Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1596
Peer-review started: November 17, 2023
First decision: December 11, 2023
Revised: December 21, 2023
Accepted: January 23, 2024
Article in press: January 23, 2024
Published online: April 15, 2024
Processing time: 145 Days and 11 Hours
Hepatitis C virus (HCV) is a blood-borne virus which globally affects around 79 million people and is associated with high morbidity and mortality. Chronic infection leads to cirrhosis in a large proportion of patients and often causes hepatocellular carcinoma (HCC) in people with cirrhosis. Of the 6 HCV genotypes (G1-G6), genotype-3 accounts for 17.9% of infections. HCV genotype-3 responds least well to directly-acting antivirals and patients with genotype-3 infection are at increased risk of HCC even if they do not have cirrhosis.
To systematically review and critically appraise all risk factors for HCC secondary to HCV-G3 in all settings. Consequently, we studied possible risk factors for HCC due to HCV-G3 in the literature from 1946 to 2023.
This systematic review aimed to synthesise existing and published studies of risk factors for HCC secondary to HCV genotype-3 and evaluate their strengths and limitations. We searched Web of Science, Medline, EMBASE, and CENTRAL for publications reporting risk factors for HCC due to HCV genotype-3 in all settings, 1946-2023.
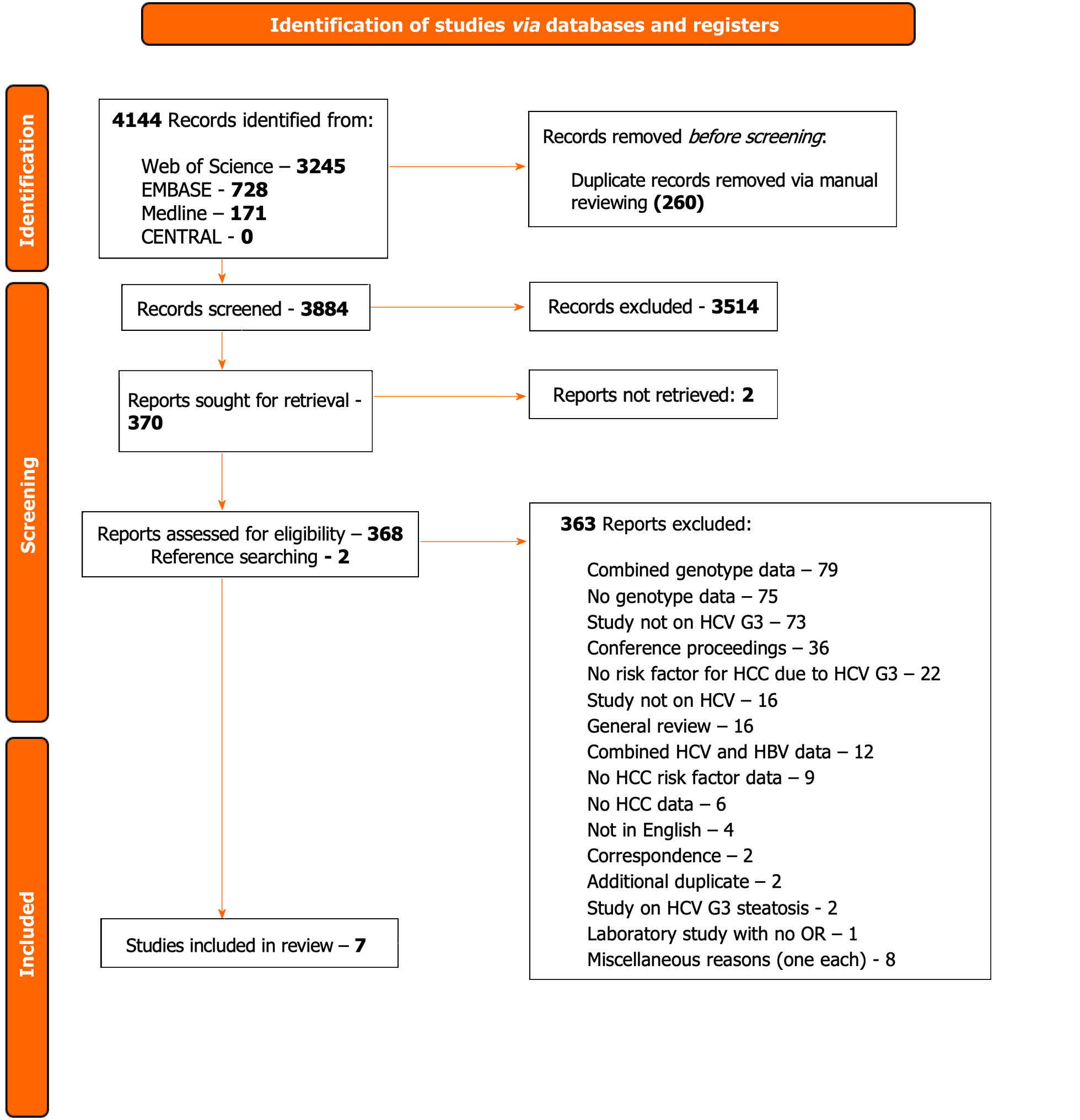
Four thousand one hundred and forty-four records were identified from the four databases with 260 records removed as duplicates. Three thousand eight hundred and eighty-four records were screened with 3514 excluded. Three hundred and seventy-one full-texts were assessed for eligibility with seven studies included for analysis. Of the seven studies, three studies were retrospective case-control trials, two retrospective cohort studies, one a prospective cohort study and one a cross-sectional study design. All were based in hospital settings with four in Pakistan, two in South Korea and one in the United States. The total number of participants were 9621 of which 167 developed HCC (1.7%). All seven studies found cirrhosis to be a risk factor for HCC secondary to HCV genotype-3 followed by higher age (five-studies), with two studies each showing male sex, high alpha feto-protein, directly-acting antivirals treatment and achievement of sustained virologic response as risk factors for developing HCC.
Although, studies have shown that HCV genotype-3 infection is an independent risk factor for end-stage liver disease, HCC, and liver-related death, there is a lack of evidence for specific risk factors for HCC secondary to HCV genotype-3. Only cirrhosis and age have demonstrated an association; however, the number of studies is very small, and more research is required to investigate risk factors for HCC secondary to HCV genotype-3.
Core Tip: Hepatitis C virus (HCV) genotype-3 accounts for 17.9% of HCV infections with an increased risk of hepatocellular carcinoma (HCC) globally. In this systematic review and meta-analysis, we screened 4144 records to find only seven studies which study risk factors for HCC. Conducted primarily in Global South hospital settings, the studies encompassed 9621 participants, revealing cirrhosis and age as consistent risk factors for HCC. While cirrhosis and age emerge as contributors, the scarcity of studies underscores the urgent need for expanded research. Limited evidence exists on other factors, emphasising the need for further research to understand specific risk contributors to HCC secondary to HCV Genotype-3.
- Citation: Farooq HZ, James M, Abbott J, Oyibo P, Divall P, Choudhry N, Foster GR. Risk factors for hepatocellular carcinoma associated with hepatitis C genotype 3 infection: A systematic review. World J Gastrointest Oncol 2024; 16(4): 1596-1612
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1596.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1596
Hepatitis C virus (HCV) is a blood-borne virus which globally affects around 79 million people[1] and is associated with high morbidity and mortality. Chronic infection leads to cirrhosis in a large proportion of patients after 30 years of asymptomatic infection and often causes hepatocellular carcinoma (HCC) in people with cirrhosis. HCV has six genotypes (G1-G6) globally with G1 accounting for 49.1% of all HCV infections, followed by G3 (17.9%), G4 (16.8%), G2 (11.0%), G5 (2.0%), and G6 (1.4%)[2].
