Published online Feb 15, 2024. doi: 10.4251/wjgo.v16.i2.493
Peer-review started: November 9, 2023
First decision: November 23, 2023
Revised: December 5, 2023
Accepted: January 12, 2024
Article in press: January 12, 2024
Published online: February 15, 2024
Processing time: 84 Days and 16.6 Hours
Gastric cancer (GC) is one of the most aggressive malignancies with limited therapeutic options and a poor prognosis. Resveratrol, a non-flavonoid poly
To identify the effects of resveratrol on GC progression and explore the related molecular mechanisms.
Action targets of resveratrol and GC-related targets were screened from public databases. The overlapping targets between the two were confirmed using a Venn diagram, and a “Resveratrol-Target-GC” network was constructed using Cyto
A total of 378 resveratrol action targets and 2154 GC disease targets were obtained from public databases, and 181 intersection targets between the two were screened by Venn diagram. The top 20 core targets were identified by PPI network analysis of the overlapping targets. GO function analysis mainly involved protein binding, identical protein binding, cytoplasm, nucleus, negative regulation of apoptotic process and response to xenobiotic stimulus. KEGG enrichment analysis suggested that the involved signaling pathways mainly included PI3K-AKT signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, TNF signaling pathway, ErbB signaling pathway, etc. FBJ murine osteosarcoma viral oncogene homolog (FOS) and matrix metallopeptidase 9 (MMP9) were selected by differential expression analysis, and they were closely associated with immune infiltration. Molecular docking results showed that resveratrol docked well with these two targets. Resveratrol treatment arrested the cell cycle at the S phase, induced apoptosis, and weakened viability, migration and invasion in a dose-dependent manner. Furthermore, resveratrol could exhibit anti-GC effect by regulating FOS and MMP9 expression.
The anti-GC effects of resveratrol are related to the inhibition of cell proliferation, migration, invasion and induction of cell cycle arrest and apoptosis by targeting FOS and MMP9.
Core Tip: Based on network pharmacology and bioinformatics, the molecular targets and signaling pathways of resveratrol on gastric cancer (GC) were explored, and experiments were used to validate the network analysis. Network analysis results suggested that the action of resveratrol on GC involved multiple targets and pathways. In vitro experiments suggested that the anti-GC effects of resveratrol were related to the inhibition of cell proliferation, migration, invasion and induction of cell cycle arrest and apoptosis by targeting FBJ murine osteosarcoma viral oncogene homolog (FOS) and matrix metallopeptidase 9 (MMP9). This study confirmed anti-GC effects of resveratrol and highlighted that FOS and MMP9 might be the effective biotargets for GC treatment.
- Citation: Ma YQ, Zhang M, Sun ZH, Tang HY, Wang Y, Liu JX, Zhang ZX, Wang C. Identification of anti-gastric cancer effects and molecular mechanisms of resveratrol: From network pharmacology and bioinformatics to experimental validation. World J Gastrointest Oncol 2024; 16(2): 493-513
- URL: https://www.wjgnet.com/1948-5204/full/v16/i2/493.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i2.493
Gastric cancer (GC) is one of the most common gastrointestinal malignancies. According to global cancer statistics, GC remains a major global cancer and is responsible for more than one million new cases (5.6% of all sites) and nearly 770000 deaths (7.7% of all sites) in 2020, ranking fifth in incidence and fourth in mortality[1]. Current therapeutic methods for GC include endoscopic resection, surgical resection, chemoradiotherapy, immunotherapy, and targeted therapy, with combination therapy being more effective than single therapy[2]. Although the application of multidisciplinary models and combination therapy in the diagnosis and treatment of GC has made certain achievements, GC remains a major contributor to the burden of global cancer incidence and mortality[3]. This is because the onset of GC is occult, and patients are often in the advanced stage of the disease when they are diagnosed, losing the optimal treatment period. The pathogenesis of GC is a complex and multistage process; it is not solely caused by mutation or dysfunction in a single gene but often involves an imbalance in the whole regulatory network[4]. Moreover, the therapeutic effect of medication is limited due to adjoint toxicity and side effects, drug resistance and difficulties in personalized medication[5]. Therefore, it is crucial to find multiple effects and rapid therapeutic strategies with low toxicity and minimal side effects to treat GC.
Traditional Chinese medicine (TCM) monomers have always played a crucial role in the drug treatment of diseases, especially in the field of oncology. TCM monomers have been widely used in clinical treatment because of their advantages of low toxicity, low cost, high efficiency and multiple targets[6,7]. Resveratrol, a natural polyphenol compound mainly extracted from Veratrum album (white hellebore), Polygonum cuspidatum and peanut, has been reported to be potentially involved in regulating the molecular pathways of malignancy to inhibit various cancers[8-10]. At present, the antitumor effects of resveratrol are mainly reflected in regulating the cell cycle, promoting apoptosis, obstructing metastasis, inhibiting epithelial-mesenchymal transformation, and sensitizing cells to chemotherapy[11-13]. In recent years, research on resveratrol in GC has developed rapidly, providing a reasonable direction for overcoming the inherent challenges in the treatment of GC. Previous studies have demonstrated that resveratrol can inhibit the proliferation of GC cells, reduce the invasion potential and inhibit epithelial-mesenchymal transition (EMT)[14-16]. In addition, the combination of resveratrol and cisplatin induced G2/M phase cell cycle arrest and apoptosis in GC cells, suggesting that resveratrol is a promising adjuvant to enhance the sensitivity of GC to standard chemotherapy[17]. A prospective open-label phase II single-arm study between October 2019 and April 2021 showed that the administration of resveratrol-copper reduced the toxicity of docetaxel-based multiagent chemotherapy and improved patients’ chemotherapy tolerance[18]. However, there are limited reports on the effects and molecular mechanisms of resveratrol in GC.
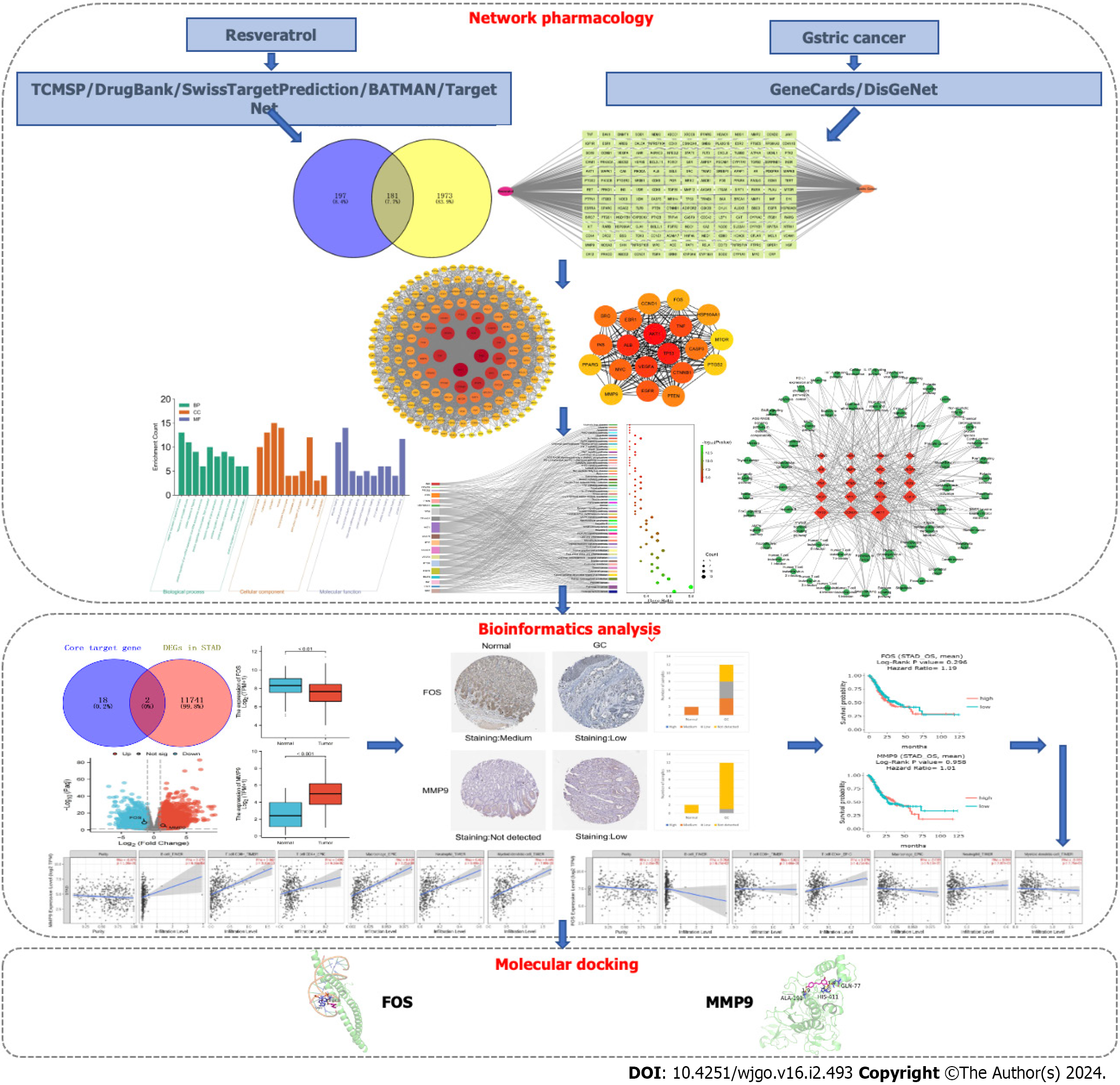
Using multiple biological information databases to analyze the correlation between drugs and diseases has become an important method for studying drug action mechanisms. Network pharmacology, a new subject based on the theory of systems biology, utilizes biological networks to mechanically link drugs and diseases, revealing multiple-targets and multiple-mechanisms of drug action[19]. In this study, the molecular biological mechanisms of resveratrol in GC were explored based on network pharmacology and bioinformatics, and experiments were used to confirm the network analysis. This study workflow is illustrated in Figure 1.
The molecular structure of resveratrol (SDF format) was derived from the PubChem database (available online: https://pubchem.ncbi.nlm.nih.gov/). The predicted targets for resveratrol were filtered from five public databases: TCM Systems Pharmacology Database and Analysis Platform[20] (TCMSP, https://old.tcmsp-e.com/tcmsp.php), DrugBank Online website[21] (https://go.drugbank.com/), SwissTargetPrediction[22] (http://swisstargetprediction.ch/), Bioinformation Analysis Tool for Molecular Mechanism of TCM[23] (BATMAN-TCM, available online: http://bionet.ncpsb.org.cn/batman-tcm/index.php), and TargetNet (http://targetnet.scbdd.com/). By using the protein database UniProt (https://www.uniprot.org/), the target protein obtained from the public database was converted into a uniform gene name, and the species was restricted to Homo sapiens.
Disease-related target information was obtained by searching the keyword “gastric cancer” in the GeneCards (http://www.genecards.org) and DisGeNet (https://www.disgenet.org/) databases. In the GeneCards database, a relevance score > 1 was set, and twice the median value to screen biological targets for GC.
The online site Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/) was used to analyze resveratrol and GC targets. The compound targets were mapped to disease targets to obtain predicted targets for resveratrol in the treatment of GC. The “resveratrol-target-GC” network was constructed by Cytoscape software version 3.9.1. Nodes in the network represent the compound, intersection targets and the disease, while edges represent the connection among the three.
The protein-protein interaction (PPI) network of common targets of resveratrol and GC was constructed by STRING version 11.5 (http://string-db.org). Selecting the appropriate filter criteria for the PPI network in the settings window: The needed minimum interaction score was set as the “medium confidence (0.400)”, and the disconnected nodes in the network were hidden. The constructed PPI network was then imported into Cytoscape software in tab separated values format, analyzing the complicated relationships among the intersection targets visually. Core targets were recognized by using the degree value.
