Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2673
Revised: February 23, 2024
Accepted: April 12, 2024
Published online: June 15, 2024
Processing time: 193 Days and 17 Hours
RAS, BRAF, and mismatch repair (MMR)/microsatellite instability (MSI) are crucial biomarkers recommended by clinical practice guidelines for colorectal cancer (CRC). However, their characteristics and influencing factors in Chinese patients have not been thoroughly described.
To analyze the clinicopathological features of KRAS, NRAS, BRAF, and PIK3CA mutations and the DNA MMR status in CRC.
We enrolled 2271 Chinese CRC patients at the China-Japan Friendship Hospital. MMR proteins were tested using immunohistochemical analysis, and the KRAS/NRAS/BRAF/PIK3CA mutations were determined using quantitative polymerase chain reaction. Microsatellite status was determined using an MSI detection kit. Statistical analyses were conducted using SPSS software and logistic regression.
The KRAS, NRAS, BRAF, and PIK3CA mutations were detected in 44.6%, 3.4%, 3.7%, and 3.9% of CRC patients, respectively. KRAS mutations were more likely to occur in patients with moderate-to-high differentiation. BRAF mutations were more likely to occur in patients with right-sided CRC, poorly differentiated, or no perineural invasion. Deficient MMR (dMMR) was detected in 7.9% of all patients and 16.8% of those with mucinous adenocarcinomas. KRAS, NRAS, BRAF, and PIK3CA mutations were detected in 29.6%, 1.1%, 8.1%, and 22.3% of patients with dMMR, respectively. The dMMR was more likely to occur in patients with a family history of CRC, aged < 50 years, right-sided CRC, poorly differentiated histology, no perineural invasion, and with carcinoma in situ, stage I, or stage II tumors.
This study analyzed the molecular profiles of KRAS, NRAS, BRAF, PIK3CA, and MMR/MSI in CRC, identifying key influencing factors, with implications for clinical management of CRC.
Core Tip: This study presents a comprehensive analysis of KRAS, NRAS, BRAF, PIK3CA mutations, and mismatch repair (MMR)/microsatellite instability status in 2271 Chinese patients with colorectal cancer (CRC). Key findings include mutation frequencies, their association with clinicopathological characteristics, and the influence of MMR deficiency. These findings have significant implications for the clinical management of CRC, providing a foundation for personalized therapy and precision medicine in CRC.
- Citation: Chen H, Jiang RY, Hua Z, Wang XW, Shi XL, Wang Y, Feng QQ, Luo J, Ning W, Shi YF, Zhang DK, Wang B, Jie JZ, Zhong DR. Comprehensive analysis of gene mutations and mismatch repair in Chinese colorectal cancer patients. World J Gastrointest Oncol 2024; 16(6): 2673-2682
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2673.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2673
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide and the second leading cause of cancer-related mortality[1]. In China, CRC ranks fifth in terms of both incidence and mortality among all malignant tumors, according to the China Cancer Statistics report in 2018[2]. Advances in understanding the pathogenesis of CRC and in gene detection technologies have led to the identification of an increasing number of genes associated with CRC development and treatment. Notably, guidelines and expert consensus worldwide now recommend the detection of CRC-related genes such as microsatellite instability (MSI)/mismatch repair (MMR), KRAS, NRAS, and BRAF[3]. Furthermore, there is a growing interest among clinical investigators in exploring the clinical significance of other genes, such as HER2, PIK3CA, and NTRK, which have shown potential implications in the management of CRC.
According to relevant studies, approximately 40%-50% of CRC patients exhibit KRAS mutations, whereas NRAS mutations account for 3.8% of CRC cases. Patients with KRAS or NRAS mutations tend to exhibit poorer prognoses than those of patients with wild-type KRAS or NRAS, and they do not benefit from EGFR inhibitors[4,5]. The frequency of BRAF mutations in Asian CRC patients ranges from 5.4% to 6.7%[6]. In CRC, BRAF V600E mutations are associated with reduced overall survival, poor treatment response, and distinct patterns of metastasis and diffusion compared with those in wild-type BRAF tumors. Importantly, BRAF mutations are mutually exclusive of KRAS/NRAS mutations[4,7]. Additionally, PIK3CA mutations occur in a subset of the Chinese population, with a mutation rate of approximately 3.5%. PIK3CA mutations coexist with RAS mutations, unlike BRAF mutations. Furthermore, studies have suggested that PIK3CA mutations may serve as predictive markers for aspirin therapy[8]. In addition, approximately 10%-15% of CRC patients exhibit high MSI (MSI-H) or deficient MMR (dMMR). MSI-H is a crucial molecular pathway involved in CRC development. Early-stage CRC with the MSI-H/dMMR phenotype is associated with a more favorable prognosis, whereas MSI-H/dMMR is considered a poor prognostic factor for advanced CRC[9]. Notably, dMMR/MSI-H has emerged as a biomarker for predicting response to immune checkpoint blockade, representing a significant advancement in the treatment of patients with metastatic CRC who do not benefit from fluorouracil-based adjuvant chemotherapy[10-12].