The highest prevalence of G3 in Western Europe is Norway (50%), England (47%), and Finland (43%) with 10% in North America (22% in Canada) with 26.9% in South America[3-5]. However, the greatest burden of G3 is in South and Central Asia with 71.6% of HCV infections being of this genotype which is very common in Pakistan and India[2,6-8].
G3 infection is not susceptible to the first generation of direct-acting antiviral (DAA) protease inhibitors and has reduced susceptibility to Sofosbuvir[6,9-13], particularly in patients with cirrhosis. The efficacy of next generation protease inhibitor-based regimens (glecaprevir/pibrentasvir) may also be reduced[14-16]. However even in patients with this genotype viral clearance rates are well over 90% and these effective, affordable oral antiviral treatments are widely available. However, in patients with HCV induced cirrhosis, viral clearance does not abolish the risk of HCC[17].
HCC is a feared complication of HCV and of all the genotypes, patients with G3 infection have the highest incidence[18,19]. In most patients, cancer is linked to cirrhosis but in subjects infected with G3 even those without cirrhosis are at increased risk[20,21]. The only effective strategy to manage liver cancer is early detection of asymptomatic tumours by screening followed by loco-regional or immunomodulatory/kinase inhibitor combination therapies. Current recommendations are to screen all cirrhotic patients by ultrasound 6-monthly. However, in G3, where cirrhosis is not an adequate risk factor, we need to screen more subjects[22] and require epidemiological risk assessment tools to determine which subjects require surveillance.
There is therefore a need to identify and evaluate risk factors for HCC secondary to HCV-G3 to assist in identification of people at high risk. However, although there are risk factors identified for the most common genotype, G1; this is not the case for G3.
To address this, we aimed to systematically review and critically appraise all risk factors for HCC secondary to HCV-G3 in all settings. Consequently, we studied possible risk factors for HCC due to HCV-G3 in the literature from 1946 to 2023.
Literature search: We searched the following four databases for articles: Web of Science, Medline, EMBASE, and CENTRAL; utilising the search strategy pre-defined by an expert librarian (Supplementary Table 1) for studies published between 1st January 1946 to 17th December 2022.
The search aimed to include all relevant studies reporting original data for the comparison of HCC risk of patients with HCV G3, from inception up until December 2022. The following keywords: “Hepatocellular carcinoma”, “hepatitis C”, and “genotype 3” were combined with other search terms, using Boolean operators and truncation. Secondly, the reference lists of all included articles were manually reviewed to identify any unidentified publications and grey literature was searched. No restrictions were set for publication year and status, or geographical area.
Selection criteria: We applied the following inclusion criteria to studies: (1) Participants/population: Patients in primary care, hospital settings and national databases; (2) exposure: Risk factor for HCC secondary to HCV-G3; (3) comparison: Risk factor for HCC secondary to non-HCV-G3 or control; and (4) outcome: Development of HCC.
We included randomized control trials and observational studies (case-control, cohort, and cross-sectional) and excluded any studies which did not fit the above criteria, mathematical modelling studies or were not published in English.
Studies were eligible for the meta-analysis if they fulfilled the following criteria: (1) Study design: Cohort studies, case-control studies or randomized controlled trials based on original data; (2) study population and exposure: For cohort studies, both HCV G3 infected group and a comparison group of HCV non-G3 infected patients in the same study, with at least 10 patients in each group, and for case-control studies at least 10 patients in each group of HCV-G3 HCC cases and non-HCC as controls. In studies where there were all HCV genotypes, we included those which had data (in the main results or supplementary appendices) of individual patients with HCV-G3 who developed HCC; (3) methods: Studies reporting odds ratios (OR), relative risks (RR), or hazard ratios (HR), or sufficient data to calculate the effect size (ES); (4) outcome: The number of HCC in each patient group is stated; and (5) the manuscript is published as a full paper in a peer-reviewed journal.
The following studies were excluded: (1) Animal or in vitro studies; (2) studies without clearly reported control or comparison group; (3) studies with unclear HCC outcome; and (4) letters to the editor, review articles, guidelines, and conference abstracts (not peer-reviewed) were excluded.
To ensure the exposure (HCV G3 infection) was present prior to the development of HCC, we excluded all studies where there was combined data in HCV genotypes and where we could not extract data (in the main results or supplementary appendices) of individual patients with HCV-G3 who developed HCC.
Screening process: We planned for two reviewers (HZF and MJ) to screen all abstracts to ensure a robust screening process with each abstract reviewed by at least two reviewers utilising the Rayyan QCRI programme. Any conflicting decisions were discussed and referred to a third reviewer if required.
Post primary screening, two reviewers (HZF and MJ) screened the full texts to ensure the papers fully fit the criteria with conflicting decisions discussed and referred to a third reviewer if needed.
Data extraction (selection and coding): Two reviewers (HZF and MJ) independently screened the full text of the included papers and extracted the following data for each included study: (1) Setting of study: Country and whether primary or secondary care; (2) characteristics of study population: Age and sex; (3) study design; (4) number of study participants in study; (5) type of HCV; (6) number participants who developed HCC; (7) risk factors identified for HCC; (8) proportion of participants with particular risk factor: Number and percentage; (9) odds ratio of risk factor; (10) hazards ratio of risk factor; and (11) number of participants who cleared HCV or were actively infected (Supplementary Table 2).
Study characteristics, context, quality, and findings were captured and summarized with similarities and differences compared across the studies in a tabular form, using appropriate subgroup analysis with comparison of the performance of different risk factors. All data were captured with a spreadsheet (MS Excel) and validated by an independent reviewer (MJ).
Risk of bias (quality) assessment: Two reviewers (HZF and MJ) utilised a standardised data extraction form based on the criteria for assessing the quality of risk factor studies. We utilised the Newcastle-Ottawa Scale (NOS) to assess the quality of the studies, judging studies based on points awarded for selection of study groups, comparability of groups and exposure/outcome ascertainment (Supplementary material). Any conflicting decisions were discussed and referred to a third reviewer if required. Studies with scores of < 5, 5-7, and > 7 points were considered to be of low, sufficient, and high quality, respectively. Any conflicting decisions were discussed and referred to a third reviewer (JA) if required.
Statistical analysis: We manually extracted the crude number of patients who developed HCC in patients with HCV-G3 and utilised these data for pooled ES and 95% confidence intervals (CIs) were estimated. As the outcome of HCV G3 HCC is rare with the worldwide HCC incidence of 9.5 cases per 100000 person-years[23] odds ratios (ORs), relative risk (RR), and hazard ratios (HRs) were deemed to be equivalent. For studies which calculated HRs, we captured these for analysis. Correspondingly, and for those that had no calculated HRs we extracted the crude number of patients who developed HCC and calculated HRs.
Meta-analysis and assessment of heterogeneity: We carried out meta-analysis of hazard ratios in Jamovi version 2.2.5 using the “meta-analysis” package minimally adjusting for age and sex reported in the studies.
We calculated pooled summary effect estimates using the restricted-maximum likelihood model (random effects model) weighting of HRs on the natural logarithmic scale and quantified between-study heterogeneity using the I2 statistic; significance of heterogeneity was investigated using Cochran’s Q test (P threshold = 0.05). Where I2 was > 0 and heterogeneity were significant, we present random-effects summary estimates. We undertook multiple sensitivity analyses whereby analyses were restricted to studies adjusting for various additional confounders, and stratified by percentage of G3, to investigate robustness of observed associations.
Publication bias: Funnel plots were utilised to assess for publication bias with Egger’s regression for small-study effects used to assess the degree of asymmetry, with statistical significance level of P < 0.05.
Funding: The funders had no role in the study design, data collection, data analysis, data interpretation, or writing of this report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit it for publication.
We identified 4144 records from the four databases with 260 records removed as duplicates via manual reviewing, 3884 records were screened with 3514 excluded (Figure 1).
Three hundred and seventy-one full texts were retrieved and assessed for eligibility of which 2 reports could not be retrieved even with contacting the study authors. Of these, 15 studies were initially included with 348 excluded. Post-preliminary analysis, seven studies were included with 363 excluded due to the defined reasons (Figure 1; Supplementary Figure 1).
