Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2716
Revised: March 18, 2024
Accepted: April 15, 2024
Published online: June 15, 2024
Processing time: 170 Days and 10.5 Hours
The role of Sm-like 5 (LSM5) in colon cancer has not been determined. In this study, we investigated the role of LSM5 in progression of colon cancer and the potential underlying mechanism involved.
To determine the role of LSM5 in the progression of colon cancer and the potential underlying mechanism involved.
The Gene Expression Profiling Interactive Analysis database and the Human Protein Atlas website were used for LSM5 expression analysis and prognosis analysis. Real-time quantitative polymerase chain reaction and Western blotting were utilized to detect the expression of mRNAs and proteins. A lentivirus tar
LSM5 was highly expressed in tumor tissue and colon cancer cells. A high ex
LSM5 knockdown inhibited the proliferation and facilitated the apoptosis of colon cancer cells by upregulating p53, CDKN1A and TNFRSF10B.
Core Tip: Sm-like 5 (LSM5) was highly expressed in tumor tissue and cells of colon cancer. Our study indicated that knockdown of LSM5 had an anti-tumor effect in colon cancer by up-regulating p53, cyclin-dependent kinase inhibitor 1A and tumor necrosis factor receptor superfamily 10B, which providing an important insight on the role of LSM5 in colon cancer development.
- Citation: Mo CJ, Deng XY, Ma RL, Zhu K, Shi L, Li K. Sm-like 5 knockdown inhibits proliferation and promotes apoptosis of colon cancer cells by upregulating p53, CDKN1A and TNFRSF10B. World J Gastrointest Oncol 2024; 16(6): 2716-2726
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2716.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2716
Colon cancer is one of the commonest malignancies in the worldwide, and ranks fifth among all tumors in morbidity and mortality[1]. Although the 5-year survival rate of patients with colon cancer has significantly improved since the 1970s[2], the survival of patients with advanced and recurrent colon cancer has still been unsatisfactory. Thus, finding new therapeutic targets and possible treatment strategies is still an urgent need.
The Sm-like (LSM) family has been found in variety of organisms. Studies have shown that LSM proteins can form stable heteromers in tri-snRNP particles and are involved in the splicing and degradation of cytoplasmic mRNA[3]. For example, the nuclear LSM2-LSM8 complex can bind to U6 snRNA and participate in pre-mRNA splicing[4,5]. The cy
Therefore, the objective of this research is to determine the effect of LSM5 in the progression of colon cancer and the underlying mechanism involved.
The following antibodies were used in this study: Anti-LSM5 (Abcam, ab151717, 1:500), anti-GAPDH (Santa-Cruz, sc-32233, 1:2000), anti-rabbit IgG (Santa-Cruz, sc-2004, 1:2000), anti-mouse IgG (Santa-Cruz, sc-2005, 1:2000), anti-p53(CST, #2527, 1:1000), anti-IRF1(Abcam, ab186384, 1:1000), anti-cyclin-dependent kinase inhibitor 1A (CDKN1A) (CST, #2947, 1:500), anti-tumor necrosis factor receptor superfamily 10B (TNFRSF10B) (Abcam, ab8416, 1:250) and anti-MDM2(Abcam, ab38618, 1:500).
Human colon cell lines (HT-29, HCT-116, RKO and SW480) (obtained from the Cell Bank of the Chinese Academy of Sciences) were cultured in the corresponding media supplemented with 10% Fetal Bovine Serum (Ausbian, A11-102) at 37 °C with 5% CO2. The culture medium was changed according to the degree of cell growth.
The different expression level of LSM5 in colon cancer tissue and the adjacent normal tissue was investigated via the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/index.html).
Immunohistochemical staining images of LSM5 in tumor tissue and adjacent normal tissue from patients with colon cancer were obtained from the Human Protein Atlas website. The differences in survival among patients with different levels of LSM5 expression in colon cancer were analyzed via the Human Protein Atlas website. The results of the prog
The real-time analysis has been previously reported[17]. Briefly, TRIzol reagent (Shanghai Pufei Biotech Co., Ltd, 3101-100) was used to obtain total RNA from colon cancer cells. The cDNA was reverse transcribed via an M-MLV kit ac
| Genes | Upstream | Downstream | Size (bp) |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA | 121 |
| LSM5 | CTAACGCTACTACCAACCCGT | TCCTTCTCCTCCAGGAACCAG | 257 |
| CDK1 | GGATGTGCTTATGCAGGATTCC | CATGTACTGACCAGGAGGGATAG | 100 |
| MDM2 | GAATCATCGGACTCAGGTACATC | TCTGTCTCACTAATTGCTCTCCT | 167 |
| CCNE2 | GGAACCACAGATGAGGTCCAT | CCATCAGTGACGTAAGCAAACT | 237 |
| RALB | TCATCAGGAAAGGAGCACT | GAGGGGATACAGGATTGTT | 189 |
| CDKN1A | CTGTCACTGTCTTGTACCCTTGT | AAATCTGTCATGCTGGTCTGC | 113 |
| TNFRSF10B | GCAGACTTGGTGCCCTTTG | CCCACTTTATCAGCATCGTGT | 133 |
| AKT2 | GCGGAAGGAAGTCATCATTG | GTGGGTCTGGAAGGCATAC | 118 |
| TP53 | CCTCCTCAGCATCTTATCC | ACAAACACGCACCTCAAA | 258 |
| IRF1 | GATGCTTCCACCTCTCACC | TGCTCCACCTCCAAGTCC | 201 |
The cells were lysed by RIPA lysis buffer (Beyotime, P0013B/). The lysates were washed with PBS and centrifuged. The abundance of proteins were detected by the BCA Protein Assay Kit (Beyotime, P0010S). The samples were separated via SDS-PAGE and transferred to PVDF membranes (Millipore, IPVH00010). After that, the antibodies were added to in
The cells were seeded into 96-well plates (2000 cells/well). MTT and Celigo cell counting assays were subsequently performed according to published protocols[20].
