Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1135
Peer-review started: February 3, 2023
First decision: March 14, 2023
Revised: March 28, 2023
Accepted: April 23, 2023
Article in press: April 23, 2023
Published online: July 15, 2023
Processing time: 158 Days and 21.1 Hours
Colorectal cancer (CRC) is a common malignancy that has the second highest incidence and mortality rate. Although there are many personalized treatment options for CRC, the therapeutic effects are ultimately limited by drug resistance. Studies have aimed to block the initiation and progression of CRC by inducing cell death to overcome this obstacle. Substantial evidence has indicated that both autophagy and ferroptosis play important regulatory roles in CRC. Autophagy, a lysosome-dependent process by which cellular proteins and organelles are degraded, is the basic mechanism for maintaining cell homeostasis. The duality and complexity of autophagy in cancer therapy is a hot topic of discussion. Ferro
Core Tip: Ferroptosis is a mode of cell death centered on iron accumulation and lipid peroxidation that plays a crucial role in colorectal cancer (CRC). Recently, an increasing number of studies have found that autophagy and ferroptosis have a cross-talk relationship in CRC. Enhancing the antitumor effect through autophagy-dependent ferroptosis will become a hot topic in medical biology. This review describes the mechanisms of autophagy and ferroptosis and their interactions in CRC with a goal of providing new strategies for the treatment of CRC.
- Citation: Zeng XY, Qiu XZ, Wu JN, Liang SM, Huang JA, Liu SQ. Interaction mechanisms between autophagy and ferroptosis: Potential role in colorectal cancer. World J Gastrointest Oncol 2023; 15(7): 1135-1148
- URL: https://www.wjgnet.com/1948-5204/full/v15/i7/1135.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i7.1135
Colorectal cancer (CRC) is the second most common cause of cancer-related death in both men and women. According to global cancer statistics, 52580 people in the United States will die from CRC in 2022[1]. At present, surgery is still the basis of CRC radical therapy. For unresectable metastatic CRC, systemic therapy, including cytotoxic chemotherapy, biologic therapy, immunotherapy, and their combinations, is the mainstay[2]. However, drug resistance in CRC significantly reduces the effectiveness of systemic treatment. Recently, an increasing number of studies have investigated methods to hinder the occurrence and development of CRC by targeted induction of different modes of cell death, such as apoptosis, necroptosis, pyroptosis, and ferroptosis[3].
Autophagy is a lysosome-dependent process for the degradation of proteins and organelles that involves the interaction of multiple protein complexes encoded by highly conserved autophagy-related (ATG) genes. Autophagy plays a central role in the maintenance of cellular homeostasis and is closely associated with human health and various diseases[4]. Current studies have shown that autophagy has a dual role in cancer, promotion or inhibition depending on the type of tumor cells, genetic background, stage of tumor progression and tumor microenvironment[5]. Nevertheless, the complex regulatory mechanisms still need further exploration. Thus, targeted autophagy has great potential in cancer therapy.
Ferroptosis is a mode of regulated cell death (RCD) centered on iron accumulation and lipid peroxidation. Originally in 2012, Dixon et al[6] discovered that erastin-induced death in RAS-mutant cancer cells could be prevented by iron chelators and antioxidants, and this iron-dependent mode of cell death was named ferroptosis. Unlike autophagy and other RCDs, such as necroptosis, the process of ferroptosis is lipid peroxidation-mediated plasma membrane damage catalyzed by iron accumulation. Morphologically, it is represented by obvious shrinkage of the mitochondria with increased membrane density, fewer or no mitochondrial cristae, and outer mitochondrial membrane rupture[7]. Because of its unique pathophysiological features, ferroptosis plays an important regulatory role in many diseases[8]. With the in-depth exploration of ferroptosis, relevant studies have indicated significant crosstalk between autophagy and ferroptosis[9-11]. The discovery of autophagy-dependent ferroptosis opens up new insights into cell death and holds great promise in the treatment of disease.
In this review, we summarized the mechanisms of autophagy and ferroptosis, as well as their roles in CRC. Next, we focused on the interaction between autophagy and ferroptosis in the context of CRC, aiming to provide new targets for clinical treatment.
Depending on how cargo is delivered to lysosomes, autophagy can be classified as the following three types: Macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). Macroautophagy transports a bulk of cytoplasm or specific cargoes to lysosomes for degradation by forming autophagosomes with a double-membrane structure[12]. Microautophagy sequesters cell cytosolic components for lysosomal degradation through lysosomal membrane invagination[13,14]. CMA occurs only in mammalian cells and involves the crucial steps of chaperone-mediated identification and targeting of specific proteins to lysosomes[15]. Among the three different forms of autophagy, macroautophagy is the major regulatory mechanism that responds to environmental and physiological cues, so it is commonly referred to as ‘autophagy’[16].
Autophagy is a dynamic process regulated by ATG proteins and can be divided into the following five distinct stages: Initiation, nucleation, elongation of the autophagosome membrane and autophagosome maturation, fusion of autophagosomes with lysosomes, and degradation of the vesicular contents[17]. The Unc-51-like kinase-1 (ULK1) complex, which includes the ULK1/FAK family–interacting protein of 200 kD (FIP200)/ATG13, is responsible for integrating nutrient and energy signals and controls the autophagy switch[18,19]. Upon receiving a starvation signal, the ULK1 complex becomes activated and phosphorylates beclin1 (BECN1) at Ser 14, progressing to the second stage of autophagy[20]. The class III phosphatidylinositol 3-kinase (PtdIns3K) complex is composed of VPS34 (phosphatidylinositol 3-kinase), VPS15 (the adaptor of VPS34), BECN1 and ATG14 to generate phosphatidylinositol 3-phosphate (PI3P), which facilitates the nucleation of autophagosomal membranes[20,21]. Following nucleation, elongation of the autophagosomal membrane involves two ubiquitin-like (Ubl) conjugation systems: ATG12-ATG5-ATG16L and LC3-phosphatidylethanolamine (PE)[22]. The C-terminus of microtubule-associated protein one light chain 3 (LC3), a mammalian ortholog of yeast Atg8, is cleaved by ATG4 to form LC3-I[23,24]. LC3-I is conjugated to PE to generate LC3-II in the presence of ATG3, ATG7, and the ATG12-ATG5-ATG16L complex[24-26]. The autophagosomal membrane is continuously elongated, eventually forming an autophagosome with closed bilayer membrane structures. Subsequently, mature autophagosomes fuse with lysosomes to form autolysosomes, which decompose the contents of vesicles.
Whether autophagy is enhanced or inhibited is a topic of discussion in cancer treatment. The mechanisms of autophagy in cancer are complex and diverse, involving numerous genes, proteins, and pathways. On the one hand, autophagy prevents cancer by removing intracellular damaged organelles or toxic substances, which helps maintain the integrity of cells and genes[27,28]. On the other hand, autophagy can provide energy and rich nutrients for tumor cells to promote their proliferation and progression[28,29]. In this way, the impact of autophagy in different stages of CRC and the mechanisms of tumor initiation and progression vary[30]. Enhancing or inhibiting autophagy may result in different effects at different stages of CRC. We now briefly introduce the roles of autophagy and several key autophagy regulatory proteins in CRC (Table 1).