Data were initially extracted from 15 selected studies[18,20,21,22-35] which provided information from a total of 12674 participants (Supplementary Table 3). Preliminary analysis of the studies showed that these studies combined genotype data even with their primary focus on G3. To ensure robustness of the data with particular reference to G3, the decision was made to exclude those which did not fully categorise G3 (i.e., inability to extract individualised G3 participant data) and thus only seven studies were included in the final data analysis[18,26,28,29,31,33,34] (Table 1).
| Ref. | Country | Journal | SJR ranking quartile | Study design | Enrolment period | Study setting | Average follow-up (months) | HCV diagnosis | HCC diagnosis | Risk estimates of HCC | Covariates adjusted for |
| Aziz et al[27], 2019 | Pakistan | Pak J Med Sc | Q3 | Cross-sectional | June 2016 to January 2018 | Hospital | 6.0 | HCV Ab, HCV RNA, and genotyping | USS abdomen, serum AFP, CT abdomen | Crude numbers | Exclusions: HBV, HIV, age < 18 yr or > 70 yr, pregnancy, previous liver lesion, “extremely fragile”, low bodyweight (not defined), known mental health issues, patients who were taking phenytoin, rifampicin, carbamazepine, patients with pancytopenia |
| Cha et al[29], 2016 | Korea | Medicine | Q3 | Retrospective case-control | January 2005 to December 2014 | Hospital | 59.6 | HCV Ab, HCV RNA, and genotyping | USS abdomen, serum AFP, CT abdomen, histological examination | HR with 95% confidence interval | Patients with < 6 months of follow-up or patients with HCC diagnosed within 6 months of enrolment in the study |
| Khan et al[30], 2009 | Pakistan | Journal of Medical Virology | Q1 | Retrospective case-control | January 2006 to September 2007 | Hospital | 6.0 | HCV Ab, HCV RNA, and genotyping | 2 of 3 criteria: Serum AFP > 400 IU/mL, CT/MRI or liver biopsy | Crude numbers | Co-infection with HBV or HDV |
| Kanwal et al[19], 2014 | United States | Journal of Hepatology | Q1 | Retrospective cohort study | October 1999 to September 2009 | Hospital | 12.0 | HCV Ab, HCV RNA, and genotyping | HCC (ICD-9 code 155.1) | HR with 95% confidence interval | < 1 yr of follow-up |
| Maryam et al[32], 2018 | Pakistan | Journal of Medical Virology | Q1 | Retrospective case-control | ND | Hospital | ND | HCV RNA and genotyping | Liver biopsy | Crude numbers | Nil |
| Park et al[35], 2019 | Korea | BMC Cancer | Q1, Q2 | Retrospective cohort study | January 2005 to December 2016 | Hospital | 24.0 | HCV Ab, HCV RNA, and genotyping | USS abdomen, serum AFP, CT Abdomen, histological examination | Crude numbers | People with HIV and/or HBV, < 6 months of follow-up |
| Tayyab et al[34], 2020 | Pakistan | BMC Gastroenterology | Q2 | Prospective cohort | October 2014 to March 2017 | Hospital | 12.0 | HCV Ab, HCV RNA, and genotyping | USS abdomen, serum AFP, CT abdomen | HR with 95% confidence interval | HBV co-infection |
Of these seven studies, two studies reported on only G3, two studies had participants which were > 90% G3 and two studies had > 5% G3 participants for which de-aggregated individual data could be collected[18,26,28,29,31,33,34] (Table 2).
| Ref. | Total number of participants | Percentage genotype 3, n (%) | Number of HCV GT3 participants | Age, yr, median or mean | Sex (M/F) | Number of HCC, n (%) | Patients without HCC, n (%) | Risk factor | Number with risk factor who developed HCC, n (%) | Cirrhosis (%) | Active HCV (%) | Cleared HCV, n (%) | HIV, n (%) | HBV, n (%) | Hazards ratio of risk factor | OR/HR/RR calculation (in study or calculated independently) |
| Aziz et al[27], 2019 | 300 | 300 (100.00) | 300 | 55.08 +/-5.62 | 179/121 | 10 (3.33) | 290 (96.67) | DAA treatment (SOF + DAC +/-RBV) | 10 (3.33) | 100 | 100 | 276 (92.00) | 0 (0) | 0 (0) | Independent calculation | |
| Child Pugh A (compensated cirrhosis) and SVR not achieved | 2 (0.67) | 100 | 100 | 276 (92.00) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Child Pugh B (compensated cirrhosis) and SVR achieved | 5 (1.67) | 100 | 100 | 276 (92.00) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Child Pugh B (Decompensated cirrhosis) and SVR not achieved | 3 (1.00) | 100 | 100 | 276 (92.00) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Male | 7 (2.33) | 100 | 100 | 276 (92.00) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Female | 3 (1.00) | 100 | 100 | 276 (92.00) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Cha et al[29], 2016 | 1335 | 98 (7.30) | 98 | 41.8 +/-10.5 | 79/19 | 4 (4.10) | 94 (95.92) | Age > 40 yr | ND | 25.50 | 100 | 34 (34.70) | 0 (0) | 4 (4.1) | 2.697 (0.436-16.683), P = 0.286 | Calculated in study |
| Cirrhosis at enrolment | 25 (25.50) | 25.50 | 100 | 34 (34.70) | 0 (0) | 4 (4.1) | 33.834 (2.088-548.269), P = 0.013 | Calculated in study | ||||||||
| Alcohol intake > 40 g/d | 53 (54.60) | 25.50 | 100 | 34 (34.70) | 0 (0) | 4 (4.1) | 8.556 (0.693-105.623), P = 0.094 | Calculated in study | ||||||||
| SVR | 34 (34.70) | 25.50 | 100 | 34 (34.70) | 0 (0) | 4 (4.1) | 0.848 (0.063-11.445), P = 0.901 | Calculated in study | ||||||||
| Decompensated cirrhosis and achieved SVR* | 1 | 25.50 | 100 | 34 (34.70) | 0 (0) | 4 (4.1) | Independent calculation | |||||||||
| Did not achieve SVR* | 1 | 25.50 | 100 | 34 (34.70) | 0 (0) | 4 (4.1) | Independent calculation | |||||||||
| Low platelet count | ND | 25.50 | 100 | 34 (34.70) | 0 (0) | 4 (4.1) | 1.00 (1.00- 1.00), P = 0.872 | Calculated in study | ||||||||
| Khan et al[30], 2009 | 158 | 147 (93.00) | 147 | 47.3 +/-12.5 | 102/56 | 65 (44.20) | 82 (55.78) | Male | 51 | 17.69 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |
| Female | 14 | 17.69 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |||||||||
| Age > 46.9 yr | 65 | 17.69 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |||||||||
| High AFP | 65 | 17.69 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |||||||||
| High HCV VL | 65 | 17.69 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |||||||||
| ALP > 68 | 65 | 17.69 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |||||||||
| Anti-HBc* | 46 | 17.69 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |||||||||
| HCV viraemia* | 58 | 17.6 | 87.10 | 30 (18.99) | 0 (0) | 5 | Independent calculation | |||||||||
| Kanwal et al[19], 2014 | 110484 | 8337 (7.54) | 8337 | 50.2 +/-6.4 | 8095/242 | ND | ND | Cirrhosis | ND | 12 | 86 | 1167 (14.00) | 242 (2.9) | 0 (0) | 1.44 (1.23-1.68) | Calculated in study |
| Diabetes | ND | 12 | 86 | 1167 (14.00) | 242 (2.9) | 0 (0) | 1.30 (1.88-1.90) | Calculated in study | ||||||||
| Age > 50 yr | ND | 12 | 86 | 1167 (14.00) | 242 (2.9) | 0 (0) | 1.79 (1.53-2.11) | Calculated in study | ||||||||
| Age < 50 yr | ND | 12 | 86 | 1167 (14.00) | 242 (2.9) | 0 (0) | 1.86 (1.56-2.22) | Calculated in study | ||||||||
| Maryam et al[32], 2018 | 50 | 50 (100.00) | 50 | 58 (47-73) | 37/23 | 27 (54.00) | 23 (46.00) | NRAS oncogene | 27 (54.00) | ND | 100 | 0 | ND | ND | Independent calculation | |
| Male* | 22 | ND | 100 | 0 | ND | ND | Independent calculation | |||||||||
| Female* | 5 | ND | 100 | 0 | ND | ND | Independent calculation | |||||||||
| Park et al[35], 2019 | 180 | 16 (8.88) | 16 | 46 (40-53) | 45306 | 16 (100.00) | 0 (0) | Male | 15 (93.80) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |
| Diabetes | 6 (40.00) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Cirrhosis | 16 (100.00) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Alcohol intake > 60 g/d | 3 (18.80) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| High HCV VL | 6 (37.70) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| MELD-score > 9.5 | 16 (100.00) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Female | 1 (6.25) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| High AFP | 16 (100.00) | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Not achieved SVR* | 2 | 100 | 100 | 2 (12.50) | 0 (0) | 0 (0) | Independent calculation | |||||||||
| Tayyab et al[34], 2020 | 653 | 593 (90.81) | 593 | 50 (41-56) | 319/334 | 40 (6.13) | 613 (93.87) | Age, per 10-yr increase | ND | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | 1.71 (1.25-2.33), P = 0.001 | Calculated in study |
| Use of SOF/DCV/RBV | 9 (22.50) | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | 17.05 (2.09-139.47), P = 0.01 | Calculated in study | ||||||||
| Cirrhosis | 40 (6.13) | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| Male* | 18 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| Female* | 22 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| High BMI* | 5 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| Hypertension* | 3 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| Diabetes* | 19 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| HBV Co-infection* | 12 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| Achieved SVR* | 35 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| Not achieved SVR* | 5 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| SOF/RBV use* | 29 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| SOF/RBV/PEG-IFN use* | 1 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation | ||||||||
| SOF/DCV use* | 1 | 49.31 | 54 (8.27) | 599 (91.78) | ND | 0 (0) | ND | Independent calculation |
Of the seven studies, three studies were retrospective case-control trials, two retrospective cohort studies, one a prospective cohort study and one a cross-sectional study design (Table 2). All were based in hospital settings with four in Pakistan, two in South Korea and one in the United States. All studies required HCV RNA sequencing with genotyping via ISO accredited standards to demonstrate HCV infection with diagnosis of HCC based on a combination of serum alpha feto-protein (AFP) and either imaging and/or histological classification. Only one study utilised ICD-coding which was based in the Global North[18].
One study involved a national database cohort (Military Veterans) with the other six studies involving single-centre hospital centres with participants enrolled between October 1999 and December 2016 (Tables 1 and 2).
The total number of participants included in the analysis were 113160 with 9541 HCV-G3 of which 162 developed HCC (Table 2). There were 8826 male participants with 796 female participants showing a preponderance of male (91.7%) participants. The demographics of participants are shown in Table 2. The mean duration of follow-up was 19.93 months ranging from six to 59.6 months. Four studies enrolled Pakistani participants (100.0%)[26,29,31,33], with two Korean (100.0%)[28,34] and one study recruiting primarily White and African-American participants (85.8%)[18].
The mean age of participants was 49.77 across the seven studies. The prevalence of cirrhosis was shown in 6 out of the 7 studies with an average of 51% ranging from 12% to 100%. The majority of the studies ensured the removal of the confounding effect of co-infection with HIV and/or hepatitis B virus (HBV) infection due to their exclusion criteria as part of their protocol with only one study (Kanwal et al[19]) including 242 out of 8337 HCV-G3 participants (2.9%) and two studies (Cha et al[29] and Khan et al[30]) including participants with HBV and HCV-G3 [4/98 (4.1%) and 5/147 (3.4%), respectively].
All seven studies demonstrated data of HCV infection status with an average of 83.05% participants having active HCV-G3 infection (ranging from 8.27% to 100.00%) with 37.71% participants clearing HCV-G3 (range 0-92%).
The majority of the studies were published in Q1 or Q2 quartile journals as per SJR with only one published in a Q3 journal. The quality of the studies was moderate with majority of studies NOS scores ranging from 6 to 8 (out of maximum score of 8) with only one study scoring a very low score of two (Table 3). The study with a low score of two primarily investigated a unique genomic marker for HCC and had a small sample size and thus was included in the analysis. Five of the studies ensured good methodological quality with two of relatively low quality. All but one study had a specified enrolment period with good data on follow-up of participants.
| Ref. | Type of Study | Selection | Comparability | Exposure | Total score | ||||||||||||
| Adequate case defini | Represe | Selection of controls1 | Definition of controls1 | Represe | Selection of non-exposed cohort2 | Ascertai | Demonst | Compara | Ascertai | Same method of ascertai | Nonres | Assess | Was follow-up long enough for out | Adequ | |||
| Aziz et al[27], 2019 | Cross-Sectional | NA | NA | NA | NA | * | * | * | * | * | NA | NA | NA | * | * | 7 | |
| Cha et al[29], 2016 | Cohort | NA | NA | NA | NA | * | * | * | * | * | NA | NA | NA | * | * | * | 8 |
| Khan et al[30], 2009 | Case-control | * | * | NA | NA | NA | NA | * | * | * | * | NA | NA | NA | 6 | ||
| Kanwal et al[19], 2014 | Cohort | NA | NA | NA | NA | * | * | * | * | * | NA | NA | NA | * | * | * | 8 |
| Maryam et al[32], 2018 | Case-control | * | NA | NA | NA | NA | * | * | NA | NA | NA | 3 | |||||
| Park et al[35], 2019 | Cohort | NA | NA | NA | NA | * | * | * | * | * | NA | NA | NA | * | * | * | 8 |
| Tayyab et al[34], 2020 | Cohort | NA | NA | NA | NA | * | * | * | * | * | NA | NA | NA | * | * | * | 8 |
Overall, 162 participants (1.7%) developed HCC during the follow-up period. The risk factors studied by the seven studies can be categorised as either participant background factors, biochemical factors or treatment factors (Supplementary Figure 1). The majority of the studies investigated the potential risk factors of gender at birth (male/female = five studies), cirrhosis (seven), age (five) from a participant background perspective. For treatment factors, the risk factors studied related to achievement of SVR (seven) or use of DAAs (five) with studied biochemical risk factors of high AFP (two), high HCV viral load (two) with one study each on ALP, low platelets levels, Child-Pugh Score (B or C), and high Model for End-Stage Liver Disease (MELD) score.
From their primary analysis, a total of seven studies assessed demonstrated cirrhosis to be a risk factor for HCC secondary to HCV-G3 followed by higher age (5), with two studies each showing male sex, high AFP, DAA treatment and achievement of SVR as risk factors for developing HCC.
A total of seven studies assessed demonstrated cirrhosis to be a risk factor for HCC secondary to HCV-G3 followed by higher age (5), with two studies each showing male sex, high AFP, DAA treatment and achievement of SVR as risk factors for developing HCC.