The cells were seeded into 6-well plates (800 cells/well) and cultured for 11 d and then washed with PBS. Then, 4% paraformaldehyde (1 mL) was used to fix the cells for 30 min-60 min. After that, Giemsa dye (500 µL) was used to stain the cells for 10 min-20 min. ddH2O was used to wash the cells several times, after which the cells were allowed to dry at 24 °C. Then, a microscope was used to scan the cell colonies[18,19].
An apoptosis detection kit (eBioscience, 88-8007) was used in this study. The digested cells were centrifuged and fixed with 70% ethanol. Flow cytometry was used to identify apoptotic cells after those were dyed with annexin-V-APC[21].
The template was set by target gene and assessed by design software (http://tools.genome-engineering.org) to find the interference target sequence of LSM5 which was 5’-TGGTACTCTTCTAGGATTT-3’. The sequence of the negative control was 5’-TTCTCCGAACGTGTCACGT-3’. Then, lentiviral construction and cell transfection were conducted according to a published protocol[18,20].
The Human GeneChip assay was performed with the 3’ IVT Plus Kit (Affymetrix, 901838). The double-stranded cDNA of the target gene was transcribed to synthesize cRNA. The cRNAs were purified, quantified and labelled. Pre hybridization mix was added, and the samples were reacted with the GeneChip. Then, Hybridization Master Mix was mixed with the labelled cRNA, and the mixture was added and reacted with the GeneChip. A GeneChip Scanner and software were used to scan and analyze the GeneChip, respectively. The raw data were filtered and standardized for further bioinformatic analysis. Then, Ingenuity Pathway Analysis (IPA) based on differentially expressed genes (DEGs) was performed. IPA software was used for bioinformatic analysis[22] and to find the potential targets after LSM5 knockdown.
GraphPad Prism 8.3.0 was used to analyze the data. The data are expressed as the mean ± SD. T tests and two-way repeated-measures ANOVA were performed to compare the difference between the two groups. A P value < 0.05 in
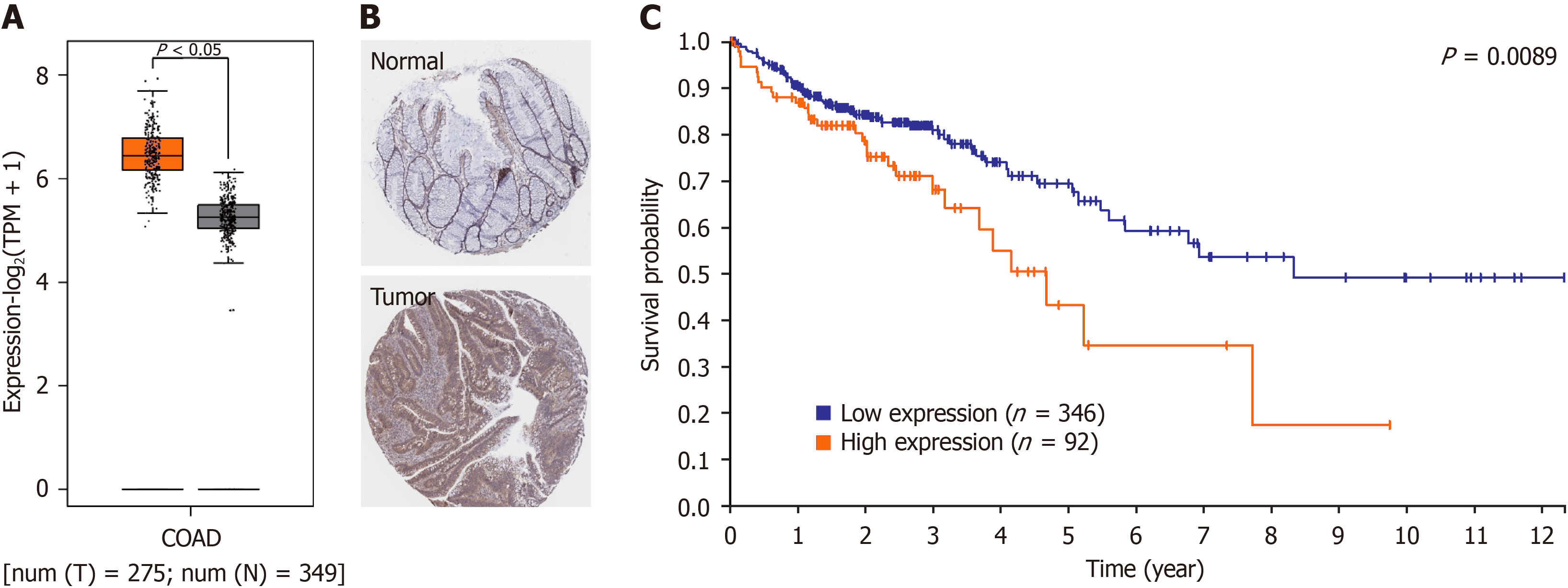
First, we used the GEPIA website to confirm the difference in LSM5 expression between tumor tissue and normal tissue from patients with colon cancer. We found that LSM5 expression was greater in tumor tissue than in paired normal tissue (Figure 1A). Immunohistochemistry of LSM5 from the Human Protein Atlas database also confirmed high expression of LSM5 in tumor tissue (Figure 1B). Further prognostic analysis revealed that higher LSM5 expression was strongly related to poor survival in patients with colon cancer (Figure 1C).
