Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2757
Revised: March 12, 2024
Accepted: April 12, 2024
Published online: June 15, 2024
Processing time: 137 Days and 9.8 Hours
Gastric cancer (GC) has a high mortality rate, and robust diagnostic biomarkers are currently lacking. However, the clinical relevance of circular RNAs (circRNAs) as GC biomarkers remains largely unexplored.
To evaluate the potential of novel circRNA circ_0004592 in the early screening and prognosis of GC.
High-throughput sequencing of circRNAs was performed to screen for potential target molecules. Circ_0004592 expression was examined in GC tissues, cells, and plasma. Plasma samples were collected from healthy subjects’ patients, as well as from patients with benign lesions, precancerous lesions, and GC, whereafter the diagnostic accuracy of circ_0004592 was evaluated. The correlation between circ_0004592 levels in plasma and clinicopathological data of patients with GC was further analyzed.
Circ_0004592 was upregulated in both the tissue and plasma of patients with GC. Further, circ_0004592 expression was higher in patients with precancerous lesions than in healthy controls while being highest in patients with GC. In the same patient, the postoperative plasma level of circ_0004592 was lower than that in the preoperative period. Moreover, circ_0004592 level was significantly correlated with tumor differentiation, tumor depth, and lymph node metastasis. The area under the curve (AUC) of plasma circ_0004592 exhibited high sensitivity and specificity for differentiating patients with GC from healthy donors. Diagnosis based on circ_0004592, carcinoembryonic antigen, and cancer antigen 199 achieved a superior AUC and was highly sensitive.
Plasma circ_0004592 may represent a potential non-invasive auxiliary diagnostic biomarker for patients with GC.
Core Tip: Circ_0004592 was significantly overexpressed in gastric cancer (GC) tissues and plasma. Reverse transcription and real-time fluorescent quantitative polymerase chain reaction was validated as a robust approach for the detection of circ_0004592. The expression level of circ_0004592 in the plasma of patients with precancerous lesions was increased relative to that in healthy controls, suggesting that plasma circ_0004592 may serve as an early diagnostic biomarker. Further, its levels in plasma decreased following surgical resection of gastric tumors. High circ_0004592 expression was associated with the histological type, depth of tumor invasion, and lymph node metastasis of GC. These observations highlight the potential of circ_0004592 as a valuable diagnostic and prognostic biomarker.
- Citation: Kong S, Xu YH, Zheng M, Ju SQ, Shi HC. Circ_0004592: An auxiliary diagnostic biomarker for gastric cancer. World J Gastrointest Oncol 2024; 16(6): 2757-2768
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2757.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2757
Gastric cancer (GC) is a malignant tumor originating in the gastric mucosal epithelium. Approximately 400000 new cases of GC occur in China every year, accounting for approximately 42% of the global cases. GC ranks first among all digestive tract tumors regarding mortality[1,2]. GC can occur in any part of the stomach, most commonly arising in the sinus (48.8%-52.5%). The greater curvature, lesser curvature, anterior wall, and posterior wall can all be affected, followed by the cardia (16.1%-20.6%), while the gastric body and the whole stomach are rarely affected (7.0%-16.6%). GC usually manifests as a single tumor, but can manifest as multiple cancerous lesions[3]. Most GC cases are of early-stage adenocarcinomas with no noticeable symptoms. The diagnosis of GC is based on upper gastrointestinal endoscopy. However, in most countries, upper gastrointestinal endoscopy is not a routine physical examination. Further, the conventional tumor biomarkers carcinoembryonic antigen (CEA) and cancer antigen 199 (CA199) are not specific or sensitive enough[4]. As a result, most patients with stomach cancer are diagnosed at an advanced stage[5]. Therefore, the search for stable and efficient diagnostic biomarkers is crucial for improving the early detection rate of GC.
Circular RNAs (circRNAs) are a new class of non-cording RNA molecules with a circular structure. Unlike other linear RNAs, circRNAs do not possess poly A tails[6]. Depending on their origin, circRNAs can be classified into exon, intron, and exon-intron circRNAs[6]. The covalently closed ring structure of circRNAs renders them more resistant to RNA exonucleases and results in a longer half-life[7-10]. Some studies have shown that circRNAs are abundant in body fluids[11-13]. Therefore, in recent years, an increasing number of studies have highlighted circRNAs as potential tumor biomarkers[14]. For example, hsa_circ_0001715 is upregulated in lung tissue and plasma, representing a potential noninvasive diagnostic biomarker and prognostic predictor for lung adenocarcinoma[15]. Hsa_circ_0065149 was significantly downregulated in the plasma exosomes of patients with early GC, exhibiting higher sensitivity and specificity than traditional clinical biomarkers for early GC diagnosis[16].
In addition, new evidence suggests that dysregulated circRNA expression is implicated in tumor development and progression[17,18]. Functionally, circRNAs can serve as sponges for miRNAs or effective competitive endogenous RNA to regulate the expression of downstream target proteins[19]. In addition, circRNAs can be used as baits to lure proteins away from their sites of function[20]. CircRNAs can also bind directly to transcription factors to regulate classical RNA splicing and protein translation[21]. For example, by serving as a miRNA sponge, hsa_circ_0004872 was shown to bind miR-224, inhibiting the proliferation, invasion, and migration of GC cells, thus increasing the expression of endogenous miR-224 target proteins p21 and Smad4[22]. Circ-FOXO3 exhibited a high affinity for the anti-aging ID-1, transcription factor E2F1, anti-stress protein FAK, and HIF1α, retaining these factors in the cytoplasm to aggravate cellular aging[23]. In addition, a recent study revealed that circMAPK1 encodes the protein MAPK1-109aa and inhibits the malignant behavior of GC cells by suppressing the activation of MAPK signaling[21].
In the present study, we detected the expression levels of plasma circ_0004592 in patients with primary GC, gastric benign lesions, and precancerous lesions to explore its potential as a biomarker for clinical diagnosis and prognosis of GC.
