignat J, Ferlay J, Murphy N, Bray F. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 926] [Article Influence: 463.0] [Reference Citation Analysis (1)] |
| 15. | Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-349.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 1233] [Article Influence: 246.6] [Reference Citation Analysis (0)] |
| 16. | Jin G, Lv J, Yang M, Wang M, Zhu M, Wang T, Yan C, Yu C, Ding Y, Li G, Ren C, Ni J, Zhang R, Guo Y, Bian Z, Zheng Y, Zhang N, Jiang Y, Chen J, Wang Y, Xu D, Zheng H, Yang L, Chen Y, Walters R, Millwood IY, Dai J, Ma H, Chen K, Chen Z, Hu Z, Wei Q, Shen H, Li L. Genetic risk, incident gastric cancer, and healthy lifestyle: a meta-analysis of genome-wide association studies and prospective cohort study. Lancet Oncol. 2020;21:1378-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 17. | Song M, Chan AT, Sun J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology. 2020;158:322-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 464] [Cited by in RCA: 514] [Article Influence: 102.8] [Reference Citation Analysis (2)] |
| 18. | Global Burden of Disease 2019 Cancer Collaboration, Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, Henrikson HJ, Lu D, Pennini A, Xu R, Ababneh E, Abbasi-Kangevari M, Abbastabar H, Abd-Elsalam SM, Abdoli A, Abedi A, Abidi H, Abolhassani H, Adedeji IA, Adnani QES, Advani SM, Afzal MS, Aghaali M, Ahinkorah BO, Ahmad S, Ahmad T, Ahmadi A, Ahmadi S, Ahmed Rashid T, Ahmed Salih Y, Akalu GT, Aklilu A, Akram T, Akunna CJ, Al Hamad H, Alahdab F, Al-Aly Z, Ali S, Alimohamadi Y, Alipour V, Aljunid SM, Alkhayyat M, Almasi-Hashiani A, Almasri NA, Al-Maweri SAA, Almustanyir S, Alonso N, Alvis-Guzman N, Amu H, Anbesu EW, Ancuceanu R, Ansari F, Ansari-Moghaddam A, Antwi MH, Anvari D, Anyasodor AE, Aqeel M, Arabloo J, Arab-Zozani M, Aremu O, Ariffin H, Aripov T, Arshad M, Artaman A, Arulappan J, Asemi Z, Asghari Jafarabadi M, Ashraf T, Atorkey P, Aujayeb A, Ausloos M, Awedew AF, Ayala Quintanilla BP, Ayenew T, Azab MA, Azadnajafabad S, Azari Jafari A, Azarian G, Azzam AY, Badiye AD, Bahadory S, Baig AA, Baker JL, Balakrishnan S, Banach M, Bärnighausen TW, Barone-Adesi F, Barra F, Barrow A, Behzadifar M, Belgaumi UI, Bezabhe WMM, Bezabih YM, Bhagat DS, Bhagavathula AS, Bhardwaj N, Bhardwaj P, Bhaskar S, Bhattacharyya K, Bhojaraja VS, Bibi S, Bijani A, Biondi A, Bisignano C, Bjørge T, Bleyer A, Blyuss O, Bolarinwa OA, Bolla SR, Braithwaite D, Brar A, Brenner H, Bustamante-Teixeira MT, Butt NS, Butt ZA, Caetano Dos Santos FL, Cao Y, Carreras G, Catalá-López F, Cembranel F, Cerin E, Cernigliaro A, Chakinala RC, Chattu SK, Chattu VK, Chaturvedi P, Chimed-Ochir O, Cho DY, Christopher DJ, Chu DT, Chung MT, Conde J, Cortés S, Cortesi PA, Costa VM, Cunha AR, Dadras O, Dagnew AB, Dahlawi SMA, Dai X, Dandona L, Dandona R, Darwesh AM, das Neves J, De la Hoz FP, Demis AB, Denova-Gutiérrez E, Dhamnetiya D, Dhimal ML, Dhimal M, Dianatinasab M, Diaz D, Djalalinia S, Do HP, Doaei S, Dorostkar F, Dos Santos Figueiredo FW, Driscoll TR, Ebrahimi H, Eftekharzadeh S, El Tantawi M, El-Abid H, Elbarazi I, Elhabashy HR, Elhadi M, El-Jaafary SI, Eshrati B, Eskandarieh S, Esmaeilzadeh F, Etemadi A, Ezzikouri S, Faisaluddin M, Faraon EJA, Fares J, Farzadfar F, Feroze AH, Ferrero S, Ferro Desideri L, Filip I, Fischer F, Fisher JL, Foroutan M, Fukumoto T, Gaal PA, Gad MM, Gadanya MA, Gallus S, Gaspar Fonseca M, Getachew Obsa A, Ghafourifard M, Ghashghaee