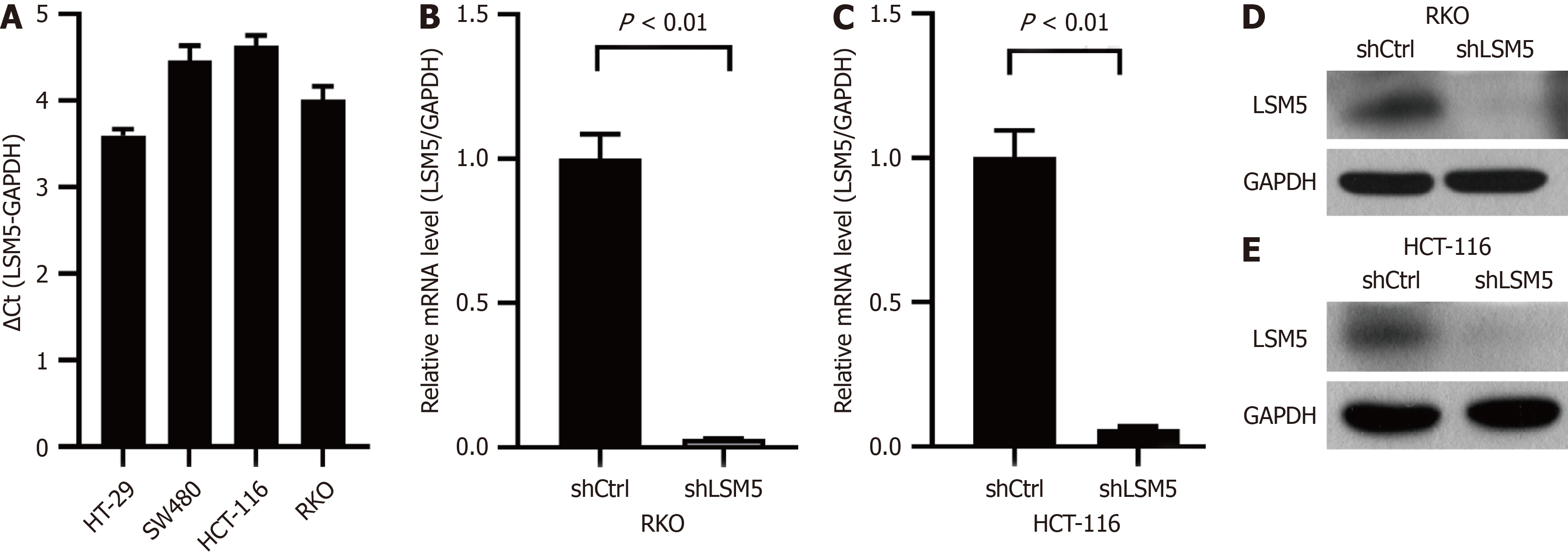
To explore the role of LSM5 in colon cancer, we confirmed that LSM5 was highly expressed in 4 colon cancer cell lines (Figure 2A). Then, we constructed a lentivirus to downregulate the expression level of LSM5, and selected the RKO and HCT-116 cell lines for further experiments. After transfection of the targeted lentivirus, the expression of LSM5 in colon cancer cells was significantly downregulated (Figure 2B-E).
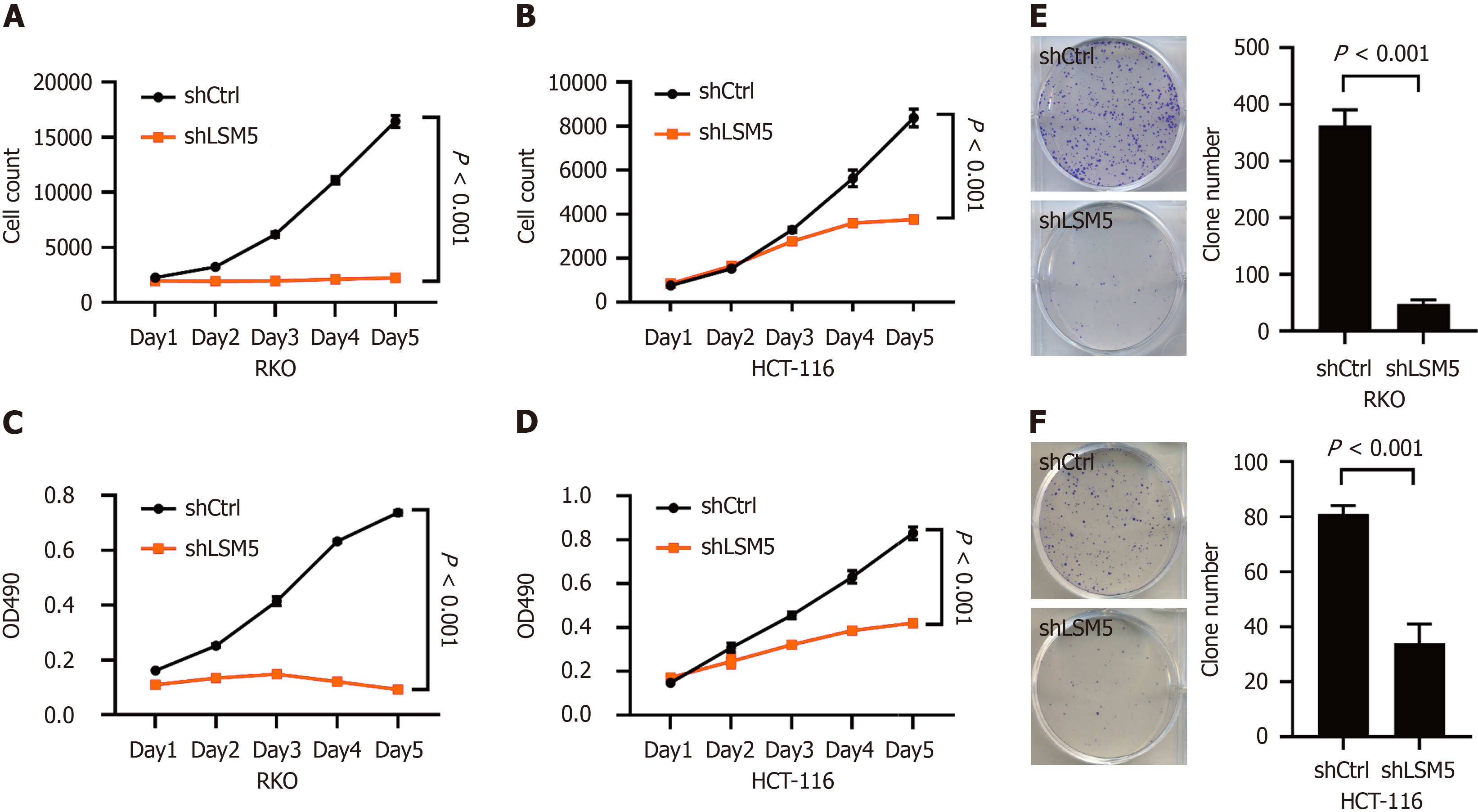
The impact of LSM5 on the growth of colon cancer was further investigated. Cell counting and MTT assays showed that knockdown of LSM5 inhibited the proliferation of colon cancer cells (Figure 3A-D). LSM5 knockdown also restrained the colony-forming ability of colon cancer cells (Figure 3E and F).