Total RNA was reverse-transcribed into complementary DNA (cDNA) using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, United States). The 20 μL RT reactions were incubated at 25 °C for 5 min, 42 °C for 1 h, and 70 °C for 5 min. The reverse transcription and real-time fluorescent quantitative polymerase chain reaction (qRT-PCR) program consisted of 40 cycles, each of which included 95 °C pre-amplification for 10 min, 95 °C denaturation for 15 s, annealing at 58 °C for 34 s, and extension at 72 °C for 30 s, performed on a Roche 480 (Roche, Germany). Primers were synthesized by Sangon Biotech (Shanghai, China). Relative expression of target genes was determined via the 2−∆∆Ct method.
Three pairs of GC tissues and corresponding paracancerous tissues were selected for RNA extraction. A Kapa RNA HyperPrep Kit with RiboErase (HMR; Illumina) was used to prepare the sequencing library: (1) Ribosomal RNA was removed from the total RNA. The ribosome-depleted RNA was incubated for 30 min at 37 °C with 10 units RNase R and purified with VAHTS RNA Clean Beads; (2) the RiboMinus RNase R(+) RNA was fragmented, whereafter, first-strand and directional second-strand syntheses were performed; (3) a tailing/adapter ligation approach was performed with the purified cDNA; and (4) the purified, adapter-ligated DNA was amplified. The library quality and concentration were assessed using a DNA 1000 chip on an Agilent 2100 Bioanalyzer. Accurate quantification for sequencing applications was determined using the qPCR-based KAPA Biosystems Library Quantification kit. Each library was diluted to 10 nM and pooled equimolar prior to clustering. Paired-end (PE150) sequencing was performed on all samples.
As summarized in Table 1, plasma was collected from 100 patients with primary GC who underwent tumor resection after the initial diagnosis and had not received any radiotherapy or chemotherapy prior to surgery between June 2020 and December 2022. These patients included 39 female (age range: 44-85 years, mean age: 65 years) and 61 male patients (age range: 42-85 years, mean age: 65 years). The age distribution of healthy males and females was matched to that of patients with GC (age range: 40-88 years, mean age: 63 years), and all physical examination indicators were normal. In addition to the 100 patients with GC and 100 healthy subjects, we included 15 patients with benign gastric lesions and 15 patients with precancerous gastric lesions. The 15 patients with benign gastric lesions included patients with gastric polyps and patients with gastric ulcers; all 15 patients with precancerous lesions had chronic atrophic gastritis with gastric mucosal dysplasia and intestinal epithelial hyperplasia. Preoperative and postoperative specimens were collected from 21 patients with GC. Twenty pairs of tissue samples were immediately fixed with RNAlater after resection in the operating room, marked in a cryopreservation tube, and stored in a refrigerator at -80 °C after quick-freezing with liquid nitrogen. Ethylenediaminetetraacetic acid anticoagulated blood was collected from all subjects in EP tubes, and samples were centrifuged at 1000 g for 10 min within 4 h of isolation. Thereafter, 300 μL of supernatant was aspirated and stored at -80 °C in EP tubes. This study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University, and all participants signed informed consent forms (Table 1).
| Specimen type | Specimen origin | Specimen number |
| Plasma | Patients with primary GC | 39 (female, 44-85 yr) |
| 61 (male, 42-85 yr) | ||
| Healthy subjects | 100 (40-88 yr) | |
| Preoperative and postoperative paired samples | 21 | |
| Patients with benign gastric lesions | 15 | |
| Patients with precancerous lesions | 15 | |
| Tissue | Gastric cancer tissues | 20 |
| Matching para-cancerous tissues | 20 |
Total RNA was extracted from tissues and cells using TRIzol Reagent (Invitrogen). Total RNA in each sample was quantified on NanoDropTM One (Thermo Fisher Scientific, United States). The NanoDrop microvolume quantification system uses a combination of fiber optic technology and natural surface tension characteristics to determine samples, not only to maximize the retention of trace samples, but also to measure a wider range of nucleic acid concentrations, essentially eliminating the need for dilution. The purine and pyrimidine rings of nucleic acid contain conjugated double bonds, which have strong absorption of ultraviolet light at about 260 nm. The concentration of RNA can be calculated by measuring the absorption value of 260 nm. The purity of the sample was determined by 260-280 nm spectrophotometry. The A260/A280 ratio ranges from 1.8-2.2. A low value indicates protein/peptide/phenol contamination. The high value, for RNA samples, may be the degradation of the sample into oligonucleotides or the presence of guanidine isothiocyanate contamination.
Human GC cell lines (HGC-27, BGC-823, MKN-1, and MKN-45) and gastric epithelial cell line were purchased from the Stem Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The composition of the culture medium for all cell lines was 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, United States) and RPMI-1640 medium (Corning, Manassas, VA, United States) supplemented with penicillin/streptomycin. Cells were cultured at 37 °C in an incubator containing 5% CO2.
Next, 10 μg of total RNA was incubated with 3-4 U/μg ribonuclease (RNase R) (Geneseed Biotech, Guangzhou, Guangdong Province, China) at 37 °C for 45 min, adding 5 μL 10-fold reaction buffer and RNase-free water to a total volume of 50 μL. The enzyme was inactivated by the reaction mixture at 70 °C for 10 min and then reverse transcribed.
One mg/mL actinomycin D was diluted to 2.5 μg/mL using the complete medium, and the complete medium was used to replace the common medium in a six-well plate. RNA was extracted from cells at 0 h, 2 h, 4 h, 8 h, 12 h, and 24 h.
Statistical analysis was carried out using GraphPad Prism 7.0 and SPSS 20.0. Heat and volcano maps were created using R version 3.5.1. The two groups of data were compared using the Student’s t-test, and multi-group comparisons were performed via one-way ANOVA. The receiver operating characteristic (ROC) curve was established to analyze the diagnostic value, and the Youden index (also known as the correct index; Youden index = specificity + sensitivity-1) was calculated to evaluate the authenticity of the screening test.