A, Ghith N, Gholamalizadeh M, Gilani SA, Ginindza TG, Gizaw ATT, Glasbey JC, Golechha M, Goleij P, Gomez RS, Gopalani SV, Gorini G, Goudarzi H, Grosso G, Gubari MIM, Guerra MR, Guha A, Gunasekera DS, Gupta B, Gupta VB, Gupta VK, Gutiérrez RA, Hafezi-Nejad N, Haider MR, Haj-Mirzaian A, Halwani R, Hamadeh RR, Hameed S, Hamidi S, Hanif A, Haque S, Harlianto NI, Haro JM, Hasaballah AI, Hassanipour S, Hay RJ, Hay SI, Hayat K, Heidari G, Heidari M, Herrera-Serna BY, Herteliu C, Hezam K, Holla R, Hossain MM, Hossain MBH, Hosseini MS, Hosseini M, Hosseinzadeh M, Hostiuc M, Hostiuc S, Househ M, Hsairi M, Huang J, Hugo FN, Hussain R, Hussein NR, Hwang BF, Iavicoli I, Ibitoye SE, Ida F, Ikuta KS, Ilesanmi OS, Ilic IM, Ilic MD, Irham LM, Islam JY, Islam RM, Islam SMS, Ismail NE, Isola G, Iwagami M, Jacob L, Jain V, Jakovljevic MB, Javaheri T, Jayaram S, Jazayeri SB, Jha RP, Jonas JB, Joo T, Joseph N, Joukar F, Jürisson M, Kabir A, Kahrizi D, Kalankesh LR, Kalhor R, Kaliyadan F, Kalkonde Y, Kamath A, Kameran Al-Salihi N, Kandel H, Kapoor N, Karch A, Kasa AS, Katikireddi SV, Kauppila JH, Kavetskyy T, Kebede SA, Keshavarz P, Keykhaei M, Khader YS, Khalilov R, Khan G, Khan M, Khan MN, Khan MAB, Khang YH, Khater AM, Khayamzadeh M, Kim GR, Kim YJ, Kisa A, Kisa S, Kissimova-Skarbek K, Kopec JA, Koteeswaran R, Koul PA, Koulmane Laxminarayana SL, Koyanagi A, Kucuk Bicer B, Kugbey N, Kumar GA, Kumar N, Kurmi OP, Kutluk T, La Vecchia C, Lami FH, Landires I, Lauriola P, Lee SW, Lee SWH, Lee WC, Lee YH, Leigh J, Leong E, Li J, Li MC, Liu X, Loureiro JA, Lunevicius R, Magdy Abd El Razek M, Majeed A, Makki A, Male S, Malik AA, Mansournia MA, Martini S, Masoumi SZ, Mathur P, McKee M, Mehrotra R, Mendoza W, Menezes RG, Mengesha EW, Mesregah MK, Mestrovic T, Miao Jonasson J, Miazgowski B, Miazgowski T, Michalek IM, Miller TR, Mirzaei H, Mirzaei HR, Misra S, Mithra P, Moghadaszadeh M, Mohammad KA, Mohammad Y, Mohammadi M, Mohammadi SM, Mohammadian-Hafshejani A, Mohammed S, Moka N, Mokdad AH, Molokhia M, Monasta L, Moni MA, Moosavi MA, Moradi Y, Moraga P, Morgado-da-Costa J, Morrison SD, Mosapour A, Mubarik S, Mwanri L, Nagarajan AJ, Nagaraju SP, Nagata C, Naimzada MD, Nangia V, Naqvi AA, Narasimha Swamy S, Ndejjo R, Nduaguba SO, Negoi I, Negru SM, Neupane Kandel S, Nguyen CT, Nguyen HLT, Niazi RK, Nnaji CA, Noor NM, Nuñez-Samudio V, Nzoputam CI, Oancea B, Ochir C, Odukoya OO, Ogbo FA, Olagunju AT, Olakunde BO, Omar E, Omar Bali A, Omonisi AEE, Ong S, Onwujekwe OE, Orru H, Ortega-Altamirano DV, Otstavnov N, Otstavnov SS, Owolabi MO, P A M, Padubidri JR, Pakshir K, Pana A, Panagiotakos D, Panda-Jonas S, Pardhan S, Park EC, Park EK, Pashazadeh Kan F, Patel HK, Patel JR, Pati S, Pattanshetty SM, Paudel U, Pereira DM, Pereira RB, Perianayagam A, Pillay JD, Pirouzpanah S, Pishgar F, Podder I, Postma MJ, Pourjafar H, Prashant A, Preotescu L, Rabiee M, Rabiee N, Radfar A, Radhakrishnan RA, Radhakrishnan V, Rafiee A, Rahim F, Rahimzadeh S, Rahman M, Rahman MA, Rahmani AM, Rajai N, Rajesh A, Rakovac I, Ram P, Ramezanzadeh K, Ranabhat K, Ranasinghe P, Rao CR, Rao SJ, Rawassizadeh R, Razeghinia MS, Renzaho AMN, Rezaei N, Rezapour A, Roberts TJ, Rodriguez JAB, Rohloff P, Romoli M, Ronfani L, Roshandel G, Rwegerera GM, S M, Sabour S, Saddik B, Saeed U, Sahebkar A, Sahoo H, Salehi S, Salem MR, Salimzadeh H, Samaei M, Samy AM, Sanabria J, Sankararaman S, Santric-Milicevic MM, Sardiwalla Y, Sarveazad A, Sathian B, Sawhney M, Saylan