Thus, downregulating the expression of LSM5 could significantly inhibit the proliferation of colon cancer cells.
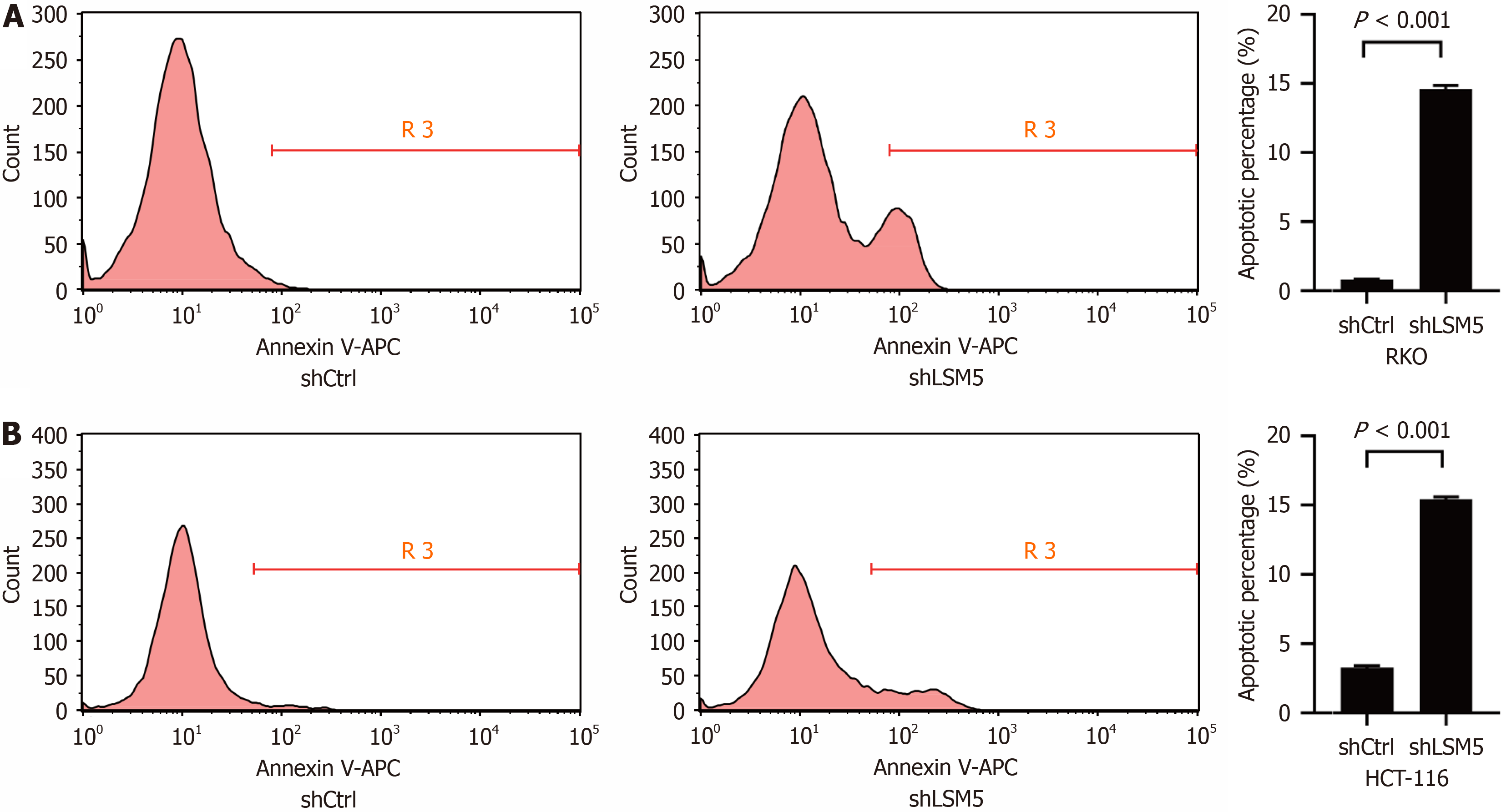
We further detected the impact of LSM5 on the apoptosis of colon cancer cells by using flow cytometry. The results indicated that reducing LSM5 expression promoted the apoptosis of LSM5 cells (Figure 4).