The Database for Annotation, Visualization and Integrated Discovery version 6.8 (https://david.ncifcrf.gov/), which provides a comprehensive set of functional annotation tools, was used to analyze the biological functions and pathways enrichment of resveratrol against GC. Cellular component (CC), molecular function (MF), and biological process (BP) were the main categories of Gene Ontology (GO) enrichment analysis. GO terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways with the highest correlation with resveratrol anti-GC were screened in ascending order according to P < 0.05, and the results were represented by a scale histogram or Sankey + dot plot. The “target-pathway” network was constructed by using Cytoscape to visualize the important functional mechanisms of resveratrol in the treatment of GC. In the network, nodes represent targets and pathways, while edges represent the interactions between them.
The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) and The Human Protein Atlas (HPA, https://www.proteinatlas.org/) databases were used to analyze the differential expression of core target genes at the RNA level and protein level, respectively. The source data for GC were from stomach adenocarcinoma (STAD) in TCGA, and gene expression data for 407 samples were obtained, including 32 normal samples and 375 GC samples. Based on the DESeq2 software package in version 4.2.1 of the R platform, normalization and difference analysis were carried out on downloaded data. |log2-fold change| > 1 and P < 0.05 were the filter conditions to identify differentially expressed genes (DEGs) between GC and normal samples. The core target genes were verified by DEG analysis, and the differential expression results are shown by volcano maps and box plots. The HPA database uses immunohistochemical techniques to map the position of proteins encoded by genes expressed in human tissues and cells and is read and indexed by professionals. The protein expression of the core target genes verified by TCGA in tumor tissues and corresponding normal tissues was compared in the HPA database.
DriverDBv3 (http://driverdb.tms.cmu.edu.tw/) is a cancer omics database where the “Gene Survival” function provides visualizations to illustrate the survival probability of selected genes across multiple cancer types. The function focuses on the relationship between the expression of target candidates verified by differential expression and overall survival (OS) and produces Kaplan-Meier plots. Hazard ratios and P value were calculated in survival analysis. If the P value of OS < 0.05, it was considered that target candidates were associated with the survival of GC. TIMER2.0 (http://timer.cistrome.org/) was used to analyze the relationship between the expression of target candidates and immune cell infiltration.
Molecular docking between small molecule ligands and protein receptors was mainly accomplished by PyMOL and AutoDock software. The three-dimensional structure of resveratrol was downloaded from PubChem and saved in SDF format. Then, Chem 3D converted the “SDF” format into a “mol2” structure to complete the small molecule ligand file preparation. The crystal structure of target proteins was downloaded from the RCSB Protein Data Bank (RCSB PDB, https://www.rcsb.org/) according to the resolution and correlation ranking and saved in “PDB” format. AutoDockTools version 1.5.6 was used for molecular docking and calculation of the binding energy of each conformation. The docking result was visualized and analyzed in PyMOL 2.6.0 The binding energy < -5 kcal/mol indicated that the compound had good binding interactions with the targets.
Adeno gastro carcinoma (AGS) cells were cultured in RPMI-1640 medium (Gibco, United States) containing 10% fetal bovine serum (FBS, ExCell Bio, China) and 1% penicillin-streptomycin in a 5% CO2 humidified incubator at 37 °C. Resveratrol was dissolved in Dimethyl sulfoxide (DMSO), configured with a gradient concentration in RPMI-1640 (final DMSO concentration did not exceed 0.1%). The effect of resveratrol on GC cell proliferation was detected by the cell counting kit 8 (CCK-8) assay. AGS cells in logarithmic growth phase were seeded at a density of 8 × 103 cells/ well in 96-well plates and cultured for 24 h. Then, cells were exposed to different concentrations of resveratrol (0, 12.5, 25, 50, 100, 200, and 400 µM) and further incubated for 24 h. The old medium was replaced with 100 µL fresh medium containing 10% CCK8 and incubated at 37 °C for another 2 h. The absorbance was measured at 450 nm by a scanning porous spectrophotometer (Thermo Scientific, China).
The cells were seeded at a density of 1 × 103 cells/ well in 6-well plates. After adhering to the wall, the cells were treated with 0, 12.5, 25 or 50 μM resveratrol for 24 h. Then, the old medium was replaced with fresh complete medium without resveratrol, and the cells were further cultured for 1 wk. The plate was fixed with 4% paraformaldehyde and stained with 0.1% crystal violet to observe cell colony formation.
The cell cycle distribution and apoptotic rate were detected by flow cytometry (BD FACSCanto, United States) and analyzed using ModFitLT software version 5.0. AGS cells were incubated in 6-well plates and treated with resveratrol at different concentrations (0, 12.5, 25, and 50 μM.) after adherent growth. After treatment for 24 h, cells were collected and then fixed with precooled 70% ethanol at 4 °C for at least 2 h. The cell cycle assay was performed with a DNA Content Quantitation Assay Kit (Solarbio, China) according to the manufacturer’s protocol. After treatment, cells were digested and collected with 0.25% trypsin digestion solutions without thylenediamine tetraacetic acid (EDTA), and cell apoptosis was detected using the fluorescein isothiocyanate-conjugated Annexin V Apoptosis Detection Kit I (BD Biosciences, United States). Finally, the samples were transferred to a flow cytometer and measured.