M, Schneider IJC, Sekerija M, Seylani A, Shafaat O, Shaghaghi Z, Shaikh MA, Shamsoddin E, Shannawaz M, Sharma R, Sheikh A, Sheikhbahaei S, Shetty A, Shetty JK, Shetty PH, Shibuya K, Shirkoohi R, Shivakumar KM, Shivarov V, Siabani S, Siddappa Malleshappa SK, Silva DAS, Singh JA, Sintayehu Y, Skryabin VY, Skryabina AA, Soeberg MJ, Sofi-Mahmudi A, Sotoudeh H, Steiropoulos P, Straif K, Subedi R, Sufiyan MB, Sultan I, Sultana S, Sur D, Szerencsés V, Szócska M, Tabarés-Seisdedos R, Tabuchi T, Tadbiri H, Taherkhani A, Takahashi K, Talaat IM, Tan KK, Tat VY, Tedla BAA, Tefera YG, Tehrani-Banihashemi A, Temsah MH, Tesfay FH, Tessema GA, Thapar R, Thavamani A, Thoguluva Chandrasekar V, Thomas N, Tohidinik HR, Touvier M, Tovani-Palone MR, Traini E, Tran BX, Tran KB, Tran MTN, Tripathy JP, Tusa BS, Ullah I, Ullah S, Umapathi KK, Unnikrishnan B, Upadhyay E, Vacante M, Vaezi M, Valadan Tahbaz S, Velazquez DZ, Veroux M, Violante FS, Vlassov V, Vo B, Volovici V, Vu GT, Waheed Y, Wamai RG, Ward P, Wen YF, Westerman R, Winkler AS, Yadav L, Yahyazadeh Jabbari SH, Yang L, Yaya S, Yazie TSY, Yeshaw Y, Yonemoto N, Younis MZ, Yousefi Z, Yu C, Yuce D, Yunusa I, Zadnik V, Zare F, Zastrozhin MS, Zastrozhina A, Zhang J, Zhong C, Zhou L, Zhu C, Ziapour A, Zimmermann IR, Fitzmaurice C, Murray CJL, Force LM. Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life Years for 29 Cancer Groups From 2010 to 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 2022;8:420-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1180] [Cited by in RCA: 1137] [Article Influence: 379.0] [Reference Citation Analysis (0)] |
| 19. | Cardoso R, Guo F, Heisser T, Hackl M, Ihle P, De Schutter H, Van Damme N, Valerianova Z, Atanasov T, Májek O, Mužík J, Nilbert MC, Tybjerg AJ, Innos K, Mägi M, Malila N, Bouvier AM, Bouvier V, Launoy G, Woronoff AS, Cariou M, Robaszkiewicz M, Delafosse P, Poncet F, Katalinic A, Walsh PM, Senore C, Rosso S, Vincerževskienė I, Lemmens VEPP, Elferink MAG, Johannesen TB, Kørner H, Pfeffer F, Bento MJ, Rodrigues J, Alves da Costa F, Miranda A, Zadnik V, Žagar T, Lopez de Munain Marques A, Marcos-Gragera R, Puigdemont M, Galceran J, Carulla M, Chirlaque MD, Ballesta M, Sundquist K, Sundquist J, Weber M, Jordan A, Herrmann C, Mousavi M, Ryzhov A, Hoffmeister M, Brenner H. Colorectal cancer incidence, mortality, and stage distribution in European countries in the colorectal cancer screening era: an international population-based study. Lancet Oncol. 2021;22:1002-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 260] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 20. | Zheng RS, Zhang SW, Sun KX, Chen R, Wang SM, Li L, Zeng HM, Wei WW, He J. [Cancer statistics in China, 2016]. Zhonghua Zhong Liu Za Zhi. 2023;45:212-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 63] [Reference Citation Analysis (0)] |
| 21. | He Y, Wang Y, Luan F, Yu Z, Feng H, Chen B, Chen W. Chinese and global burdens of gastric cancer from 1990 to 2019. Cancer Med. 2021;10:3461-3473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 70] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 22. | Qin Y, Tong X, Fan J, Liu Z, Zhao R, Zhang T, Suo C, Chen X, Zhao G. Global Burden and Trends in Incidence, Mortality, and Disability of Stomach Cancer From 1990 to 2017. Clin Transl Gastroenterol. 