| Intervention | Target | Effects and mechanism | Ref. |
| Aspirin | mTOR↓, AMPK↑ | Inhibits mTOR signal transduction and activates AMPK to induce autophagy of CRC | [33] |
| TBK1 | mTORC1↓ | TBK1 initiates mTORC1 inhibition and induces autophagy to promote CRC progression | [139] |
| ABHD5 | BECN1↑ | ABHD5 prevents CASP3 from cleaving BECN1 and enhances autophagy flux to inhibit CRC | [38] |
| SOX2 | BECN1↑ | SOX2-β-catenin/Beclin1/autophagy signaling axis promotes chemoresistance of CRC | [41] |
| FIRRE | Stabilize BECN1 | Stabilizes BECN1 and promotes autophagy in a PTBP1 mediated manner to stimulate the development of CRC | [42] |
| IL-6 | BECN1↑ | IL-6/BECN1 pathway activates autophagy and promotes chemotherapy resistance of CRC | [39] |
| Pelareorep | BECN1↑ | Upregulates BECN1 expression and induces autophagy to enhance CRC proliferation | [140] |
| Fusobacterium nucleatum | BECN1↑, CARD3↑ | Upregulates CARD3 expression and activates autophagy signal to promote CRC metastasis | [141] |
| DCZ5248 | p62↑ | Induces lysosomal acidification and weakens lysosomal cathepsin activity to inhibit autophagy | [142] |
| Claudin 1 | p62↓ | Reduces the level of p62 and stimulates autophagy to promote CRC progression | [143] |
| DBTTS | p62↑ | Induces accumulation of p62 protein and inhibits autophagy to induce CRC cell death | [47] |
The mammalian target of rapamycin (mTOR) is a serine/threonine kinase that primarily regulates cell growth and proliferation and plays an anabolic role[31]. mTOR is recognized as the upstream negative regulator of the autophagy initiation stage. Under nutrient-rich conditions, high mTOR activity prevents ULK1 activation by phosphorylating ULK1 Ser 757, thereby inhibiting autophagy[32]. As early as 2012, aspirin was found to induce autophagy by reducing mTOR signaling in CRC cells through inhibition of the mTOR effector S6K1[33]. Moreover, oxiconazole (OXI) can downregulate the expression of the peroxiredoxin-2 (PRDX2) protein, extinguish mTOR, and initiate autophagy[34]. Moreover, OXI can block the fusion of autophagosomes with lysosomes, resulting in the extreme accumulation of autophagosomes and subsequent inhibition of CRC cell growth[34]. In addition, there are numerous compounds that target the mTOR pathway to regulate autophagy in CRC, all with promising antitumor effects[35,36]. Thus, mTOR may serve as an effective target for the treatment of CRC.
BECN1 is a core protein in autophagy nucleation. In one study, BECN1 expression was higher in CRC tissues than in normal colorectal tissues[37]. Patients with lower BECN1 expression had longer overall survival than those with high BECN1 expression[37]. ABHD5, an activator of cellular lipolysis, binds to BECN1, preventing its cleavage and conse
Sequestosome 1 (SQSTM1/p62) is both a cargo receptor for autophagy and a substrate for selective autophagy. P62 has been identified as having multiple domains, including the Phox1 and Bem1p (PB1) domain, zinc finger (ZZ), tumor necrosis factor receptor-associated factor 6 (TRAF6) binding domain, LC3-interacting region (LIR), KEAP1-interacting region (KIR) and ubiquitin-associated (UBA) domain[43]. While p62 attaches to the autophagosome by binding with LC3 via the LIR, its other domains bind the corresponding proteins, and eventually, they are degraded together in the autolysosome[44,45]. Therefore, the accumulation of p62 represents a decrease in autophagic flux. One group has shown that the expression of p62 is associated with the prognosis of CRC[46,47]. Increased autophagy leads to decreased p62 expression, which enables GATA4 to evade autophagic degradation, enhance NF-κB function, and drive the antioxidant reaction to support CRC survival[48]. Toll-like receptor (TLR) signaling acts as an immunomodulator that regulates inflammatory responses and plays a critical role in colitis-associated CRC[49]. TLR4 mediates the formation of the TRAF6-BECN1 complex, which activates autophagy, facilitating the migration and invasion of cancer cells[50]. P62 negatively regulates TLR4-induced autophagy activation and inhibits cancer cell progression[50]; thus, p62 may become a therapeutic target for CRC.
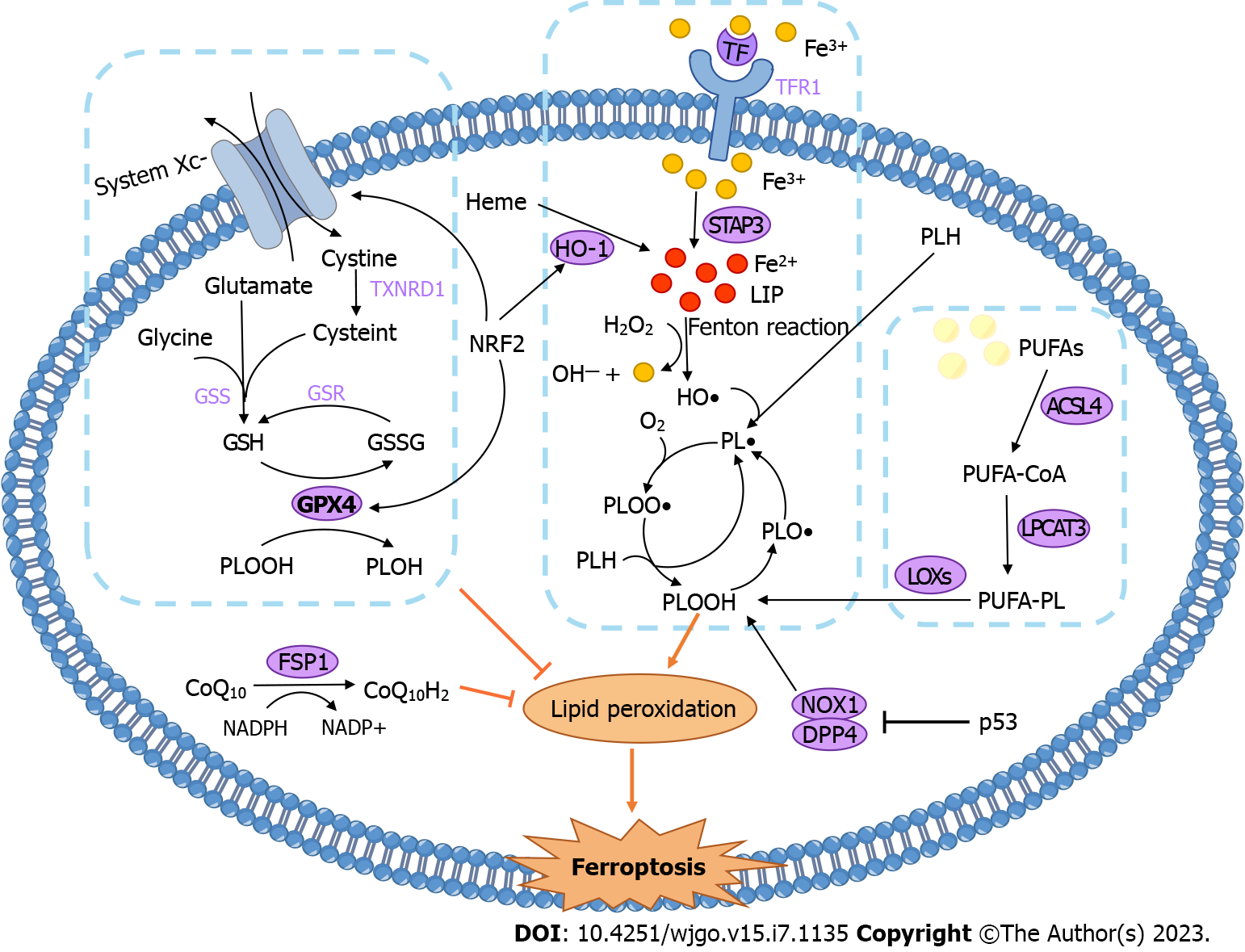
Ferroptosis is a form of cell death caused by iron-dependent lipid peroxidation. In general, the regulatory mechanism of ferroptosis can be divided into three main pathways: iron metabolism, lipid metabolism, and the antioxidant system. The three pathways are inseparable, and an imbalance in any one of the pathways drives ferroptosis. The mechanisms of ferroptosis and its effect on CRC are summarized in Figure 1 and Table 2.