Given the clinical implications of the MSI/dMMR, KRAS, NRAS, BRAF, and PIK3CA status, it is crucial for clinicians to accurately identify these molecular characteristics in patients with CRC to facilitate appropriate management decisions for patients and their at-risk family members. Therefore, this study aimed to evaluate the relationship between clinicopathological features and the prevalence of MMR/MSI, KRAS, NRAS, BRAF, and PIK3CA mutations, thereby enhancing our understanding of the clinical relevance of these genes in guiding treatment decisions.
We conducted a retrospective analysis of the pathology reports from consecutively treated patients with CRC who underwent surgical resection at the China-Japan Friendship Hospital between January 2015 and December 2020. A total of 2271 eligible patients with CRC were included in this study. The clinical stage of each case was reassessed using the 8th edition of the American Joint Committee on Cancer Tumor Node Metastasis staging system. Detailed data on clinicopathological characteristics of the patients were collected and analyzed. This study was conducted in accordance with the ethical guidelines and regulations approved by the Ethics Committee of China-Japan Friendship Hospital (Approval No. QX2021-001-01). Given the retrospective nature of this study, the requirement for written informed consent was waived.
For each case, 4 μm thick sections were obtained from a representative block of formalin-fixed, paraffin-embedded tumor tissue. Immunohistochemistry (IHC) was performed using monoclonal antibodies against MLH1 (clone ES05, 1:20), MSH2 (clone FE11, 1:20), MSH6 (clone EP49, 1:20), and PMS2 (clone EPR3947, 1:20) sourced from Beijing Zhongshan Golden Bridge Biotechnology, China. IHC staining was carried out using an automated slide stainer (Ventana Benchmark) in combination with an OptiView DAB IHC Detection Kit (Ventana Medical Systems, AZ, United States). Evaluation of protein expression relied on the absence of nuclear staining in tumor cells or very faint nuclear staining in focal tumor cells, indicating a loss of protein expression (abnormal staining). The positive internal controls consisted of stromal/lymphoid cells and the nearby normal glandular epithelium of the bowel. To maintain consistency and accuracy, two specialized pathologists assessed the IHC results, and only samples with concordant interpretations were included in the present study.
Genomic DNA was extracted from formalin-fixed and paraffin-embedded (FFPE) tissues, and subsequent quantitative polymerase chain reaction (qPCR) analysis was conducted using the human KRAS/NRAS/PIK3CA/BRAF mutation detection kit according to the manufacturer’s instructions (AmoyDx, Xiamen, China). Briefly, a total volume of 25 μL was prepared, including a complex mixture comprising 0.3 μM primers and Taqman probes, 200 μM deoxynucleotide triphosphates, 200 μM Taq polymerase, and 90 ng of DNA. qPCR amplification was performed carried out using the ABI 7500 instrument (Thermo Fisher Scientific, Massachusetts, United States) according to the manufacturer’s instructions. The resulting data were analyzed using 7500 software ver 2.3 (Applied Biosystems, Foster City, CA, United States).
CRC tissue samples and adjacent normal tissue samples were collected from patients. A pathologist reviewed hematoxylin and eosin-stained slides of the tumors, and areas containing < 10% viable tumor cells were marked for macrodissection to enrich the tumor DNA. Genomic DNA was then extracted from all paired tumor-normal samples using the AmoyDx® FFPE DNA Kit (Xiamen, China). Microsatellite status was determined using six microsatellite markers (NR21, NR24, NR27, BAT25, BAT26, and MONO-27) and was detected using an MSI detection kit (Microread, Beijing, China). The reaction complex was subsequently analyzed by DNA fragmentation assays using an Applied Biosystems 3500DX instrument (Massachusetts, United States). Further details on the specific practices for MSI detection can be found in references[13]. Allelic sizes were evaluated using GeneMapper software ver 4.1 (Thermo Fisher Scientific, Waltham, MA, United States). MSI status was classified as microsatellite stable (MSS), MSI-low (MSI-L, with instability in one marker), and MSI-H (with instability in more than two markers).
Statistical analyses were conducted using SPSS (version 22; SPSS Inc., Chicago, IL, United States) or Prism software (version 7; San Diego, CA, United States). Data that did not meet the criteria for the χ2 test were combined into appropriate groups to meet the required standards. Logistic regression analysis was used to determine the factors influencing driver gene mutations. P values < 0.05 were considered statistically significant.
A total of 2271 patients diagnosed with CRC were included in the analysis. Among these patients, 1361 (59.9%) were male and 910 (40.1%) were female. Of the total number of patients, 1983 (87.3%) were over 50 years of age, 300 (13.2%) had a family history of CRC, 1762 (77.6%) had no history of smoking, and 1848 (81.4%) had no history of alcohol consumption. Histologically, 2150 (99.1%) patients had adenocarcinomas, including 113 (5.0%) diagnosed with mucinous adenocarcinomas. Tumor grading, according to the World Health Organization (WHO) criteria (Fourth Edition of the WHO Classification of Tumors of the Digestive System), revealed that 226 (10.0%) tumors were poorly differentiated, while 1878 (82.7%) were moderately or well differentiated. Most tumors were in stages II and III (36.8% and 41.2%, respectively). Right-sided colon cancer was diagnosed in 551 (24.3%) patients, while left-sided colon cancer was diagnosed in 1718 (75.6%) patients. Additionally, 489 (21.5%), 786 (34.6%), and 486 (21.4%) patients had cancerous nodules, perineural invasion, and vascular invasion, respectively. Detailed clinicopathological characteristics of the patients are summarized in Table 1.
| Characteristics | All cases (n = 2271) | Characteristics | All cases (n = 2271) |
| Sex | Perineural invasion | ||
| Male | 1361 (59.9) | No | 1484 (65.3) |
| Female | 910 (40.1) | Yes | 786 (34.6) |
| Age category (yr) | NA | 1 (< 0.01) | |
| ≤ 50 | 288 (12.7) | Vascular invasion | |
| > 50 | 1983 (87.3) | No | 1779 (78.3) |
| Smoking status | Yes | 486 (21.4) | |
| No | 1762 (77.6) | NA | 6 (0.3) |
| Yes | 509 (22.4) | Histological type | |
| Drinking status | Adenocarcinoma | 2250 (99.1) | |
| No | 1848 (81.4) | Mucinous adenocarcinoma | 113 (5.0) |
| Yes | 423 (18.6) | Others | 21 (0.9) |
| Family history | Histological differentiation grade | ||
| No | 1971 (86.8) | Poorly differentiated | 226 (10.0) |
| Yes | 300 (13.2) | Moderately/well differentiated | 1878 (82.7) |
| Tumor location | NA | 167 (7.4) | |
| Right colon | 551 (24.3) | Stage | |
| Left colon | 1718 (75.6) | Tis | 18 (0.8) |
| Liver | 2 (0.1) | I | 304 (13.4) |
| Cancer nodules | II | 836 (36.8) | |
| No | 1782 (78.5) | III | 935 (41.2) |
| Yes | 489 (21.5) | IV | 178 (7.8) |
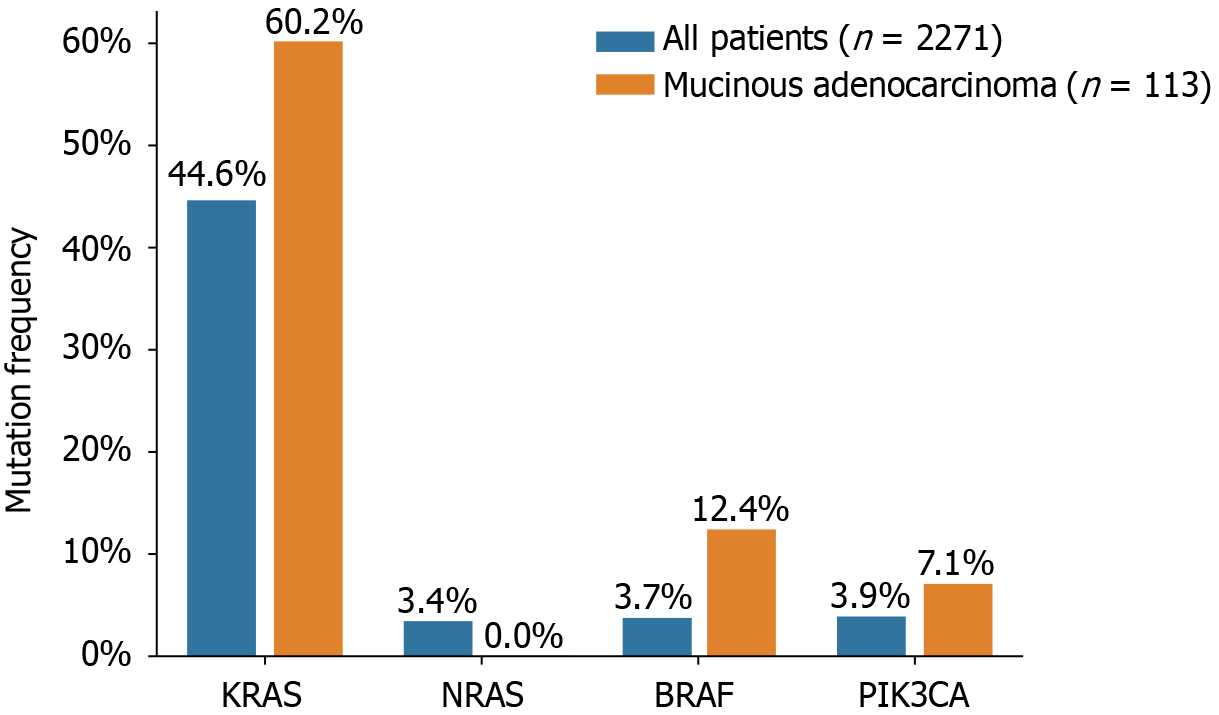
The status of KRAS, NRAS, BRAF, and PIK3CA mutations in 2271 Chinese patients with CRC is presented in Figure 1 and Table 2. KRAS mutations were detected in 44.6% (1014/2271) of all patients and 44.2% (50/113) of patients with mucinous adenocarcinomas. Among the overall patient population, 28.8% (653/2271) of the KRAS mutations occurred in exon 2, 1.5% (34/2271) in exon 3, 2.1% (48/2271) in exon 4, and 0.2% (6/2271) were indistinguishable between exons 2/3 or 2/4. In patients with mucinous adenocarcinoma, 37.2% (42/113), 2.7% (3/113), and 4.4% (5/113) of the mutations were in exons 2, 3, and 4, respectively. Of the analyzed samples, 3.4% (78/2271) harbored NRAS mutations, of which 1.5% (33/2271) occurred in exon 2 and 1.1% (25/2271) in exon 3. No NRAS mutations were detected in patients with mucinous adenocarcinoma. Interestingly, six patients displayed concurrent mutations in both KRAS and NRAS. PIK3CA mutations were detected in 88 patients (3.9%, 88/2271), of whom 43 (48.8%, 43/88) had KRAS mutations as well. PIK3CA mutations were present in 43 of 1014 (4.2%) patients with KRAS mutations, compared to 45 of 1256 (3.6%) patients with wild-type KRAS. Notably, in mucinous adenocarcinoma, PIK3CA exon 20 mutations accounted for 7.1% (8/113) of the cases. Furthermore, BRAF V600E mutations were detected in 85 of the 2271 patients with CRC (3.7%) and in 14 of the 113 patients with mucinous adenocarcinoma (12.4%). The mutual exclusivity of KRAS (exons 2, 3, and 4), NRAS (exons 2, 3, and 4), and BRAF mutations was confirmed, as none of the patients with KRAS or NRAS mutations displayed simultaneous BRAF mutations. However, three tumors exhibited concurrent mutations in BRAF and PIK3CA.
| Mutation genes | All patients (n = 2271) | Mucinous adenocarcinoma (n = 113) |
| KRAS | ||
| Exon 2 | 653 (28.8) | 42 (37.2) |
| Exon 3 | 34 (1.5) | 3 (2.7) |
| Exon 4 | 48 (2.1) | 5 (4.4) |
| Exon 2/3 | 3 (0.1) | 0 |
| Exon 2/4 | 3 (0.1) | 0 |
| NRAS | ||
| Exon 2 | 33 (1.5) | 0 |
| Exon 3 | 25 (1.1) | 0 |
| BRAF | ||
| Exon 15 | 85 (3.7) | 14 (12.4) |
| PIK3CA | ||
| Exon 20 | 88 (3.9) | 8 (7.1) |
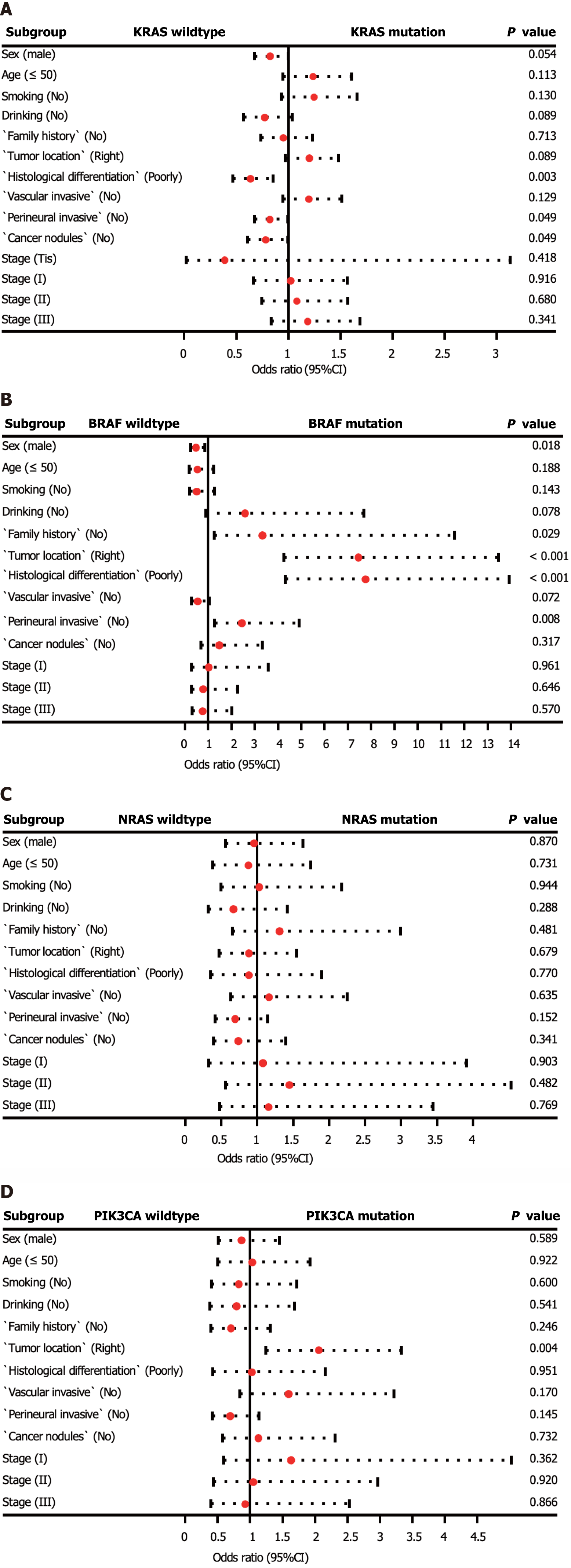
In addition, we investigated the factors affecting the mutation status of KRAS, NRAS, BRAF, and PIK3CA (Supplementary Table 1, Figure 2). Logistic regression analysis revealed that KRAS mutations were more prevalent in patients with moderately and highly differentiated tumors (P = 0.003). BRAF mutations were more frequently observed in patients with right-sided CRC, poorly differentiated tumors, and no perineural invasion (P < 0.01). Additionally, PIK3CA mutations were more common in patients with right-sided CRC (P = 0.004). No statistically significant differences were observed between patients with wild-type NRAS and those with mutations.
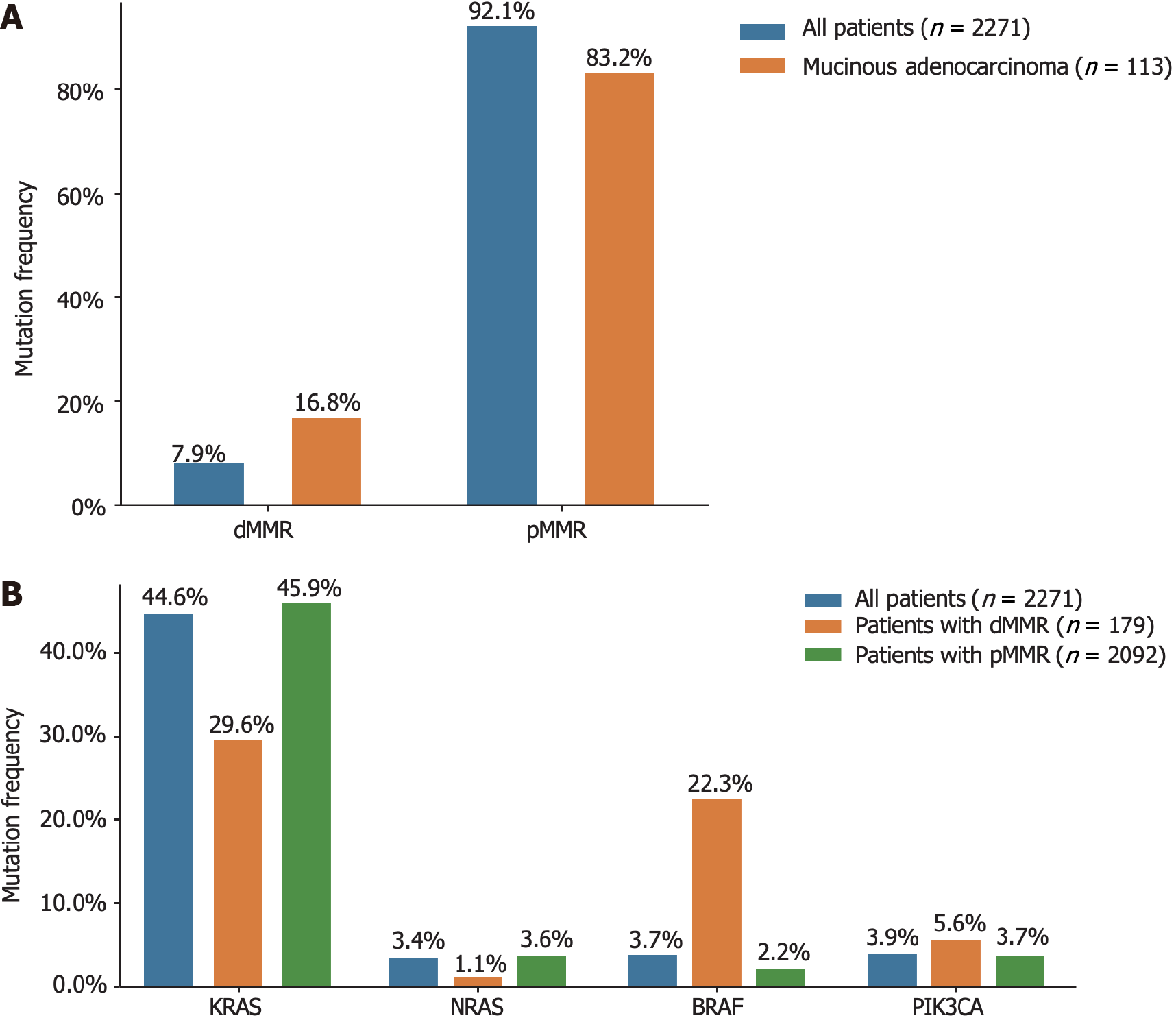
All 2271 patients with CRC included in our study underwent both MMR-IHC and MSI-PCR analyses. Abnormal MMR IHC results were detected in 179 tumors (7.9% of patients). Specifically, 2, 3, 16, and 28 patients exhibited loss of MLH1, MSH2, MSH6, and PMS2 expression, respectively. Furthermore, 96, 28, and 6 patients showed simultaneous loss of MLH1/PMS2, MSH2/MSH6, and MLH1/PMS2/MSH6 expression, respectively. In terms of MSI status, 186 patients (8.2% of the total) were classified as having MSI, including seven patients (3.8%) with MSI-L and 179 patients (96.2%) with MSI-H. The concordance between MSI-H cases and those exhibiting abnormal MMR IHC was remarkably high, with 95.5% of cases with abnormal MMR IHC showing the MSI-H status. However, eight patients (0.4%) with MSI-H showed no abnormalities in MMR IHC, and six patients (3.4%) with abnormal MMR IHC were found to be MSS upon initial assessment (Table 3).
| Test results of MSI | MMR IHC normal (n = 2092) | MMR IHC abnormal (n = 179) |
| MSS | 2079 (99.4) | 6 (3.4) |
| MSI-L | 5 (0.2) | 2 (1.1) |
| MSI-H | 8 (0.4) | 171 (95.5) |
The relationships between clinicopathological features and MSI or MMR status are shown in Supplementary Table 2. The incidence of dMMR in mucinous adenocarcinomas was as high as 16.8%. Interestingly, the prevalence of KRAS mutations in patients with dMMR was 29.6% (53 out of 179), which was lower than that observed in the general population. Additionally, the frequency of BRAF mutations among patients with dMMR was 22.3% (40 out of 179), as depicted in Figure 3.
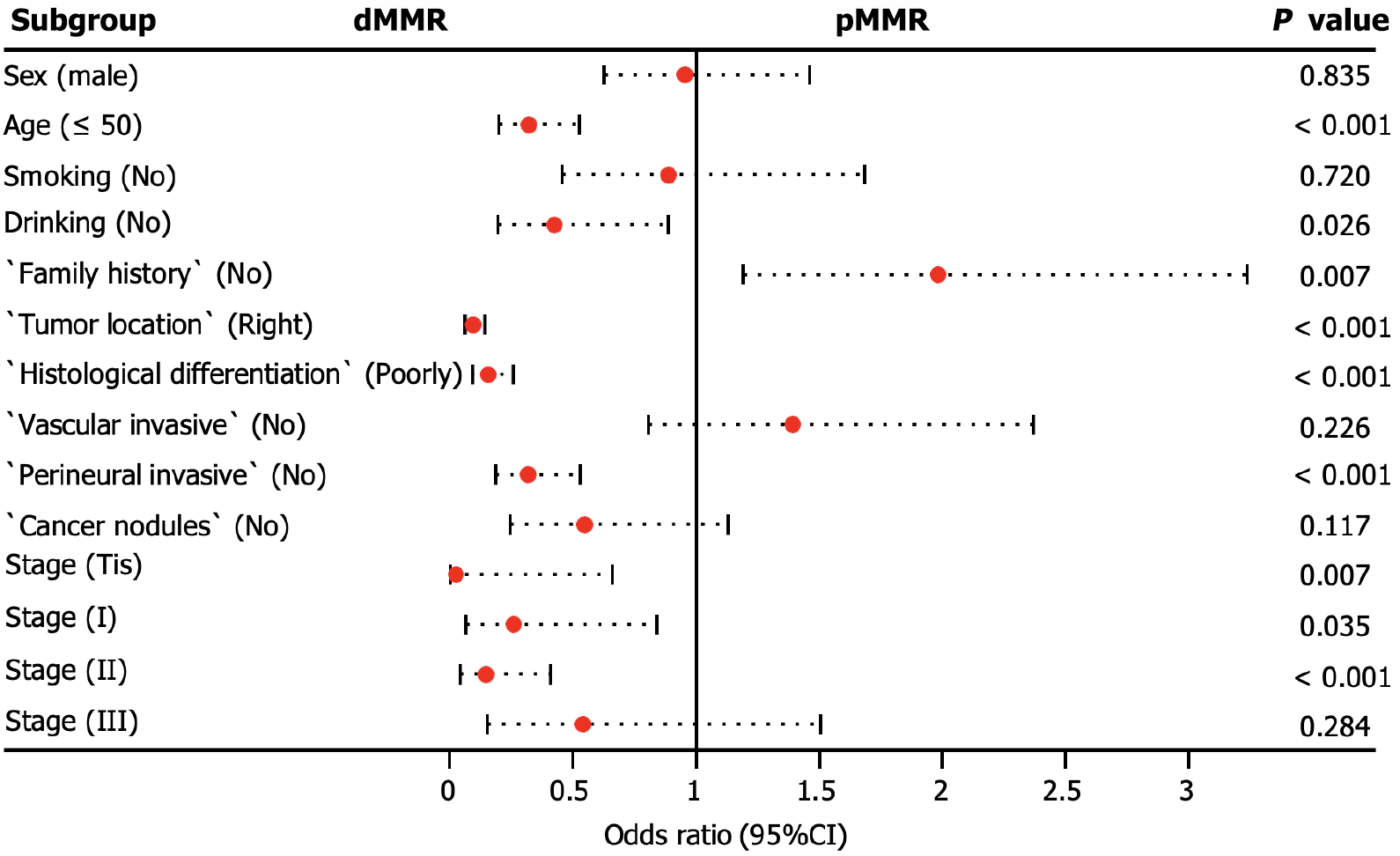
We conducted additional analyses to assess the clinicopathological factors associated with dMMR. Our findings revealed that dMMR was more likely to accumulate in patients aged < 50 years, with a positive family history, and with tumors located in the right colorectal site (P < 0.001). In addition, dMMR tumors were significantly associated with poorly differentiated histology (P < 0.001) and the absence of perineural invasion (P < 0.001), particularly in stage carcinoma in situ (Tis; P = 0.007) or stage II (P = 0.001), as illustrated in Figure 4.
Mutations in KRAS, NRAS, BRAF, and PIK3CA and the assessment of MMR/MSI status play crucial roles in evaluating patients with CRC. In this real-world study, we conducted a retrospective analysis of KRAS, NRAS, BRAF, and PIK3CA mutated genes, as well as MMR protein status, in a cohort of 2271 patients with CRC from northern China. The analysis of this large-scale dataset aimed to elucidate the clinicopathological characteristics and clinical relevance of these significant driver genes.
The mutation rates and profiles of KRAS, NRAS, BRAF, and PIK3CA in our study were consistent with those of previous reports[14,15]. Mucinous colorectal adenocarcinoma, although a rare histological type accounting for 5%-15% of CRC cases, is associated with worse survival than conventional adenocarcinoma[16,17]. Furthermore, mucinous adenocarcinoma exhibits higher frequencies of KRAS, BRAF, and PIK3CA mutations when compared to those in all CRC types[18]. In our study, we observed mutation rates of 60.2%, 12.4%, and 7.1% for KRAS, BRAF, and PIK3CA, respectively, in mucinous adenocarcinoma, compared to 44.6%, 3.7%, and 3.9%, respectively, in all CRC cases. These findings are consistent with those reported by Li et al[16].
To provide meaningful insights for clinical practice, we analyzed the clinicopathological factors associated with driver gene mutations. Our multivariate logistic regression analysis revealed that tumor differentiation grade was a key factor related to KRAS mutations. Specifically, patients with moderately to highly differentiated tumors were more likely to harbor KRAS mutations, as reported by Imamura et al[19]. Interestingly, no significant clinicopathological differences were found between NRAS mutated tumors and wild-type tumors in our study, as well as in other studies. Additionally, we found that high BRAF mutation rates were associated with low differentiation, the absence of perineural invasion, and right-sided colon cancer, which is consistent with the results of previous research[20,21]. Moreover, our study identified cancer location as the only relevant factor for PIK3CA mutations, with these mutations being enriched in right-sided CRC.
MSI is a molecular manifestation of a defective MMR system[22]. PCR-based MSI detection is commonly employed because it can provide insight into the status of the MMR system and exhibits high concordance with MMR protein detection through immunohistochemical analysis. Multiple clinical trials have demonstrated a strong correlation between molecular MSI and protein-based MMR detection[23]. In the present study, the consistency between MSI-PCR and MMR-IHC detection was 99.6%, which surpassed that reported in previous clinical studies. In addition, we found a higher incidence of dMMR in patients with mucinous adenocarcinoma (16.8%) than in all patients (7.9%), which is consistent with previous reports[18]. Among patients with dMMR, there was an enrichment of BRAF (22.3% vs 3.7% in all CRC) and PIK3CA mutations (5.6% vs 3.9% in all CRC), as well as a lower frequency of KRAS (29.6% vs 44.6% in all CRC) and NRAS mutations (1.1% vs 3.4% in all CRC). BRAF V600E has been strongly correlated with MLH1 promoter methylation, leading to dMMR[24]. Additionally, KRAS/NRAS mutations have been shown to be mutually exclusive of dMMR[25]. In our study, factors associated with a higher incidence of dMMR included age (≤ 50 years), right-sided CRC, poorly differentiated histology, and early stages (Tis, I, and II), which were similar to the influencing factors reported by Li et al[26] and Guo et al[27]. Notably, our study demonstrated a trend of younger patients with CRC, which may have arisen from the smaller age stratification in our cohort. It has been reported that dMMR is a good prognostic indicator for stage II CRC and that patients do not benefit from 5-fluorouracil-based adjuvant chemotherapy[28]. In this study, we collected clinical information from 15 patients who experienced relapse following surgery at stage II, with 12 patients exhibiting the proficient MMR (pMMR) status and experiencing a disease-free survival of less than 30 months. Limited data indicate that patients with pMMR stage II are prone to relapse, necessitating the collection of additional patient data for a more comprehensive comparative analysis during follow-up.
We conducted a comprehensive analysis of the clinicopathological characteristics and factors influencing KRAS, NRAS, BRAF, PIK3CA, and MMR/MSI in CRC. The results revealed that patients with mucinous adenocarcinoma exhibited higher mutation rates in KRAS, BRAF, PIK3CA, and dMMR. Further validation is required to confirm the clinicopathological features and prognostic value of these markers, particularly in relation to different treatment regimens. Nonetheless, the findings obtained from this large-scale, real-world dataset undoubtedly contribute to a better understanding of the significance of genetic testing and provide valuable guidance for clinical practice.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country of origin: China
Peer-review report’s classification
Scientific Quality: Grade A
Novelty: Grade A
Creativity or Innovation: Grade A
Scientific Significance: Grade A
P-Reviewer: Ampollini L, Italy S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD
| 1. | Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16:690-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 812] [Article Influence: 135.3] [Reference Citation Analysis (0)] |
| 2. | Zheng RS, Zhang SW, Sun KX, Chen R, Wang SM, Li L, Zeng HM, Wei WW, He J. [Cancer statistics in China, 2016]. Zhonghua Zhong Liu Za Zhi. 2023;45:212-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 63] [Reference Citation Analysis (0)] |