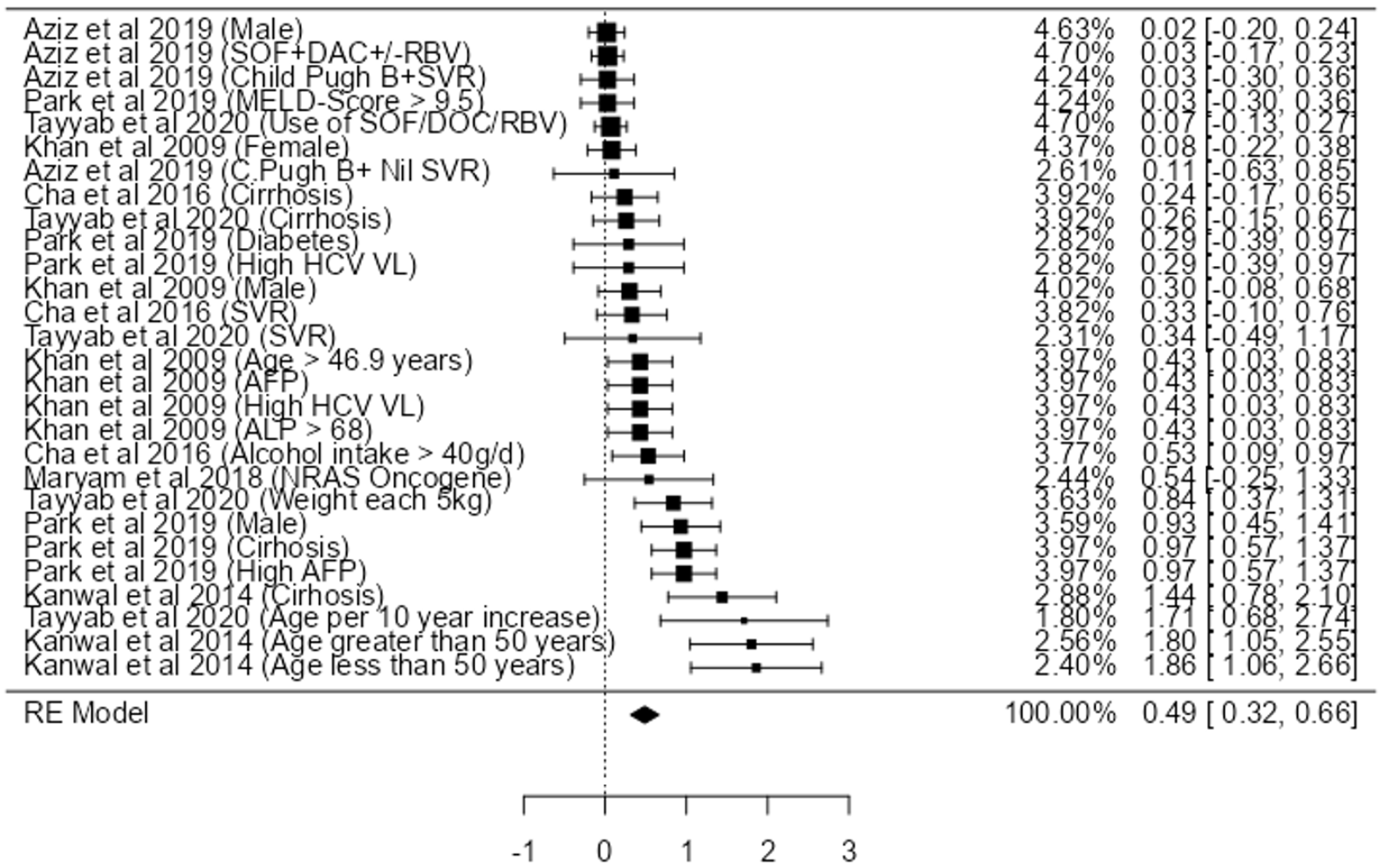
Utilising the individual participant data (Table 4) and pooling the data from the seven studies, we only found a strong association between age > 50 (HR: 1.86, 95%CI: 1.05-2.55). We also found a relatively moderate association with cirrhosis (HR: 1.44, 95%CI: 0.78-2.10), high AFP (HR: 0.97, 95%CI: 0.57-1.37), male gender (HR: 0.93, 95%CI: 0.45-1.41) and weight gain (HR: 0.84, 95%CI: 0.37-1.31), high HCV VL (HR: 0.43, 95%CI: 0.03-0.83), ALP > 68 (HR: 0.43, 95%CI: 0.03-0.83), and alcohol intake > 40 g/dL (HR: 0.24, 95%CI: 0.17-0.34) (Figure 2).
| Risk factor | Number of participants |
| Patient-dependent factors | |
| Cirrhosis | 66 |
| Male | 118 |
| Female | 62 |
| Age > 40 yr | 65 |
| Alcohol intake > 40 g/d | 56 |
| Anti-HBc | 46 |
| Diabetes | 25 |
| Age < 50 yr | 0 |
| NRAS oncogene | 27 |
| Age, per 10-yr increase | 0 |
| High BMI | 5 |
| Hypertension | 3 |
| HBV co-infection | 12 |
| Treatment dependent factors | |
| DAA treatment | 66 |
| SVR achieved | 74 |
| SVR not achieved | 13 |
| Decompensated cirrhosis and achieved SVR | 1 |
| Use of SOF/DCV/RBV | 9 |
| Biochemical factors | |
| Low platelet count | 16 |
| High AFP | 65 |
| High HCV VL | 71 |
| ALP > 68 | 65 |
| HCV viraemia | 58 |
| MELD-score > 9.5 | 16 |
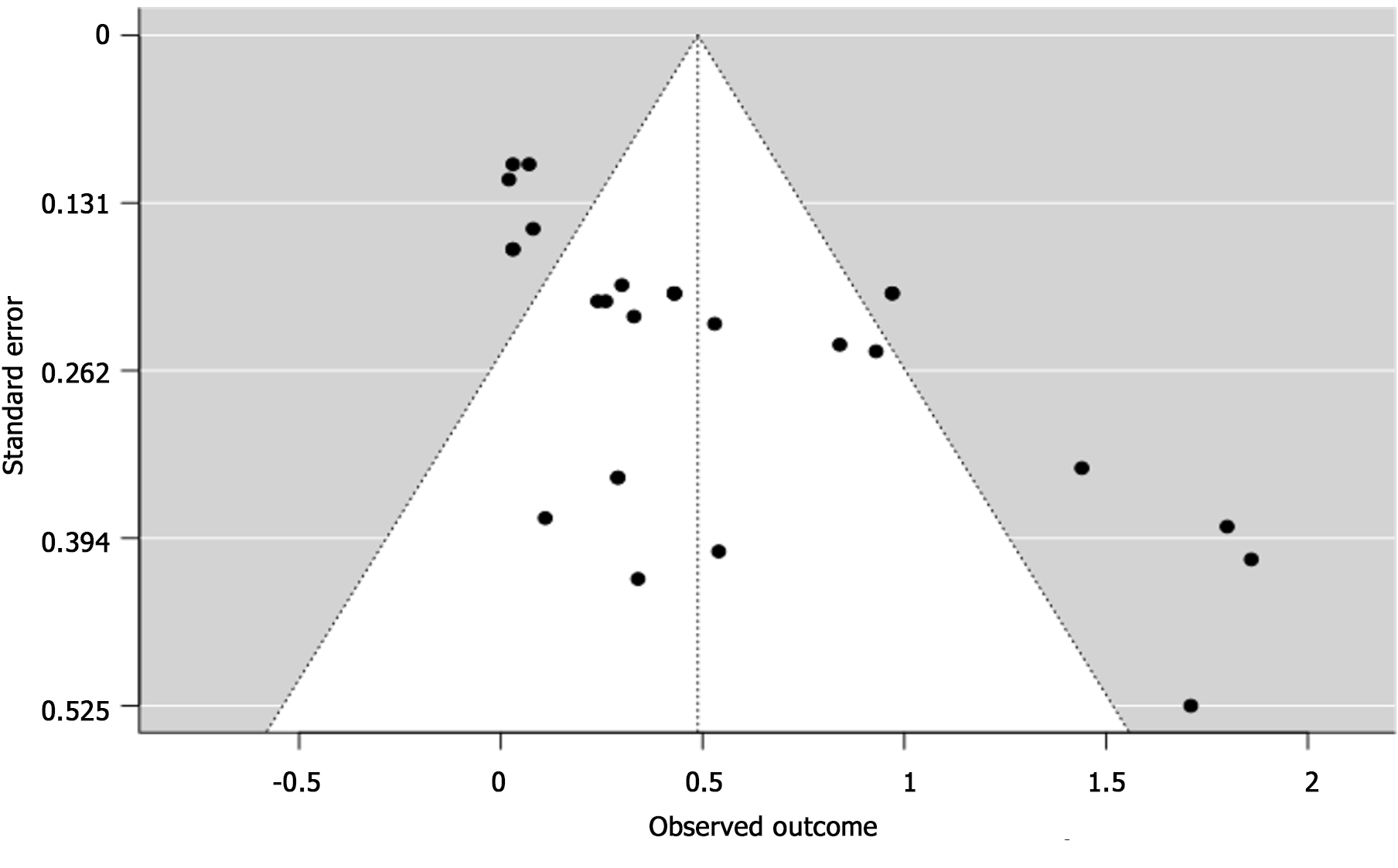
Some studies also showed an association between DAA use, MELD Score >9.5, Female, Diabetes, NRAS Oncogene (Figure 2). However, high statistical heterogeneity (I2 = 79.84%, with P < 0.001) was observed. As the heterogeneity was high, the factors were not fully combined for a pooled HR: to demonstrate an appropriate view of the data. The funnel plot did demonstrate asymmetry (Egger’s test = 4.936, P < 0.001) did not indicate for small-study effects (Figure 3).