AGS cells at the logarithmic growth stage were adjusted to a density of 2.5 × 105 cells/ mL of cell suspension. Six-well plates were inoculated with 2 mL of cell suspension per well. After 24 h of culture, resveratrol was added at final concentrations of 0, 12.5, 25, and 50 μM. The plate was fixed with 4% paraformaldehyde at room temperature for 15 min. Hoechst 33258 was labeled by conventional methods. Apoptosis was observed under a fluorescence microscope and photographed.
The cells (6 × 105 cells/ well) were seeded into 6-well plates, cultured overnight to 100% confluence, scratched using a sterile pipette tip to create an incision-like gap, and washed 3 times with phosphate-buffered saline. The cells were treated with resveratrol at different concentrations (0, 12.5, 25, and 50 μM). The scratch areas were photographed at 0 h and 24 h after wounding. Cell migration was quantified by ImageJ software and expressed as the average percentage of the closure of the scratched area.
Cell migration and invasion were detected by the Transwell method. Cells were collected with 0.25% trypsin-EDTA solution (Solarbio, China) and centrifuged. Cells were resuspended at a density of 5 × 105 cells/ mL in RPMI-1640 medium and treated with resveratrol at different concentrations (0, 12.5, 25, and 50 μM). Then, 100 μL of treated cells was inoculated into the upper compartment of the cell culture chamber (Corning, United States), and 700 μL of medium containing 10% FBS was added to the lower compartment for further culture for 24 h at 37 °C. After incubation, the cells on the upper surface of the membrane were removed. Cells below the membrane were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet ammonium oxalate solution. In the cell invasion assay, the Transwell chamber was coated with 1 mg/ mL Matrigel (Corning, United States) before the experiment, and the other steps were basically the same as in the migration assay. Cell migration and invasion were photographed with an inverted microscope.
The total RNA components of cells were extracted by using TRNzol Universal Reagent (TIANGEN, China) according the manufacturer’s instructions and quantified using a NanoDrop Spectrophotometer (Thermo Scientific, United States). cDNA was obtained by reverse transcription using the FastKing RT Kit (With gDNase, TIANGEN, China) and then stored in a refrigerator at -20 °C until use. The polymerase chain reaction (PCR) system was prepared using the SuperReal PreMix Plus Kit (SYBR Green, TIANGEN, China). Glyceraldehyde-3-phosphate dehydrogenase was used as an internal reference, and the 2−ΔΔCt method was used to analyze the relative expression of the target genes. The primer sequences are shown in Table 1.
| Gene name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
| FOS | TCTTACTACCACTCACCCGC | GGGAATGAAGTTGGCACTG |
| MMP9 | TTTGGAAACGCAGATGGC | ATTGCCGTCCTGGGTGTAG |
| GAPDH | CAAGAAGGTGGTGAAGCAGG | AAAGGTGGAGGAGTGGGTGT |
In this study, R 4.2.1 and GraphPad Prism 8.0 were used for data analysis. A t test was used to determine differences between the two groups. Analysis of Variance was used to compare multiple groups of independent data. All experiments were repeated three times, and differences were considered statistically significant when P < 0.05.
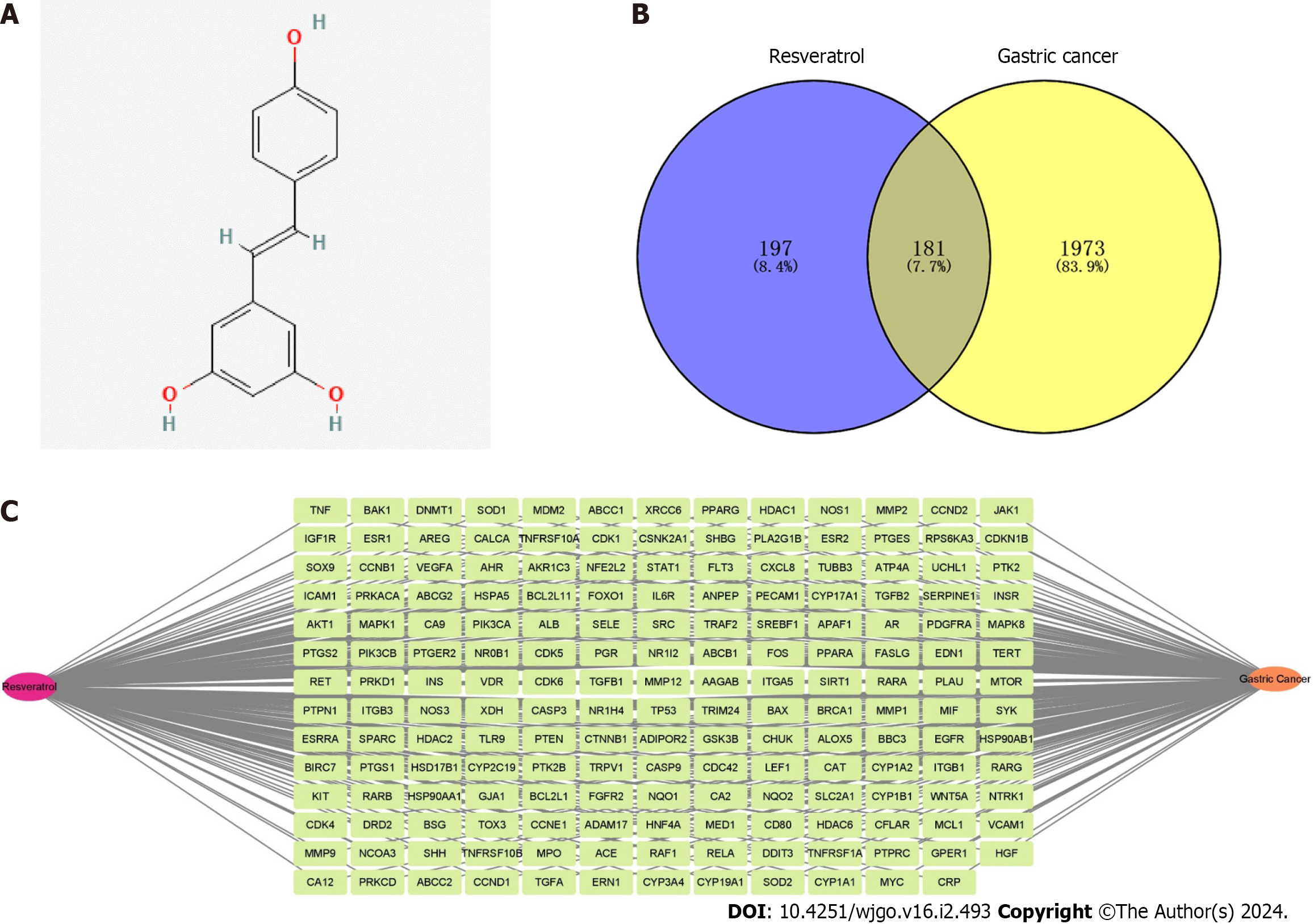
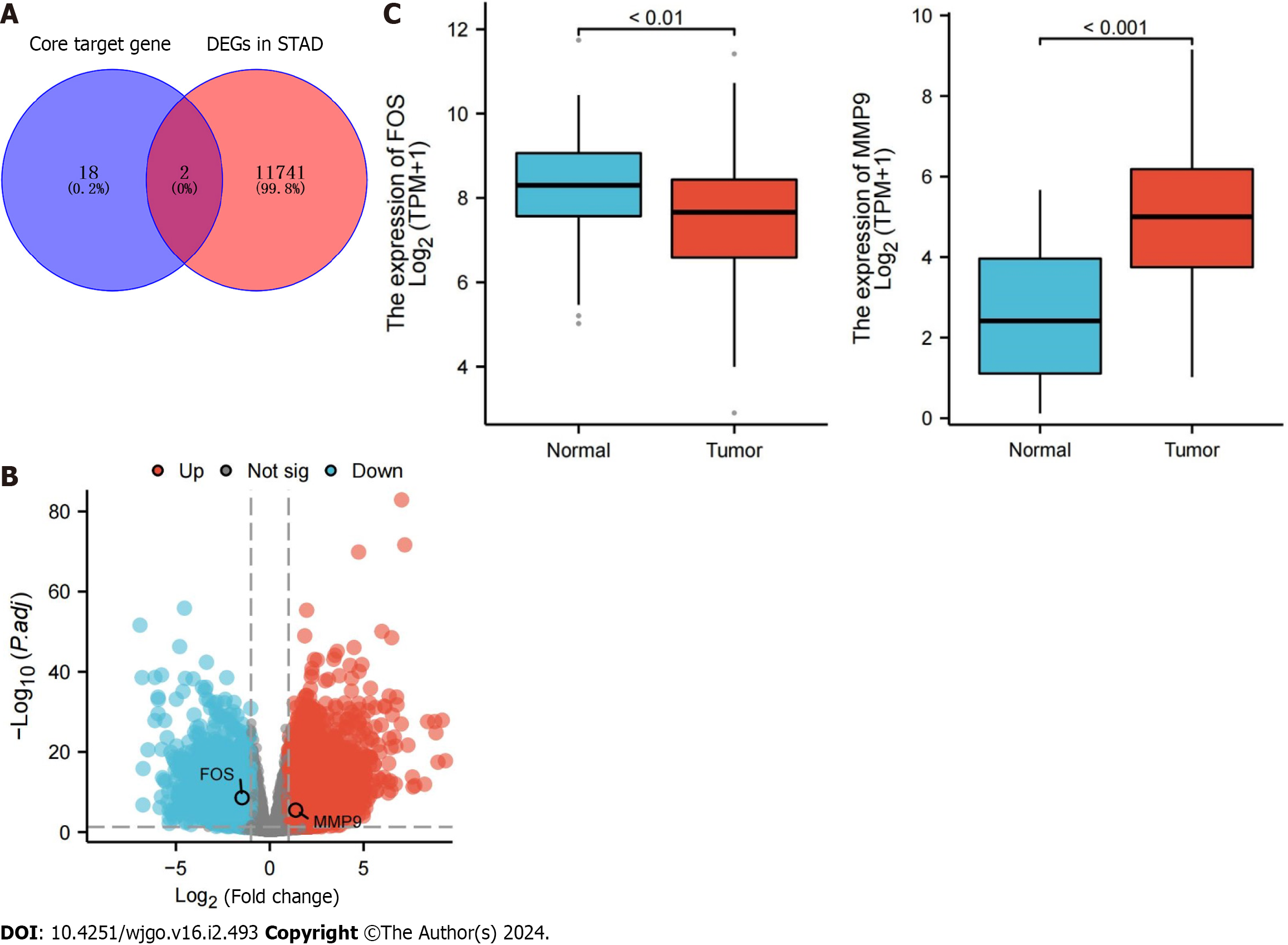
The molecular structure of resveratrol is shown in Figure 2A, and its molecular formula is C14H12O3. A total of 378 potential targets were identified for resveratrol based on the TCMSP, DrugBank Online, BATMAN-TCM, Swiss Target Prediction and TargetNet databases. GC-related targets were mainly obtained from the GeneCards and DisGeNet databases, in which a total of 2154 targets were selected. There were 181 common targets of resveratrol and GC by using a Venn diagram for the overlap analysis, which predicted active targets of resveratrol acting on GC (Figure 2B). These common targets were imported into Cytocsape 3.9.1 software to construct the “resveratrol-target-GC” network (Figure 2C).
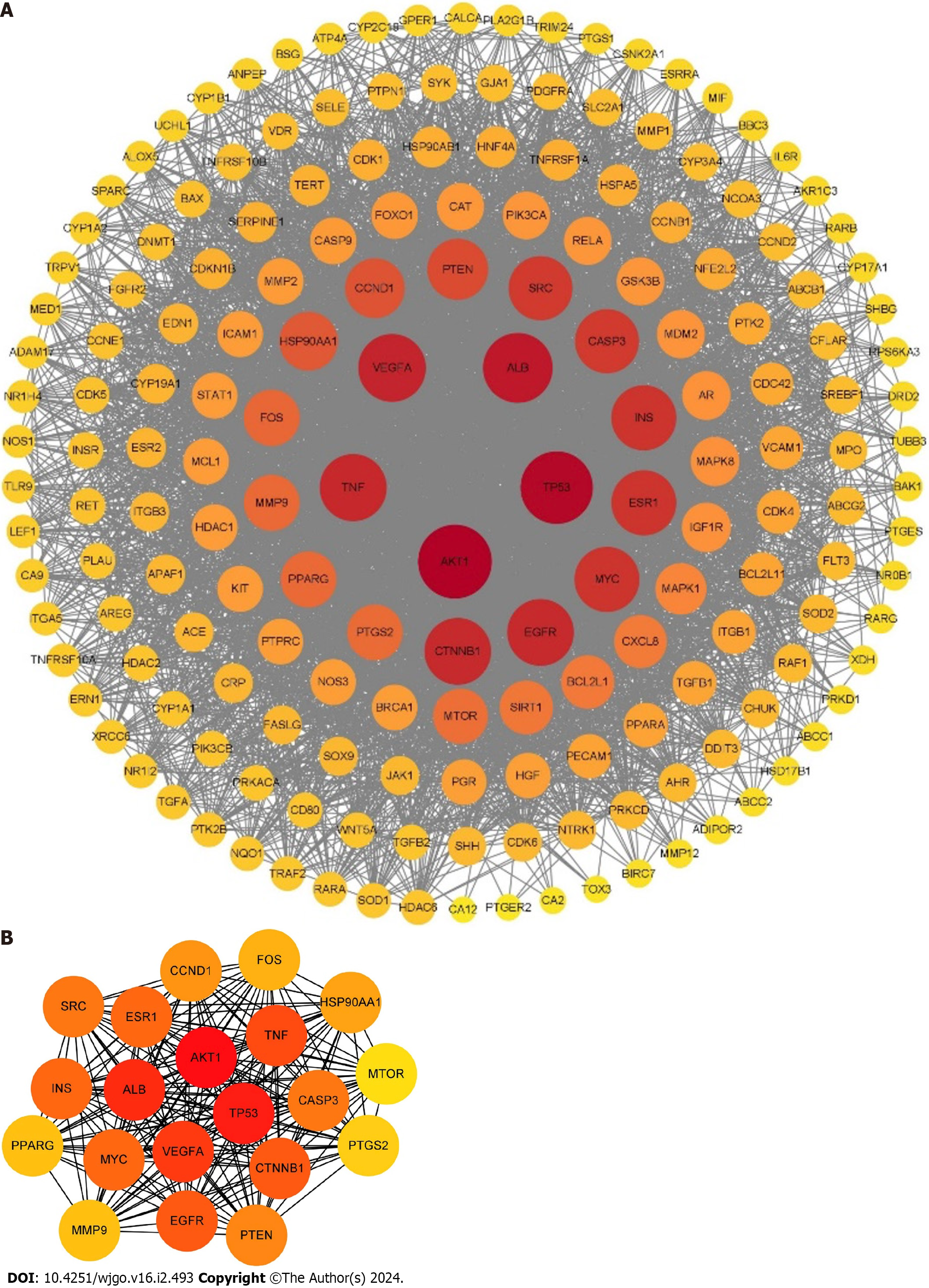
A total of 181 targets of resveratrol anti-GC were submitted to the STRING database to obtain the PPI network. Then, the PPI network was analyzed by Cytoscape 3.9.1, which contained 179 nodes and 3991 edges (Figure 3A). The top 20 core targets were selected based on the degree value: AKT1, TP53, ALB, VEGFA, TNF, CTNNB1, EGFR, INS, ESR1, MYC, CASP3, SRC, PTEN, CCND1, HSP90AA1, FBJ murine osteosarcoma viral oncogene homolog (FOS), PPARG, matrix metallopeptidase 9 (MMP9), PTGS2, and MTOR (Figure 3B).
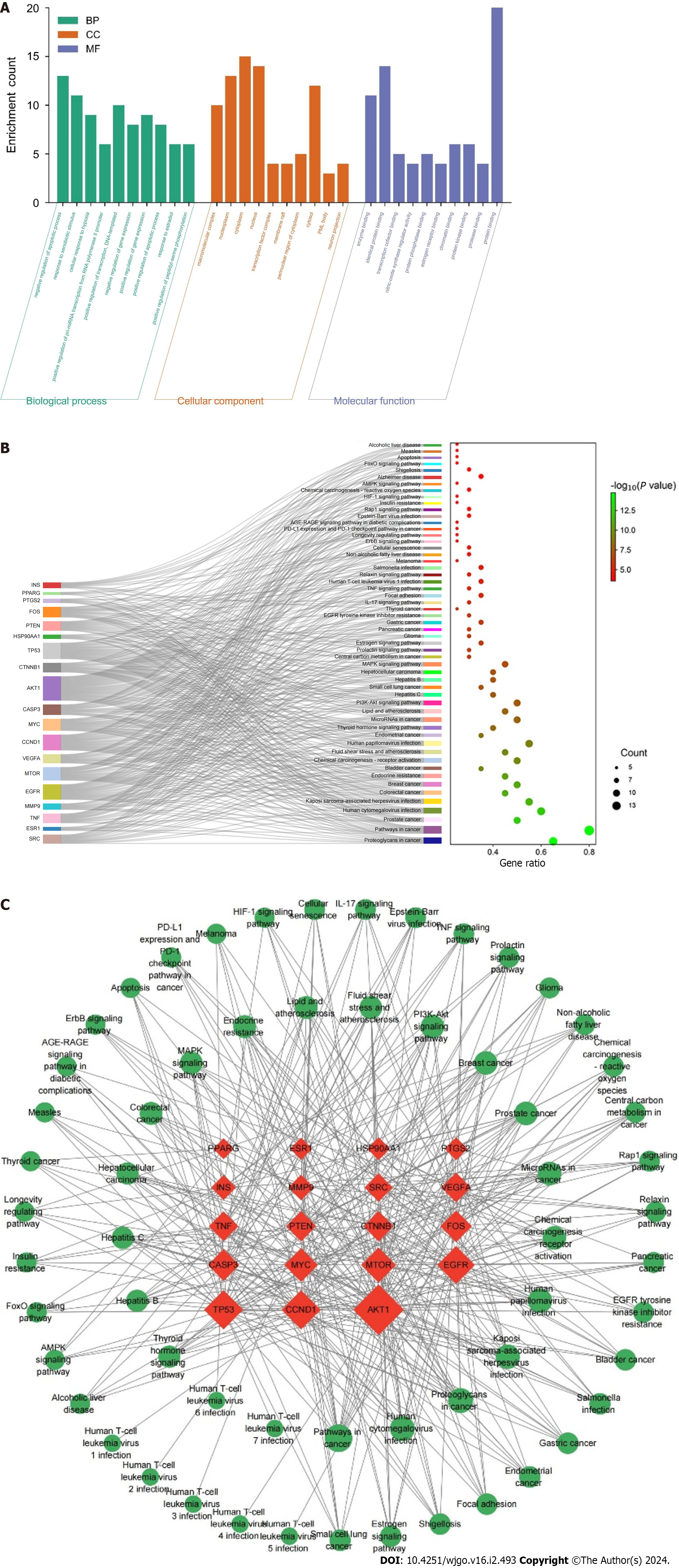
GO and KEGG enrichment analyses were performed to further explore the biological functions and signaling pathways of 20 core targets of resveratrol against GC. The GO enrichment results revealed 264 BPs, 31 CCs and 47 MFs. The top 10 enrichment terms of BP, CC and MF were plotted as a scale histogram (Figure 4A). BPs mainly involved negative regulation of apoptotic process, response to xenobiotic stimulus, and positive regulation of transcription, DNA-templated. CCs were primarily associated with cytoplasm, nucleus and nucleoplasm. MFs were enriched in protein binding, identical protein binding and enzyme binding. It was found that 110 pathways were identified in KEGG pathway analysis, and the top 55 enrichment terms were plotted as a Sankey + dot diagram to observe the relationship between signaling pathways and core targets (Figure 4B). These signaling pathways are closely related to the inhibitory effects of resveratrol on GC, including PI3K-AKT signaling pathway, MAPK signaling pathway, IL-17 signaling pathway, TNF signaling pathway, and ErbB signaling pathway. The “target- signaling pathway” network was drawn by Cytoscape 3.9.1 to further investigate the mechanisms of resveratrol anti-GC (Figure 4C). The high enrichment of the PI3K-Akt and MAPK signaling pathways suggested that resveratrol might play a role in the treatment of GC through these two pathways.