2021;12:e00406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 23. | Arnold M, Rutherford MJ, Bardot A, Ferlay J, Andersson TM, Myklebust TÅ, Tervonen H, Thursfield V, Ransom D, Shack L, Woods RR, Turner D, Leonfellner S, Ryan S, Saint-Jacques N, De P, McClure C, Ramanakumar AV, Stuart-Panko H, Engholm G, Walsh PM, Jackson C, Vernon S, Morgan E, Gavin A, Morrison DS, Huws DW, Porter G, Butler J, Bryant H, Currow DC, Hiom S, Parkin DM, Sasieni P, Lambert PC, Møller B, Soerjomataram I, Bray F. Progress in cancer survival, mortality, and incidence in seven high-income countries 1995-2014 (ICBP SURVMARK-2): a population-based study. Lancet Oncol. 2019;20:1493-1505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 734] [Article Influence: 122.3] [Reference Citation Analysis (0)] |
| 24. | Aggarwal A, Han L, van der Geest S, Lewis D, Lievens Y, Borras J, Jayne D, Sullivan R, Varkevisser M, van der Meulen J. Health service planning to assess the expected impact of centralising specialist cancer services on travel times, equity, and outcomes: a national population-based modelling study. Lancet Oncol. 2022;23:1211-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 25. | Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond). 2021;41:1037-1048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 584] [Article Influence: 146.0] [Reference Citation Analysis (0)] |
| 26. | Maomao C, He L, Dianqin S, Siyi H, Xinxin Y, Fan Y, Shaoli Z, Changfa X, Lin L, Ji P, Wanqing C. Current cancer burden in China: epidemiology, etiology, and prevention. Cancer Biol Med. 2022;19:1121-1138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 164] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 27. | Lu L, Mullins CS, Schafmayer C, Zeißig S, Linnebacher M. A global assessment of recent trends in gastrointestinal cancer and lifestyle-associated risk factors. Cancer Commun (Lond). 2021;41:1137-1151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 187] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 28. | Luo XJ, Zhao Q, Liu J, Zheng JB, Qiu MZ, Ju HQ, Xu RH. Novel Genetic and Epigenetic Biomarkers of Prognostic and Predictive Significance in Stage II/III Colorectal Cancer. Mol Ther. 2021;29:587-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 29. | Kastrinos F, Samadder NJ, Burt RW. Use of Family History and Genetic Testing to Determine Risk of Colorectal Cancer. Gastroenterology. 2020;158:389-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 30. | Cullinane CM, Creavin B, O'Connell EP, Kelly L, O'Sullivan MJ, Corrigan MA, Redmond HP. Risk of colorectal cancer associated with BRCA1 and/or BRCA2 mutation carriers: systematic review and meta-analysis. Br J Surg. 2020;107:951-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 31. | Oh M, McBride A, Yun S, Bhattacharjee S, Slack M, Martin JR, Jeter J, Abraham I. BRCA1 and BRCA2 Gene Mutations and Colorectal Cancer Risk: Systematic Review and Meta-analysis. J Natl Cancer Inst. 2018;110:1178-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (1)] |
| 32. | Heide T, Househam J, Cresswell GD, Spiteri I, Lynn C, Mossner M, Kimberley C, Fernandez-Mateos J, Chen B, Zapata L, James C, Barozzi I, Chkhaidze K, Nichol D, Gunasri V, Berner A, Schmidt M, Lakatos E, Baker AM, Costa H, Mitchinson M, Piazza R, Jansen M, Caravagna G, Ramazzotti D, Shibata D, Bridgewater J, Rodriguez-Justo M, Magnani L, Graham TA, Sottoriva A. The co-evolution of the genome and epigenome in colorectal cancer. Nature. 