| Intervention | Target | Effects and mechanism | Ref. |
| SRSF9 | GPX4↓ | Inhibition of SRSF9 increases erastin-induced iron death by downregulation of GPX4 level | [74] |
| IMCA | SLC7A11↓ | IMCA induces SLC7A11 mediated ferroptosis through AMPK/mTOR pathway | [77] |
| TalaA | ROS↑, SLC7A11↓ | TalaA induces ferroptosis to kill CRC cells | [144] |
| Lipocalin 2 | GPX4↑, system Xc−↑ | Overexpression of Lipocalin 2 inhibits ferroptosis and promotes CRC progression | [75] |
| TIGAR | GSH↓, ROS↑ | TIGAR induces ferroptosis resistance in CRC by ROS/AMPK/SCD1 signaling pathway | [145] |
| HIF-2α | Iron↑ | HIF-2α activation potentiates oxidative cell death in CRCs by increasing cellular iron | [79] |
| TP53 | Lipid peroxidation, ACSL4↑ | Restricts ferroptosis by blocking DPP4 activity in a transcription independent manner TP53 | [91] |
| Tagitinin C | NRF2/HO-1↑, lipid peroxidation | Tagitin C activates NRF2/HO-1 pathway to induce ferroptosis | [82] |
| Cetuximab | NRF2↓, ROS↑ | Cetuximab inhibits Nrf2/HO-1 pathway to promote ferroptosis in CRC | [85] |
| Beta-elemene | GSH↓, GPX4↓ | Combined treatment with beta-elemene and cetuximab induces ferroptosis in CRC | [96] |
| Vitamin C | Iron↑, ROS↑ | Vitamin C limits CRC resistance to EGFR-targeted therapies | [97] |
| FeOOH NSs | H2S↓ | FeOOH NSs eliminate endogenous H2S to induce ferroptosis | [99] |
Iron metabolism: Iron is an essential trace element in physiological metabolism, and an imbalance in iron homeostasis might lead to many pathological processes[51]. Iron mainly exists in the form of ferric ions (Fe3+) outside the cell, which are transported by transferrin (Tf). Iron-laden Tf binds to the transferrin receptor (TfR1) on the cell membrane to transport iron into the cell[52]. Intracellular Fe3+ is reduced to ferrous ions (Fe2+) by six-transmembrane epithelial antigen of prostate 3 (STEAP3), forming the labile iron pool (LIP), which is involved in various metabolic reactions[53]. In ferroptosis, the role of iron can be summarized into two crucial types as follows: (1) Iron that catalyzes the nonenzymatic lipid autoxidation chain via the Fenton reaction; and (2) iron acting as an enzyme cofactor in enzymatic reactions of lipid peroxidation (see Figure 1). Fe2+ in LIP reacts with hydrogen peroxide (H2O2) to generate hydroxyl radicals (HO•), Fe3+, and hydroxide ions, in a process known as the Fenton reaction[54]. HO• is a highly oxidative species that reacts with phospholipids to generate the phospholipid radical (PL•), initiating a lipid autoxidation chain reaction[55,56]. Ferrostatin-1 (fer-1), a ferroptosis inhibitor, can form a complex with Fe2+ to reduce LIP and scavenge alkoxy radicals produced by the lipid autoxidation chain, thereby inhibiting lipid peroxidation[57].
Lipid peroxidation: In addition to the iron-dependent Fenton reaction, another pathway of lipid peroxide production is the enzymatic reaction dominated by lipoxygenases (LOXs)[55]. Polyunsaturated fatty acids (PUFAs) are highly prone to peroxidation because of their bis-allylic carbons, and PUFAs play a central role in ferroptosis[58,59]. Polyunsaturated-fatty-acid-containing phospholipids (PUFA-PLs) are oxidized to phospholipid hydroperoxides (PLOOH) under the catalysis of LOXs[60]. The constant accumulation of lipid peroxides can destabilize membranes (membrane thinning and increased curvature), leading to pore formation, micellization, and ultimately cell death[61]. Furthermore, oxidative lipidomic analysis revealed that after cells were treated with ferroptosis inducers, the major oxidized phospholipid was PE[62]. Intracellular free PUFAs, especially arachidonic acid (AA) and adrenic acid (AdA), can be esterified to PE under the action of acyl-CoA synthetase long-chain family member 4 (ACSL4) and lysophosphatidylcholine acyltransferase 3 (LPCAT3), providing fuel for ferroptosis[58,63]. First, ACSL4 catalyzes the combination of coenzyme A (CoA) and PUFA to form a PUFA-CoA intermediate[64]. Next, LPCAT3 incorporates PUFA-CoA into membrane phospholipids[65]. The ACSL4 inhibitor rosiglitazone or LPCAT3 inhibitors can prevent ferroptosis[66,67]. Thus, ACSL4 and LPCAT3 are also important parts of the enzymatic reaction in ferroptosis.
Antioxidant defense systems: After the discovery of ferroptosis, researchers found the following common mediator for 12 ferroptosis-inducing small molecule compounds: glutathione peroxidase 4 (GPX4)[68]. GPX4 can reduce intracellular PLOOH to harmless phosphatidyl alcohol (PLOH), preventing the accumulation of lipid peroxides. This reaction requires the consumption of two molecules of glutathione (GSH) each time[69]. GSH, an important reducing substance in the body, is composed of three amino acids (glutamate, cysteine and glycine)[70]. Upon cellular oxidative stress, system Xc−, a cystine-glutamate antiporter, transports cystine into cells to provide raw materials for GSH synthesis[71]. At present, system Xc−-GSH-GPX4 is recognized as the most critical pathway by which the body resists ferroptosis. Ferroptosis inducers such as erastin inhibit system Xc− to prevent cystine uptake, resulting in the inability to synthesize GSH. GSH depletion inactivates the GPX4 enzyme, eventually triggering ferroptosis[56,68]. Another inducer, RSL3, binds directly to GPX4 to inactivate it[68]. Recently, new research has identified a GSH-independent pathway to inhibit ferroptosis. Ferroptosis suppressor protein 1 (FSP1) expression positively correlates with ferroptosis resistance[72]. Its main mode of action is that the reduced form of ubiquinone (also known as CoQ10) consumes lipid peroxyl radicals, while FSP1 uses NAD(P)H to catalyze the regeneration of ubiquinone[73].
Targeting the three pathways of ferroptosis described above can effectively impact CRC. Currently, there are more studies targeting system Xc−-GSH-GPX4 (versus the other two pathways) to regulate ferroptosis in CRC. Serine- and arginine-rich splicing factor 9 (SFRS9) is considered to be a carcinogen of cervical and bladder cancer. However, one group revealed that the expression of SFRS9 mRNA and protein was significantly higher in CRC tissues than in adjacent tissues. SFRS9 can bind to GPX4 mRNA and upregulate the expression of GPX4. Knockdown of SFRS9 inhibits CRC progression by triggering GPX4 reduction-mediated ferroptosis[74]. Lipocalin 2 has also been reported to inhibit ferroptosis by stimulating the expression of GPX4 and system Xc−[75]. Moreover, researchers isolated and purified petunidin 3-O-[rhamnopyranosyl-(trans-p-coumaroyl)]-5-O-(β-D-glucopyranoside) (Pt3R5G) from Lycium ruthenicum Murray, which inhibits RKO cell proliferation by downregulating solute carrier family 7 member 11 (SLC7A11), a subunit of system Xc−, which reduces ferroptosis[76]. In HCT116 cells, the benzopyran derivative 2-imino-6-methoxy-2H-chromene-3-carbothioamide (IMCA) downregulated SLC7A11 expression and decreased the content of cysteine and glutathione, leading to ROS accumulation and ferroptosis[77]. Therefore, inducing ferroptosis by inhibiting the system Xc−-GSH-GPX4 pathway may be an effective way to treat CRC. In addition, alterations in intracellular iron levels affect the growth of CRC cells. HIF-2α is a critical transcriptional regulator of cellular iron levels[78]. Activation of HIF-2α can lead to an increase in cellular iron and ROS levels; when this process is coupled with lipid-ROS induction by ferroptosis inducers, CRC cell death occurs[79].