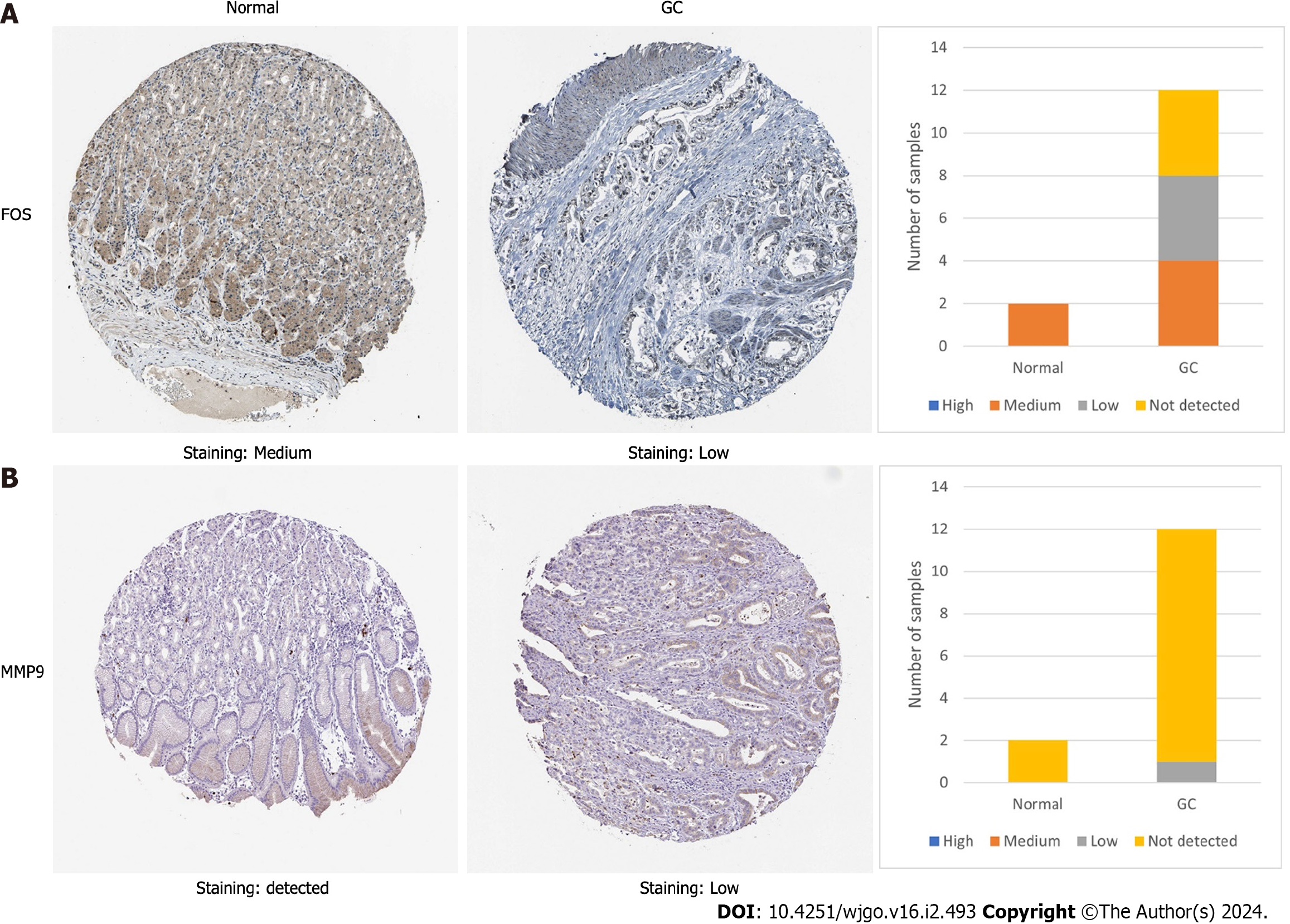
The TCGA database was used to compare the differential expression of core target genes at the transcriptome level. The results showed that FOS and MMP9 were differentially expressed among core target genes, in which FOS was downregulated and MMP9 was upregulated. (Figure 5A and B). The RNA expression of FOS was lower in GC tissues than in normal stomach tissues, while that of MMP9 was higher (Figure 5C). The HPA database was used to further compare whether the protein expression of FOS and MMP9 is different in GC tissues and the corresponding normal tissues. Immunohistochemistry results from the HPA database showed that the protein expression of FOS was lower in GC tissues than in normal gastric tissues. The protein expression of MMP9 was not detected in normal gastric tissues, but low expression of MMP9 was detected in GC tissues (Figure 6).
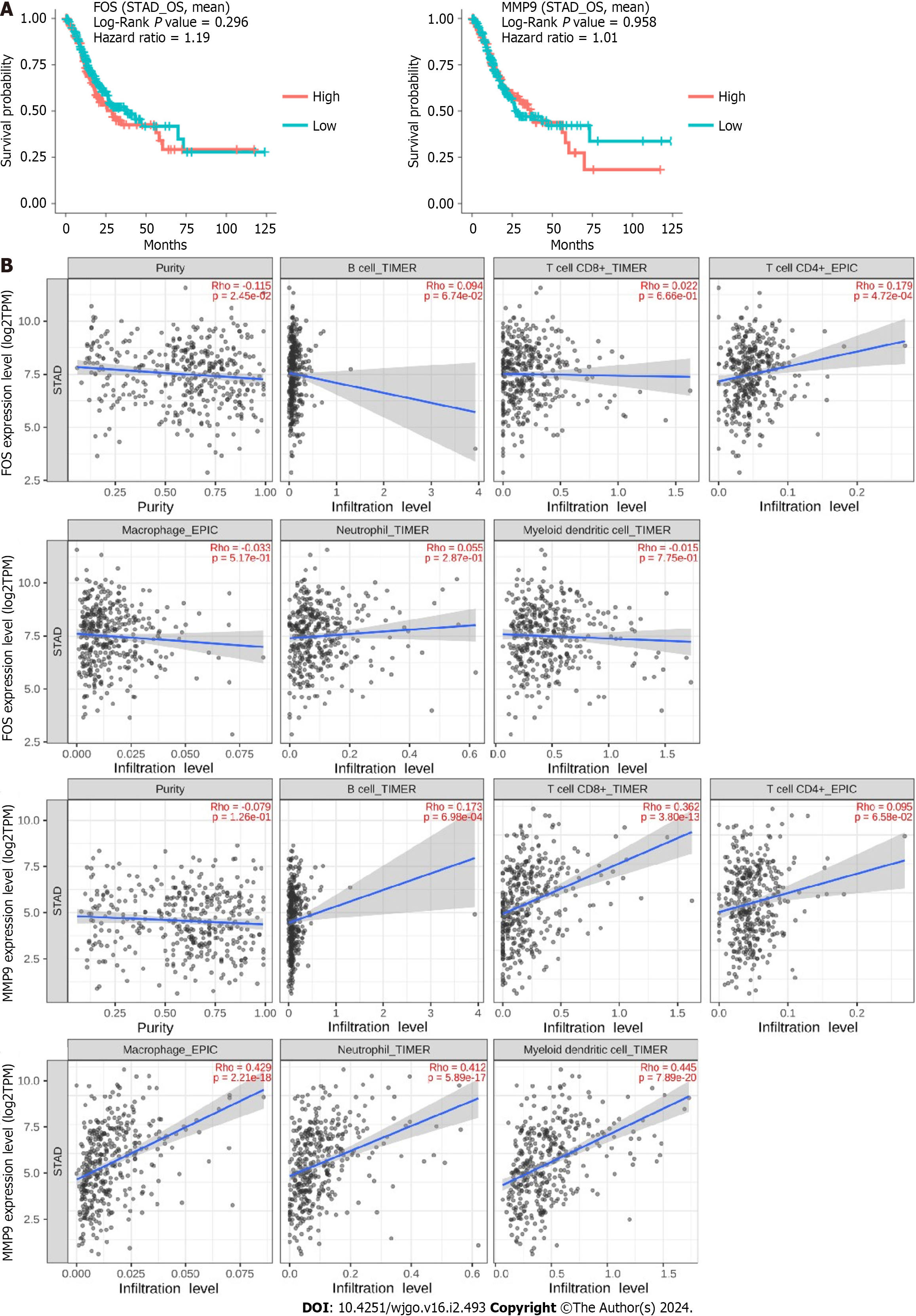
To explore the prognostic value of FOS and MMP9 with differential expression values, Kaplan-Meier survival curves were applied for survival comparison between low-expression and high-expression groups via the “Gene Survival” function in the DriverDBv3 database. The results showed that FOS and MMP9 had no significant relationship with survival prognosis (Figure 7A). The TIMER2.0 database was used to assess the association between FOS and MMP9 expression and immune cell infiltration in STAD. The results showed that FOS expression was positively correlated with the infiltration degree of CD4+ T cells (Rho = 0.179, P = 4.72e-04). MMP9 expression was positively correlated with the infiltration degree of B cells (Rho = 0.173, P = 6.98e-04), CD8+ T cells (Rho = 0.362, P = 3.80e-13), macrophages (Rho =0.429, P = 2.21e-18), neutrophils (Rho = 0.412, P = 5.89e-17), and DCs (Rho = 0.445, P = 7.89e-20) (Figure 7B).