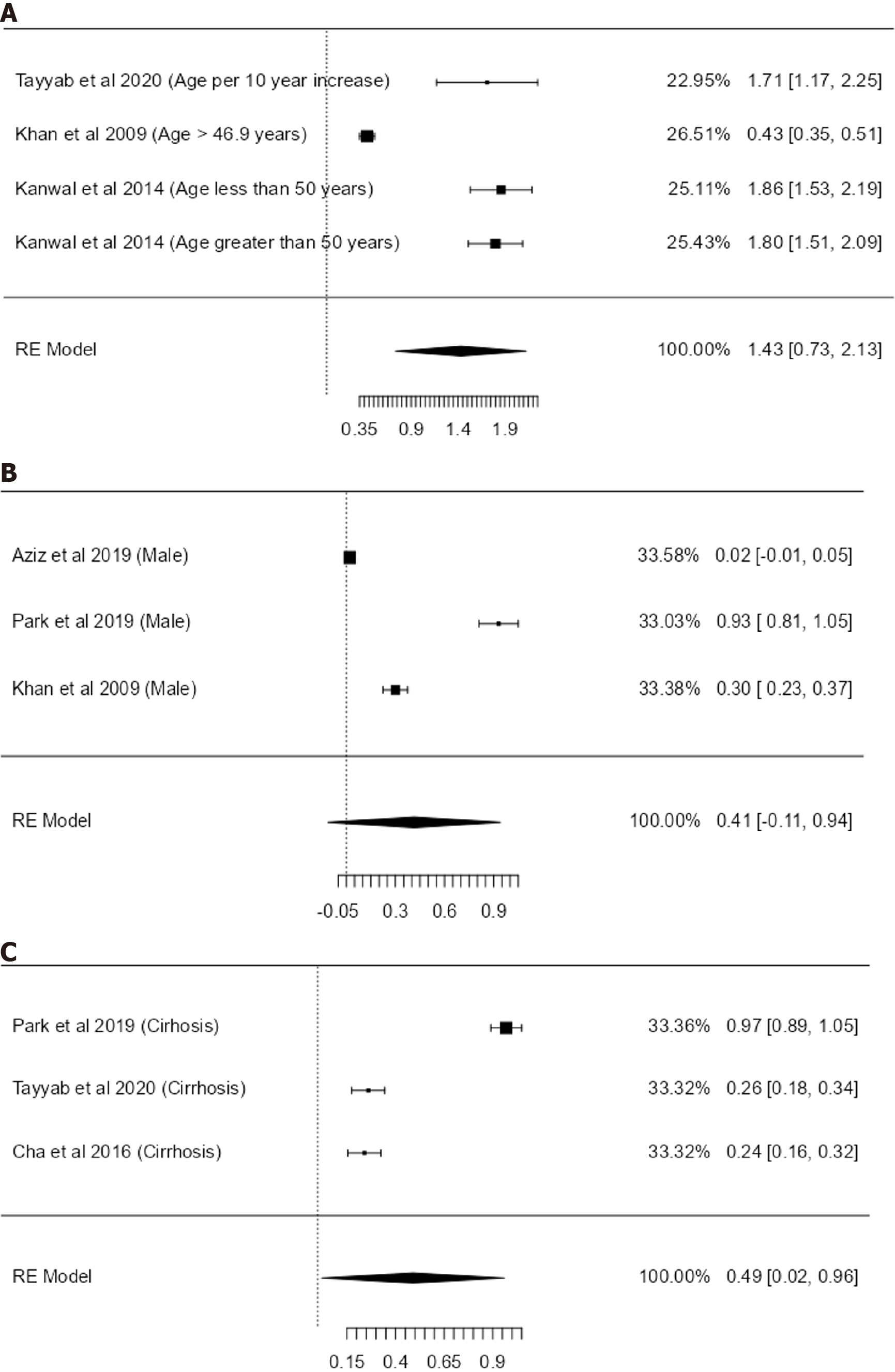
We performed a sub-analysis of risk factors where there were more than three studies studying the uniform risk factor. We pooled the HRs of to show an overall effect size, utilising the random effects model (Figure 4).
When exclusively pooling the studies, the combined HR: for cirrhosis is 0.49 (95%CI: 0.02-0.96, I2 = 98.96%, P ≤ 0.001, n = 3), for age 1.43 (95%CI: 0.73-2.13, I2 = 96.44%, n = 4) and for male gender 0.41 (95%CI: -0.11 to 0.94, I2 = 99.45%, P ≤ 0.001, n = 3).
HCV infection represents a significant global health burden, with millions of individuals affected worldwide. Considered a “viral time bomb”[36], the World Health Organization’s (WHO) ambitious target of eliminating HCV as a public health threat by 2030 has spurred unprecedented efforts to increase screening, diagnosis, and treatment access. While advances in DAA therapy have led to remarkable rates of viral clearance, the emergence of HCC in post-treatment patients has raised concerns and new challenges.
Among the various HCV genotypes, G3 has attracted particular attention due to its distinctive association with HCC development. Notably, patients infected with G3 have a higher predisposition to developing HCC, even in the absence of cirrhosis. This unique genotype’s enhanced hepatocarcinogenic potential warrants further exploration and the need to investigate risk factors associated with the development to HCC.
This systematic review of seven HCV-G3 studies with 9541 HCV-G3 participants shows that cirrhosis and age greater than 40 are principal risk factors for developing HCC in people with HCV-G3. It is the largest study focusing on HCC secondary to HCV-G3 of which 162 developed HCC.
This study shows that there are few published studies on HCV-G3 and HCC and the majority of the studies are observational studies of retrospective design which do not have the ability to fully investigate confounding factors. It also demonstrates that the data is very heterogenous in HCV-G3 studies with a lack of high-quality studies and randomised control trials with a focus on HCV-G3. Of note, there is a lack of data and association with diabetes, HBV co-infection and/or high BMI especially with the increasing prevalence of metabolic dysfunction-associated steatotic liver disease.
This may be due to the lack of G3 patients and participants in the countries where the majority of HCV and HCC clinical trials occur. The highest global prevalence is of G3 in South and Central Asia (71.6% of HCV infection), contrasting to 24.8% in Western Europe and 10%-12% in the United States[5], where the highest number of HCV and HCC clinical trials occur. Without adequate G3 participants in the Global North, it is difficult to power studies to demonstrate appropriate risk factors for HCC in HCV-G3.