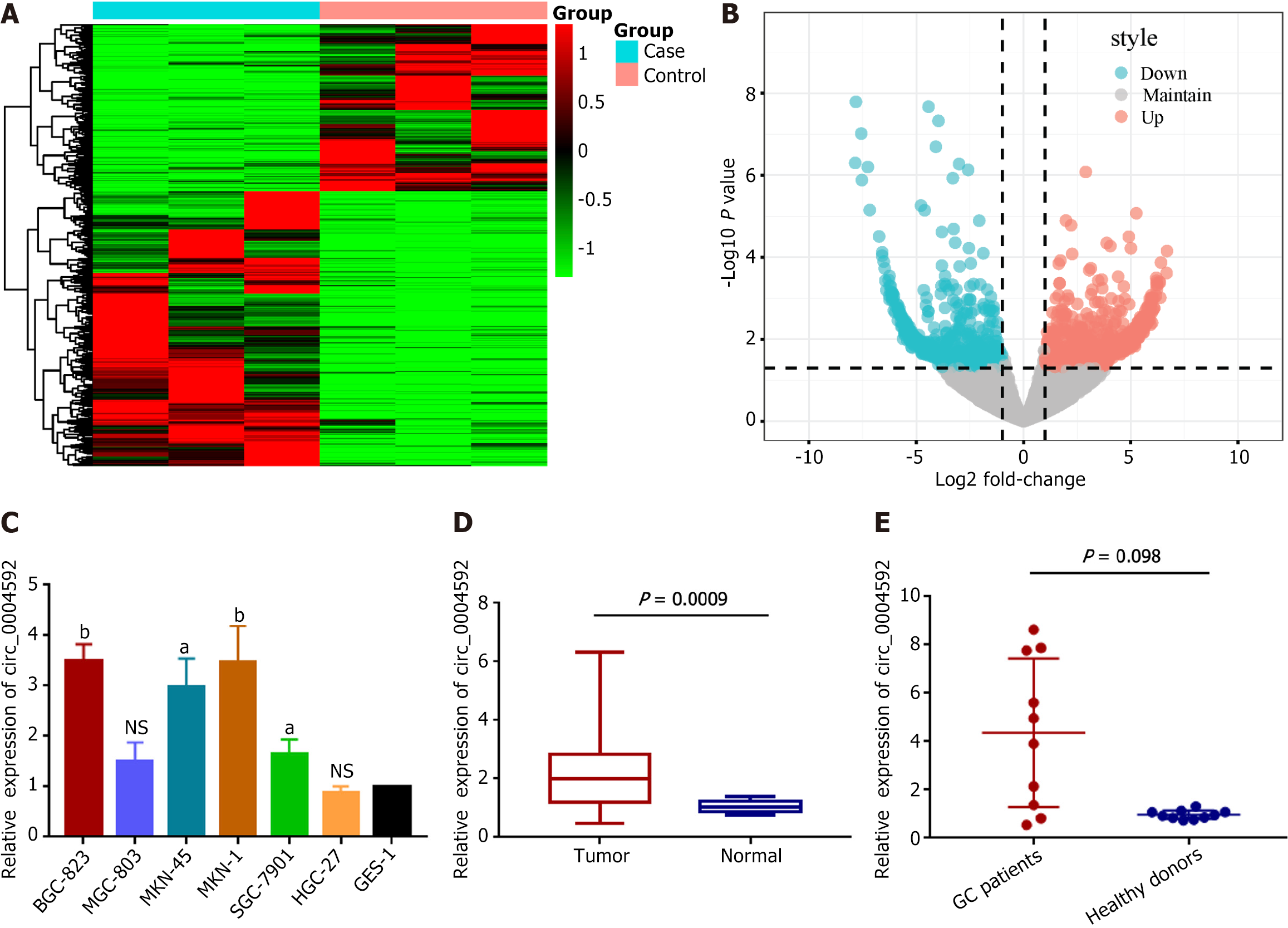
High-throughput sequencing was performed on three pairs of GC and matched normal gastric mucosal tissues to screen for abnormally expressed circRNAs. Hierarchical clustering analysis showed that circRNAs were differentially expressed between GC and adjacent normal tissues (Figure 1A). Overall, 15607 circRNAs were detected, among which 14295 circRNAs were maintained in both cancerous and para-cancerous tissues. One thousand three hundred and twelve differentially expressed circRNAs (fold-change > 2.0, P value < 0.05) were identified; of them, 815 were upregulated and 497 were downregulated in GC (Figure 1B). Combined with the CircBank database, which provides the basic characteristics of the candidate molecules, we selected hsa_circ_0004592 with noticeable expression differences. We verified expression levels in six GC cell lines (Figure 1C) and 20 pairs of GC tissues (Figure 1D). qRT-PCR indicated an upregulation of circ_0004592 in GC, which was consistent with the sequencing results. Subsequently, we detected the expression levels of circ_0004592 in the plasma of 10 patients with GC and 10 healthy subjects, observing an upregulation in the former (Figure 1E).