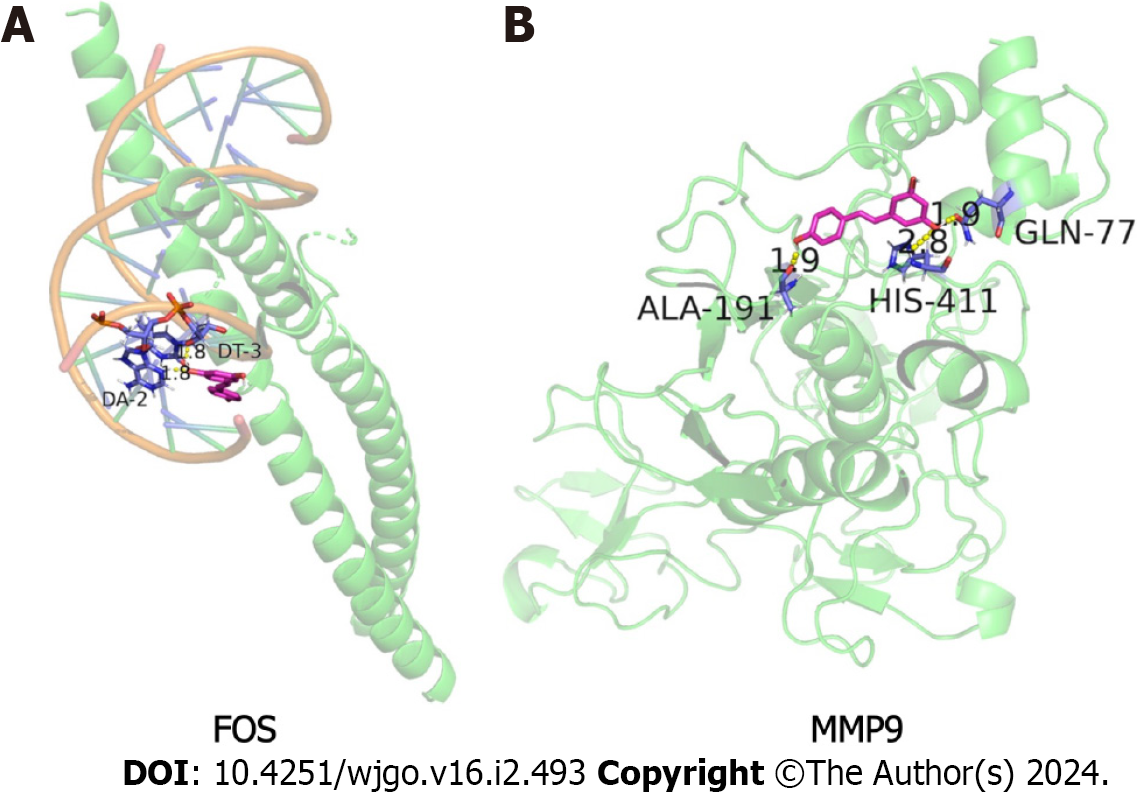
Molecular docking is mainly used to explore the interaction between molecules, as well as to predict their binding patterns and affinity. To explore the interaction between resveratrol and FOS or MMP9, a molecular docking model was constructed by AutoDock software. A binding energy < -5.0 kcal/ mol indicated good binding activity. In this study, the docking scores of FOS (PDB ID: 2WT7) and MMP9 (PDB ID: 1L6J) were -6.7 kcal/ mol and -6.13 kcal/ mol, respectively (Table 2). The visualization results of resveratrol binding to FOS and MMP9 proteins in GC revealed that resveratrol had a stable binding site in these target protein models (Figure 8). Resveratrol plays an inhibitory role in GC and may be used as an effective anti-GC drug by targeting FOS and MMP9.
| Target | PDB ID | Docking score (kcal/mol) | Hydrogen bond and other interacting residues |
| FOS | 2WT7 | -6.7 | DA-2, DT-3 |
| MMP9 | 1L6J | -6.13 | ALA-191, HIS-411, GLN-77 |
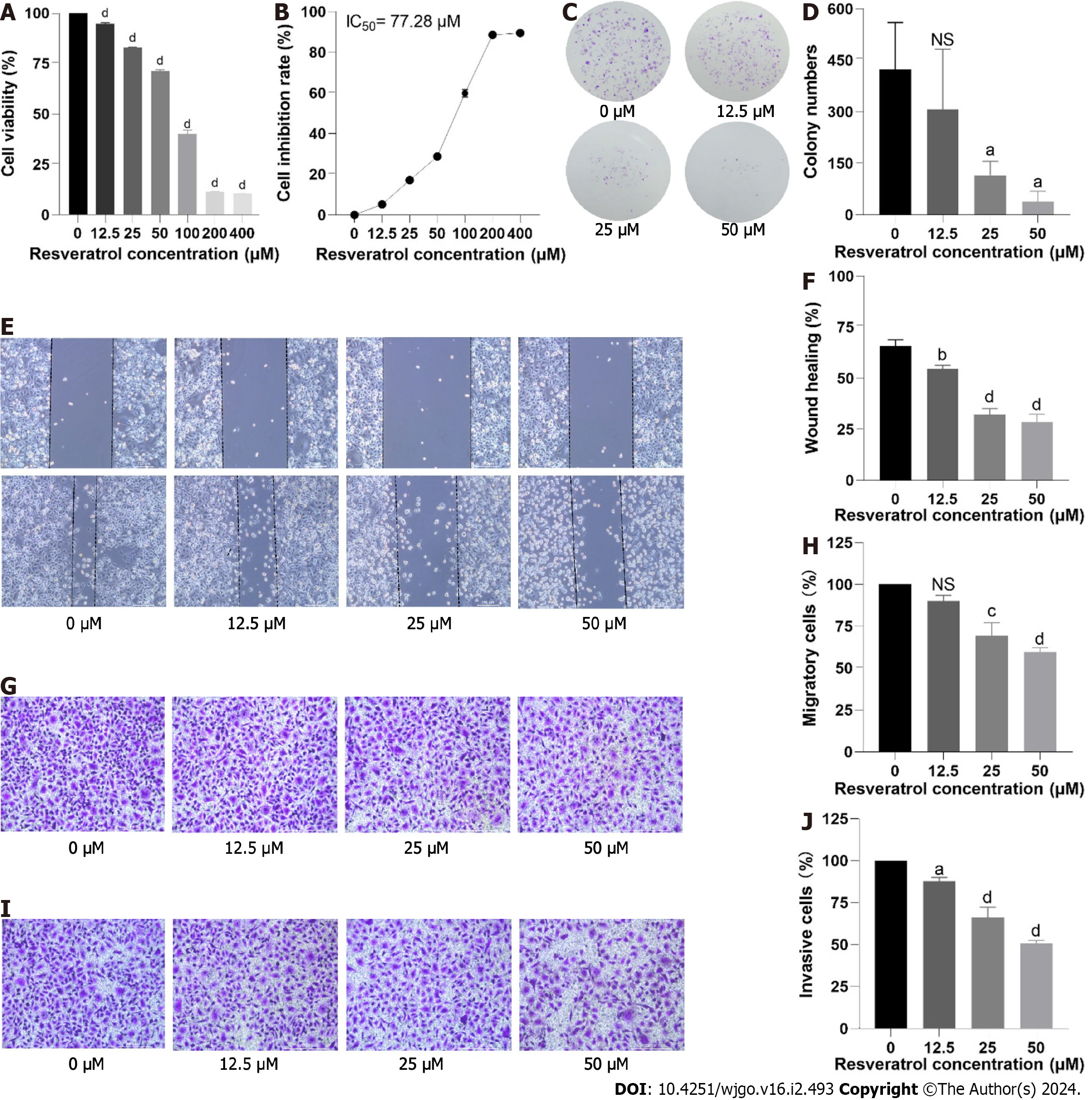
We evaluated the in vitro effects of resveratrol on the human GC cell line AGS. A CCK-8 assay was used to determine AGS cell viability after treatment with resveratrol at different concentrations (0, 12.5, 25, 50, 100, 200, and 400 µM) for 24 h. The results showed that the viability rate of AGS cells was markedly reduced in a dose-dependent manner (Figure 9A). The half maximal inhibitory concentration (IC50) value of AGS cells treated with resveratrol is shown in Figure 9B. The IC50 value of resveratrol intervention in AGS cells for 24 h was 77.28 µM. The dose of administration of subsequent experiments was adjusted according to the IC50 value. The colony formation rate reflects two important traits of cell population dependence and cell proliferation. The effect of resveratrol on cell colony formation ability is shown in Figure 9C and D. The results showed that compared with that in the control group, the cell colony number in the resveratrol intervention group was significantly reduced. The colony number of AGS cells gradually decreased with increasing resveratrol dosage. The above results indicated that resveratrol inhibited AGS cell proliferation.
The motility of cancer cells enables them to migrate and invade. The effect of resveratrol at different concentrations (0-50 μM) on the motility of AGS cells was evaluated by wound healing assay and Transwell assays. In the wounding healing assay, compared with the control group, the healing rate of cells in the resveratrol intervention group was slower after 24 h of treatment (Figure 9E and F). A similar phenomenon was observed in the Transwell migration assay, where fewer cells crossed the chamber membrane in the resveratrol intervention group (Figure 9G and H). Both the wounding healing assay and Transwell migration assay indicated that resveratrol prevented GC cell migration. A Matrigel-coated Transwell chamber mimics the extracellular matrix environment and can be used to measure cell invasion ability. Resveratrol significantly reduced the number of cells crossing the chamber membrane, suggesting that it can reduce the invasion ability of AGS cells (Figure 9 I and J). Additionally, the changes in AGS cell migration and invasion were dose dependent. These findings suggested that resveratrol had an inhibitory effect on AGS cell migration and invasion.
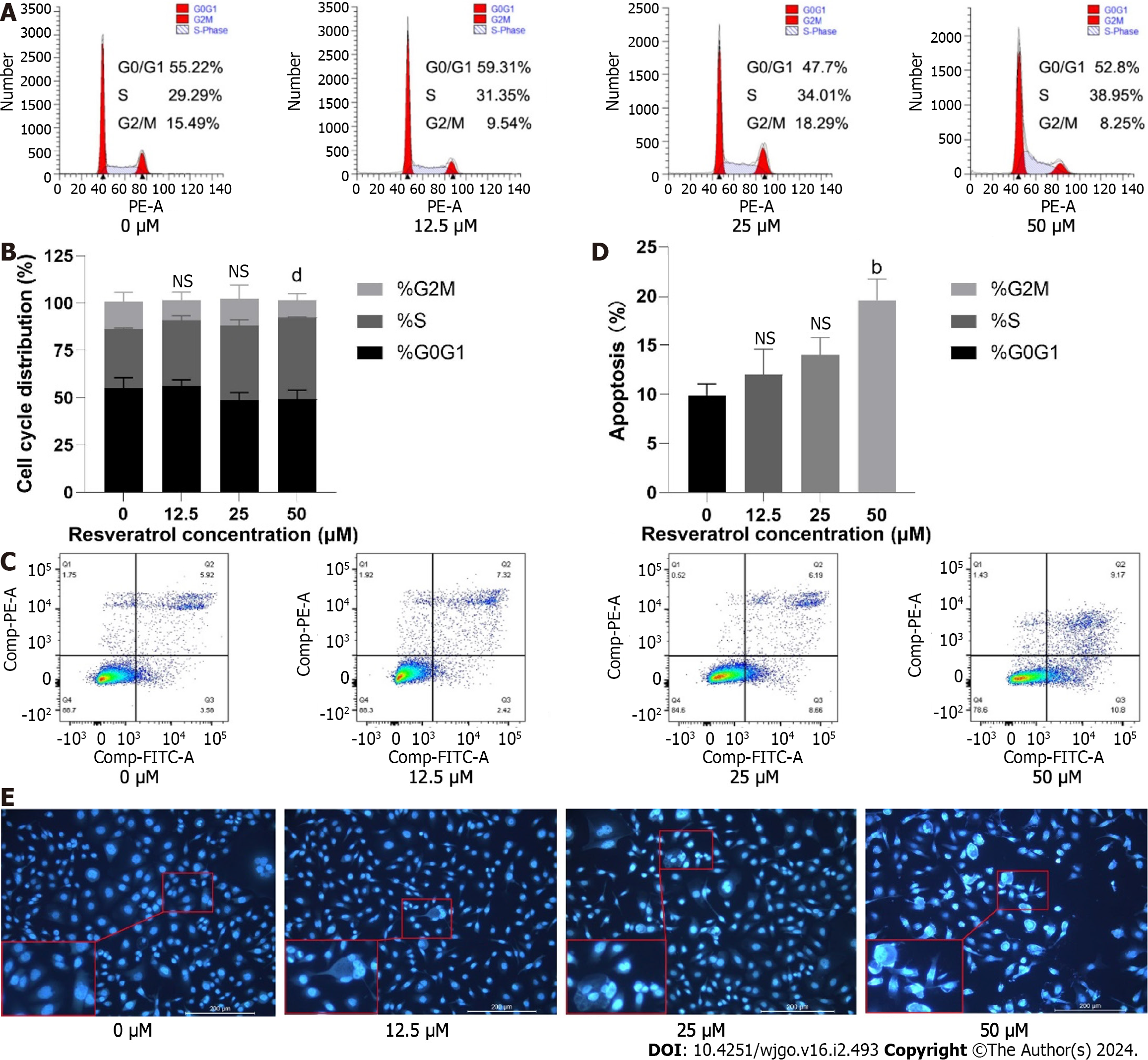
To explore the cell cycle arrest effect of resveratrol on AGS cells, AGS cells were exposed to resveratrol for 24 h and analyzed by flow cytometry. The results showed that the proportion of AGS cells in S phase increased with the increasing drug concentration (Figure 10A and B). We further determined the apoptosis of the resveratrol treated cells by Annexin V-FITC and propidium iodide staining. The results illustrated that resveratrol treatment increased the apoptosis rate compared with that of the control group (Figure 10C and D). Cell apoptosis morphology was observed by the Hoechst 33258 fluorescence method. The chromatin of the control group was uniform, the nuclear shape was regular, circular or oval, and showed uniform blue fluorescence. The morphology of cells in the administration group was changed with uneven distribution of chromatin. Some cells showed characteristic apoptotic changes, gradually appearing karyo
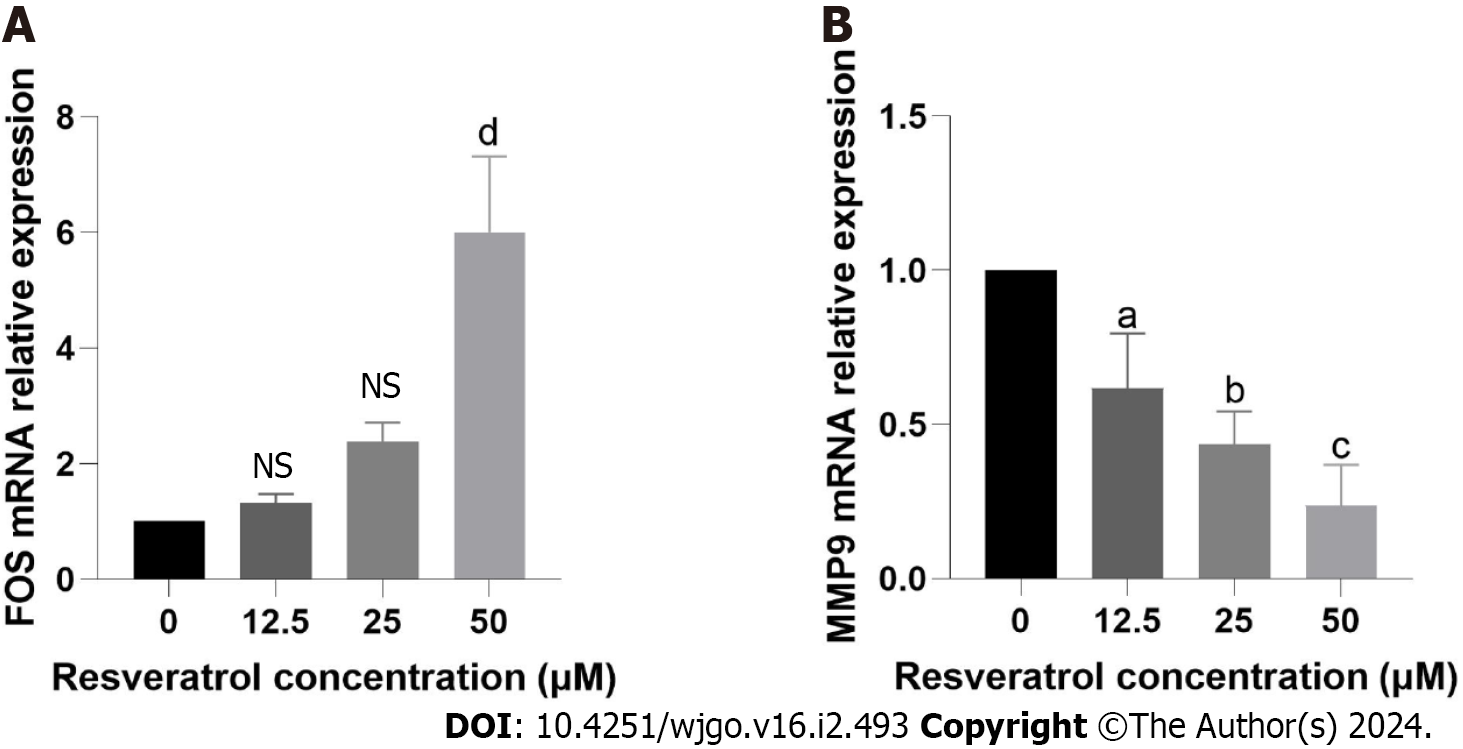