In addition to the canonical system Xc−-GSH-GPX4 pathway, the nuclear factor erythroid 2-related factor 2/heme oxygenase 1 (NRF2/HO-1) axis also plays a major role in ferroptosis. NRF2 is the master transcription factor responsible for endogenous antioxidative stress, and many of its downstream target genes are also involved in the regulation of iron metabolism, particularly HO-1[80]. HO-1 catalyzes the cleavage of heme to produce Fe2+, which increases the LIP and thus promotes ferroptosis[80,81]. For example, the natural product tagitinin C (TC), a novel inducer of ferroptosis, can inhibit the growth of erastin-insensitive HCT116 cell lines[82]. Mechanistically, tagitinin C first induces oxidative stress, which activates the NRF2/HO-1 pathway and leads to the accumulation of iron, thus driving ferroptosis. Additionally, NRF2 is involved in the regulation of lipid metabolism. Another transcriptional target of NRF2 is GPX4, which allows NRF2 to exert anti-ferroptosis effects[83,84]. One group reported that cetuximab enhanced RSL3-induced lipid ROS accumulation by inhibiting the expression of NRF2 and HO-1 and ultimately promoted ferroptosis in KRAS-mutant CRC cells[85]. There is also research showing that lysionotin (Lys, a flavonoid) promotes the degradation of Nrf2, which leads to decreased expression of GPX4 and system Xc− and subsequently promotes ferroptosis[86]. Given the two different results of the above studies, further evidence is needed to clarify the relationship between NRF2, ferroptosis, and CRC. The balance between the driving effect and the suppressive effect is the key to treatment.
P53 is one of the most widely studied tumor suppressor genes, and it is mutated in almost all human cancers[87]. P53 is involved in a wide range of regulatory processes, including DNA repair, senescence, apoptosis, cell metabolism, ROS production, and ferroptosis[88]. P53 has a dual role in the regulation of ferroptosis[89]. The most classical pathway that promotes iron-mediated death involves p53-mediated repression of the transcription of the SLC7A11 gene, which decreases the expression of SLC7A11, affects the generation of GSH, and induces ferroptosis[90]. However, p53 has an antiferroptotic effect in CRC cells. Mechanistically, p53 binds with dipeptidyl peptidase 4 (DPP4) to block the formation of the DPP4-NOXI complex, leading to a decrease in DPP4-dependent lipid peroxidation, which suppresses ferroptosis[91]. In addition, TP53 target genes, such as cytochrome c oxidase 2 (SCO2), glutaminase 2 (GLS2), and spermidine/spermine N1 acetyltransferase 1 (SAT1), are also involved in the regulation of ferroptosis, but they have not been thoroughly studied in CRC[92,93].
Cisplatin is one of the most widely used anticancer drugs in the clinic, and its most prominent mechanism of action is DNA damage and ultimately apoptosis. However, the chemotherapeutic efficacy of cisplatin has been greatly limited, as the attenuation of DNA damage-mediated apoptotic signaling leads to drug resistance[94]. However, recently, it was found that cisplatin could promote ferroptosis via GSH depletion and GPX inactivation in CRC and had a synergistic effect with erastin[95]. In addition, β-elemene, a compound isolated from the Chinese herb Curcumae Rhizoma, combined with cetuximab can induce ferroptosis in KRAS-mutant CRC cells by increasing cellular iron accumulation and lipid peroxidation, which inhibits CRC growth and metastasis[96]. Another study showed that combinatorial treatment with pharmacological doses of vitamin C and cetuximab can trigger ferroptosis, which ultimately prevents the emergence of acquired resistance to anti-EGFR targeted therapy[97]. In addition to the drugs mentioned above, some novel compounds that inhibit CRC by inducing ferroptosis have also emerged with the rapid development of nanotechnology. For example, zinc oxide-coated virus-like silica nanoparticles (VZnO) can induce ferroptosis by scavenging H2S and depleting GSH to inhibit CRC growth[98]. Additionally, iron oxide hydroxide nanospindles (FeOOH NSS) had similar effects and hold promise as therapeutic agents for CRC[99]. In summary, ferroptosis regulation has great potential for addressing the current problem of anticancer drug resistance and may provide a new strategy for the treatment of CRC.
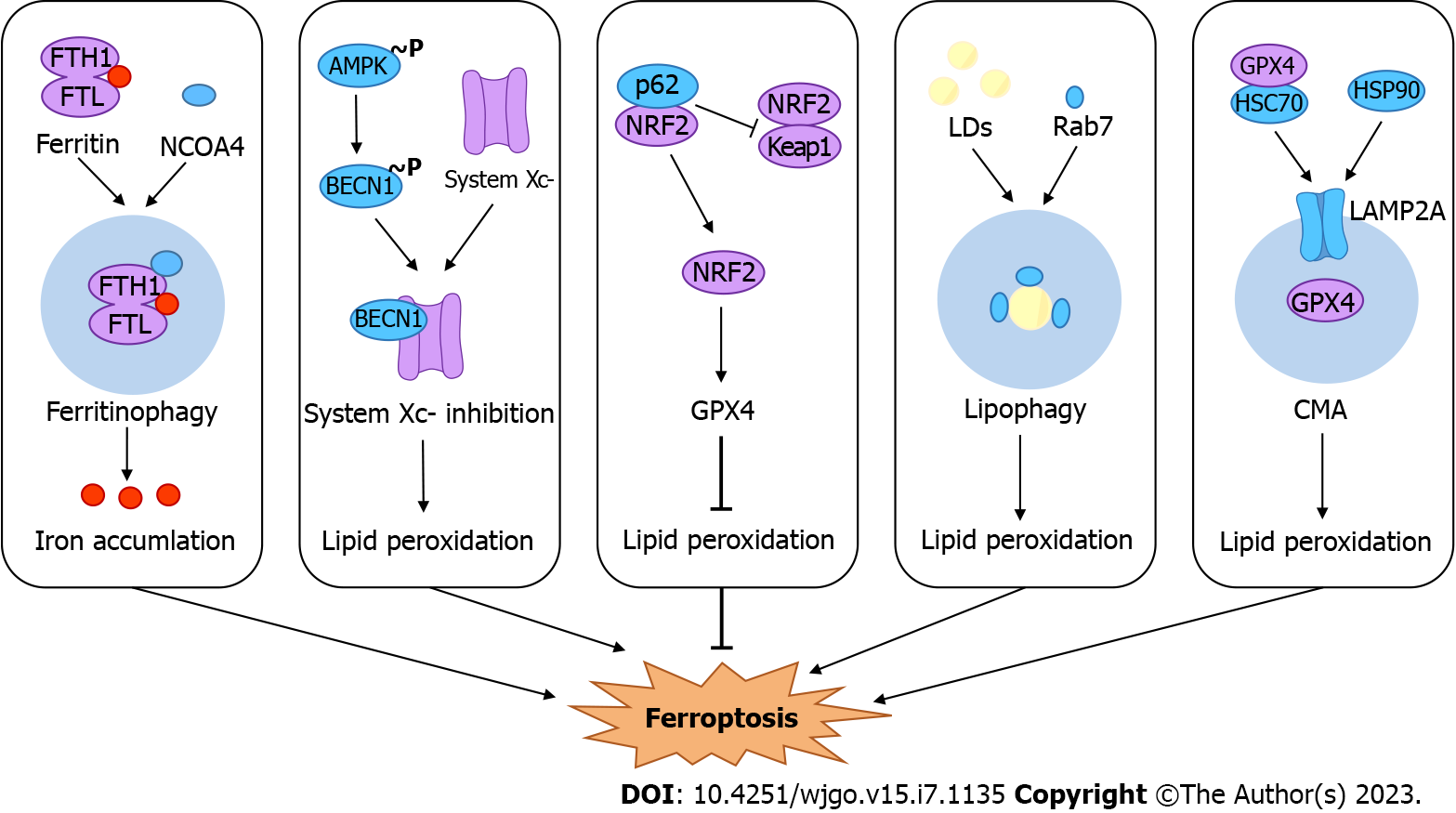
Autophagy and ferroptosis are two mechanistically distinct forms of cell death. Recent studies have revealed that autophagy inhibitors can prevent erastin-induced ferroptosis in cells and noted that ferroptosis is a form of autophagic cell death[9]. Although the interaction between autophagy and ferroptosis is not yet clear, several studies have noted the role of selective autophagy or certain autophagy factors in ferroptosis. For example, ferritinophagy, lipophagy, clockophagy, CMA and so on promote ferroptosis by inducing iron accumulation or lipid peroxidation[10,11,100]. Here, we summarize selective autophagy and possible regulatory pathways driving ferroptosis in the context of CRC (Figure 2).