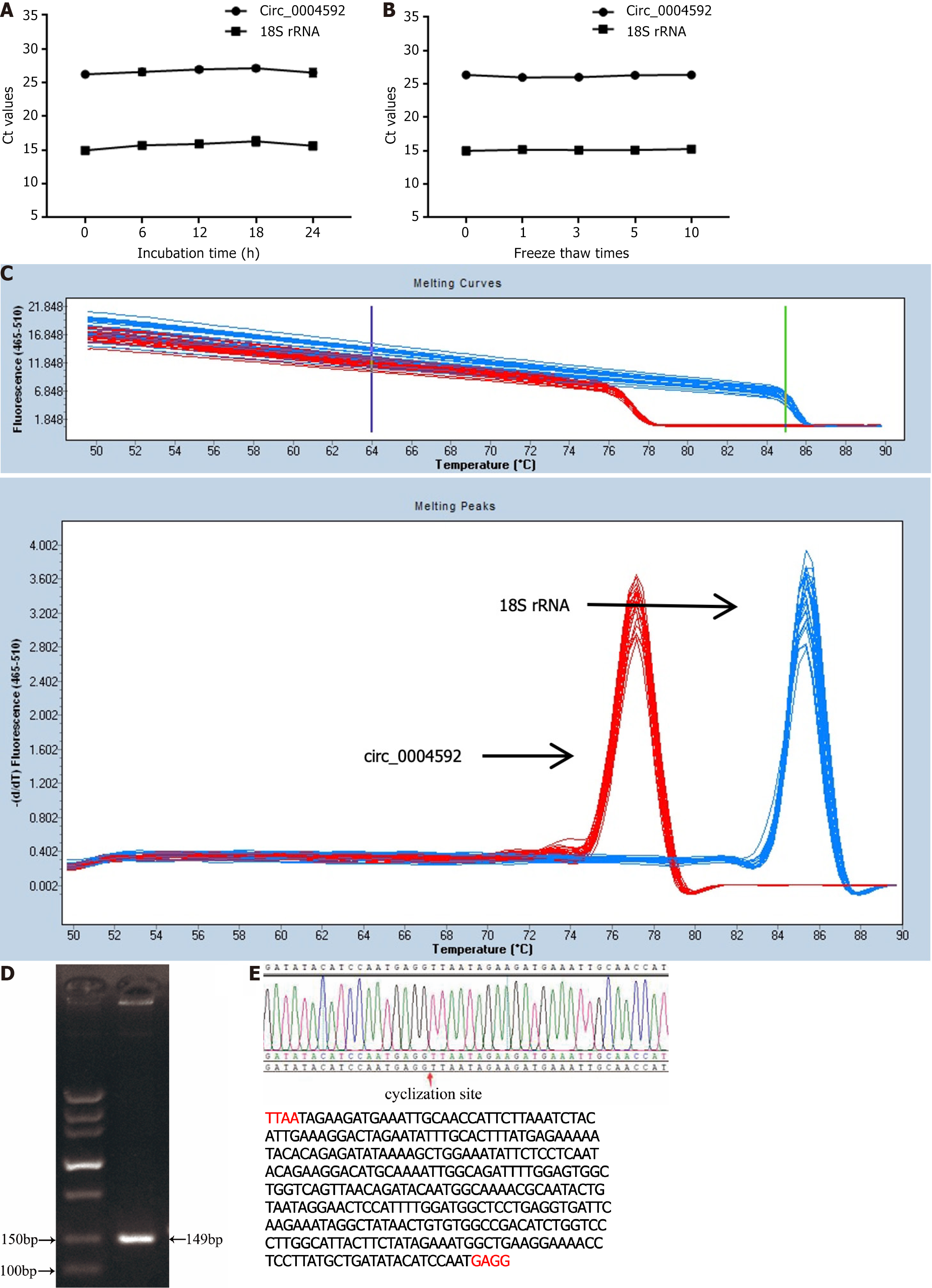
To validate the measured circ_0004592 expression differences, we selected several commonly used internal reference genes: 18S rRNA, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin (ACTB), α-tubulin (TUB), and RNA polymerase II (RP II). Internal reference gene expression was detected in the plasma of 10 healthy donors and 10 patients with GC, revealing the highest expression of 18S rRNA and the lowest CT value (Supplementary Table 1, Supplementary Figure 1). Furthermore, no significant difference was observed in the expression of 18S rRNA between patients with GC and normal controls (Supplementary Figure 2). We therefore used 18S in subsequent experiments. We then extracted 10 mixed plasma RNA from the same batch and detected the expression levels of circ_0004592 and 18S rRNA in the same batch. In addition, we extracted RNA from the mixed plasma in 10 batches and detected the expression levels of circ_0004592 and 18S rRNA in these batches. The inter- and intra-assay coefficients of variation (CV) of circ_0004592 and 18S rRNA were less than 5%. The repeatability met the requirements of the experiment (Table 2). In addition, the mixed plasma was placed at room temperature for 0 h, 6 h, 12 h, 18 h, and 24 h or freeze-thawed 0, 1, 3, 5, and 10 times, whereafter the results showed stable CT values for circ_0004592 and 18S rRNA (Figure 2A and B). Finally, we designed specific reverse primers for the cyclization site, and the melting curve of circ_0004592 was single peak-specific (Figure 2C). Further, the AGE band was single, and the size of the product was the same as that of the predicted target gene product (Figure 2D). Sanger sequencing results were consistent with CircBank data (Figure 2E).
| Intra-assay | Inter-assay | |
| mean ± SD, CV (%) | mean ± SD, CV (%) | |
| Circ_0004592 | 23.23 ± 0.48, 2.06 | 24.01 ± 0.91, 3.79 |
| 18S rRNA | 15.32 ± 0.28, 1.83 | 14.87 ± 0.56, 3.77 |
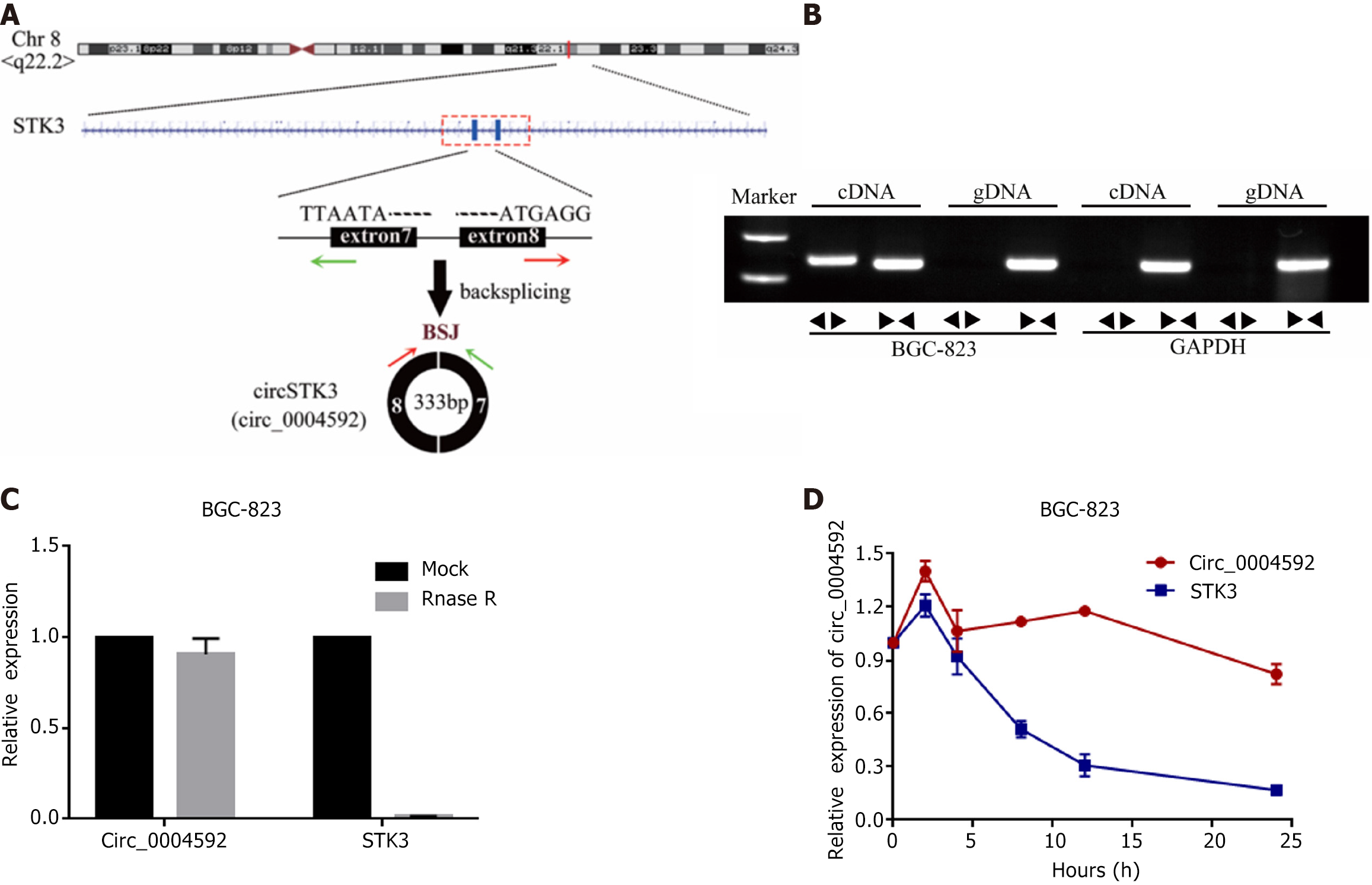