Based on the previous network analysis and screening and molecular docking simulation, FOS and MMP9 were considered to be two important targets of resveratrol against GC. We further determined whether resveratrol mediated the GC inhibition effects by targeting FOS and MMP9. The results of the reverse transcription quantitative PCR (RT-qPCR) experiment demonstrated that resveratrol increased the mRNA expression of FOS but decreased the mRNA expression of MMP9, which corresponded to the results of the previous bioinformatic analysis (Figure 11). The changes in the mRNA expression of FOS and MMP9 were dose dependent. The results indicated that resveratrol could suppress the development of AGS cells by regulating the expression of FOS and MMP9.
It has been reported that resveratrol has anti-GC effects. However, studies on the mechanisms of resveratrol in GC are not comprehensive. It is difficult to elucidate the characteristics of multitarget and multipathway mechanisms of drug action based on the single thinking of traditional pharmacology. In this study, we investigated the mechanisms of action of the natural polyphenol compound resveratrol in the treatment of GC based on network pharmacology and bioinformatics. To our knowledge, this is the first study to explore the core therapeutic targets and potential mechanisms of resveratrol against GC by combining network analysis and experimental validation.
A total of 181 anti-GC targets of resveratrol were identified from multiple public databases, and a PPI network was constructed. Core targets were selected based on the degree value in the PPI network, and then functional and pathway enrichment was performed using GO and KEGG analysis. Protein binding, identical protein binding, cytoplasm, nucleus, negative regulation of apoptotic process and response to xenobiotic stimulus were the GO terms with the most enriched core targets. The KEGG pathway analysis revealed that these targets were primarily enriched in cancer-related pathways, such as PI3K-Akt signaling pathway, MAPK signaling pathway, IL-17 signaling pathway and TNF signaling pathway. It is worth noting that the high enrichment of the PI3K-Akt and MAPK signaling pathways suggested that resveratrol was most likely involved in the treatment of GC through these two pathways. PI3K is an evolutionarily conserved family of lipid kinases that have been found to be oncogenic and linked to a variety of cancers. AKT, a key hub of the PI3K signaling pathway, can further regulate multiple downstream signaling targets and is closely correlated with tumor progression and drug resistance[24]. The PI3K/AKT signaling pathway can be abnormally activated in most cancers, promoting tumor cell proliferation, inhibiting apoptosis, and enhancing invasion and metastasis[25]. Wang et al[26] verified that monoamine oxidase A inhibits the progression of GC by reducing glycolysis through the PI3K/Akt/mTOR pathway. Inhibition of the PI3K/Akt pathway may provide a potential method for GC treatment. As the integration point of various biochemical signals, mitogen-activated protein kinases are widely involved in cell proliferation, differentiation, transcriptional regulation and development. The MAPK signaling pathway is closely related to cancers. Chen et al[27] reported that SAM-domain- and HD-domain-containing protein 1 is downregulated in GC cell lines and tissues, and negatively regulates the activation of the MAPK p38 signaling pathway, thereby inhibiting the proliferation of GC. Zhang et al[28] found that spindle and kinetochore associated complex subunit 3 (SKA3) expression can promote the proliferation, migration, invasion and epithelial-mesenchymal transformation of GC. After silencing SKA3, dual specificity phosphatase 2 negatively regulates the MAPK/ERK pathway to slow GC progression. IL-17 and TNF are potent proinflammatory cytokines involved in tumorigenesis, metastasis, and immune resistance. Targeting genes located in the IL-17 signaling pathway or the TNF signaling pathway to alter the effects of these signaling pathways in cancer is an effective strategy for cancer treatment. Downregulation of the IL-17 signaling pathway gene Lipocalin 2 inhibits the proliferation and clone formation of IL-17 treated AGS cells, partially reverses invasion and migration, and promotes apoptosis and cycle arrest[29]. Compound Kushen injection (CKI) is a TCM formula, which has achieved satisfactory results in treating GC and improving the immunity of patients. Some studies have found that it regulates the TNF signaling pathway with VCAM1 as a key target, and then affects the EMT of GC, providing new clues for the study of the complex molecular mechanism of CKI in the treatment of GC[30]. These findings suggest that the anti-GC mechanism of resveratrol may be closely related to PI3K-AKT, MAPK, IL-17 and TNF signaling pathways. It is worth noting that the high enrichment of the PI3K-Akt and MAPK signaling pathways suggested that resveratrol was most likely involved in the treatment of GC through these two pathways.