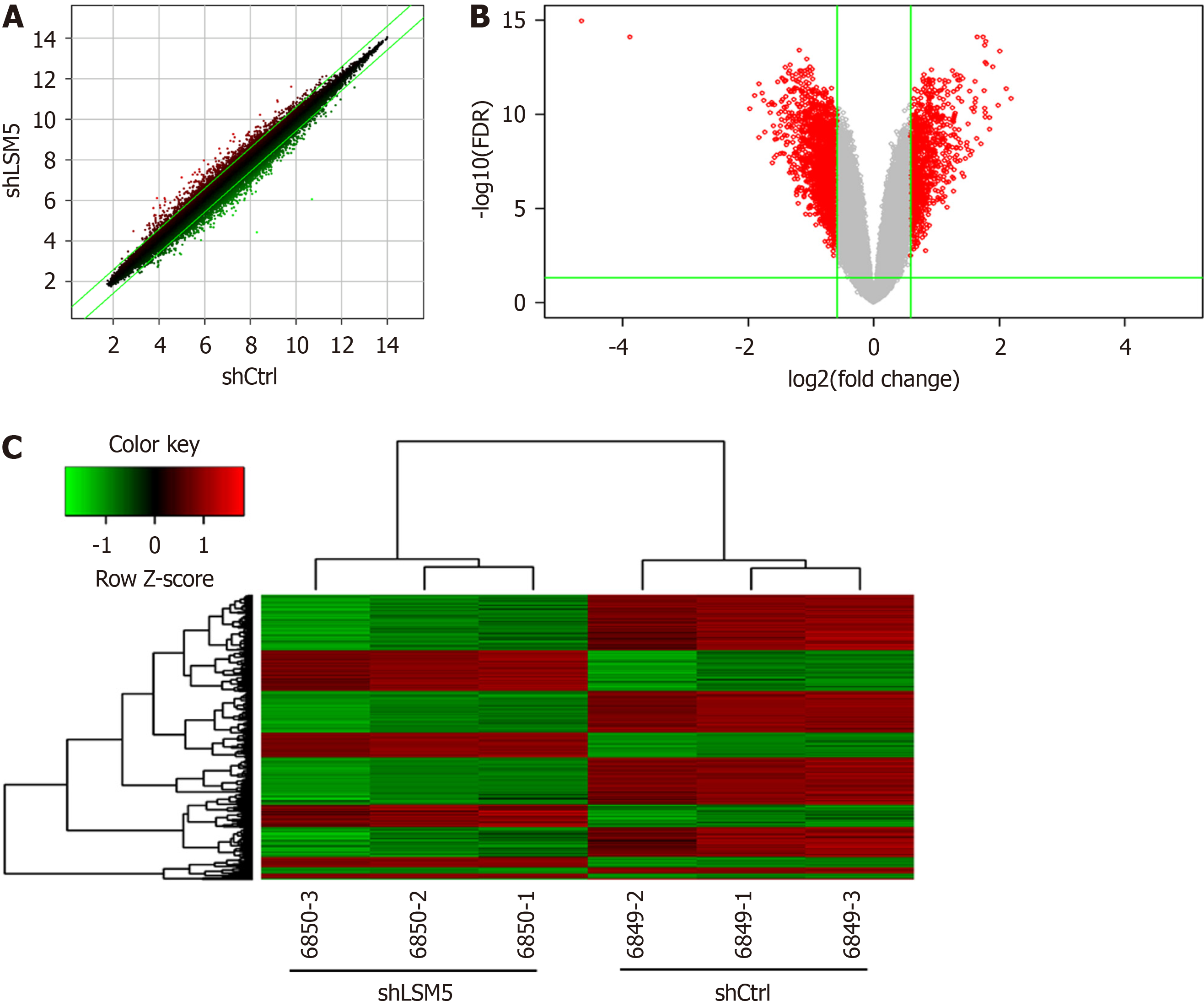
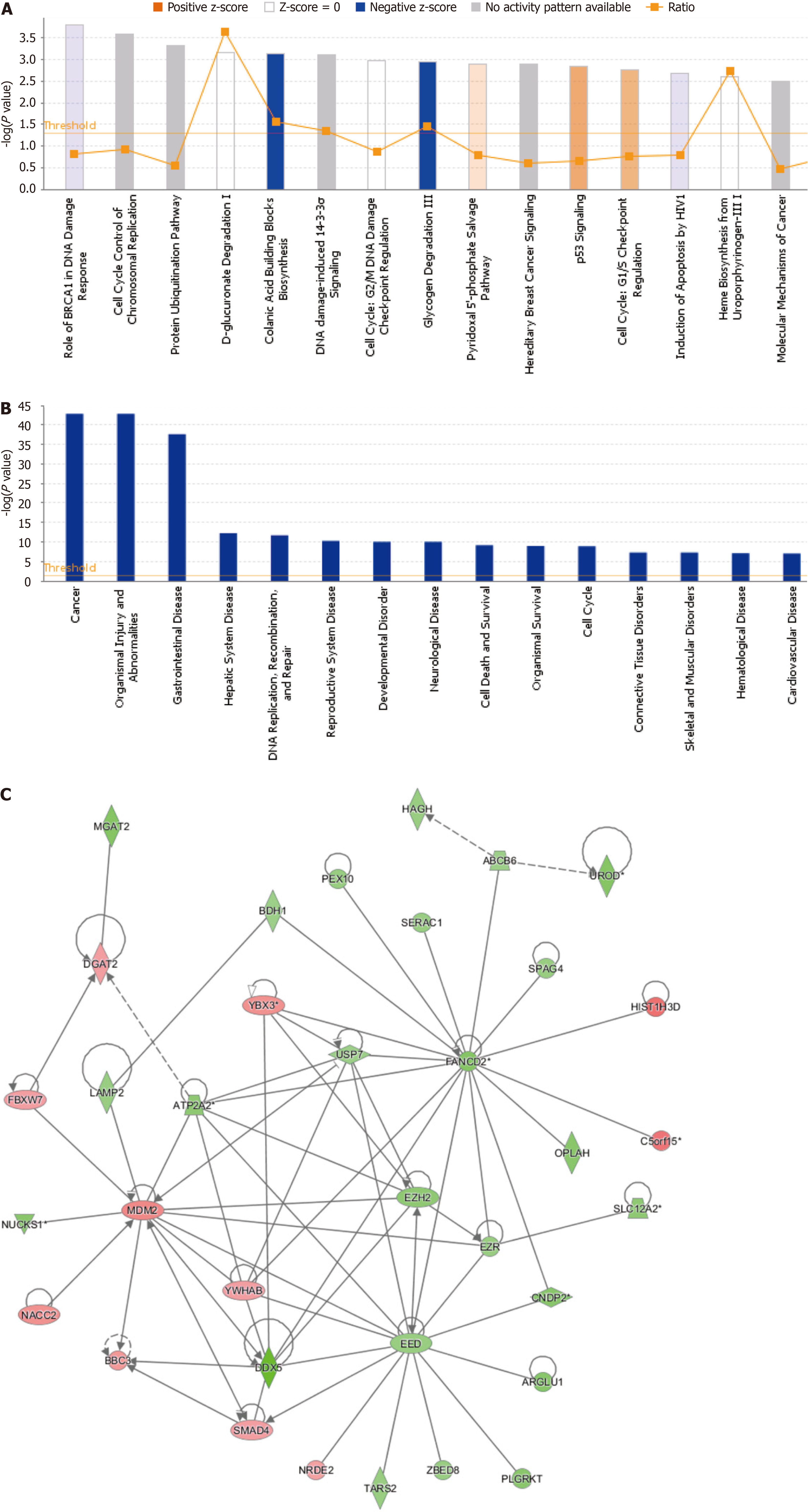
To investigate the possible mechanism by which LSM5 knockdown inhibited the growth of colon cancer cells, RKO cells were transfected with lentivirus targeting LSM5, and a GeneChip assay was conducted. The quality of the raw microarray data was evaluated, and the results suggested that the data met the criteria for further analysis (Table 2 and Supplementary Figure 1). Then, bioinformatics analysis of the filtered data was performed. Differential gene expression analysis revealed that 757 genes were upregulated and 1318 genes were downregulated following LSM5 knockdown (Figure 5). The results of the classical pathway analysis of the DEGs are shown in the top 15 signalling pathways in Figure 6A. As shown in Figure 6A, “Colanic Acid Building Blocks Biosynthesis” was significantly restrained, while “p53 Signalling” was activated. In addition, Figure 6B shows the 15 diseases and functions associated with the DEGs according to disease and functional analysis. DEGs were enriched mainly in “cancer” (Figure 6B). Finally, we analyzed the interaction network of these DEGs, and the interactions between DEGs after LSM5 knockdown are shown in Figure 6C.
| Group | Sample name | Thermo NanoDrop 2000 | 2100 Result | Quality inspection results | ||
| Concentration (ng/μL) | A260/A280 | RIN | 28S/18S | |||
| ShCtrl | 6849-1 | 663.7 | 1.98 | 10 | 2.1 | Qualified |
| 6849-2 | 663.7 | 1.98 | 10 | 2.5 | Qualified | |
| 6849-3 | 663.7 | 1.98 | 10 | 2.2 | Qualified | |