Under normal physiological conditions, the LIP in cells maintains a dynamic balance, and excess Fe2+ is stored by ferritin[53]. Ferritin, composed of ferritin heavy chain 1 (FTH1) and ferritin light chain (FTL), protects against harmful oxidative stress under the condition of free iron overload. When cells are iron deficient, iron is released through an autophagy-related mechanism, known as "ferritinophagy"[101]. Nuclear receptor coactivator 4 (NCOA4), as a selective cargo receptor for ferritinophagy, transports ferritin for lysosomal degradation by binding to FTH1[102,103]. Ferritinophagy enhances cellular susceptibility to ferroptosis by controlling the size of the LIP[104,105]. New research has identified a novel ferroptosis inhibitor, 9a, that acts by disrupting the NCOA4-FTH1 interaction to reduce the amount of intracellular Fe2+[106].
Immunohistochemistry of specimens from patients with colon adenocarcinoma showed that the expression of the NCOA4 protein in tumor tissues was lower than that in peritumoral neighboring tissues. Moreover, the NCOA4 expression level was highly correlated with overall survival, and patients with low protein expression had a worse prognosis[107]. One study showed that in CRC cells, inhibition of the GTP cyclohydrolase-1/tetrahydrobiopterin (GCH1/BH4) pathway resulted in increased levels of NCOA4 protein, decreased levels of FTH1, and the accumulation of free iron. This phenomenon can be reversed with the use of autophagy inhibitors. Altogether, inhibition of the GCH1/BH4 pathway promoted erastin-induced ferroptosis by activating ferritinophagy[108]. In contrast, another study showed that the knockdown of NCOA4 disrupted ferritinophagy and had no significant effect on erastin-induced ferroptosis in HCT116 cells[109]. We should consider whether there is another mechanism by which erastin-induced ferroptosis in CRC cells copes with iron reduction caused by decreased ferritinophagy. Therefore, we need more evidence to validate the role of ferritinophagy in CRC. Studies on ferritinophagy, which involves two major mechanisms, autophagy and ferroptosis, will provide new insights into the treatment of CRC.
BECN1 is a key factor in autophagy initiation, and its role in CRC is complex, as it can promote or inhibit autophagy. Thus far, in a variety of cancers, such as hepatocellular carcinoma, lung cancer, head and neck cancer, and others, there is substantial evidence that BECN1 regulates autophagy-dependent ferroptosis[110,111]. In several studies, BECN1 merely plays the classical role of activating autophagy, promoting the degradation of autophagic ferritin, which in turn leads to ferroptosis. For example, ELAV-like RNA binding protein 1 can bind to the 3’-untranslated region of BECN1, allow intracellular iron accumulation, and eventually lead to ferroptosis[112]. However, one study found that in CRC, BECN1 plays a direct role in regulating ferroptosis[113]. The transporter system Xc− in the anti-ferroptosis system is another agonist of this pathway. System Xc− consists of the following two core components: SLC7A11 and solute carrier family 3 member 2 (SLC3A2)[114]. BECN1 can block the activity of system Xc− by directly binding to SLC7A11, thereby promoting ferroptosis[113]. Adenosine monophosphate-activated protein kinase (AMPK) is upstream of this pathway, and its phosphorylation of BECN1 at Ser90/93/96 could promote the formation of a complex of BECN1 with system Xc−[113]. Taken together, these results indicate that BECN1 could be a regulatory target for ferroptosis, and its detailed regulatory pathways require further investigation.
NRF2 acts as an important defense factor against oxidative stress, and its negative regulator is Kelch-like ECH-associated protein 1 (KEAP1)[115]. Under normal physiological conditions, NRF2 binds to KEAP1, which is constantly ubiquitinated and degraded by the proteasome so that it has no function. When the organism undergoes oxidative stress, the site of KEAP1 binding to NRF2 changes so that NRF2 can translocate to the nucleus and activate the transcription of the antioxidant response element[116,117]. P62 is a selective cargo receptor for autophagy, and its regulation of NRF2-KEAP1 was revealed as early as 2010[118]. Upon autophagy deficiency and p62 accumulation, p62 competes with KEAP1 for the binding site of NRF2, exempting NRF2 from degradation and enabling the transcriptional activation of its target genes[119,120]. Recently, with the uncovering of new mechanistic insights into the regulation of ferroptosis by NRF2, a growing number of studies have demonstrated the role of the p62-KEAP1-NRF2 pathway in the regulation of ferroptosis[121,122]. A study showed that CRC cells could be treated with RH4 (the primary pharmacologically active component of ginseng) and found an increase in Beclin1, LC3B, and NRF2 and a decrease in p62, which could ultimately induce ferroptosis. Treatment with the autophagy inhibitor 3-MA could reverse RH4-induced ferroptosis[123]. In another study, silencing NRF2 decreased the expression of p62, which improved the antitumor effects of tributyltin (IV) ferulate (TBT-F)[124]. These results establish a basis for the crosstalk between autophagy and ferroptosis and suggest that the p62-KEAP1-NRF2 pathway influences ferroptosis, which may be an important topic for future research.
In addition to the above, there are other potential mechanisms by which autophagy regulates ferroptosis in CRC. Lipophagy is a process in which intracellular lipid droplets (LDs) are targeted for transport into lysosomes for breakdown. LD is a dynamic organelle that stores neutral fatty acids and is involved in maintaining energy and redox homeostasis[125]. In hepatocytes, the small GTPase Rab7 recruits autophagosomes and lysosomes to the surface of LDs, resulting in lipophagy[126]. Tumor protein D52 (TPD52) or knockdown of Rab7 increased lipid storage, reduced lipid peroxidation, and suppressed RSL3-induced ferroptosis[127]. The results show that lipophagy is closely related to lipid peroxidation in ferroptosis. In addition, the accumulation of LDs contributes to chemoresistance in CRC[128]. Therefore, driving lipophagy, which leads to an increase in the occurrence of lipid peroxidation and promotes ferroptosis, may emerge as a novel treatment strategy for CRC. Most importantly, we must find the specific receptor of lipophagy in CRC.
CMA, unlike macroautophagy and microautophagy, is a type of selective autophagy that degrades only a specific subset of soluble proteins[129]. Heat shock cognate 71 kDa protein (HSC70) detects cytoplasmic proteins containing a KFERQ-like motif and then docks with lysosomes via lysosome-associated membrane protein type 2A (LAMP2A) to send the target proteins to lysosomes for degradation[130]. One research team found that GPX4 contains pentapeptide sequences (124 NVKFD 128, 169 LIDKN 173, and 187 QVIEK 191) consistent with a KFERQ-like motif, which is one of the substrates of CMA[131]. Heat shock protein 90 (HSP90) increases the levels of LAMP2A, mediating the degradation of GPX4 and leading to ferroptosis[131]. Antimony (sb) can upregulate the expression of HSP90, HSC70, and LAMP2A, which increases the rate of formation of the chaperone-GPX4 complex to mediate ferroptosis via CMA[132]. In addition, ACSL4 can also be recognized by HSC70 as a substrate for CMA-mediated ferroptosis[133]. In HCT116 cells, the lack of sorting nexin 10 (SNX10) promotes the proliferation of cancer cells by enhancing the degradation of the CMA substrate p21Cip1/WAF1[134]. The above studies laid the foundation for CMA-mediated ferroptosis of CRC cells. Future studies should focus on finding ferroptosis-related proteins containing a KFERQ-like motif and identifying the targets that drive CMA to degrade ferroptosis-related proteins.
Furthermore, hippocalcin-like 1 (HPCAL1), a neuronal calcium sensor, was identified as an important negative regulator of lipid synthesis and mTOR signaling activation, thereby blunting lipid metabolism to suppress tumorigenesis in the liver[135]. A recent study showed that HPCAL1 selectively degrades cadherin 2 and promotes lipid peroxidation to induce ferroptosis[136]. This phenomenon has been confirmed in a variety of cancer cells, including pancreatic cancer, non-small cell lung cancer, and bladder cancer cells, but this trend needs to be explored further in CRC.