2022;611:733-743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 33. | Tong Y, Gao H, Qi Q, Liu X, Li J, Gao J, Li P, Wang Y, Du L, Wang C. High fat diet, gut microbiome and gastrointestinal cancer. Theranostics. 2021;11:5889-5910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 127] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 34. | Huang T, Braun D, Lynch HT, Parmigiani G. Variation in cancer risk among families with genetic susceptibility. Genet Epidemiol. 2021;45:209-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 35. | Hryhorowicz S, Kaczmarek-Ryś M, Lis-Tanaś E, Porowski J, Szuman M, Grot N, Kryszczyńska A, Paszkowski J, Banasiewicz T, Pławski A. Strong Hereditary Predispositions to Colorectal Cancer. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 36. | Samadder NJ, Baffy N, Giridhar KV, Couch FJ, Riegert-Johnson D. Hereditary Cancer Syndromes-A Primer on Diagnosis and Management, Part 2: Gastrointestinal Cancer Syndromes. Mayo Clin Proc. 2019;94:1099-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 37. | Holowatyj AN, Washington MK, Tavtigian SV, Eng C, Horton C. Inherited Cancer Susceptibility Gene Sequence Variations Among Patients With Appendix Cancer. JAMA Oncol. 2022;9:95-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Ten Broeke SW, van Bavel TC, Jansen AML, Gómez-García E, Hes FJ, van Hest LP, Letteboer TGW, Olderode-Berends MJW, Ruano D, Spruijt L, Suerink M, Tops CM, van Eijk R, Morreau H, van Wezel T, Nielsen M. Molecular Background of Colorectal Tumors From Patients With Lynch Syndrome Associated With Germline Variants in PMS2. Gastroenterology. 2018;155:844-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Valle L, de Voer RM, Goldberg Y, Sjursen W, Försti A, Ruiz-Ponte C, Caldés T, Garré P, Olsen MF, Nordling M, Castellvi-Bel S, Hemminki K. Update on genetic predisposition to colorectal cancer and polyposis. Mol Aspects Med. 2019;69:10-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 118] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 40. | Thomas M, Sakoda LC, Hoffmeister M, Rosenthal EA, Lee JK, van Duijnhoven FJB, Platz EA, Wu AH, Dampier CH, de la Chapelle A, Wolk A, Joshi AD, Burnett-Hartman A, Gsur A, Lindblom A, Castells A, Win AK, Namjou B, Van Guelpen B, Tangen CM, He Q, Li CI, Schafmayer C, Joshu CE, Ulrich CM, Bishop DT, Buchanan DD, Schaid D, Drew DA, Muller DC, Duggan D, Crosslin DR, Albanes D, Giovannucci EL, Larson E, Qu F, Mentch F, Giles GG, Hakonarson H, Hampel H, Stanaway IB, Figueiredo JC, Huyghe JR, Minnier J, Chang-Claude J, Hampe J, Harley JB, Visvanathan K, Curtis KR, Offit K, Li L, Le Marchand L, Vodickova L, Gunter MJ, Jenkins MA, Slattery ML, Lemire M, Woods MO, Song M, Murphy N, Lindor NM, Dikilitas O, Pharoah PDP, Campbell PT, Newcomb PA, Milne RL, MacInnis RJ, Castellví-Bel S, Ogino S, Berndt SI, Bézieau S, Thibodeau SN, Gallinger SJ, Zaidi SH, Harrison TA, Keku TO, Hudson TJ, Vymetalkova V, Moreno V, Martín V, Arndt V, Wei WQ, Chung W, Su YR, Hayes RB, White E, Vodicka P, Casey G, Gruber SB, Schoen RE, Chan AT, Potter JD, Brenner H, Jarvik GP, Corley DA, Peters U, Hsu L. Genome-wide Modeling of Polygenic Risk Score in Colorectal Cancer Risk. Am J Hum Genet. 2020;107:432-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 41. | Xu LL, Lin Y, Han LY, Wang Y, Li JJ, Dai XY. Development and validation of a prediction model for early screening of people at high risk for colorectal cancer. World J Gastroenterol. 2024;30:450-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (1)] |