Correspondingly, in this study we have noted that there is only a moderate association of HCV-G3 with cirrhosis leading to HCC. This contrasts to G1 where there has been established a high association of cirrhosis with HCC[37-39] with some studies demonstrating a significant HR of 6.686 (4.319-10.350)[40]. Similar significant associations with cirrhosis and HCC were noted in G4[24] and G6[41], with a lack of data for G5 due to its low global prevalence. Majority of HCC predictive scores aim to quantify HCC risk in the presence of cirrhosis due to the high association with HCC[42-45]. However, these scores have been developed and validated on a predominance of G1 and G2 participants with a low percentage of G3 participants, warranting further studies for HCC in G3-predominant populations.
Efforts to eliminate HCV, especially in regions with high endemicity of G3, such as in India and Pakistan, face substantial challenges. The efficacy of treatment strategies in curbing HCV transmission must be supported by surveillance for potential risk of subsequent HCC development in patients with G3 mono-infection and those with co-infection with HBV and/or HIV. The evolving epidemiological landscape demands careful surveillance and long-term follow-up of patients treated for HCV, particularly those belonging to high-risk populations.
To supplement the WHO’s ambitious HCV elimination goals and reduce the burden of associated disease, it is imperative to implement proactive measures for identifying and managing HCC risk in patients post viral clearance. Strategies may include intensified surveillance, targeted risk stratification, and tailored treatment approaches based on HCV genotype and individual patient characteristics.
There is currently a lack of data in the literature regarding the risk factors for HCC secondary to hepatitis HCV-G3. Yet, no confirmed risk factors have been identified. To better understand the risk factors for HCC secondary to HCV-G3, a case-control trial is needed. Such a trial would allow for a more in-depth investigation of the risk factors associated with this condition.
The global initiative to eliminate HCV by 2030 represents a remarkable public health undertaking. However, the emergence of HCC as a significant concern in patients post viral clearance, particularly in HCV-G3 infections, demands careful consideration. Collaborative efforts between healthcare providers, researchers, and policymakers are essential to develop effective risk mitigation strategies while ensuring the successful elimination of HCV on a global scale. Continued research into the mechanistic basis of HCC development in HCV-G3 infections will be crucial in shaping preventive and therapeutic interventions to safeguard the progress made towards an HCV-free future.
Neglected hepatitis C genotype 3 (HCV-G3) is a global health concern as it is more oncogenic than other genotypes.
It leads to hepatocellular carcinoma (HCC) in people without cirrhosis and HCV-G3 HCC risk factors are currently unknown with no validated risk assessment tools.
To systematically review and critically appraise all risk factors for HCC secondary to HCV-G3 in all settings. Consequently, we studied possible risk factors for HCC due to HCV-G3 in the literature from 1946 to 2023.
We searched the following four databases for articles: Web of Science, Medline, EMBASE, and CENTRAL; for studies published between 1st January 1946 to 17th December 2022.
Cirrhosis, higher age, and male gender were found to be strongly associated with HCC due to HCV-G3.
There is currently a lack of data in the literature regarding the risk factors for HCC secondary to HCV-G3. As of yet, no confirmed risk factors have been identified.
With limited studies on HCV-G3 and HCC, further research is needed to provide a risk assessment tool for HCC secondary to HCV-G3.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Du H, China S-Editor: Chen YL L-Editor: A P-Editor: Zhang XD
| 1. | Mahmud S, Al Kanaani Z, Abu-Raddad LJ. Characterization of the hepatitis C virus epidemic in Pakistan. BMC Infect Dis. 2019;19:809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 2. | Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22:7824-7840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 514] [Cited by in RCA: 568] [Article Influence: 63.1] [Reference Citation Analysis (7)] |
| 3. | Chan CP, Uemura H, Kwan TH, Wong NS, Oka S, Chan DPC, Lee SS. Review on the molecular epidemiology of sexually acquired hepatitis C virus infection in the Asia-Pacific region. J Int AIDS Soc. 2020;23:e25618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 4. | Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61:S45-S57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1325] [Cited by in RCA: 1361] [Article Influence: 123.7] [Reference Citation Analysis (0)] |
| 5. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1145] [Article Influence: 114.5] [Reference Citation Analysis (0)] |
| 6. | Chan A, Patel K, Naggie S. Genotype 3 Infection: The Last Stand of Hepatitis C Virus. Drugs. 2017;77:131-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Goossens N, Negro F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology. 2014;59:2403-2412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 8. | Erman Daloğlu A, Parkan ÖM, Erdoğan A, Peker BO, Can Sarınoğlu R, Sağlık İ, İnan D, Kuloğlu MM, Mutlu D, Öngüt G, Çolak D. [Distribution of Hepatitis C Virus (HCV) Genotypes Among Intravenous Drug and Non-Drug User Patients]. Mikrobiyol Bul. 2021;55:30-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Reference Citation Analysis (0)] |
| 9. | Butt AS, Abbas Z, Jafri W. Hepatocellular carcinoma in pakistan: where do we stand? Hepat Mon. 2012;12:e6023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Young J, Wong S, Janjua NZ, Klein MB. Comparing direct acting antivirals for hepatitis C using observational data - Why and how? Pharmacol Res Perspect. 2020;8:e00650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Afzal MS. Hepatitis C Virus and Interferon-Free Antiviral Therapeutics Revolution: Implications for Pakistan. Viral Immunol. 2017;30:252-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Akhtar S, Moatter T, Azam SI, Rahbar MH, Adil S. Prevalence and risk factors for intrafamilial transmission of hepatitis C virus in Karachi, Pakistan. J Viral Hepat. 2002;9:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Amougou-Atsama M, Jean Adrien Atangana P, Noah Noah D, Fewou Moundipa P, Pineau P, Njouom R. The role of hepatitis C virus genotypes and core mutations in hepatocellular carcinoma in Cameroon. J Viral Hepat. 2020;27:880-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 14. | Chhatwal J, Chen Q, Ayer T, Bethea ED, Kanwal F, Kowdley KV, Wang X, Roberts MS, Gordon SC. Hepatitis C virus re-treatment in the era of direct-acting antivirals: projections in the USA. Aliment Pharmacol Ther. 2018;47:1023-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Zeuzem S, Foster GR, Wang S, Asatryan A, Gane E, Feld JJ, Asselah T, Bourlière M, Ruane PJ, Wedemeyer H, Pol S, Flisiak R, Poordad F, Chuang WL, Stedman CA, Flamm S, Kwo P, Dore GJ, Sepulveda-Arzola G, Roberts SK, Soto-Malave R, Kaita K, Puoti M, Vierling J, Tam E, Vargas HE, Bruck R, Fuster F, Paik SW, Felizarta F, Kort J, Fu B, Liu R, Ng TI, Pilot-Matias T, Lin CW, Trinh R, Mensa FJ. Glecaprevir-Pibrentasvir for 8 or 12 Weeks in HCV Genotype 1 or 3 Infection. N Engl J Med. 2018;378:354-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 314] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 16. | Wei L, Wang G, Alami NN, Xie W, Heo J, Xie Q, Zhang M, Kim YJ, Lim SG, Fredrick LM, Lu W, Liu W, Kalluri HV, Krishnan P, Tripathi R, Mobashery N, Burroughs M, Asatryan A, Jia J, Hou J. Glecaprevir-pibrentasvir to treat chronic hepatitis C virus infection in Asia: two multicentre, phase 3 studies- a randomised, double-blind study (VOYAGE-1) and an open-label, single-arm study (VOYAGE-2). Lancet Gastroenterol Hepatol. 