In recent years, ferroptosis has consistently been under the spotlight in medical research. Ferroptosis can be used as a new treatment to clear cancer cells. For example, sorafenib itself is a ferroptosis inducer and ferroptosis inducers combined with chemotherapy drugs can overcome drug resistance; in addition, nanoparticulate anticancer drug delivery systems based on ferroptosis have emerged[137,138]. Although many studies are still in the experimental stage, these results have revealed the great potential of ferroptosis in cancer treatment. Furthermore, with a deeper understanding of the mechanisms of ferroptosis, an increasing number of studies have demonstrated crosstalk between ferroptosis and other types of RCD. Thus, autophagy-dependent ferroptosis takes the stage. Clarification of the crosstalk between autophagy and ferroptosis would not only provide a comprehensive understanding of the mechanisms of cell death but could also provide new insights for cancer treatment. Although much progress has been made, research on autophagy-dependent ferroptosis in CRC is still at an early stage. In this review, we summarized the mechanisms of autophagy and ferroptosis and their roles in CRC and focused on the possible pathways of crosstalk between them. While ferritinophagy, the BECN1–system Xc− pathway, and the p62-KEAP1-NRF2 pathway play a significant role in ferroptosis, the roles of lipophagy, CMA, or other regulators have not been validated in CRC. The mechanisms involved in the two different types of cell death are complex but also build a broader platform for subsequent research. Defining targets that regulate autophagy-dependent ferroptosis might lead to the discovery of novel therapeutic strategies for CRC.
We are very grateful to the other members of the research team for their careful review and suggestions.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bordonaro M, United States; Chen X, China; Sipos F, Hungary; Tzeng IS, Taiwan S-Editor: Yan JP L-Editor: A P-Editor: Yuan YY
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11438] [Article Influence: 3812.7] [Reference Citation Analysis (4)] |
| 2. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1442] [Article Influence: 360.5] [Reference Citation Analysis (0)] |
| 3. | Patankar JV, Becker C. Cell death in the gut epithelium and implications for chronic inflammation. Nat Rev Gastroenterol Hepatol. 2020;17:543-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 246] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 4. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 1914] [Article Influence: 159.5] [Reference Citation Analysis (0)] |
| 5. | Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1271] [Cited by in RCA: 2041] [Article Influence: 340.2] [Reference Citation Analysis (0)] |
| 6. | Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4711] [Cited by in RCA: 11765] [Article Influence: 905.0] [Reference Citation Analysis (1)] |
| 7. | Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, Sun B, Wang G. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 639] [Cited by in RCA: 2536] [Article Influence: 507.2] [Reference Citation Analysis (0)] |
| 8. | Yan HF, Zou T, Tuo QZ, Xu S, Li H, Belaidi AA, Lei P. Ferroptosis: mechanisms and links with diseases. Signal Transduct Target Ther. 2021;6:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 382] [Cited by in RCA: 834] [Article Influence: 208.5] [Reference Citation Analysis (0)] |
| 9. | Gao M, Monian P, Pan Q, Zhang W, Xiang J, Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 1328] [Article Influence: 147.6] [Reference Citation Analysis (2)] |
| 10. | Zhou B, Liu J, Kang R, Klionsky DJ, Kroemer G, Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 718] [Article Influence: 119.7] [Reference Citation Analysis (0)] |
| 11. | Liu J, Kuang F, Kroemer G, Klionsky DJ, Kang R, Tang D. Autophagy-Dependent Ferroptosis: Machinery and Regulation. Cell Chem Biol. 2020;27:420-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 576] [Article Influence: 115.2] [Reference Citation Analysis (0)] |
| 12. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5293] [Article Influence: 311.4] [Reference Citation Analysis (0)] |
| 13. | Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 378] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 14. | Shpilka T, Elazar Z. Shedding light on mammalian microautophagy. Dev Cell. 2011;20:1-2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 635] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 16. | Mizushima N, Levine B. Autophagy in Human Diseases. N Engl J Med. 2020;383:1564-1576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 758] [Article Influence: 151.6] [Reference Citation Analysis (0)] |
| 17. | Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11:28-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 253] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 18. | Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 390] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 19. | Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497-510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 676] [Cited by in RCA: 768] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 20. | Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 Lipid kinase. Nat Cell Biol. 2013;15:741-750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1001] [Cited by in RCA: 1234] [Article Influence: 102.8] [Reference Citation Analysis (0)] |
| 21. | Wirth M, Joachim J, Tooze SA. Autophagosome formation--the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 216] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 22. | Kim KH, Lee MS. Autophagy--a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10:322-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 741] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 23. | Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720-5728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4962] [Cited by in RCA: 5534] [Article Influence: 221.4] [Reference Citation Analysis (0)] |
| 24. | Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341-1350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 306] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 25. | Don Wai Luu L, Kaakoush NO, Castaño-Rodríguez N. The role of ATG16L2 in autophagy and disease. Autophagy. 2022;18:2537-2546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 26. | Frudd K, Burgoyne T, Burgoyne JR. Oxidation of Atg3 and Atg7 mediates inhibition of autophagy. Nat Commun. 2018;9:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 27. | Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913-1930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 635] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 28. | Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 1091] [Article Influence: 218.2] [Reference Citation Analysis (0)] |
| 29. | Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 475] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 30. | Wen X, Klionsky DJ. At a glance: A history of autophagy and cancer. Semin Cancer Biol. 2020;66:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 31. | Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1577] [Article Influence: 157.7] [Reference Citation Analysis (0)] |
| 32. | Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4208] [Cited by in RCA: 5539] [Article Influence: 395.6] [Reference Citation Analysis (0)] |
| 33. | Din FV, Valanciute A, Houde VP, Zibrova D, Green KA, Sakamoto K, Alessi DR, Dunlop MG. Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology. 2012;142:1504-15.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 334] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 34. | Shi J, Zhou L, Huang HS, Peng L, Xie N, Nice E, Fu L, Jiang C, Huang C. Repurposing Oxiconazole against Colorectal Cancer via PRDX2-mediated Autophagy Arrest. Int J Biol Sci. 2022;18:3747-3761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 35. | Xiang X, Tian Y, Hu J, Xiong R, Bautista M, Deng L, Yue Q, Li Y, Kuang W, Li J, Liu K, Yu C, Feng G. Fangchinoline exerts anticancer effects on colorectal cancer by inducing autophagy via regulation AMPK/mTOR/ULK1 pathway. Biochem Pharmacol. 2021;186:114475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Shen J, Dong J, Shao F, Zhao J, Gong L, Wang H, Chen W, Zhang Y, Cai Y. Graphene oxide induces autophagy and apoptosis via the ROS-dependent AMPK/mTOR/ULK-1 pathway in colorectal cancer cells. Nanomedicine (Lond). 2022;17:591-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 37. | Yang Z, Ghoorun RA, Fan X, Wu P, Bai Y, Li J, Chen H, Wang L, Wang J. High expression of Beclin-1 predicts favorable prognosis for patients with colorectal cancer. Clin Res Hepatol Gastroenterol. 2015;39:98-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 38. | Peng Y, Miao H, Wu S, Yang W, Zhang Y, Xie G, Xie X, Li J, Shi C, Ye L, Sun W, Wang L, Liang H, Ou J. ABHD5 interacts with BECN1 to regulate autophagy and tumorigenesis of colon cancer independent of PNPLA2. Autophagy. 