| ShLSM5 | 6850-1 | 418.1 | 1.91 | 9.7 | 1.2 | Qualified |
| 6850-2 | 418.1 | 1.91 | 9.8 | 1.1 | Qualified | |
| 6850-3 | 418.1 | 1.91 | 9.5 | 1.2 | Qualified | |
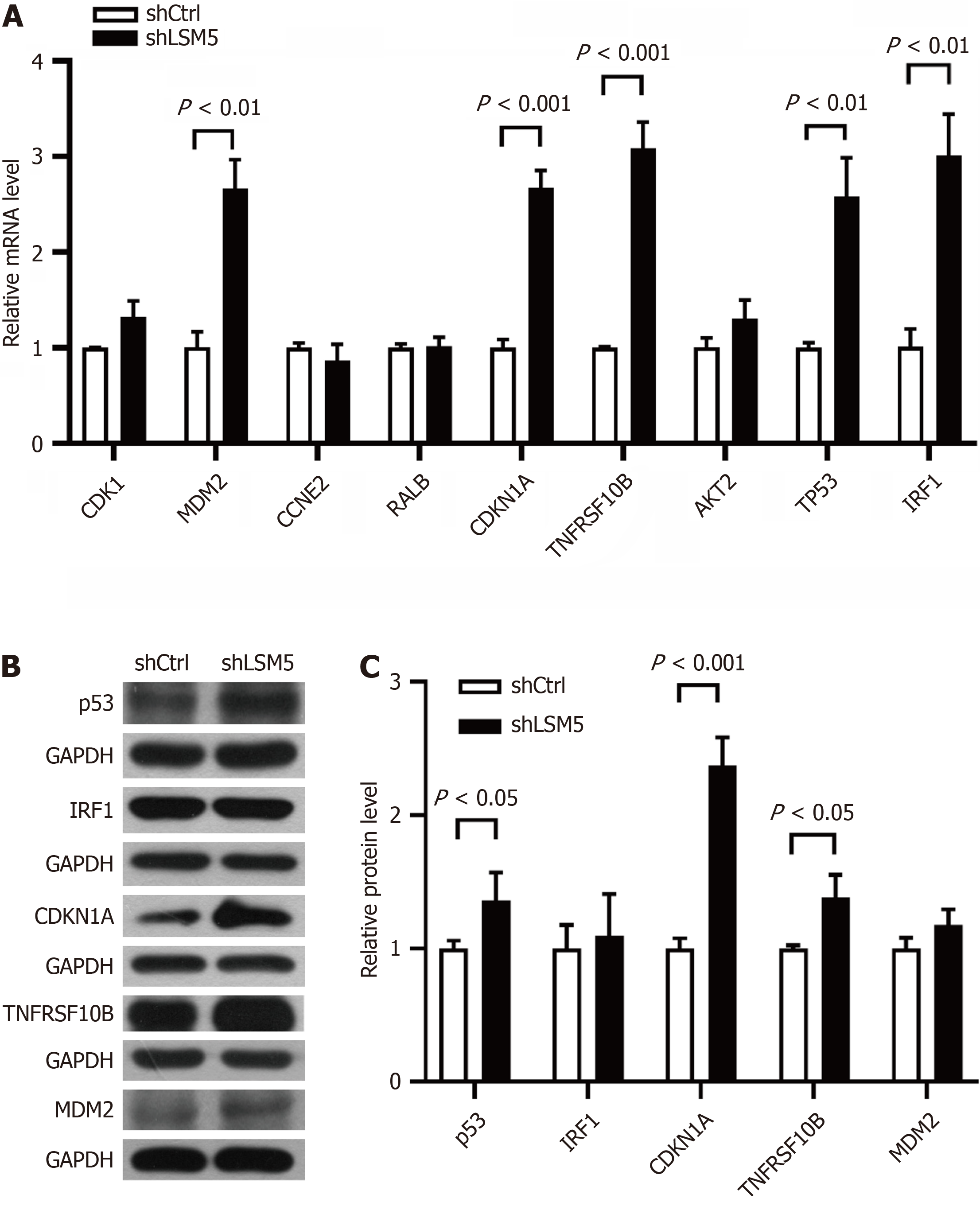
RT–qPCR showed that the expression levels of MDM2, CDKN1A, TNFRSF10B, TP53 and IRF1 were significantly increased following LSM5 knockdown (Figure 7A). Further Western blotting confirmed that LSM5 knockdown caused the upregulation of p53, CDKN1A and TNFRSF10B (Figure 7B and C).
Although the survival rate of patients with colorectal cancer has significantly improved in the past five decades[2], identifying new available therapeutic targets for patients with advanced metastasis or recurrence is still warranted. On the Human Atlas website, we found that colon cancer patients with low LSM5 expression had a greater survival rate than patients with high LSM5 expression (69% vs 43%), suggesting that LSM5 may play a role in the occurrence and de
Indeed, scientists have focused mainly on the role of the LSM family in the splicing and degradation of cytoplasmic mRNA[3-6]. Only a few studies have reported the expression level and role of LSM family members in tumors[7-14]. For example, LSM1 was usually over expressed in tumor tissues and cells, and it was closely associated with tumor growth[7,9-11]. Similarly, we found that LSM5 was highly expressed in colon cancer tissues and cells. Colon cancer patients with high LSM5 expression also had a lower survival rate, indicating the adverse effect of LSM5 on tumor development. Therefore, we knocked down LSM5 to determine whether targeting LSM5 