We analyzed the clinical significance of the core targets using bioinformatics databases to further screen target genes. FOS and MMP9 were screened out by differential analysis in TGCA and HPA databases. The results of differential expression of FOS and MMP9 at the RNA and protein levels were basically consistent, which indicated that they might be the most favorable targets for resveratrol action on GC. Their associations with survival and immune infiltration were subsequently analyzed using DriverDBv3 and TIMER2.0. The analysis showed that the differential expression of FOS and MMP9 had no significant relationship with the OS of patients with GC. The results of immune infiltration showed a strong correlation between FOS and MMP9 and immune cell infiltration, suggesting that they might influence GC progression by regulating the immune microenvironment. Subsequently, molecular docking was used to analyze the affinity between FOS, MMP9 and resveratrol. The results showed that the binding energies between FOS, MMP9 and resveratrol were all lower than -5 kcal/ mol, indicating that these two targets had a strong affinity for resveratrol. It is well known that the major hallmarks of malignant tumors include rapid growth, poorly defined borders, strong invasiveness, and easy metastasis. Behind these characteristics, multiple molecular events are found to be involved. Traditional pathological classification is mainly based on the morphological structure of tumor tissues and cell biological features. Due to the differences in subjective judgment, the low diagnosis rate of early-stage cancers, and the lack of scientific evidence at the molecular level, it is difficult to objectively reflect the intrinsic characteristics of tumors and guide the subsequent individualized treatment. With the rapid development of molecular detection technology, molecular classification based on various molecules shows unique genomic features, which can make up for the shortcomings of traditional pathological classification and is conducive to the discovery of important biomarkers and new therapeutic targets, thus providing more precise assistance in the clinical diagnosis and treatment of GC[31]. However, the identification of molecular subtypes of GC is currently incomplete, and the clinical application is still very limited. Therefore, it is urgently needed to identify more valuable and accurate novel biomolecules for early diagnosis and personalized therapy of GC. The expression of Fos (c-Fos) is closely related to clinicopathological variables and survival of GC. Some studies have shown that the tumor size of GC patients with positive c-Fos expression is significantly smaller than that of GC patients with negative c-Fos expression. The loss of c-Fos expression is positively correlated with the depth of tumor infiltration, lymph node metastasis and advanced stage of GC. In survival analysis, the OS of GC patients with positive c-Fos expression is significantly better than that of GC patients with negative c-Fos expression[32,33]. These findings suggest that the loss of c-Fos expression may accelerate the progression of GC and lead to poor prognosis. Helicobacter pylori (HP) is an important predisposing factor for several gastric lesions and plays a key role in the development and spread of gastric diseases. Investigating the relationship between HP infection and GC has been the focus of GC research, but previous studies focused on the virulence of HP strains and environmental risk factors. However, the occurrence and progression of GC is very complex, often involving multiple genes, pathways, and stages. With the in-depth exploration of the molecular level of GC, researchers have further elucidated the pathogenesis of GC from the aspects of epigenetics and molecular features, and HP-related genes are of great significance in the diagnosis and treatment of GC. Chronic HP infection can develop into chronic atrophic gastritis and GC, and MMP9 is significantly expressed in this process, which is a potential biomarker for the early stage of GC development[34]. The expression of MMP9 is closely associated with HP infection in GC patients, so the quantification of MMP9 expression level in vivo is beneficial for the diagnosis of HP-infected GC. Moreover, it is directly involved in the progression of GC, showing great potential in providing new molecular targets for GC treatment and prognosis[35]. At present, the treatment of progressive GC is in the bottleneck period due to the high heterogeneity of GC, few available chemotherapy drugs and the lag of targeted drugs. It is gratifying that genomics technology has been continuously improved in recent years, especially the development of high-throughput sequencing and gene chip technology allows the study of GC at high resolution and molecular level. The role of FOS and MMP9 as therapeutic targets for anticancer drugs in cancer has also been investigated in previous studies. For example, Zhang et al[36] reported that c-Fos plays a key role in the inhibition of human bladder cancer T24 cells by the purified natural product brazilin. The overexpression of c-Fos induces specific alterations in the morphology and viability of tumor cells, suggesting that brazilin may inhibit the proliferation of T24 cells by targeting c-Fos. c-Fos is involved in cell proliferation, differentiation, invasiveness and apoptosis as an inducible transcription factor. P53 is a tumor suppressor protein, and its transcriptional activity is regulated by the transcription factor activator protein-1 (AP-1), which is mainly composed of Jun and Fos in mammalian cells[37,38]. Sun et al[39] revealed that the competitive binding of AS3MT and c-Jun with c-Fos could inhibit the formation of AP-1 complexes, thereby reducing the activity of p53, inhibiting the apoptosis of tumor cells and inducing proliferation. FOS/JUN may be biomarkers for the predeteriorated epithelial cell subpopulation in colorectal cancer, as their expression is reduced with epithelial cell degeneration and promotes epithelial cell deterioration by interfering with the p53 signaling pathway[40]. Data mining has found that decreased FOS expression is closely associated with the occurrence, metastasis and poor prognosis of prostate cancer. Moreover, in vitro and in vivo experiments have shown that the lack of FOS can activate cancer signaling pathways related to aggressiveness, proving that FOS can be used as a tumor suppressor in prostate cancer initiation and the progression to invasive disease[41]. FOS also plays an important role in the photodynamic therapy of melanoma. Photodynamic treatment significantly inhibits the growth of melanoma, which is related to the increased expression of FOS and the induction of autophagy in tumor cells. Photodynamic therapy can induce cell autophagy by elevating FOS expression in melanoma, inhibit cancer progression and improve patient prognosis[42]. Matrix metalloproteinases (MMPs) play an important role in the degradation of tumor extracellular matrix components, among which MMP9 overexpression can accelerate the metastasis of different types of cancer and is considered a marker of tumor invasion and metastasis[43]. Liang et al[44] predicted the core therapeutic targets of dihydroartemisinin in GC by using network pharmacology and revealed that dihydroartemisinin suppressed the tumorigenesis and invasion of GC by regulating STAT1/KDR/MMP9. It has been reported that the anticancer properties of melatonin are related to its ability to suppress EMT in cancer cells. Melatonin downregulates MMP2 and MMP9 expression and reduces cell invasion and migration by suppressing EMT and nuclear factor-kappa B activity[45]. IL-17 has been found to trigger the metastasis of non-small cell lung cancer. In vitro experiments showed that the migration and invasion of cancer cells are mainly related to the up-regulation of the downstream gene MMP9, and in vivo experiments found that the number of metastatic nodules in lung tissues is significantly reduced after interfering with MMP9 expression. Specific mechanism exploration suggested that IL-17 regulation of non-small cell lung cancer metastasis is closely related to the GCN5-SOX4-MMP9 axis[46]. Metastasis is a major cause of cancer development and death, and the production of neutrophil extracellular traps (NETs) have an influential role in the pathogenesis of the metastatic cascade. Studies have shown that NETs promote the proliferation and migration of mouse colon cancer MC38 cells, while silencing MMP9 inhibits this promotion. The results suggested that NETs can promote the proliferation and migration of colon cancer by upregulating MMP9 expression and activating FAK signaling pathway[47]. The above findings provide clues for our current research that FOS and MMP9 may be the most promising anticancer targets for GC. This leads us to speculate that the inhibition of GC may be achieved by regulating the expression of FOS and MMP9, which requires to be confirmed by extra experiments. Tumor immunotherapy has made breakthrough progress in recent years, and with the in-depth study of GC at the molecular level, personalized immunotherapy has become one of the most promising directions in GC treatment. Here, our results exhibited FOS and MMP9 expression notably related to immune cell infiltration in GC. FOS and MMP9 may be immunomodulators in the tumor microenvironment of GC. The important role of FOS and MMP9 in GC immunotherapy still needs more evidence to prove. The study of TCGA is of landmark significance, which opens a new era of molecular classification of GC. In this study, FOS and MMP9, two molecular targets with differential expression significance, were screened based on the TCGA database, and their relationship with the clinical aspects of GC was discussed. Therefore, FOS and MMP9 contribute to the early detection and targeted treatment of GC, and may be used as important biomarkers for the diagnosis and intervention of GC.
Many studies have suggested that the FOS and MMP9 are associated with tumor cell proliferation, migration, invasion and apoptosis[48-51]. Our results also indicated that resveratrol inhibited the proliferation of AGS cells using CCK-8 and colony formation assays and suppressed the migration and invasion of AGS cells by wound healing and Transwell assays. By flow cytometry, we verified the changes in the proportion of each phase of the cell cycle and the number of apoptotic cells after resveratrol treatment. In addition, apoptotic morphological changes were observed after resveratrol treatment in the Hoechst 33258 experiment. The cell cycle refers to the whole process that a cell goes through from the completion of one division to the completion of the next[52]. Abnormalities in cell cycle progression are one of the basic mechanisms of tumorigenesis, and the regulation of the cell cycle is expected to be a reasonable method of tumor treatment[53]. Apoptosis is a programmed cell death mode that plays an important role in the early development of organisms and the maintenance of homeostasis under pathophysiological conditions[54,55]. Studies have shown that apoptosis is one of the important mechanisms inducing tumor cell death and a key step in anticancer therapy[56,57]. Our results indicated that resveratrol could block the cell cycle in the S phase and induce cell apoptosis to inhibit AGS cells. Based on the previous network prediction and analysis, we selected FOS and MMP9 as the most promising targets of resveratrol against GC. Our RT-qPCR results also demonstrated this. With an increase in resveratrol, the expression level of FOS significantly increased, while the expression level of MMP9 gradually decreased. The in vitro experiments showed that resveratrol inhibited AGS cell proliferation, migration, and invasion and induced S phase arrest and apoptosis by targeting FOS and MMP9.
However, there are some limitations to this study. First, the pathways that may play critical roles in the anti-GC effects of resveratrol have not been fully validated and should be further studied. Second, this study only verified the effects of resveratrol on AGS cells, and its interaction with other GC cell lines needs further study. Animal and in vivo studies are needed to validate these results and support the function of resveratrol in the treatment of GC in future clinical practice.
In conclusion, the mechanisms of action of resveratrol against GC were systematically elucidated based on network pharmacology and bioinformatics. Our results demonstrate that resveratrol inhibited proliferation, migration, and invasion and induced cell cycle arrest and apoptosis in GC cells by targeting FOS and MMP9, thereby playing an important role in regulating the progression of GC.
Gastric cancer (GC) is a malignant tumor of digestive tract with high incidence and mortality. The treatment of GC is more difficult due to its characteristics such as rapid invasive growth, difficulties in personalized medication and high risk of recurrence. Resveratrol, as a traditional Chinese medicine (TCM) monomer, plays an outstanding anticancer role in a variety of cancers.
In this study, network pharmacology, bioinformatics, molecular docking technology and experimental verification were used to explore the important effects and key targets of resveratrol in anti-GC. This discovery is of great clinical significance for identifying potential novel biomarkers and therapeutic targets for GC treatment.
The main objective of this study was to explore the mechanism of resveratrol based on network analysis. In this study, through network analysis and in vitro experiments, we verified the anti-cancer effects of resveratrol on GC cells, including inhibiting proliferation, invasion and migration, inducing cycle arrest and apoptosis, as well as important action targets. Our results suggested that resveratrol has a great clinical value as an anti-GC drug, providing a new direction for the drug treatment of GC.
Network pharmacology, bioinformatics, molecular docking technology and experiments were used in this study. These methods utilize biological networks to mechanically link drugs and diseases, comprehensively elucidate the roles and molecular mechanisms of resveratrol in GC.
In the research, FBJ murine osteosarcoma viral oncogene homolog (FOS) and matrix metallopeptidase 9 (MMP9) were screened as the most important targets of resveratrol against GC by using multiple biological information databases. The experiments verified the anti-GC effect of resveratrol by targeting FOS and MMP9. These findings provide a scientific basis for GC treatment of resveratrol, and also provide a new method for the study of the mechanism of TCM monomer or compound in the diseases However, the pathways which may play critical roles in the anti-GC effect of resveratrol have not been fully validated and should be further studied.
In the research, the key biotargets and biological effects of resveratrol against GC were explored by molecular network analysis and experimental verification for the first time.