Circ_0004592 is located on chromosome 8, derived from exon 7 and 8. Mature transcripts were 333 bp in length (Figure 3A). Genomic DNA (gDNA) and cDNA were used as templates for PCR. The AGE of PCR products indicated that circ_0004592 could be amplified from PCR products with cDNA as a template, but no noticeable band was found in the control group with gDNA as a template (Figure 3B). CircRNAs are more stable than linear RNA. After RNA treatment with the exonuclease, the expression level of circ_0004592 remained almost unchanged (Figure 3C). Circ _0004592 expression was detected 24 h after RNA cells were treated with transcription inhibitor actinomycin D. Compared with linear RNA, circ_0004592 had a longer half-life (Figure 3D).
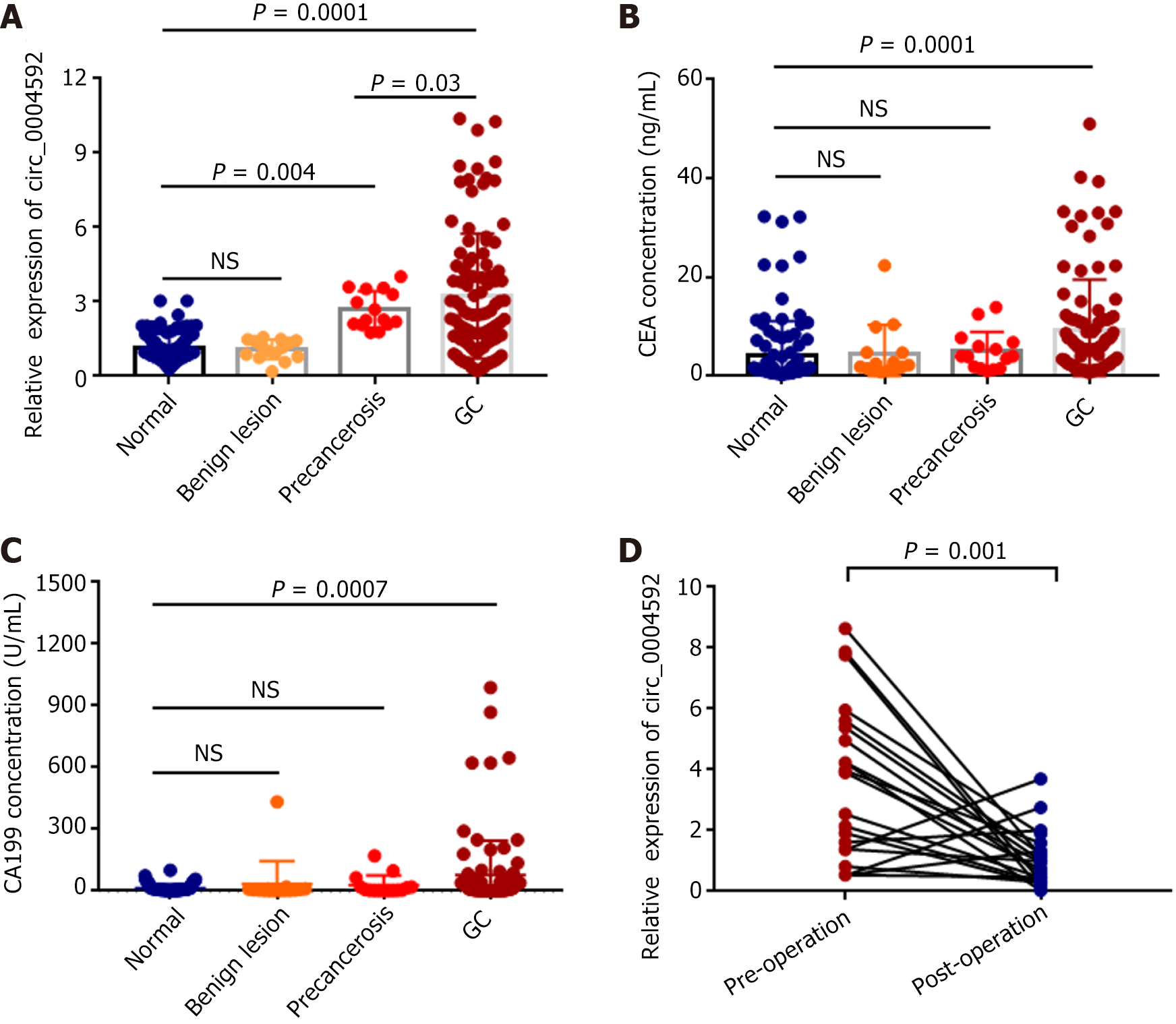
Based on these results, we explored the potential of circ_0004592 as a diagnostic biomarker in GC. Plasma samples were collected from 100 patients with primary GC, 15 with benign gastric lesions, 15 with precancerous gastric lesions, and 100 healthy controls. The expression levels of circ_0004592 in the plasma of subjects were detected using qRT-PCR. The results showed that circ_0004592 levels were significantly higher in the plasma of patients with primary GC than in healthy controls and in patients with benign gastric lesions (P = 0.0001). Expression of circ_0004592 in the plasma of patients with benign lesions was not significantly different from that in the plasma of healthy controls. In addition, the plasma circ_0004592 expression level was higher in patients with precancerous lesions than in healthy donors (P = 0.004). The results also indicated that circ_0004592 expression levels were higher in patients with GC than in patients with precancerous lesions (P = 0.03) (Figure 4A). The levels of CEA (P = 0.0001) and CA199 (P = 0.0007) were also upregulated in patients with GC (Figure 4B and C). We also collected plasma samples from 21 patients with primary GC who underwent radical gastrectomy and measured circ_0004592 expression levels. Plasma circ_0004592 levels in the same patient decreased after surgery (P = 0.001) (Figure 4D).