affects the growth of colon cancer cells. As expected, we found that LSM5 knockdown inhibited the proliferation and colony formation ability of colon cancer cells and promoted cell apoptosis, revealing that targeting LSM5 could have an antitumor effect on colon cancer.
We further explored the possible mechanism by which targeting LSM5 inhibits colon cancer. A human GeneChip assay was also conducted, and bioinformatics analysis was subsequently performed to identify the downstream molecules of LSM5. The results showed that the DEGs between the control group and the LSM5 knockdown group were related to “p53 signalling” and “cancers”. In addition, RT-qPCR and Western blotting of the genes among these DEGs confirmed that the expression levels of p53, CDKN1A and TNFRSF10B were significantly upregulated following LSM5 knockdown. TP53 is a tumor suppressor gene whose inactivation drives invasion, proliferation, and cell survival, thereby facilitating cancer progression and metastasis[23]. High expression of the TP53 gene and p53 protein can significantly inhibit the occurrence and development of colon cancer[24]. In addition, CDKN1A encodes a potent cyclin-dependent kinase in
This study has several limitations. First, we explored the role of LSM5 in only colon cancer at the cellular level. No experiments have been carried out on animals. Second, we did not further investigate the interaction between p53, CDKN1A and TNFRSF10B in colon cancer. Therefore, in the next step, we explored the potential mechanism of the LSM5-p53-CDKN1A/TNFRSF10B axis in colon cancer.