2016;12:2167-2182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 39. | Hu F, Song D, Yan Y, Huang C, Shen C, Lan J, Chen Y, Liu A, Wu Q, Sun L, Xu F, Hu F, Chen L, Luo X, Feng Y, Huang S, Hu J, Wang G. IL-6 regulates autophagy and chemotherapy resistance by promoting BECN1 phosphorylation. Nat Commun. 2021;12:3651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 162] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 40. | Pan Y, Zhao Z, Li J, Luo Y, Li W, You W, Zhang Y, Li Z, Yang J, Xiao ZJ, Wang Y. Nuclear Beclin 1 Destabilizes Retinoblastoma Protein to Promote Cell Cycle Progression and Colorectal Cancer Growth. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 41. | Zhu Y, Huang S, Chen S, Chen J, Wang Z, Wang Y, Zheng H. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis. 2021;12:449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 42. | Wang Y, Li Z, Xu S, Li W, Chen M, Jiang M, Fan X. LncRNA FIRRE functions as a tumor promoter by interaction with PTBP1 to stabilize BECN1 mRNA and facilitate autophagy. Cell Death Dis. 2022;13:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 43. | Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672-4678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 643] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 44. | Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131-24145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3082] [Cited by in RCA: 3631] [Article Influence: 201.7] [Reference Citation Analysis (0)] |
| 45. | Islam MA, Sooro MA, Zhang P. Autophagic Regulation of p62 is Critical for Cancer Therapy. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 189] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 46. | Schmitz KJ, Ademi C, Bertram S, Schmid KW, Baba HA. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J Surg Oncol. 2016;14:189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 114] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 47. | Yagdi Efe E, Mazumder A, Lee JY, Gaigneaux A, Radogna F, Nasim MJ, Christov C, Jacob C, Kim KW, Dicato M, Chaimbault P, Cerella C, Diederich M. Tubulin-binding anticancer polysulfides induce cell death via mitotic arrest and autophagic interference in colorectal cancer. Cancer Lett. 2017;410:139-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 48. | Liu X, Zhao M, Sun X, Meng Z, Bai X, Gong Y, Xu L, Hao X, Yang T, Wei Z, Zhang X, Guo H, Li P, Liu Q, Shi Y, Shao C. Autophagic Flux Unleashes GATA4-NF-κB Axis to Promote Antioxidant Defense-Dependent Survival of Colorectal Cancer Cells under Chronic Acidosis. Oxid Med Cell Longev. 2021;2021:8189485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 49. | Moradi-Marjaneh R, Hassanian SM, Fiuji H, Soleimanpour S, Ferns GA, Avan A, Khazaei M. Toll like receptor signaling pathway as a potential therapeutic target in colorectal cancer. J Cell Physiol. 2018;233:5613-5622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 50. | Kim MJ, Min Y, Im JS, Son J, Lee JS, Lee KY. p62 is Negatively Implicated in the TRAF6-BECN1 Signaling Axis for Autophagy Activation and Cancer Progression by Toll-Like Receptor 4 (TLR4). Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Koleini N, Shapiro JS, Geier J, Ardehali H. Ironing out mechanisms of iron homeostasis and disorders of iron deficiency. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 52. | Gammella E, Buratti P, Cairo G, Recalcati S. The transferrin receptor: the cellular iron gate. Metallomics. 2017;9:1367-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 198] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 53. | Lv H, Shang P. The significance, trafficking and determination of labile iron in cytosol, mitochondria and lysosomes. Metallomics. 2018;10:899-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 54. | Winterbourn CC. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett. 1995;82-83:969-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 813] [Cited by in RCA: 932] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 55. | Gaschler MM, Stockwell BR. Lipid peroxidation in cell death. Biochem Biophys Res Commun. 2017;482:419-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 717] [Cited by in RCA: 1192] [Article Influence: 149.0] [Reference Citation Analysis (0)] |
| 56. | Zhang S, Xin W, Anderson GJ, Li R, Gao L, Chen S, Zhao J, Liu S. Double-edge sword roles of iron in driving energy production vs instigating ferroptosis. Cell Death Dis. 2022;13:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 123] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 57. | Miotto G, Rossetto M, Di Paolo ML, Orian L, Venerando R, Roveri A, Vučković AM, Bosello Travain V, Zaccarin M, Zennaro L, Maiorino M, Toppo S, Ursini F, Cozza G. Insight into the mechanism of ferroptosis inhibition by ferrostatin-1. Redox Biol. 2020;28:101328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 573] [Article Influence: 114.6] [Reference Citation Analysis (0)] |
| 58. | Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, Stockwell BR. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci U S A. 2016;113:E4966-E4975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 699] [Cited by in RCA: 1622] [Article Influence: 180.2] [Reference Citation Analysis (0)] |
| 59. | Feng H, Stockwell BR. Unsolved mysteries: How does lipid peroxidation cause ferroptosis? PLoS Biol. 2018;16:e2006203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 567] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 60. | Shintoku R, Takigawa Y, Yamada K, Kubota C, Yoshimoto Y, Takeuchi T, Koshiishi I, Torii S. Lipoxygenase-mediated generation of lipid peroxides enhances ferroptosis induced by erastin and RSL3. Cancer Sci. 2017;108:2187-2194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 343] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 61. | Agmon E, Solon J, Bassereau P, Stockwell BR. Modeling the effects of lipid peroxidation during ferroptosis on membrane properties. Sci Rep. 2018;8:5155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 251] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 62. | Wiernicki B, Dubois H, Tyurina YY, Hassannia B, Bayir H, Kagan VE, Vandenabeele P, Wullaert A, Vanden Berghe T. Excessive phospholipid peroxidation distinguishes ferroptosis from other cell death modes including pyroptosis. Cell Death Dis. 2020;11:922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 184] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 63. | Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, Prokisch H, Trümbach D, Mao G, Qu F, Bayir H, Füllekrug J, Scheel CH, Wurst W, Schick JA, Kagan VE, Angeli JP, Conrad M. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1074] [Cited by in RCA: 2722] [Article Influence: 302.4] [Reference Citation Analysis (0)] |
| 64. | Kuwata H, Nakatani E, Shimbara-Matsubayashi S, Ishikawa F, Shibanuma M, Sasaki Y, Yoda E, Nakatani Y, Hara S. Long-chain acyl-CoA synthetase 4 participates in the formation of highly unsaturated fatty acid-containing phospholipids in murine macrophages. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:1606-1618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 65. | Jalil A, Bourgeois T, Ménégaut L, Lagrost L, Thomas C, Masson D. Revisiting the Role of LXRs in PUFA Metabolism and Phospholipid Homeostasis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 66. | Wang Y, Zhang M, Bi R, Su Y, Quan F, Lin Y, Yue C, Cui X, Zhao Q, Liu S, Yang Y, Zhang D, Cao Q, Gao X. ACSL4 deficiency confers protection against ferroptosis-mediated acute kidney injury. Redox Biol. 2022;51:102262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 263] [Article Influence: 87.7] [Reference Citation Analysis (0)] |
| 67. | Reed A, Ichu TA, Milosevich N, Melillo B, Schafroth MA, Otsuka Y, Scampavia L, Spicer TP, Cravatt BF. LPCAT3 Inhibitors Remodel the Polyunsaturated Phospholipid Content of Human Cells and Protect from Ferroptosis. ACS Chem Biol. 2022;17:1607-1618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 91] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 68. | Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, Brown LM, Girotti AW, Cornish VW, Schreiber SL, Stockwell BR. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2388] [Cited by in RCA: 5203] [Article Influence: 473.0] [Reference Citation Analysis (0)] |
| 69. | Forcina GC, Dixon SJ. GPX4 at the Crossroads of Lipid Homeostasis and Ferroptosis. Proteomics. 2019;19:e1800311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 656] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 70. | Wang L, Ahn YJ, Asmis R. Sexual dimorphism in glutathione metabolism and glutathione-dependent responses. Redox Biol. 2020;31:101410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 71. | Parker JL, Deme JC, Kolokouris D, Kuteyi G, Biggin PC, Lea SM, Newstead S. Molecular basis for redox control by the human cystine/glutamate antiporter system xc(). Nat Commun. 2021;12:7147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 141] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 72. | Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, Bassik MC, Nomura DK, Dixon SJ, Olzmann JA. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688-692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1498] [Cited by in RCA: 2431] [Article Influence: 405.2] [Reference Citation Analysis (0)] |