Patients with GC were divided into high and low expression groups according to median plasma circ_0004592 expression. Chi-square or Fisher’s exact tests indicated that high levels of circ_0004592 were associated with histological classification (P = 0.002), depth of tumor invasion (P = 0.004), and lymph node metastasis (P = 0.022). Of note, correlation existed between the levels of plasma circ_0004592 and CEA in plasma (Table 3), indicating the potential of circ_0004592 as a biomarker of GC. However, no significant correlation was found between plasma circ_0004592 and other clinicopathological features, such as sex, age, tumor size, and serum CA199.
| Characteristics | No. of patients | Low expression | High expression | P value |
| Gender | ||||
| Male | 61 | 33 | 28 | 0.206 |
| Female | 39 | 17 | 22 | |
| Age | ||||
| ≤ 60 | 34 | 14 | 20 | 0.146 |
| > 60 | 66 | 36 | 30 | |
| Tumor size, cm | ||||
| < 5 | 72 | 36 | 36 | 0.176 |
| ≥ 5 | 28 | 14 | 14 | |
| Pathological differentiation | ||||
| Poor-undifferentiation | 59 | 22 | 37 | 0.002b |
| Well-moderate | 41 | 28 | 13 | |
| Tumor depth | ||||
| T1-T2 | 50 | 32 | 18 | 0.004b |
| T3-T4 | 50 | 18 | 32 | |
| Lymph node metastasis | ||||
| Yes | 49 | 19 | 30 | 0.022a |
| No | 51 | 31 | 20 | |
| Nerve/vascular invasion | ||||
| Positive | 48 | 22 | 26 | 0.274 |
| Negative | 52 | 28 | 24 | |
| CEA | ||||
| Positive | 60 | 36 | 24 | 0.012a |
| Negative | 40 | 14 | 26 | |
| CA199 | ||||
| Positive | 51 | 25 | 26 | 0.500 |
| Negative | 49 | 25 | 24 |
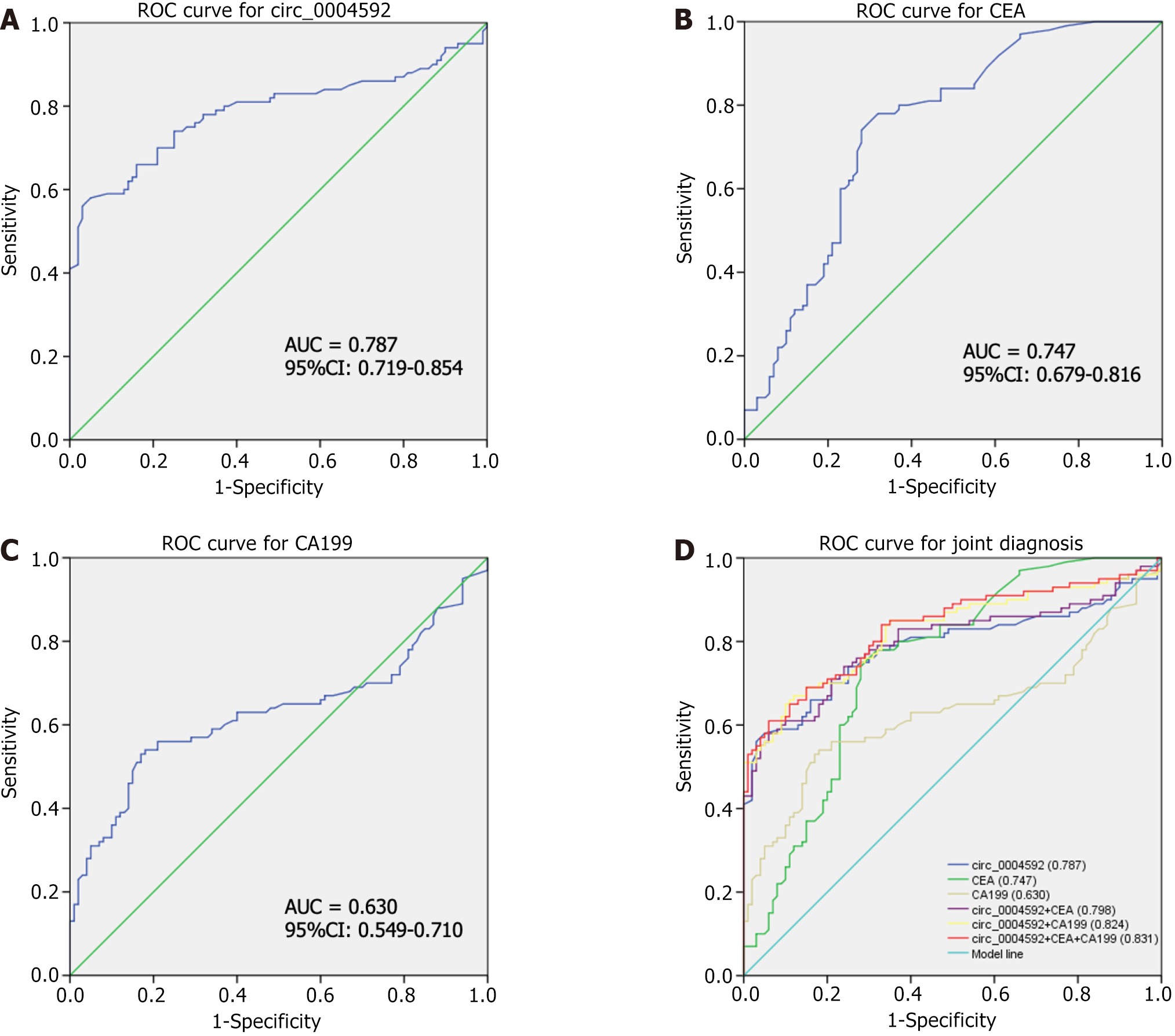
We further explored the potential of circ_0004592 as a diagnostic biomarker for GC. A ROC curve was drawn according to the expression levels of circ_0004592, CEA, and CA199 in 100 patients with primary GC and 100 healthy controls. The area under the curve (AUC) was used to evaluate diagnostic efficiency. The results showed that the AUC of circ_0004592 for differentiating patients with GC from healthy donors was 0.787 [95% confidence interval (95%CI): 0.719-0.854] (Figure 5A). The AUC for CEA was 0.747 (95%CI: 0.679-0.816) (Figure 5B), and the AUC of CA199 was 0.630 (95%CI: 0.549-0.710) (Figure 5C). Circ_0004592 was therefore more effective than CA199 for the diagnosis of GC. When the cut-off value was defined at 1.700, the Youden index (sensitivity + specificity-1) was 0.500. Sensitivity (66.00%), specificity (84.00%), positive predictive value (80.49%), negative predictive value (71.19%), and accuracy (75.00%) of circ_0004592 were higher than those of CEA and CA199. An increasing number of reports have shown that, compared with a single diagnostic index, joint diagnosis significantly improves diagnostic efficiency[24,25]. Therefore, we explored whether the combined use of circ_0004592, CEA, and CA199 could improve diagnostic accuracy. The diagnostic efficacy of circ_0004592 combined with CEA was 0.798 (95%CI: 0.733-0.863), and that of circ_0004592 combined with CA199 was 0.824 (95%CI: 0.764-0.883). Compared with that of single indexes and pairwise combinations, the diagnostic efficiency of combining all three was the highest, at 0.831 (95%CI: 0.773-0.889) (Figure 5D). Simultaneously, the sensitivity for GC diagnosis increased to 85.00%, which greatly improved detection rate (Table 4).