Our study indicated that knockdown of LSM5 has an antitumor effect on colon cancer by upregulating p53, CDKN1A and TNFRSF10B, which provides important insight into the role of LSM5 in colon cancer development.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s classification
Scientific Quality: Grade C
Novelty: Grade B
Creativity or Innovation: Grade B
Scientific Significance: Grade B
P-Reviewer: Hegazy AA, Egypt S-Editor: Zhang L L-Editor: A P-Editor: Xu ZH
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64442] [Article Influence: 16110.5] [Reference Citation Analysis (176)] |
| 2. | Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1548] [Reference Citation Analysis (3)] |
| 3. | Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661-1671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 310] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 4. | Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3'-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789-5802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 256] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 5. | Pannone BK, Kim SD, Noe DA, Wolin SL. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics. 2001;158:187-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 61] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 338] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 7. | Zhuang H, Chen B, Tang C, Chen X, Tan W, Yang L, Xie Z, Ma X, Wang Q, Zhang C, Shang C, Chen Y. Identification of LSM Family Members as Novel Unfavorable Biomarkers in Hepatocellular Carcinoma. Front Oncol. 2022;12:871771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 8. | Liu Q, Lian Q, Song Y, Yang S, Jia C, Fang J. Identification of LSM family members as potential chemoresistance predictive and therapeutic biomarkers for gastric cancer. Front Oncol. 2023;13:1119945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 9. | Little EC, Camp ER, Wang C, Watson PM, Watson DK, Cole DJ. The CaSm (LSm1) oncogene promotes transformation, chemoresistance and metastasis of pancreatic cancer cells. Oncogenesis. 2016;5:e182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Yan Y, Rubinchik S, Watson PM, Kelley JR, Fraser MM, Wood AL, Dong JY, Gillanders WE, Boylan AM, Watson DK, Cole DJ. Establishing a murine pancreatic cancer CaSm model: up-regulation of CaSm is required for the transformed phenotype of murine pancreatic adenocarcinoma. Mol Ther. 2005;11:363-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Streicher KL, Yang ZQ, Draghici S, Ethier SP. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene. 2007;26:2104-2114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Sun X, Zhang J, Hu J, Han Q, Ge Z. LSM2 is associated with a poor prognosis and promotes cell proliferation, migration, and invasion in skin cutaneous melanoma. BMC Med Genomics. 2023;16:129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 13. | Sun X, Zhang J, Xiao C, Ge Z. Expression profile and prognostic values of LSM family in skin cutaneous melanoma. BMC Med Genomics. 2022;15:238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 14. | Dong Y, Xue L, Zhang Y, Liu C, Jiang N, Ma X, Chen F, Li L, Yu L, Liu X, Shao S, Guan S, Zhang J, Xiao Q, Li H, Dong A, Huang L, Shi C, Wang Y, Fu M, Lv N, Zhan Q. Identification of RNA-splicing factor Lsm12 as a novel tumor-associated gene and a potent biomarker in Oral Squamous Cell Carcinoma (OSCC). J Exp Clin Cancer Res. 2022;41:150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Zhong X, Ni J, Jia Z, Yan H, Zhang Y, Liu Y. CBX3 is associated with metastasis and glutathione/glycosphingolipid metabolism in colon adenocarcinoma. J Gastrointest Oncol. 2022;13:246-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Kryczka J, Boncela J. Integrated Bioinformatics Analysis of the Hub Genes Involved in Irinotecan Resistance in Colorectal Cancer. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 17. | Ibrahim BA, Hegazy AA, Gobran MA, Zaitoun MA, Elmigdadi F, El-Gindy GA, Alashkar EM, Omar WE. Expression of microRNAs ‘let-7d and miR-195’ and Apoptotic Genes ‘BCL2 and Caspase-3’ as Potential Biomarkers of Female Breast Carcinogenesis. Biomed Pharmacol J. 2023;16:2299-2313. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Ying H, Ji L, Xu Z, Fan X, Tong Y, Liu H, Zhao J, Cai X. TRIM59 promotes tumor growth in hepatocellular carcinoma and regulates the cell cycle by degradation of protein phosphatase 1B. Cancer Lett. 2020;473:13-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 19. | Pu Q, Lv YR, Dong K, Geng WW, Gao HD. Tumor suppressor OTUD3 induces growth inhibition and apoptosis by directly deubiquitinating and stabilizing p53 in invasive breast carcinoma cells. BMC Cancer. 2020;20:583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Ma R, Zhu K, Yuan D, Gong M, Li Y, Li K, Meng L. Downregulation of the FBXO43 gene inhibits tumor growth in human breast cancer by limiting its interaction with PCNA. J Transl Med. 2021;19:425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Yuan D, Ma R, Sun T, Zhu K, Dang C, Ye H, Li K. Knockdown of RSPH14 inhibits proliferation, migration, and invasion and promotes apoptosis of hepatocellular carcinoma via RelA. Cancer Cell Int. 2022;22:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 22. | Lai CH, Xu K, Zhou J, Wang M, Zhang W, Liu X, Xiong J, Wang T, Wang Q, Wang H, Xu T, Hu H. DEPDC1B is a tumor promotor in development of bladder cancer through targeting SHC1. Cell Death Dis. 2020;11:986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Wang H, Guo M, Wei H, Chen Y. Targeting p53 pathways: mechanisms, structures, and advances in therapy. Signal Transduct Target Ther. 2023;8:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 432] [Cited by in RCA: 380] [Article Influence: 190.0] [Reference Citation Analysis (0)] |
| 24. | Marei HE, Althani A, Afifi N, Hasan A, Caceci T, Pozzoli G, Morrione A, Giordano A, Cenciarelli C. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021;21:703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 284] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 25. | Ueda M, Kono J, Sengiku A, Nagumo Y, Mathis BJ, Shimba S, Taketo MM, Kobayashi T, Ogawa O, Negoro H. Bmal1 Regulates Prostate Growth via Cell-Cycle Modulation. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 26. | Xiao BD, Zhao YJ, Jia XY, Wu J, Wang YG, Huang F. Multifaceted p21 in carcinogenesis, stemness of tumor and tumor therapy. World J Stem Cells. 2020;12:481-487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (2)] |
| 27. | Gong Y, Liu Z, Yuan Y, Yang Z, Zhang J, Lu Q, Wang W, Fang C, Lin H, Liu S. PUMILIO proteins promote colorectal cancer growth via suppressing p21. Nat Commun. 2022;13:1627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 28. | Benoit YD, Laursen KB, Witherspoon MS, Lipkin SM, Gudas LJ. Inhibition of PRC2 histone methyltransferase activity increases TRAIL-mediated apoptosis sensitivity in human colon cancer cells. J Cell Physiol. 2013;228:764-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |