Published online Jun 15, 2024. doi: 10.4251/wjgo.v16.i6.2663
Revised: February 18, 2024
Accepted: April 7, 2024
Published online: June 15, 2024
Processing time: 171 Days and 19.2 Hours
Early diagnosis of pancreatic ductal adenocarcinoma (PDAC) has been a longstanding challenge. The prognosis of patients with PDAC depends on the stage at diagnosis. It is necessary to identify biomarkers for the detection and differentiation of pancreatic tumors and optimize PDAC sample preparation procedures for DNA and RNA analysis. Most molecular studies are done using paraffin-embedded blocks; however, the integrity of DNA and RNA is often compromised in this format. Moreover, RNA isolated from human pancreatic tissue samples is generally of low quality, in part, because of the high concentration of endogenous pancreatic RNAse activity present.
To assess the potential of endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) to obtain specimens from pancreatic neoplasms for subsequent RNA molecular profiling, including next-generation sequencing (NGS).
Thirty-four EUS-FNA samples were included in this study: PDAC (n = 15), chronic pancreatitis (n = 5), pancreatic cysts (n = 14), mucinous cysts (mucinous cystic neoplasia/intraductal papillary mucinous neoplasia) n = 7, serous cystic neoplasms n = 5, and pseudocysts n = 2. Cyst material consisted of cyst fluid and cyst wall samples obtained by through-the-needle biopsy (TTNB). Samples were stored at -80 °C until analysis. RNA purity (A260/230, A260/280 ratios), concentration, and integrity (RIN) were assessed. Real-time polymerase chain reaction was conducted on all samples, and small RNA libraries were prepared from solid mass samples.
RNA was successfully extracted from 29/34 (85%) EUS-FNA samples: 100% pancreatic adenocarcinoma samples, 100% chronic pancreatitis samples, 70% pancreatic fluid cyst samples, and 50% TTNB samples. The relative expression of GAPDH and HPRT were obtained for all successfully extracted RNA samples (n = 29) including low-quality RNA specimens. Low concentration and nonoptimal RIN values (no less than 3) of RNA extracted from EUS-FNA samples did not prevent NGS library preparation. The suitability of cyst fluid samples for RNA profiling varied. The quality of RNA extracted from mucinous cyst fluid had a median RIN of 7.7 (5.0-8.2), which was compatible with that from solid neoplasms [6.2 (0-7.8)], whereas the quality of the RNA extracted from all fluids of serous cystic neoplasms and TTNB samples had a RIN of 0.
The results demonstrate the high potential of EUS-FNA material for RNA profiling of various pancreatic lesions, including low-quality RNA specimens.
Core Tip: In this study, the use of RNA from endoscopic ultrasound-guided, fine-needle aspiration (EUS-FNA) samples of cystic and solid pancreatic neoplasms, including samples of low-quality RNA, was demonstrated. The process of sample preparation from EUS-FNA material is detailed along with the necessary criteria for assessing RNA quality for effective molecular biological analysis. Issues regarding the dependence of RNA quality on the type of neoplasm and material are identified. Modification of the library preparation protocol made it possible to obtain libraries for samples with RNA integrity number equivalent values of three and above, meeting the quality criteria for purity and concentration.
- Citation: Seyfedinova SS, Freylikhman OA, Sokolnikova PS, Samochernykh KA, Kostareva AA, Kalinina OV, Solonitsyn EG. Fine-needle aspiration technique under endoscopic ultrasound guidance: A technical approach for RNA profiling of pancreatic neoplasms. World J Gastrointest Oncol 2024; 16(6): 2663-2672
- URL: https://www.wjgnet.com/1948-5204/full/v16/i6/2663.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i6.2663
Pancreatic cancer (PC) remains one of the most lethal malignancies because of its asymptomatic nature and the difficulties in differentiating it from chronic pancreatitis or benign pancreatic cysts[1]. Molecular profiling of pancreatic neoplasms is of paramount importance for identifying predictors of severe dysplasia, early malignancy, metastasis, and therapeutic targets as well as for developing personalized approaches for selecting effective chemotherapy regimens. The 2023 National Comprehensive Cancer Network (NCCN) guidelines recommend molecular profiling for all patients with locally advanced or metastatic PC[2]; however, one of the key conditions for its success is obtaining high-quality biopsy material[3]. Most studies involving the molecular genetic analysis of pancreatic neoplasms rely upon formalin-fixed paraffin-embedded (FFPE) samples derived from postoperative material[4-6]. However, preoperative diagnosis at the primary stage is important for improving diagnosis, prognosis, and selecting appropriate therapy. NCCN recommends endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) as the primary method for morphological diagnosis when PC is suspected; however, the information obtained from EUS-FNA samples may be limited because of technical difficulties, concurrent inflammation, and other conditions. One problem is that 22 G needles only allow for a small amount of tissue to be harvested. Furthermore, pancreatic adenocarcinomas are characterized by hypocellularity with a prominent desmoplastic component and often exhibit neoplastic cellularity of only 5%-20%[7]. This further reduces the amount of available information obtained from these specimens.
The suitability of EUS-FNA samples for molecular analyses has already been assessed in recent studies of FFPE blocks, not native material, and DNA informativeness[8,9]. The quality of nucleic acids extracted from formalin-fixed tissue is generally low and variable, primarily because the integrity is compromised. It remains to be determined whether this method is adequate for molecular profiling of neoplasms at the DNA and RNA levels.
RNA isolated from human pancreatic tissue samples is often characterized by low integrity, primarily because of the high concentration of endogenous pancreatic RNAses present. However, total RNA isolated from tissue representing various RNA classes, including messenger RNA (mRNA) and microRNA (miRNA), can serve as highly informative material for studying gene expression or its regulation, although significant workflow adjustments are required to overcome the limitations caused by low RNA input and integrity[10]. Therefore, optimizing EUS-FNA sample preparation method is important for the extraction of high-quality total RNA.
Particular attention has been given to the development of methods for the diagnosis of pancreatic cystic neoplasms as precancerous conditions. Mucinous cysts have a higher potential for malignancy compared with other types of cystic neoplasms, and thus require close monitoring. ESGE (the 2017 update)[11] recommends performing EUS-FNA of pancreatic cysts only for biochemical and cytological analyses. As a result, the diagnosis is usually based on a combination of such data along with clinical symptoms. The sensitivity of EUS-FNA for cystic neoplasms is only 51%-54% because of the low cell content of the fluid, with a specificity of 93%-94%[12,13]. The sensitivity and specificity for differentiating mucinous and non-mucinous cystic neoplasms are 42% and 99%, respectively[14]; however, the number of nucleic acids extracted from lysed or exfoliated cyst cavity cells is usually sufficient and informative for molecular typing[15]. Previous studies that added next-generation sequencing (NGS) data to cytology exhibited a higher sensitivity (94.1% vs 87.1%) and specificity (100% vs 50%) compared with cytology alone for the detection of mucinous neoplasms and, in part, the detection of high-grade lesions[16]. Another study reported that the addition of molecular analysis altered disease management in approximately 25% of the patients and 40% of the cases when carcinoembryonic antigen levels were undetermined[17]. Several studies performed a molecular genetic analysis of cystic fluid based on RNA obtained using EUS-FNA used various methods of material transport and storage[4,18].
To improve the diagnostic accuracy of EUS-FNA, the through-the-needle biopsy (TTNB) method may be used to obtain tissue directly from the cyst wall. According to the literature, the sensitivity of TTNB for detecting mucinous cysts is 87%-90%, with a specificity of 94%-95%[19]. The aim of this study was to assess the potential of the EUS-FNA technique to obtain tissue specimens from pancreatic neoplasms for subsequent RNA profiling, including NGS.
Biobanking of 95 EUS-FNA pancreatic samples was done from October 2019 to May 2023. Diagnoses were established based on histological findings, whereas cytological and biochemical examinations were also conducted on cyst samples. The EUS-FNA technique is standard and performed on patients for histological verification of neoplasms. This study included 34 patients with unequivocal diagnoses and stages in case of pancreatic ductal adenocarcinoma (PDAC). The patients were categorized into the following groups: PDAC (n = 15), chronic pancreatitis (n = 5), and pancreatic cysts (n = 14). Cystic lesions were further divided into mucinous cysts [mucinous cystic neoplasia (MCN)/intraductal papillary mucinous neoplasia (IPMN)] n = 7, serous cystic neoplasms (SCN) n = 5, and pseudocysts n = 2.
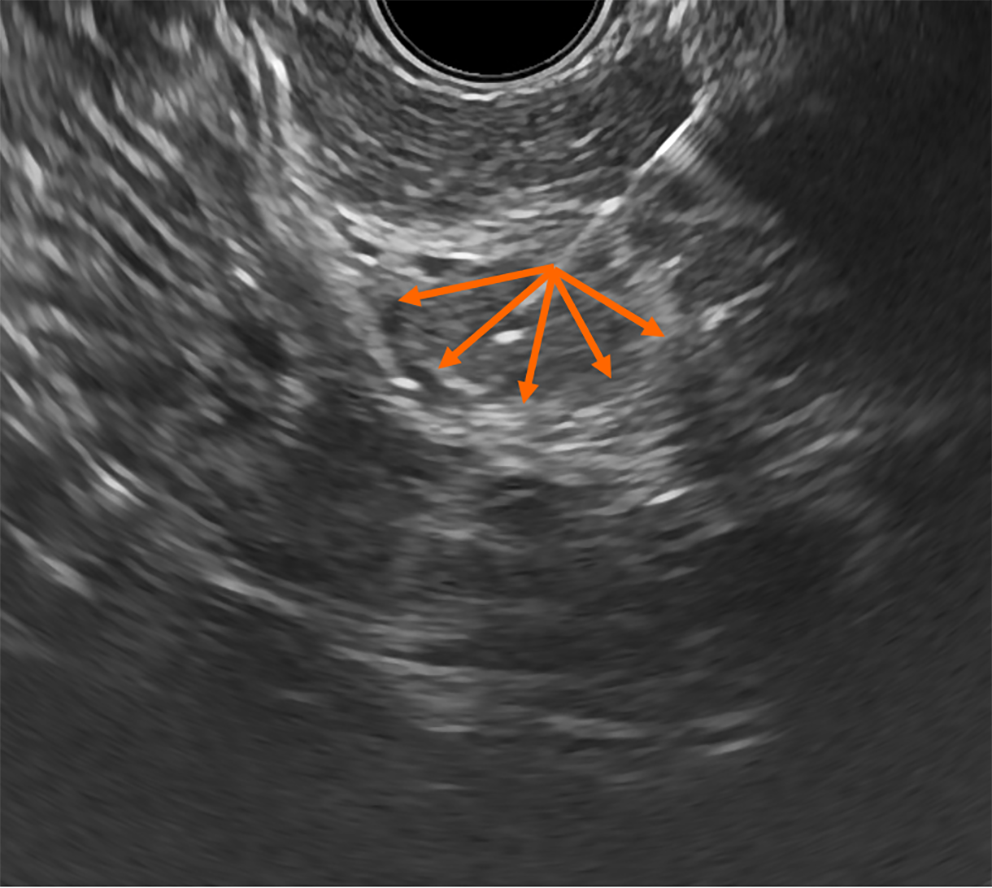
Solid lesions: EUS-FNA was performed using a 22 G needle with the wet suction technique[20]. Considering the hypocellularity and potential heterogeneity of tumors, a “fanning” technique was used during needle passes, which involved back-and-forth movements with continuous aspiration while varying the angle of approach (Figure 1)[21]. As a result, the material was sampled from the central and peripheral regions of the neoplasm to enhance diagnostic accuracy. Two to four passes were performed for each lesion until adequate material was obtained. Then, macroscopic on-site evaluation[22] of the acquired specimens was done to assess the adequacy of the sample for histological diagnosis.
Performing a puncture from sites of chronic pancreatitis was challenging because of the increased discharge of chronic inflamed tissue as well as the presence of areas of destruction, fluid collection, and calcification. Because of the absence of a dense stroma, the amount of material that could be obtained from a single pass varied and was generally smaller compared with that obtained from neoplastic changes. In this study, the EUS-FNA technique did not differ from that used for solid neoplasms; however, obtaining adequate material required a larger number of passes.
Cystic lesions: EUS-FNA of pancreatic cysts was done using a 19 G or 25 G needle, depending on the localization of the cyst and the vascularity of its septations. Initially, cyst fluid was aspirated and submitted for biochemical and cytological analyses. Approximately 0.5-2 mL of fluid was placed in a cryovial for genetic analysis. EUS-guided TTNB was performed for histological examination of neoplastic cysts, which enabled us to obtain tissue samples from the cyst wall and/or internal septations. Typically, the resulting material was macroscopically visible as small tissue fragments up to 1 mm in size, with 1-2 such fragments placed into a cryovial.
Biobanking the EUS-FNA samples: The complexity of standardizing the procedure for biological material sample preparation can negatively impact quality, subsequently leading to a decrease in the quantity and integrity of the extracted RNA. The most standardized sample preparation stage is the immediate placement of the biopsy in liquid nitrogen after collection and its further storage in liquid nitrogen vapors, which reduces the opportunity for tissue lysis over time[23,24].
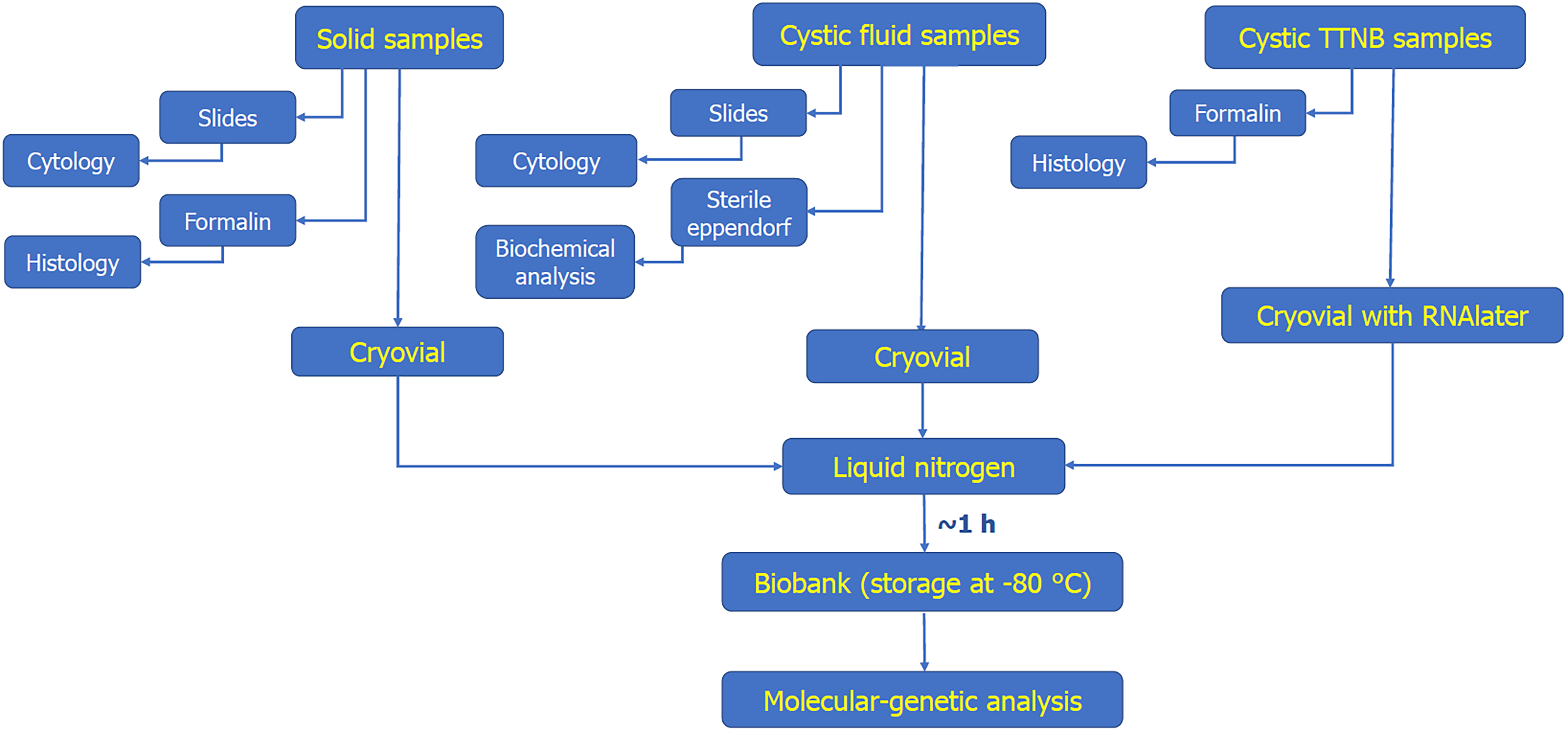
After each pass, the needle was withdrawn from the echoendoscope, and the initial portion of the material from the distal end of the needle was placed onto a slide. When visually identifiable “worm-like” fragments were obtained, the remaining material was transferred into a 2 mL cryovial, immediately placed into a container filled with liquid nitrogen, and delivered to the sample processing room. Solid samples as well as cyst fluid were placed into vials in their native form, whereas TTNB tissue fragments were transferred to a vial filled with RNAlater Stabilization Solution (Thermo Fisher Scientific, United States) to prevent potential loss during defrosting because of its extremely small size. The samples were then immediately transported to the biobank for storage in a cryogenic storage depot until RNA extraction. The entire process is presented as a diagram in Figure 2.
RNA isolation: RNA was isolated from EUS-FNA biopsy specimens, which had the following average sample weights: 0.45 g for PDAC, 0.35 g for chronic pancreatitis; and for pancreatic cystic neoplasia, 0.75 g for liquid biopsy, and 0.001 g for forceps biopsy. Total RNA isolation from all samples was performed using a TRIzol Reagent (Invitrogen, United States) based on the manufacturer’s instructions. RNA was resuspended in 15 μL of nuclease-free H2O (Invitrogen, United States) and stored at -80 °C until use.
Assessment of RNA quality and quantity: RNA purity and concentration were assessed by spectrophotometry (NanoDrop ND-1000 spectrophotometer, Thermo Fisher Scientific, United States). The RNA concentration was measured in ng/μL and the purity was assessed based on the absorbance ratios, A260/280 and A260/230. The optimal A260/A280 ratio for pure RNA is in the range of 1.8-2.2. A value less than 1.8 may indicate contamination of the sample with protein, whereas a greater than 2 may indicate possible degradation and the presence of free nucleotides. The A260/A230 ratio should be approximately 2. Lower values indicate contamination with components that remain after the RNA isolation procedure (phenol, isopropanol). RNA integrity was analyzed based on the RNA integrity index, RNA integrity number equivalent (RIN) which ranged from 1 to 10 using a 4150 Tape Station (Agilent Technologies, United States), a cartridge, and an RNA ScreenTape reagent kit (Agilent Technologies, United States). A value of zero was beyond the detection capability of the device, 1 was considered the most degraded, and 10 was mostly intact based on the RIN algorithm.
Real-time reverse transcriptase-polymerase chain reaction: Reverse transcription of the RNA (100 ng of each sample) was performed using random primers and the MMLV RT kit reagents (Evrogen, Russia) according to the manufacturer’s instructions. Quantitative polymerase chain reaction (PCR) reactions were performed in technical duplicate using specific primers designed to amplify the HPRT and GAPDH genes[25] and a qPCRmix-HS SYBR low ROX reaction mixture (Evrogen, Russia) following the manufacturer’s instructions. All samples were amplified on an ABI 7500 apparatus (Applied Biosystems, United States) under the following reaction conditions: denaturation at 95 °C for 5 min, followed by 40 cycles consisting of 95 °C for 15 s, 60 °C for 30 s, and 70 °C for 30 s. Following amplification, the reaction efficiency and the average threshold cycle (Ct) value were determined. To confirm product specificity, a melting curve analysis was performed.
Preparation of libraries for small RNA sequencing using NGS: To prepare small RNA libraries, we used 500 ng of total RNA from each sample and the TruSeq Small RNA Preparation Kit (Illumina, United States) according to the manufacturer’s instructions or with some modifications. Briefly, during the modification, the number of amplification cycles was increased from 11 to 16, T4 RNA ligase was used instead of T4 RNA ligase 2, deletion mutant, and the initial RNA concentration was increased from 500 ng to 1 μg. The quality and concentration of the libraries were assessed using a Bioanalyzer 2100 instrument (Agilent Technologies, United States) with high-sensitivity DNA chips (Agilent Technologies, United States). The optimal library concentration was 10000-30000 pM/L.
Statistical analysis was conducted using IBM SPSS Statistics software. For descriptive statistics, continuous data were presented as means with standard deviation as well as median and range. Differences between groups were analyzed using an analysis of variance. The significance of statistical differences between the two groups was assessed using the independent samples t-test and Welch’s criterion for t-tests with different variances. Pearson’s coefficients and P values were used to determine the correlation between variables.
The most informative approach involves a comprehensive analysis of quantitative and qualitative RNA sample parameters[26]. Statistical analysis of each parameter individually enables us to identify the most significant factors relevant to a specific research method.
RNA assessment was conducted for all 34 samples (Table 1). Extraction was considered successful when the sample concentration exceeded 20 ng/μL, regardless of the RIN values and A260/A280 and A260/A230 ratios. RNA concentrations exceeded 20 ng/μL in 29 out of 34 samples: 15/15 (100%) pancreatic adenocarcinoma samples, 5/5 (100%) chronic pancreatitis samples, 7/10 (70%) pancreatic fluid cyst samples, and 2/4 (50%) TTNB samples.
| EUS-FNA samples | Total, n | Lesion diameter, mm | RNA indicators | |||||
| RIN | Concentration, ng/μL | A260/A230 ratio | A260/A280 ratio | |||||
| Ductal adenocarcinoma | 15 | 45 | 3.9 | 29 | 1.1 | 1.6 | ||
| 32 | 0 | 156 | 1.3 | 1.8 | ||||
| 39 | 5 | 618 | 1.9 | 1.9 | ||||
| 42 | 0 | 387 | 1.8 | 1.8 | ||||
| 35 | 7 | 248 | 2.1 | 1.9 | ||||
| 24 | 7 | 895 | 2.2 | 1.95 | ||||
| 38 | 6.9 | 1378 | 2.2 | 1.99 | ||||
| 35 | 7 | 227 | 1.94 | 1.83 | ||||
| 35 | 6.3 | 780 | 2.24 | 1.98 | ||||
| 40 | 5.5 | 1081 | 1.91 | 1.92 | ||||
| 38 | 5.1 | 360 | 1.82 | 1.93 | ||||
| 40 | 6.2 | 2044 | 2.13 | 1.98 | ||||
| 50 | 5.7 | 432 | 2.23 | 1.96 | ||||
| 40 | 7.8 | 912 | 2.2 | 1.96 | ||||
| 32 | 7.6 | 374 | 1.62 | 1.86 | ||||
| Median (range) | 38 (24-50) | 6.2 (0-7.8) | 432 (29-2044) | 1.9 (1.1-2.2) | 1.92 (1.6-2.0) | |||
| Chronic pancreatitis | 5 | Size not estimated | 5.8 | 109 | 2.15 | 1.72 | ||
| - | 5.1 | 294 | 2.3 | 1.88 | ||||
| - | 2.5 | 543 | 2.26 | 1.84 | ||||
| - | 6.4 | 68 | 1.9 | 1.84 | ||||
| - | 7.8 | 71 | 2.4 | 1.69 | ||||
| Median (range) | - | 5.8 (2.5-7.8) | 109 (68-543) | 2.3 (1.9-2.4) | 1.8 (1.7-1.9) | |||
| Cysts | Fluid | MCN, IPMN | 4 | 22 | 5 | 304 | 1.94 | 1.95 |
| 30 | 7.5 | 201 | 2.21 | 1.86 | ||||
| 27 | 8.2 | 203 | 1.48 | 1.87 | ||||
| 19 | 7.9 | 170 | 2.05 | 1.87 | ||||
| Median (range) | 24.5 (19-30) | 7.7 (5.0-8.2) | 202 (170-304) | 1.9 (1.5-2.2) | 1.9 (1.9-2.0) | |||
| SCN | 4 | 70 | 0 | 21 | 0.9 | 1.75 | ||
| 30 | 0 | 0 | 0 | 0 | ||||
| 57 | 0 | 0 | 0 | 0 | ||||
| 24 | 0 | 0 | 0 | 0 | ||||
| Median (range) | 44 (24-70) | 0 | 0 (0-21) | 0 (0-0.9) | 0 (0-1.8) | |||
| Pseudocyst | 2 | 21 | 6.5 | 33.6 | 1.87 | 1.73 | ||
| 35 | 3.5 | 1000 | 1.6 | 1.8 | ||||
| Median (range) | 28 (21-35) | 5 (3.5-6.5) | 516.8 (33.6-1000) | 1.7 (1.6-1.9) | 1.7 (1.7-1.8) | |||
| TTNB | MCN, IPMN | 3 | 20 | 0 | 33 | 1.3 | 2.0 | |
| 45 | 0 | 0 | 1.9 | 1.9 | ||||
| 38 | 0 | 23 | 0.27 | 1.9 | ||||
| Median (range) | 34 (20-45) | 0 | 11.5 (0-33) | 0.8 (0-1.9) | 1.9 (0-1.97) | |||
| SCN | 1 | 30 | 0 | 0 | 0 | 0 | ||
The concentration of successfully extracted RNA samples ranged from 21 ng/μL to 2.0 μg/μL (mean 272.5 ng/μL). The concentration depended on the material origin and it was positively correlated with purity and RIN. Concentration, as a single factor, was not the key characteristic for using RNA for qPCR analysis or for preparing small RNA libraries. The working concentrations are all values presented for the set of samples in the study. The optimal concentration was within a range > 100 ng/μL.
In the group of ductal adenocarcinoma samples (n = 15) the RIN values were the highest with a median of 6.2. Two samples had a RIN of 0, whereas the other values ranged from 3.8 to 7.8. There was no correlation observed when assessing the effect of tumor diameter on RIN and RNA concentration (r = -0.18, P = 0.5, and r = -0.01, P = 0.9, respectively). Therefore, the differences were likely the result of structural characteristics and heterogeneity of the tumor.
The range of RIN values in the patient group with chronic pancreatitis ranged from 2.5 to 7.8; however, the median RIN in this group did not significantly differ compared with that of the ductal adenocarcinoma samples (5.8 vs 6.2) (P = 0.9), although the samples from the chronic pancreatitis group had an expectedly significantly lower concentration (median 109 ng/μL vs 432 ng/μL) (P = 0.02).
The indicator of RNA sample purity was characterized by the 260/280 and 260/230 ratios, which showed low significance for RT-qPCR efficiency and the preparation of small RNA libraries. Analysis of the distribution of 260/280 ratio values as a fundamental indicator of purity revealed that it varied within a range of 0-2.0 for all samples (median 1.9). The median values for the adenocarcinoma (median 1.92) and chronic pancreatitis (median 1.8) specimens were close to the optimal range. The MCN/IPMN and pseudocyst subgroups also had optimal values (median 1.9 and 1.7, respectively), whereas the median value in the SCN group was expectedly zero. Interestingly, in the TTNB subgroup, the 260/280 ratio value was satisfactory (median 1.9), despite low RIN and concentration values. The minimum effective values for qPCR and small RNA library preparation were established as 1.6 and 1.1 for 260-280 and 260-230, respectively. Values lower than these thresholds can lead to erroneous results in some molecular analyses.
RT-qPCR was performed to indirectly evaluate the quality and suitability of RNA from all types of EUS-FNA samples by analyzing the relative abundance of the GAPDH and HPRT1 gene transcripts. Efficiency, threshold cycle (Ct), and melting curve analysis indicated the specificity of the amplified product, the efficiency of the reaction, and the reproducibility of the results. Although the dependence of the Ct values and reproducibility of the results as indicators of RNA integrity, purity, and concentration was observed, analysis by qPCR was successful for all 29 samples with successfully isolated RNA, including the samples with undetectable RIN.
The original protocol was used successfully for library preparation on the first attempt for only 10 of 17 small RNA libraries. Presumably, this is due to a compromised RNA integrity for some samples, as evidenced by the RIN value. A modified library preparation protocol was developed, which enabled the generation of libraries that met the quality criteria for purity and concentration (with a total concentration of two target peaks ranging from 10000 to 70000 pM/L) for samples with an RIN of 3 or higher and purity ratios of 260/230 ≥ 1.1 and 260/280 ≥ 1.6. Samples with RIN values between 2.5 and 3 may be used under conditions that relax library quality standards (reduced concentration or the presence of additional peaks). No libraries were obtained for the two adenocarcinoma samples with RIN values of zero.
RNA integrity was characterized by RIN ranging from 0 to 8.2. Among the solid samples, adenocarcinoma had the highest RIN (median 6.2), whereas the chronic pancreatitis subgroup exhibited lower values (median 5.8). Among the samples of cystic fluid, RIN exhibited pronounced heterogeneity, with relatively high values in the mucinous cyst subgroup (median 7.7), low values in the pseudocyst subgroup (median 5.0), and extremely low values in the serous cyst subgroup (RIN below the detection limit for all samples).
For RNA samples used in modern molecular biology, including NGS, the recommended RIN is ≥ 8[27,28]. However, RNA samples extracted from pathological pancreatic tumor material generally exhibit lower RIN compared not only with healthy tissue, but also with RNA extracted from other organ tumors, which typically achieve a value near 7[29,30]. Nevertheless, the RIN of RNA extracted from EUS-FNA specimens usually falls within even lower ranges, from 2.7 to 5.15[25]. Our findings largely confirm these data and characterize the variable integrity of RNA samples obtained during EUS-FNA, depending on the pathological type and collection method.
RNA quality in the cyst group was separately evaluated in fluid and TTNB samples. In the fluid samples (n = 10), significant differences in RNA integrity and concentration were observed based on cyst morphology. Mucinous cysts (n = 4) exhibited high RIN values (median 7.7) and a relatively high concentration (median 202 ng/μL). Among pseudocysts, the RIN was lower (median 5), but the RNA concentration was higher (median 516.8 ng/μL), which is likely due to the presence of tissue fragments, detritus, and areas of discharged pancreatic parenchyma within the cyst cavity. Interestingly, all SCN fluid samples were characterized by unsuccessful RNA extraction with RIN values of 0, and the median concentration was also < 1 ng/μL (range 0.1-21). These results may be associated with the low cellularity of the serous cyst fluid. Cytological studies of SCN fluid confirm the cellular depletion and variability of cuboidal epithelial cells compared with mucinous cysts[31]. In addition, the average diameter of serous cysts in our study was larger compared with that in other groups [52.3 mm vs 24.5 mm (MCN) and 28 mm (pseudocysts)], which could further reduce cellularity. In larger cysts, the concentration of exfoliated cells and free RNA in the fluid is lower compared with that in smaller cysts.
RNA extraction from samples obtained by TTNB, both from serous and mucinous cysts, proved to be less effective (median RIN 0, median concentration 11.5 ng/μL). Presumably, this is related to the insufficient size of the sample, which is sufficient for histological examination, but not sufficient for RNA extraction. A recent study revealed high sensitivity and specificity for the diagnosis of mucinous cysts and IPMNs using TTNB samples by NGS. However, TTNB samples in this study were presented by cell blocks, and targeted NGS was performed[32]. Typically, 2-3 samples were obtained when performing 4-5 TTN biopsies of the cyst wall. Given the high accuracy of morphological diagnosis using TTNB samples, their value for both clinical and research studies, and the extremely small amount of biopsy material, we preferred histological analysis as the most effective approach in such cases.
The average Ct value, with a reaction efficiency of 95%-100%, for GAPDH amplification varied within a range of 20.4-33.5 (median 25.4), and for HPRT1 it was 27.3-34.1 (median 29.4), which also indicates satisfactory RNA quality to obtain reproducible results when measuring gene expression. Significant dispersion in the average Ct values for GAPDH was observed, although this may be associated with differential expression of the GAPDH gene mRNA for this group of samples[33,34]. Thus, the use of GAPDH as a reference gene for this group is questionable, but this assumption needs to be confirmed with a larger set of samples.
The original protocol was used successfully for library preparation on the first attempt for only 10 of 17 small RNA libraries. Presumably, this is due to a compromised RNA integrity for some samples, as evidenced by the RIN value. A modified library preparation protocol was developed, which enabled the generation of libraries that met the quality criteria for purity and concentration (with a total concentration of two target peaks ranging from 10000 to 70000 pM/L) for samples with an RIN of 3 or higher and purity ratios of 260/230 ≥ 1.1 and 260/280 ≥ 1.6. Samples with RIN values between 2.5 and 3 may be used under conditions that relax library quality standards (reduced concentration or the presence of additional peaks). No libraries were obtained for the two adenocarcinoma samples with RIN values of zero.
Given the small volume of EUS-FNA samples, their heterogeneity in quality, and rapid autolysis due to the abundance of pancreatic endogenous RNases, DNases, and proteases, a major factor for increasing the accuracy of molecular analyses is the standardization of the process of RNA sample preparation.
In this study, we demonstrated the potential of EUS-FNA in various pancreatic specimens for RNA profiling, including low-quality RNA samples. However, the suitability of EUS-FNA samples for RNA extraction for molecular studies varied significantly depending on the technique used to collect the biological material. In particular, the TTNB technique yielded the lowest resolution among the samples studied with respect to the quality and quantity of samples for RNA profiling. The suitability of cyst fluid samples for RNA profiling also varied. The quality of RNA extracted from mucinous cysts was comparable to that of solid neoplasms, whereas the fluid of serous cystic neoplasms exhibited low RNA quality. There is a trend of inverse dependence between cyst diameter and RNA suitability. The samples containing RNA with a RIN value less than 3 and a concentration of approximately 20 ng/mL can be informative for qPCR assays. The samples containing RNA with RIN values greater than 3, concentrations of approximately 20 ng/mL, and purity ratios of 260/230 ≥ 1.1 and 260/280 ≥ 1.6 are sufficient for NGS library preparation.
Taken together, the EUS-FNA technique is a promising tool for identifying RNA markers for the early diagnosis of pancreatic neoplasms and the prediction of carcinogenesis. Further studies using a larger sample size will be useful for estimating the sensitivity and specificity of RNA profiling for diagnostic assays. Considering that biochemical and cytological examination of EUS-FNA cyst samples does not provide sufficient sensitivity and specificity, and the diagnosis of malignant forms requires a more thorough approach, molecular studies should provide greater efficiency and significantly increase the sensitivity and specificity of the diagnosis of pancreatic neoplasms.
The authors would like to thank Sorokina M for the kindly provided human HPRT1 primers and Ignatieva E for the support and recommendations for library preparation.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Russia
Peer-review report’s classification
Scientific Quality: Grade C
Novelty: Grade C
Creativity or Innovation: Grade C
Scientific Significance: Grade B
P-Reviewer: Tang Y, China S-Editor: Wang JJ L-Editor: A P-Editor: Li X
| 1. | Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY, Wang L, Zhan HX. Early detection of pancreatic cancer: Where are we now and where are we going? Int J Cancer. 2017;141:231-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 2. | National Comprehensive Cancer Network®. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Pancreatic Adenocarcinoma. version 1. [cited 4 May 2023]. Available from: https://www2.tri-kobe.org/nccn/guideline/archive/pancreas2020/english/pancreatic.pdf. |
| 3. | Chen W, Ahmed N, Krishna SG. Pancreatic Cystic Lesions: A Focused Review on Cyst Clinicopathological Features and Advanced Diagnostics. Diagnostics (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Matthaei H, Wylie D, Lloyd MB, Dal Molin M, Kemppainen J, Mayo SC, Wolfgang CL, Schulick RD, Langfield L, Andruss BF, Adai AT, Hruban RH, Szafranska-Schwarzbach AE, Maitra A. miRNA biomarkers in cyst fluid augment the diagnosis and management of pancreatic cysts. Clin Cancer Res. 2012;18:4713-4724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 5. | Nishiwada S, Sho M, Banwait JK, Yamamura K, Akahori T, Nakamura K, Baba H, Goel A. A MicroRNA Signature Identifies Pancreatic Ductal Adenocarcinoma Patients at Risk for Lymph Node Metastases. Gastroenterology. 2020;159:562-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 6. | Bang JY, Jhala N, Seth A, Krall K, Navaneethan U, Hawes R, Wilcox CM, Varadarajulu S. Standardisation of EUS-guided FNB technique for molecular profiling in pancreatic cancer: results of a randomised trial. Gut. 2023;72:1255-1257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 7. | Ren B, Cui M, Yang G, Wang H, Feng M, You L, Zhao Y. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol Cancer. 2018;17:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 409] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 8. | Ohyama H, Mikata R, Hirotsu Y, Amemiya K, Miura Y, Hirose S, Oyama T, Takano A, Iimuro Y, Kojima Y, Mochizuki H, Ikeda J, Kato N, Omata M. Genomic profiling amplifies the utility of endoscopic ultrasound-guided fine needle biopsy by identifying clinically applicable druggable mutations in pancreatic cancer. Ann Diagn Pathol. 2022;60:152016. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 9. | Redegalli M, Grassini G, Magliacane G, Pecciarini L, Schiavo Lena M, Smart CE, Johnston RL, Waddell N, Maestro R, Macchini M, Orsi G, Petrone MC, Rossi G, Balzano G, Falconi M, Arcidiacono PG, Reni M, Doglioni C, Cangi MG. Routine Molecular Profiling in Both Resectable and Unresectable Pancreatic Adenocarcinoma: Relevance of Cytologic Samples. Clin Gastroenterol Hepatol. 2023;21:2825-2833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Andreou I, Storbeck M, Hahn P, Rulli S, Lader E. Optimized Workflow for Whole Genome and Transcriptome Next-Generation Sequencing of Single Cells or Limited Nucleic Acid Samples. Curr Protoc. 2023;3:e753. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 11. | Dumonceau JM, Deprez PH, Jenssen C, Iglesias-Garcia J, Larghi A, Vanbiervliet G, Aithal GP, Arcidiacono PG, Bastos P, Carrara S, Czakó L, Fernández-Esparrach G, Fockens P, Ginès À, Havre RF, Hassan C, Vilmann P, van Hooft JE, Polkowski M. Indications, results, and clinical impact of endoscopic ultrasound (EUS)-guided sampling in gastroenterology: European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline - Updated January 2017. Endoscopy. 2017;49:695-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 233] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 12. | Wang QX, Xiao J, Orange M, Zhang H, Zhu YQ. EUS-Guided FNA for Diagnosis of Pancreatic Cystic Lesions: a Meta-Analysis. Cell Physiol Biochem. 2015;36:1197-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Thornton GD, McPhail MJ, Nayagam S, Hewitt MJ, Vlavianos P, Monahan KJ. Endoscopic ultrasound guided fine needle aspiration for the diagnosis of pancreatic cystic neoplasms: a meta-analysis. Pancreatology. 2013;13:48-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 187] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 14. | Gillis A, Cipollone I, Cousins G, Conlon K. Does EUS-FNA molecular analysis carry additional value when compared to cytology in the diagnosis of pancreatic cystic neoplasm? A systematic review. HPB (Oxford). 2015;17:377-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Turner RC, Melnychuk JT, Chen W, Jones D, Krishna SG. Molecular Analysis of Pancreatic Cyst Fluid for the Management of Intraductal Papillary Mucinous Neoplasms. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 16. | Haeberle L, Schramm M, Goering W, Frohn L, Driescher C, Hartwig W, Preissinger-Heinzel HK, Beyna T, Neuhaus H, Fuchs K, Keitel-Anselmino V, Knoefel WT, Esposito I. Molecular analysis of cyst fluids improves the diagnostic accuracy of pre-operative assessment of pancreatic cystic lesions. Sci Rep. 2021;11:2901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | Arner DM, Corning BE, Ahmed AM, Ho HC, Weinbaum BJ, Siddiqui U, Aslanian H, Adams RB, Bauer TW, Wang AY, Shami VM, Sauer BG. Molecular analysis of pancreatic cyst fluid changes clinical management. Endosc Ultrasound. 2018;7:29-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Shirakami Y, Iwashita T, Uemura S, Imai H, Murase K, Shimizu M. Micro-RNA Analysis of Pancreatic Cyst Fluid for Diagnosing Malignant Transformation of Intraductal Papillary Mucinous Neoplasm by Comparing Intraductal Papillary Mucinous Adenoma and Carcinoma. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Tacelli M, Celsa C, Magro B, Barchiesi M, Barresi L, Capurso G, Arcidiacono PG, Cammà C, Crinò SF. Diagnostic performance of endoscopic ultrasound through-the-needle microforceps biopsy of pancreatic cystic lesions: Systematic review with meta-analysis. Dig Endosc. 2020;32:1018-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 20. | Villa NA, Berzosa M, Wallace MB, Raijman I. Endoscopic ultrasound-guided fine needle aspiration: The wet suction technique. Endosc Ultrasound. 2016;5:17-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Bang JY, Magee SH, Ramesh J, Trevino JM, Varadarajulu S. Randomized trial comparing fanning with standard technique for endoscopic ultrasound-guided fine-needle aspiration of solid pancreatic mass lesions. Endoscopy. 2013;45:445-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 190] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 22. | Gaia S, Rizza S, Bruno M, Ribaldone DG, Maletta F, Sacco M, Pacchioni D, Rizzi F, Saracco GM, Fagoonee S, De Angelis CG. Impact of Macroscopic On-Site Evaluation (MOSE) on Accuracy of Endoscopic Ultrasound-Guided Fine-Needle Biopsy (EUS-FNB) of Pancreatic and Extrapancreatic Solid Lesions: A Prospective Study. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Reference Citation Analysis (0)] |
| 23. | From the American Association of Neurological Surgeons (AANS); American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE); Canadian Interventional Radiology Association (CIRA); Congress of Neurological Surgeons (CNS), European Society of Minimally Invasive Neurological Therapy (ESMINT), European Society of Neuroradiology (ESNR); European Stroke Organization (ESO); Society for Cardiovascular Angiography and Interventions (SCAI), Society of Interventional Radiology (SIR), Society of NeuroInterventional Surgery (SNIS); and World Stroke Organization (WSO); Sacks D, Baxter B, Campbell BCV, Carpenter JS, Cognard C, Dippel D, Eesa M, Fischer U, Hausegger K, Hirsch JA, Hussain MS, Jansen O, Jayaraman MV, Khalessi AA, Kluck BW, Lavine S, Meyers PM, Ramee S, Rüfenacht DA, Schirmer CM, Vorwerk D. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. AJNR Am J Neuroradiol. 2018;39:E61-E76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 24. | Archibugi L, Ruta V, Panzeri V, Redegalli M, Testoni SGG, Petrone MC, Rossi G, Falconi M, Reni M, Doglioni C, Sette C, Arcidiacono PG, Capurso G. RNA Extraction from Endoscopic Ultrasound-Acquired Tissue of Pancreatic Cancer Is Feasible and Allows Investigation of Molecular Features. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Dmitrieva RI, Lelyavina TA, Komarova MY, Galenko VL, Ivanova OA, Tikanova PA, Khromova NV, Golovkin AS, Bortsova MA, Sergushichev A, Sitnikova MY, Kostareva AA. Skeletal Muscle Resident Progenitor Cells Coexpress Mesenchymal and Myogenic Markers and Are Not Affected by Chronic Heart Failure-Induced Dysregulations. Stem Cells Int. 2019;2019:5690345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Caixeiro NJ, Lai K, Lee CS. Quality assessment and preservation of RNA from biobank tissue specimens: a systematic review. J Clin Pathol. 2016;69:260-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Kap M, Oomen M, Arshad S, de Jong B, Riegman P. Fit for purpose frozen tissue collections by RNA integrity number-based quality control assurance at the Erasmus MC tissue bank. Biopreserv Biobank. 2014;12:81-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Augereau C, Lemaigre FP, Jacquemin P. Extraction of high-quality RNA from pancreatic tissues for gene expression studies. Anal Biochem. 2016;500:60-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Butler AE, Matveyenko AV, Kirakossian D, Park J, Gurlo T, Butler PC. Recovery of high-quality RNA from laser capture microdissected human and rodent pancreas. J Histotechnol. 2016;39:59-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Rudloff U, Bhanot U, Gerald W, Klimstra DS, Jarnagin WR, Brennan MF, Allen PJ. Biobanking of human pancreas cancer tissue: impact of ex-vivo procurement times on RNA quality. Ann Surg Oncol. 2010;17:2229-2236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 31. | Abdelkader A, Hunt B, Hartley CP, Panarelli NC, Giorgadze T. Cystic Lesions of the Pancreas: Differential Diagnosis and Cytologic-Histologic Correlation. Arch Pathol Lab Med. 2020;144:47-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (1)] |
| 32. | Rift CV, Melchior LC, Kovacevic B, Klausen P, Toxværd A, Grossjohann H, Karstensen JG, Brink L, Hassan H, Kalaitzakis E, Storkholm J, Scheie D, Hansen CP, Lund EL, Vilmann P, Hasselby JP. Targeted next-generation sequencing of EUS-guided through-the-needle-biopsy sampling from pancreatic cystic lesions. Gastrointest Endosc. 2023;97:50-58.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 33. | Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem. 2001;295:17-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 291] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 34. | Zhong H, Simons JW. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun. 1999;259:523-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 357] [Article Influence: 13.7] [Reference Citation Analysis (0)] |