| SEN, % | SPE, % | ACCU, % | PPV, % | NPV, % | |
| Circ_0004592 | 66.00 (66/100) | 84.00 (84/100) | 75.00 (150/200) | 80.49 (66/82) | 71.19 (84/118) |
| CEA | 60.00 (60/100) | 77.00 (77/100) | 68.50 (137/200) | 72.29 (60/83) | 65.81 (77/117) |
| CA199 | 51.00 (51/100) | 84.00 (84/100) | 67.50 (135/200) | 76.12 (51/67) | 63.16 (84/133) |
| Circ_0004592 + CEA | 74.00 (74/100) | 76.00 (76/100) | 75.00 (150/200) | 75.51 (74/98) | 74.51 (76/102) |
| Circ_0004592 + CA199 | 67.00 (67/100) | 88.00 (88/100) | 77.50 (155/200) | 84.81 (67/79) | 72.73 (88/121) |
| Circ_0004592 + CEA + CA199 | 85.00 (85/100) | 65.00 (65/100) | 75.00 (150/200) | 70.83 (85/120) | 81.25 (65/80) |
Advances in medicine have led to great progress in the diagnosis and treatment of GC. However, the mortality rate of GC remains high, with invasion and metastasis posing a significant challenge to clinical diagnosis and treatment[26]. The key factors leading to high GC mortality include late diagnosis and tumor heterogeneity. The current treatment options for GC include endoscopy, gastrectomy, chemotherapy, or chemoradiotherapy[27]. In recent decades, studies into the molecular mechanism of GC pathogenesis have revealed various roles for non-coding RNAs (miRNAs, lncRNAs, and circRNAs) in the occurrence and development of GC[28-30].
In recent years, circRNAs have become a focus of research owing to their remarkable characteristics. They are highly conserved, cannot be degraded, and are widely expressed in human cells, sometimes at levels higher than linear RNA[31]. Therefore, circRNAs may be superior to long non-coding RNAs and miRNAs for the purpose of cancer diagnosis. While many studies have focused on the functional exploration of circRNAs, their diagnostic value remains underexplored[32].
In this study, circRNA sequencing and GEO database analysis were used to screen out circRNAs with abnormal expression in GC. Circ_0004592 was significantly overexpressed in GC tissues and plasma. qRT-PCR was validated as a robust approach for the detection of circ_0004592. The expression level of circ_0004592 in the plasma of patients with precancerous lesions was increased relative to that in healthy controls, suggesting that plasma circ_0004592 may serve as an early diagnostic biomarker. Further, its levels in plasma decreased following surgical resection of gastric tumors. These observations highlight the potential of circ_0004592 as a valuable diagnostic and prognostic biomarker. In addition, high circ_0004592 expression was associated with the histological type, depth of tumor invasion, and lymph node metastasis of GC.
CEA and CA199 are the most commonly used serum markers for GC[33]. Nevertheless, the positive rate during early GC screening is extremely low[34-36]. Yang et al[37] found that plasma circ-LDLRAD3 content was significantly increased in patients with pancreatic cancer, and combined detection with CA19-9 could significantly improve diagnostic efficiency. When the AUC was used to evaluate the diagnostic efficiency of circ_0004592, the sensitivity and specificity of the AUC of circ_0004592 were higher than those of CEA and CA199. When the three indices were combined, the AUC was the largest, and the sensitivity was the highest.