Development and validation of biomarkers for classifying and treating diseases through bioinformatics and machine learning.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chen J, China; Emran TB, Bangladesh S-Editor: Qu XL L-Editor: A P-Editor: Zhang XD
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64702] [Article Influence: 16175.5] [Reference Citation Analysis (177)] |
| 2. | Eom SS, Choi W, Eom BW, Park SH, Kim SJ, Kim YI, Yoon HM, Lee JY, Kim CG, Kim HK, Kook MC, Choi IJ, Kim YW, Park YI, Ryu KW. A Comprehensive and Comparative Review of Global Gastric Cancer Treatment Guidelines. J Gastric Cancer. 2022;22:3-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 49] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 3. | Thrift AP, El-Serag HB. Burden of Gastric Cancer. Clin Gastroenterol Hepatol. 2020;18:534-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 983] [Article Influence: 196.6] [Reference Citation Analysis (1)] |
| 4. | Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 897] [Cited by in RCA: 860] [Article Influence: 172.0] [Reference Citation Analysis (0)] |
| 5. | Alsina M, Arrazubi V, Diez M, Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol. 2023;20:155-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 184] [Article Influence: 92.0] [Reference Citation Analysis (1)] |
| 6. | Wang K, Chen Q, Shao Y, Yin S, Liu C, Liu Y, Wang R, Wang T, Qiu Y, Yu H. Anticancer activities of TCM and their active components against tumor metastasis. Biomed Pharmacother. 2021;133:111044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 226] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 7. | Nan Y, Su H, Zhou B, Liu S. The function of natural compounds in important anticancer mechanisms. Front Oncol. 2022;12:1049888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 8. | Zaffaroni N, Beretta GL. Resveratrol and Prostate Cancer: The Power of Phytochemicals. Curr Med Chem. 2021;28:4845-4862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 9. | Behroozaghdam M, Dehghani M, Zabolian A, Kamali D, Javanshir S, Hasani Sadi F, Hashemi M, Tabari T, Rashidi M, Mirzaei S, Zarepour A, Zarrabi A, De Greef D, Bishayee A. Resveratrol in breast cancer treatment: from cellular effects to molecular mechanisms of action. Cell Mol Life Sci. 2022;79:539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 10. | Asemi R, Rajabpoor Nikoo N, Asemi Z, Shafabakhsh R, Hajijafari M, Sharifi M, Homayoonfal M, Davoodvandi A, Hakamifard A. Modulation of long non-coding RNAs by resveratrol as a potential therapeutic approach in cancer: A comprehensive review. Pathol Res Pract. 2023;246:154507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 11. | Sun DP, Chen JT, Yang ST, Chen TH, Liu SH, Chen RM. Resveratrol triggers the ER stress-mediated intrinsic apoptosis of neuroblastoma cells coupled with suppression of Rho-dependent migration and consequently prolongs mouse survival. Chem Biol Interact. 2023;382:110645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 12. | Yu W, Wang Z, Dai P, Sun J, Li J, Han W, Li K. The activation of SIRT1 by resveratrol reduces breast cancer metastasis to lung through inhibiting neutrophil extracellular traps. J Drug Target. 2023;31:962-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Kong F, Zhang L, Zhao X, Zhao L, Wang P, Zhang R, Tian H, Ma S. Resveratrol augments paclitaxel sensitivity by modulating miR-671-5p/STOML2/PINK1/Parkin-mediated autophagy signaling in A549 cell. J Biochem Mol Toxicol. 2023;e23557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 14. | Kim S, Kim W, Kim DH, Jang JH, Kim SJ, Park SA, Hahn H, Han BW, Na HK, Chun KS, Choi BY, Surh YJ. Resveratrol suppresses gastric cancer cell proliferation and survival through inhibition of PIM-1 kinase activity. Arch Biochem Biophys. 2020;689:108413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 15. | Rojo D, Madrid A, Martín SS, Párraga M, Silva Pinhal MA, Villena J, Valenzuela-Valderrama M. Resveratrol Decreases the Invasion Potential of Gastric Cancer Cells. Molecules. 2022;27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 16. | Deng L, Zou J, Su Y, Wang M, Zhao L. Resveratrol inhibits TGF-β1-induced EMT in gastric cancer cells through Hippo-YAP signaling pathway. Clin Transl Oncol. 2022;24:2210-2221. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Ren M, Zhou X, Gu M, Jiao W, Yu M, Wang Y, Liu S, Yang J, Ji F. Resveratrol synergizes with cisplatin in antineoplastic effects against AGS gastric cancer cells by inducing endoplasmic reticulum stress‑mediated apoptosis and G2/M phase arrest. Oncol Rep. 2020;44:1605-1615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Ostwal V, Ramaswamy A, Bhargava P, Srinivas S, Mandavkar S, Chaugule D, Peelay Z, Baheti A, Tandel H, Jadhav VK, Shinde S, Jadhav S, Gota V, Mittra I. A pro-oxidant combination of resveratrol and copper reduces chemotherapy-related non-haematological toxicities in advanced gastric cancer: results of a prospective open label phase II single-arm study (RESCU III study). Med Oncol. 2022;40:17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Nogales C, Mamdouh ZM, List M, Kiel C, Casas AI, Schmidt HHHW. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol Sci. 2022;43:136-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 565] [Article Influence: 141.3] [Reference Citation Analysis (0)] |
| 20. | Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1623] [Cited by in RCA: 3148] [Article Influence: 286.2] [Reference Citation Analysis (0)] |
| 21. | Liu B, He H, Luo H, Zhang T, Jiang J. Artificial intelligence and big data facilitated targeted drug discovery. Stroke Vasc Neurol. 2019;4:206-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Daina A, Zoete V. Application of the SwissDrugDesign Online Resources in Virtual Screening. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 23. | Li X, Wei S, Niu S, Ma X, Li H, Jing M, Zhao Y. Network pharmacology prediction and molecular docking-based strategy to explore the potential mechanism of Huanglian Jiedu Decoction against sepsis. Comput Biol Med. 2022;144:105389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 210] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 24. | Huang X, You L, Nepovimova E, Psotka M, Malinak D, Valko M, Sivak L, Korabecny J, Heger Z, Adam V, Wu Q, Kuca K. Inhibitors of phosphoinositide 3-kinase (PI3K) and phosphoinositide 3-kinase-related protein kinase family (PIKK). J Enzyme Inhib Med Chem. 2023;38:2237209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 25. | Li P, Huang D, Gu X. Exploring the dual role of circRNA and PI3K/AKT pathway in tumors of the digestive system. Biomed Pharmacother. 2023;168:115694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 26. | Wang YY, Zhou YQ, Xie JX, Zhang X, Wang SC, Li Q, Hu LP, Jiang SH, Yi SQ, Xu J, Cao H, Zhao EH, Li J. MAOA suppresses the growth of gastric cancer by interacting with NDRG1 and regulating the Warburg effect through the PI3K/AKT/mTOR pathway. Cell Oncol (Dordr). 2023;46:1429-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 27. | Chen Z, Jiang Z, Meng L, Wang Y, Lin M, Wei Z, Han W, Ying S, Xu A. SAMHD1, positively regulated by KLF4, suppresses the proliferation of gastric cancer cells through MAPK p38 signaling pathway. Cell Cycle. 2022;21:2065-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Zhang C, Zhao S, Tan Y, Pan S, An W, Chen Q, Wang X, Xu H. The SKA3-DUSP2 Axis Promotes Gastric Cancer Tumorigenesis and Epithelial-Mesenchymal Transition by Activating the MAPK/ERK Pathway. Front Pharmacol. 2022;13:777612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 29. | Xu J, Lv S, Meng W, Zuo F. LCN2 Mediated by IL-17 Affects the Proliferation, Migration, Invasion and Cell Cycle of Gastric Cancer Cells by Targeting SLPI. Cancer Manag Res. 2020;12:12841-12849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 30. | Huang Z, Wu C, Zhou W, Lu S, Tan Y, Wu Z, You R, Stalin A, Guo F, Zhang J, Liu P, Wang W, Duan X, You L, Wu J. Compound Kushen Injection inhibits epithelial-mesenchymal transition of gastric carcinoma by regulating VCAM1 induced by the TNF signaling pathway. Phytomedicine. 2023;118:154984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 31. | Huo J, Xie W, Fan X, Sun P. Pyroptosis, apoptosis, and necroptosis molecular subtype derived prognostic signature universal applicable for gastric cancer-A large sample and multicenter retrospective analysis. Comput Biol Med. 2022;149:106037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 32. | Jin SP, Kim JH, Kim MA, Yang HK, Lee HE, Lee HS, Kim WH. Prognostic significance of loss of c-fos protein in gastric carcinoma. Pathol Oncol Res. 2007;13:284-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Zhou L, Zhang JS, Yu JC, Cui QC, Zhou WX, Kang WM, Ma ZQ. Negative association of c-fos expression as a favorable prognostic indicator in gastric cancer. Arch Med Res. 2010;41:201-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Rivas-Ortiz CI, Morales-Guerrero SE, Ponce-de-León-Rosales S, Gamboa-Domínguez A, Rangel-Escareño C, Uscanga-Domínguez LF, Aguilar-Gutiérrez GR, Kershenobich-Stalnikowitz D, López-Vidal Y, Castillo-Rojas G. Overview of Gene Expression Analysis in Gastric Disease Infected with Helicobacter pylori: CLDN1 and MMP9 Could Be Biomarkers for Early Diagnosis of Gastric Cancer. Processes. 2022;10:196. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 35. | Liu L, Li Y, Zhang X, Zhang H. The correlation of the miR-29a/MMP9 axis with Helicobacter pylori infection in gastric cancer. Am J Transl Res. 2021;13:10155-10162. [PubMed] |
| 36. | Zhang T, Fan X, Song L, Ren L, Ma E, Zhang S, Zheng Y, Zhang J. c-Fos is involved in inhibition of human bladder carcinoma T24 cells by brazilin. IUBMB Life. 2015;67:175-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J. 2015;469:325-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 218] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 38. | Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390-2400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 1298] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 39. | Sun M, Cheng H, Yu T, Tan J, Li M, Chen Q, Gu Y, Jiang C, Li S, He Y, Wen W. Involvement of a AS3MT/c-Fos/p53 signaling axis in arsenic-induced tumor in human lung cells. Environ Toxicol. 2023;38:615-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 40. | Huang X, Han C, Zhong J, Hu J, Jin Y, Zhang Q, Luo W, Liu R, Ling F. Low expression of the dynamic network markers FOS/JUN in pre-deteriorated epithelial cells is associated with the progression of colorectal adenoma to carcinoma. J Transl Med. 2023;21:45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 41. | Riedel M, Berthelsen MF, Cai H, Haldrup J, Borre M, Paludan SR, Hager H, Vendelbo MH, Wagner EF, Bakiri L, Thomsen MK. In vivo CRISPR inactivation of Fos promotes prostate cancer progression by altering the associated AP-1 subunit Jun. Oncogene. 2021;40:2437-2447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 42. | Shao J, Jiang F, Hu M, Mei E, Pan Z, Chen C, Lin L, Zheng T, Cai W, Li Z, Liu J. The role of FOS-mediated autophagy activation in the indocyanine green-based photodynamic therapy for treating melanoma. J Photochem Photobiol B. 2021;214:112101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 43. | Shang W, Wang Y, Liang X, Li T, Shao W, Liu F, Cui X, Lv L, Chai L, Qu L, Zheng L, Jia J. SETDB1 promotes gastric carcinogenesis and metastasis via upregulation of CCND1 and MMP9 expression. J Pathol. 2021;253:148-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 44. | Liang R, Chen W, Chen XY, Fan HN, Zhang J, Zhu JS. Dihydroartemisinin inhibits the tumorigenesis and invasion of gastric cancer by regulating STAT1/KDR/MMP9 and P53/BCL2L1/CASP3/7 pathways. Pathol Res Pract. 2021;218:153318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 45. | Wang X, Wang B, Xie J, Hou D, Zhang H, Huang H. Melatonin inhibits epithelial‑to‑mesenchymal transition in gastric cancer cells via attenuation of IL‑1β/NF‑κB/MMP2/MMP9 signaling. Int J Mol Med. 2018;42:2221-2228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 46. | Ge W, Gong Y, Li Y, Wu N, Ruan Y, Xu T, Shu Y, Qiu W, Wang Y, Zhao C. IL-17 induces non-small cell lung cancer metastasis via GCN5-dependent SOX4 acetylation enhancing MMP9 gene transcription and expression. Mol Carcinog. 2023;62:1399-1416. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 47. | He Y, Hou S, Memg C. [Neutrophil extracellular traps activates focal adhesion kinase by upregulating MMP9 expression to promote proliferation and migration of mouse colorectal cancer cells]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2023;39:416-422. [PubMed] |
| 48. | Anuj, Arivazhagan L, Venkatraman G, Rayala SK. Increased Expression of MicroRNA 551a by c-Fos Reduces Focal Adhesion Kinase Levels and Blocks Tumorigenesis. Mol Cell Biol. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 49. | Gupte R, Lin KY, Nandu T, Lea JS, Kraus WL. Combinatorial Treatment with PARP-1 Inhibitors and Cisplatin Attenuates Cervical Cancer Growth through Fos-Driven Changes in Gene Expression. Mol Cancer Res. 2022;20:1183-1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 50. | Xia Q, Du Z, Chen M, Zhou X, Bai W, Zheng X, Lin L, Zhao Y, Ding J, Wu Z, Zou H, Wang S, Xu L, Li E, Wu B. A protein complex of LCN2, LOXL2 and MMP9 facilitates tumour metastasis in oesophageal cancer. Mol Oncol. 2023;17:2451-2471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 51. | Xie QH, Wang WM, Yang JG, Xia HF, Xiao BL, Chen GH, Huang J, Li RF, Chen G. ALIX promotes cell migration and invasion of head and neck squamous cell carcinoma by regulating the expression of MMP9, MMP14, VEGF-C. Arch Oral Biol. 2023;151:105696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 52. | Gao SW, Liu F. Novel insights into cell cycle regulation of cell fate determination. J Zhejiang Univ Sci B. 2019;20:467-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 53. | Liu J, Peng Y, Wei W. Cell cycle on the crossroad of tumorigenesis and cancer therapy. Trends Cell Biol. 2022;32:30-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 243] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 54. | Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20:175-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1074] [Cited by in RCA: 1400] [Article Influence: 233.3] [Reference Citation Analysis (0)] |
| 55. | Fu Y, Sui B, Xiang L, Yan X, Wu D, Shi S, Hu X. Emerging understanding of apoptosis in mediating mesenchymal stem cell therapy. Cell Death Dis. 2021;12:596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 56. | Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17:395-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 1629] [Article Influence: 325.8] [Reference Citation Analysis (0)] |
| 57. | Kashyap D, Garg VK, Goel N. Intrinsic and extrinsic pathways of apoptosis: Role in cancer development and prognosis. Adv Protein Chem Struct Biol. 2021;125:73-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 196] [Article Influence: 49.0] [Reference Citation Analysis (0)] |