2020;5:839-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 17. | Yamana Y, Kanda T, Matsumoto N, Honda M, Kumagawa M, Sasaki R, Kanezawa S, Mizutani T, Yamagami H, Masuzaki R, Ishii T, Nirei K, Moriyama M. Efficacy of Glecaprevir/Pibrentasvir for Real-World HCV Infected Patients in the Northern Part of Tokyo, Japan. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Dash S, Aydin Y, Widmer KE, Nayak L. Hepatocellular Carcinoma Mechanisms Associated with Chronic HCV Infection and the Impact of Direct-Acting Antiviral Treatment. J Hepatocell Carcinoma. 2020;7:45-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 19. | Kanwal F, Kramer JR, Ilyas J, Duan Z, El-Serag HB. HCV genotype 3 is associated with an increased risk of cirrhosis and hepatocellular cancer in a national sample of U.S. Veterans with HCV. Hepatology. 2014;60:98-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 240] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 20. | Lee SS, Kim CY, Kim BR, Cha RR, Kim WS, Kim JJ, Lee JM, Kim HJ, Ha CY, Kim TH, Jung WT, Lee OJ. Hepatitis C virus genotype 3 was associated with the development of hepatocellular carcinoma in Korea. J Viral Hepat. 2019;26:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Nkontchou G, Ziol M, Aout M, Lhabadie M, Baazia Y, Mahmoudi A, Roulot D, Ganne-Carrie N, Grando-Lemaire V, Trinchet JC, Gordien E, Vicaut E, Baghad I, Beaugrand M. HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepat. 2011;18:e516-e522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 22. | McMahon BJ, Bruden D, Townshend-Bulson L, Simons B, Spradling P, Livingston S, Gove J, Hewitt A, Plotnik J, Homan C, Espera H, Negus S, Snowball M, Barbour Y, Bruce M, Gounder P. Infection With Hepatitis C Virus Genotype 3 Is an Independent Risk Factor for End-Stage Liver Disease, Hepatocellular Carcinoma, and Liver-Related Death. Clin Gastroenterol Hepatol. 2017;15:431-437.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77:1598-1606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 1037] [Article Influence: 345.7] [Reference Citation Analysis (0)] |
| 24. | El-Serag HB, Kanwal F, Richardson P, Kramer J. Risk of hepatocellular carcinoma after sustained virological response in Veterans with hepatitis C virus infection. Hepatology. 2016;64:130-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 312] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 25. | Adnan F, Khan NU, Iqbal A, Ali I, Petruzziello A, Sabatino R, Guzzo A, Loquercio G, Botti G, Khan S, Naeem M, Khan MI. Interleukin-6 polymorphisms in HCC patients chronically infected with HCV. Infect Agent Cancer. 2020;15:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Asim M, Sarma MP, Kar P. Etiological and molecular profile of hepatocellular cancer from India. Int J Cancer. 2013;133:437-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 27. | Aziz B, Nazar T, Akhlaq S. The frequency of occurrence of Hepatocellular Carcinoma after direct antiviral therapy in Hepatitis C virus patients. Pak J Med Sci. 2019;35:101-105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Ul Abideen Z, Siddique S, Nasrullah I, Khan JS, Rehman S, Tong Y, Qamar R, Butt AM. A 2-year retrospective study of viral and host-associated risk factors in Pakistani hepatocellular carcinoma patients. Eur J Gastroenterol Hepatol. 2019;31:1103-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Cha RR, Lee SS, Lee CM, Ji SB, Jung HC, Cho HC, Kim JJ, Lee JM, Kim HJ, Ha CY, Kim TH, Jung WT, Lee OJ. Clinical Features and Outcomes of Patients With Genotype 3 Hepatitis C Virus Infection in Korea: A Retrospective Observational Study. Medicine (Baltimore). 2016;95:e2755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 30. | Khan A, Tanaka Y, Azam Z, Abbas Z, Kurbanov F, Saleem U, Hamid S, Jafri W, Mizokami M. Epidemic spread of hepatitis C virus genotype 3a and relation to high incidence of hepatocellular carcinoma in Pakistan. J Med Virol. 2009;81:1189-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Gulnaz A, Sayyed AH, Amin F, Khan Au, Aslam MA, Shaikh RS, Ali M. Association of XRCC1, XRCC3, and XPD genetic polymorphism with an increased risk of hepatocellular carcinoma because of the hepatitis B and C virus. Eur J Gastroenterol Hepatol. 2013;25:166-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Maryam M, Idrees M. Study of promoter hypomethylation profiles of RAS oncogenes in hepatocellular carcinoma derived from hepatitis C virus genotype 3a in Pakistani population. J Med Virol. 2018;90:1516-1523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Wu N, Rao HY, Yang WB, Gao ZL, Yang RF, Fei R, Gao YH, Jin Q, Wei L. Impact of hepatitis C virus genotype 3 on liver disease progression in a Chinese national cohort. Chin Med J (Engl). 2020;133:253-261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Tayyab GUN, Rasool S, Nasir B, Rubi G, Abou-Samra AB, Butt AA. Hepatocellular carcinoma occurs frequently and early after treatment in HCV genotype 3 infected persons treated with DAA regimens. BMC Gastroenterol. 2020;20:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Park HK, Lee SS, Im CB, Im C, Cha RR, Kim WS, Cho HC, Lee JM, Kim HJ, Kim TH, Jung WT, Lee OJ. Hepatitis C virus genotype affects survival in patients with hepatocellular carcinoma. BMC Cancer. 2019;19:822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Littera R, Zamboni F, Tondolo V, Fantola G, Chessa L, Orrù N, Sanna M, Valentini D, Cappai L, Mulargia M, Caocci G, Arras M, Floris A, Orrù S, La Nasa G, Carcassi C. Absence of activating killer immunoglobulin-like receptor genes combined with hepatitis C viral genotype is predictive of hepatocellular carcinoma. Hum Immunol. 2013;74:1288-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Detonating a viral time bomb--the hepatitis C pandemic. Lancet. 2013;381:178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 38. | Silini E, Bottelli R, Asti M, Bruno S, Candusso ME, Brambilla S, Bono F, Iamoni G, Tinelli C, Mondelli MU, Ideo G. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a case-control study. Gastroenterology. 1996;111:199-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 126] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Silini E, Bono F, Cividini A, Cerino A, Bruno S, Rossi S, Belloni G, Brugnetti B, Civardi E, Salvaneschi L. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology. 1995;21:285-290. [PubMed] |
| 40. | Nousbaum JB, Pol S, Nalpas B, Landais P, Berthelot P, Bréchot C. Hepatitis C virus type 1b (II) infection in France and Italy. Collaborative Study Group. Ann Intern Med. 1995;122:161-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 348] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 41. | Shiha G, Mikhail NNH, Soliman R, Hassan A, Eslam M. Predictive performance and clinical utility of HCC risk scores in chronic hepatitis C: a comparative study. Hepatol Int. 2022;16:159-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 42. | Lee MH, Hsiao TI, Subramaniam SR, Le AK, Vu VD, Trinh HN, Zhang J, Jin M, Wong VW, Wong GL, Nguyen MH. HCV Genotype 6 Increased the Risk for Hepatocellular Carcinoma Among Asian Patients With Liver Cirrhosis. Am J Gastroenterol. 2017;112:1111-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Ioannou GN, Green PK, Beste LA, Mun EJ, Kerr KF, Berry K. Development of models estimating the risk of hepatocellular carcinoma after antiviral treatment for hepatitis C. J Hepatol. 2018;69:1088-1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 44. | Sharma SA, Kowgier M, Hansen BE, Brouwer WP, Maan R, Wong D, Shah H, Khalili K, Yim C, Heathcote EJ, Janssen HLA, Sherman M, Hirschfield GM, Feld JJ. Toronto HCC risk index: A validated scoring system to predict 10-year risk of HCC in patients with cirrhosis. J Hepatol. 2017;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 132] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 45. | Flemming JA, Yang JD, Vittinghoff E, Kim WR, Terrault NA. Risk prediction of hepatocellular carcinoma in patients with cirrhosis: the ADRESS-HCC risk model. Cancer. 2014;120:3485-3493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (1)] |