In the present study, we confirmed the high expression of circ_0004592 in GC tissues and plasma. More importantly, we demonstrated its diagnostic potential for the first time. However, the mechanisms of action of circulating RNAs in the blood are complex. Some RNAs in bodily fluids may be byproducts of cancer cell death. Recent studies have shown that circRNAs can be transferred via exosomes and taken up by other tumor cells. Thus, GC cells may not only selectively release cellular RNA in different forms but also selectively take up certain RNAs[38,39]. This suggests that circRNAs in plasma do not fully reflect their expression in tumor tissue. While the expression level of circ_0004592 was increased in the plasma of patients, this does not rule out the possibility that circ_0004592 is secreted by other tumor cells. Therefore, the potential of circ_0004592 as a non-invasive diagnostic marker needs to be further determined in larger patient cohorts.
Plasma circ_0004592 may represent a potential non-invasive auxiliary diagnostic biomarker for patients with GC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s classification
Scientific Quality: Grade B
Novelty: Grade B
Creativity or Innovation: Grade A
Scientific Significance: Grade B
P-Reviewer: Exbrayat JM, France S-Editor: Chen YL L-Editor: A P-Editor: Zhang XD
| 1. | Kim IW, Jang H, Kim JH, Kim MG, Kim S, Oh JM. Computational Drug Repositioning for Gastric Cancer using Reversal Gene Expression Profiles. Sci Rep. 2019;9:2660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55820] [Article Influence: 7974.3] [Reference Citation Analysis (132)] |
| 3. | Yan Y, Chen Y, Jia H, Liu J, Ding Y, Wang H, Hu Y, Ma J, Zhang X, Li S. Patterns of Life Lost to Cancers with High Risk of Death in China. Int J Environ Res Public Health. 2019;16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1159] [Cited by in RCA: 1327] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 5. | Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, Ogunbiyi OJ, Azevedo E Silva G, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP; CONCORD Working Group. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2711] [Cited by in RCA: 3422] [Article Influence: 488.9] [Reference Citation Analysis (1)] |
| 6. | Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71:428-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 988] [Cited by in RCA: 1484] [Article Influence: 212.0] [Reference Citation Analysis (0)] |
| 7. | Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1230] [Cited by in RCA: 1621] [Article Influence: 180.1] [Reference Citation Analysis (0)] |
| 8. | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3481] [Cited by in RCA: 3419] [Article Influence: 284.9] [Reference Citation Analysis (0)] |
| 9. | Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1370] [Cited by in RCA: 1808] [Article Influence: 180.8] [Reference Citation Analysis (0)] |
| 10. | Zhang C, Wu H, Wang Y, Zhu S, Liu J, Fang X, Chen H. Circular RNA of cattle casein genes are highly expressed in bovine mammary gland. J Dairy Sci. 2016;99:4750-4760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 11. | Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 527] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 12. | Liang Z, Guo W, Fang S, Zhang Y, Lu L, Xu W, Qian H. CircRNAs: Emerging Bladder Cancer Biomarkers and Targets. Front Oncol. 2020;10:606485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015;10:e0141214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 503] [Cited by in RCA: 517] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 14. | Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 15. | Lu GJ, Cui J, Qian Q, Hou ZB, Xie HY, Hu W, Hao KK, Xia N, Zhang Y. Overexpression of hsa_circ_0001715 is a Potential Diagnostic and Prognostic Biomarker in Lung Adenocarcinoma. Onco Targets Ther. 2020;13:10775-10783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, Ye G, Guo J. Hsa_circ_0065149 is an Indicator for Early Gastric Cancer Screening and Prognosis Prediction. Pathol Oncol Res. 2020;26:1475-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 17. | Liu J, Zhang X, Yan M, Li H. Emerging Role of Circular RNAs in Cancer. Front Oncol. 2020;10:663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 18. | Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, Li X, Sun Z. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 180] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 19. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6045] [Article Influence: 503.8] [Reference Citation Analysis (0)] |
| 20. | Xie F, Huang C, Liu F, Zhang H, Xiao X, Sun J, Zhang X, Jiang G. CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021;20:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 21. | Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, Xuan Z, Xie L, Qiu S, He Z, Wang L, Xu Z. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 151] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 22. | Ma C, Wang X, Yang F, Zang Y, Liu J, Xu X, Li W, Jia J, Liu Z. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020;19:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 23. | Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402-1412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 359] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 24. | Yang Q, Kong S, Zheng M, Hong Y, Sun J, Ming X, Gu Y, Shen X, Ju S. Long intergenic noncoding RNA LINC00173 as a potential serum biomarker for diagnosis of non-small-cell lung cancer. Cancer Biomark. 2020;29:441-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Zong W, Feng W, Jiang Y, Ju S, Cui M, Jing R. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clin Chem Lab Med. 2019;57:1063-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20512] [Article Influence: 2051.2] [Reference Citation Analysis (20)] |
| 27. | Matboli M, El-Nakeep S, Hossam N, Habieb A, Azazy AE, Ebrahim AE, Nagy Z, Abdel-Rahman O. Exploring the role of molecular biomarkers as a potential weapon against gastric cancer: A review of the literature. World J Gastroenterol. 2016;22:5896-5908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869-881.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1086] [Cited by in RCA: 1185] [Article Influence: 197.5] [Reference Citation Analysis (0)] |
| 29. | Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer. 2020;19:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 207] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 30. | Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut. 2021;70:784-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 301] [Article Influence: 75.3] [Reference Citation Analysis (0)] |
| 31. | Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1572] [Cited by in RCA: 2073] [Article Influence: 207.3] [Reference Citation Analysis (0)] |
| 32. | Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, Xu X, Liang Q, Christiani DC, Wang M, Liu L, Du M. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 210] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 33. | Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X, Xu C, Xiao B, Sun Y, Guo J. Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2016;37:1183-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 34. | Wu J, Li G, Wang Z, Yao Y, Chen R, Pu X, Wang J. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis Markers. 2015;2015:435656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 35. | Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 207] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 36. | Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G, Guo M, Lian X, Fan D, Zhang H. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 267] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 37. | Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:8345-8354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 151] [Cited by in RCA: 152] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 38. | Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, Wu J, Quan W, Yao Y, Zhou Y, Sun Z, Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 334] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 39. | Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 391] [Cited by in RCA: 402] [Article Influence: 80.4] [Reference Citation Analysis (0)] |