| 3. | Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C, Lindor NM, Minsky BD, Monzon FA, Sargent DJ, Singh VM, Willis J, Clark J, Colasacco C, Rumble RB, Temple-Smolkin R, Ventura CB, Nowak JA. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline Summary From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J Oncol Pract. 2017;13:333-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Dienstmann R, Mason MJ, Sinicrope FA, Phipps AI, Tejpar S, Nesbakken A, Danielsen SA, Sveen A, Buchanan DD, Clendenning M, Rosty C, Bot B, Alberts SR, Milburn Jessup J, Lothe RA, Delorenzi M, Newcomb PA, Sargent D, Guinney J. Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann Oncol. 2017;28:1023-1031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 168] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 5. | de Cuba EM, Snaebjornsson P, Heideman DA, van Grieken NC, Bosch LJ, Fijneman RJ, Belt E, Bril H, Stockmann HB, Hooijberg E, Punt CJ, Koopman M, Nagtegaal ID, Coupé VH, Carvalho B, Meijer GA. Prognostic value of BRAF and KRAS mutation status in stage II and III microsatellite instable colon cancers. Int J Cancer. 2016;138:1139-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 6. | Yoshino T, Arnold D, Taniguchi H, Pentheroudakis G, Yamazaki K, Xu RH, Kim TW, Ismail F, Tan IB, Yeh KH, Grothey A, Zhang S, Ahn JB, Mastura MY, Chong D, Chen LT, Kopetz S, Eguchi-Nakajima T, Ebi H, Ohtsu A, Cervantes A, Muro K, Tabernero J, Minami H, Ciardiello F, Douillard JY. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol. 2018;29:44-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 421] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 7. | Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:103-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 752] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 8. | Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, Sun R, Nosho K, Meyerhardt JA, Giovannucci E, Fuchs CS, Chan AT, Ogino S. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 651] [Cited by in RCA: 667] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 9. | Gelsomino F, Barbolini M, Spallanzani A, Pugliese G, Cascinu S. The evolving role of microsatellite instability in colorectal cancer: A review. Cancer Treat Rev. 2016;51:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 205] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 10. | Oliveira AF, Bretes L, Furtado I. Review of PD-1/PD-L1 Inhibitors in Metastatic dMMR/MSI-H Colorectal Cancer. Front Oncol. 2019;9:396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 11. | Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1114] [Cited by in RCA: 1221] [Article Influence: 203.5] [Reference Citation Analysis (0)] |
| 12. | Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz JF, Sinicrope F, Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219-3226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1132] [Cited by in RCA: 1209] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 13. | Song Y, Wang L, Ran W, Li G, Xiao Y, Wang X, Zhang L, Xing X. Effect of Tumor Location on Clinicopathological and Molecular Markers in Colorectal Cancer in Eastern China Patients: An Analysis of 2,356 Cases. Front Genet. 2020;11:96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, Moris D, Cloyd J, Spartalis E, Pawlik TM. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg Oncol. 2018;27:280-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 15. | Vacante M, Borzì AM, Basile F, Biondi A. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J Clin Cases. 2018;6:869-881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 81] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (4)] |
| 16. | Li J, Yang L, Bai F, Cai Y, Zhang J, Wu Z, Fu Y, Deng Y. Clinicopathological, molecular features and prognosis of colorectal cancer with mucinous component. Future Oncol. 2021;17:1351-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Kang H, O'Connell JB, Maggard MA, Sack J, Ko CY. A 10-year outcomes evaluation of mucinous and signet-ring cell carcinoma of the colon and rectum. Dis Colon Rectum. 2005;48:1161-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 243] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 18. | Luo C, Cen S, Ding G, Wu W. Mucinous colorectal adenocarcinoma: clinical pathology and treatment options. Cancer Commun (Lond). 2019;39:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 189] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 19. | Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X, Nishihara R, Jung S, Wu K, Nosho K, Wang YE, Peng S, Bass AJ, Haigis KM, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 119] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 20. | Fariña-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VEPP, Rutten HJT, van den Brule AJC. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396-2402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 21. | Kawazoe A, Shitara K, Fukuoka S, Kuboki Y, Bando H, Okamoto W, Kojima T, Fuse N, Yamanaka T, Doi T, Ohtsu A, Yoshino T. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer. 2015;15:258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 22. | De' Angelis GL, Bottarelli L, Azzoni C, De' Angelis N, Leandro G, Di Mario F, Gaiani F, Negri F. Microsatellite instability in colorectal cancer. Acta Biomed. 2018;89:97-101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 102] [Reference Citation Analysis (0)] |
| 23. | Takehara Y, Nagasaka T, Nyuya A, Haruma T, Haraga J, Mori Y, Nakamura K, Fujiwara T, Boland CR, Goel A. Accuracy of four mononucleotide-repeat markers for the identification of DNA mismatch-repair deficiency in solid tumors. J Transl Med. 2018;16:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49:151-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 225] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 25. | Poulsen TS, de Oliveira DVNP, Espersen MLM, Klarskov LL, Skovrider-Ruminski W, Hogdall E. Frequency and coexistence of KRAS, NRAS, BRAF and PIK3CA mutations and occurrence of MMR deficiency in Danish colorectal cancer patients. APMIS. 2021;129:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Li P, Xiao ZT, Braciak TA, Ou QJ, Chen G, Oduncu FS. Impact of age and mismatch repair status on survival in colorectal cancer. Cancer Med. 2017;6:975-981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Guo TA, Wu YC, Tan C, Jin YT, Sheng WQ, Cai SJ, Liu FQ, Xu Y. Clinicopathologic features and prognostic value of KRAS, NRAS and BRAF mutations and DNA mismatch repair status: A single-center retrospective study of 1,834 Chinese patients with Stage I-IV colorectal cancer. Int J Cancer. 2019;145:1625-1634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 28. | Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16:30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 293] [Article Influence: 32.6] [Reference Citation Analysis (0)] |