| 73. | Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, Goya Grocin A, Xavier da Silva TN, Panzilius E, Scheel CH, Mourão A, Buday K, Sato M, Wanninger J, Vignane T, Mohana V, Rehberg M, Flatley A, Schepers A, Kurz A, White D, Sauer M, Sattler M, Tate EW, Schmitz W, Schulze A, O'Donnell V, Proneth B, Popowicz GM, Pratt DA, Angeli JPF, Conrad M. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1492] [Cited by in RCA: 2137] [Article Influence: 356.2] [Reference Citation Analysis (0)] |
| 74. | Wang R, Xing R, Su Q, Yin H, Wu D, Lv C, Yan Z. Knockdown of SFRS9 Inhibits Progression of Colorectal Cancer Through Triggering Ferroptosis Mediated by GPX4 Reduction. Front Oncol. 2021;11:683589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 75. | Chaudhary N, Choudhary BS, Shah SG, Khapare N, Dwivedi N, Gaikwad A, Joshi N, Raichanna J, Basu S, Gurjar M, P K S, Saklani A, Gera P, Ramadwar M, Patil P, Thorat R, Gota V, Dhar SK, Gupta S, Das M, Dalal SN. Lipocalin 2 expression promotes tumor progression and therapy resistance by inhibiting ferroptosis in colorectal cancer. Int J Cancer. 2021;149:1495-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 76. | Han L, Yan Y, Fan M, Gao S, Zhang L, Xiong X, Li R, Xiao X, Wang X, Ni L, Tong D, Huang C, Cao Y, Yang J. Pt3R5G inhibits colon cancer cell proliferation through inducing ferroptosis by down-regulating SLC7A11. Life Sci. 2022;306:120859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 77. | Zhang L, Liu W, Liu F, Wang Q, Song M, Yu Q, Tang K, Teng T, Wu D, Wang X, Han W, Li Y. IMCA Induces Ferroptosis Mediated by SLC7A11 through the AMPK/mTOR Pathway in Colorectal Cancer. Oxid Med Cell Longev. 2020;2020:1675613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 78. | Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 339] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 79. | Singhal R, Mitta SR, Das NK, Kerk SA, Sajjakulnukit P, Solanki S, Andren A, Kumar R, Olive KP, Banerjee R, Lyssiotis CA, Shah YM. HIF-2α activation potentiates oxidative cell death in colorectal cancers by increasing cellular iron. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 80. | Kerins MJ, Ooi A. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxid Redox Signal. 2018;29:1756-1773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 532] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 81. | Anandhan A, Dodson M, Schmidlin CJ, Liu P, Zhang DD. Breakdown of an Ironclad Defense System: The Critical Role of NRF2 in Mediating Ferroptosis. Cell Chem Biol. 2020;27:436-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 82. | Wei R, Zhao Y, Wang J, Yang X, Li S, Wang Y, Fei J, Hao X, Gui L, Ding X. Tagitinin C induces ferroptosis through PERK-Nrf2-HO-1 signaling pathway in colorectal cancer cells. Int J Biol Sci. 2021;17:2703-2717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 271] [Article Influence: 67.8] [Reference Citation Analysis (1)] |
| 83. | Dodson M, Castro-Portuguez R, Zhang DD. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23:101107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1085] [Cited by in RCA: 1532] [Article Influence: 255.3] [Reference Citation Analysis (0)] |
| 84. | Dong S, Lu Y, Peng G, Li J, Li W, Li M, Wang H, Liu L, Zhao Q. Furin inhibits epithelial cell injury and alleviates experimental colitis by activating the Nrf2-Gpx4 signaling pathway. Dig Liver Dis. 2021;53:1276-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 85. | Yang J, Mo J, Dai J, Ye C, Cen W, Zheng X, Jiang L, Ye L. Cetuximab promotes RSL3-induced ferroptosis by suppressing the Nrf2/HO-1 signalling pathway in KRAS mutant colorectal cancer. Cell Death Dis. 2021;12:1079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 201] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 86. | Gao Z, Jiang J, Hou L, Ji F. Lysionotin Induces Ferroptosis to Suppress Development of Colorectal Cancer via Promoting Nrf2 Degradation. Oxid Med Cell Longev. 2022;2022:1366957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 87. | Machado-Silva A, Perrier S, Bourdon JC. p53 family members in cancer diagnosis and treatment. Semin Cancer Biol. 2010;20:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 88. | Hernández Borrero LJ, El-Deiry WS. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim Biophys Acta Rev Cancer. 2021;1876:188556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 351] [Article Influence: 87.8] [Reference Citation Analysis (0)] |
| 89. | Ji H, Wang W, Li X, Han X, Zhang X, Wang J, Liu C, Huang L, Gao W. p53: A double-edged sword in tumor ferroptosis. Pharmacol Res. 2022;177:106013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 90. | Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 2404] [Article Influence: 240.4] [Reference Citation Analysis (0)] |
| 91. | Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, Zhong M, Yuan H, Zhang L, Billiar TR, Lotze MT, Zeh HJ 3rd, Kang R, Kroemer G, Tang D. The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep. 2017;20:1692-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 688] [Article Influence: 98.3] [Reference Citation Analysis (0)] |
| 92. | Liu J, Zhang C, Wang J, Hu W, Feng Z. The Regulation of Ferroptosis by Tumor Suppressor p53 and its Pathway. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 180] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 93. | Ou Y, Wang SJ, Li D, Chu B, Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc Natl Acad Sci U S A. 2016;113:E6806-E6812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 596] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 94. | Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265-7279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2302] [Cited by in RCA: 2522] [Article Influence: 114.6] [Reference Citation Analysis (1)] |
| 95. | Guo J, Xu B, Han Q, Zhou H, Xia Y, Gong C, Dai X, Li Z, Wu G. Ferroptosis: A Novel Anti-tumor Action for Cisplatin. Cancer Res Treat. 2018;50:445-460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 554] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 96. | Chen P, Li X, Zhang R, Liu S, Xiang Y, Zhang M, Chen X, Pan T, Yan L, Feng J, Duan T, Wang D, Chen B, Jin T, Wang W, Chen L, Huang X, Zhang W, Sun Y, Li G, Kong L, Li Y, Yang Z, Zhang Q, Zhuo L, Sui X, Xie T. Combinative treatment of β-elemene and cetuximab is sensitive to KRAS mutant colorectal cancer cells by inducing ferroptosis and inhibiting epithelial-mesenchymal transformation. Theranostics. 2020;10:5107-5119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 284] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 97. | Lorenzato A, Magrì A, Matafora V, Audrito V, Arcella P, Lazzari L, Montone M, Lamba S, Deaglio S, Siena S, Bertotti A, Trusolino L, Bachi A, Di Nicolantonio F, Bardelli A, Arena S. Vitamin C Restricts the Emergence of Acquired Resistance to EGFR-Targeted Therapies in Colorectal Cancer. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 98. | Pan X, Qi Y, Du Z, He J, Yao S, Lu W, Ding K, Zhou M. Zinc oxide nanosphere for hydrogen sulfide scavenging and ferroptosis of colorectal cancer. J Nanobiotechnology. 2021;19:392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 99. | Li Y, Chen W, Qi Y, Wang S, Li L, Li W, Xie T, Zhu H, Tang Z, Zhou M. H(2) S-Scavenged and Activated Iron Oxide-Hydroxide Nanospindles for MRI-Guided Photothermal Therapy and Ferroptosis in Colon Cancer. Small. 2020;16:e2001356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 100. | Chen X, Yu C, Kang R, Kroemer G, Tang D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021;28:1135-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 392] [Article Influence: 98.0] [Reference Citation Analysis (0)] |
| 101. | Arosio P, Elia L, Poli M. Ferritin, cellular iron storage and regulation. IUBMB Life. 2017;69:414-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 165] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 102. | Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 728] [Cited by in RCA: 1445] [Article Influence: 131.4] [Reference Citation Analysis (0)] |
| 103. | Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, Cornella-Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF, Helliwell SB, D'Aquin S, Bastien J, Wang H, Wiederschain D, Kuerth J, Bergman P, Schwalb D, Thomas J, Ugwonali S, Harbinski F, Tallarico J, Wilson CJ, Myer VE, Porter JA, Bussiere DE, Finan PM, Labow MA, Mao X, Hamann LG, Manning BD, Valdez RA, Nicholson T, Schirle M, Knapp MS, Keaney EP, Murphy LO. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 587] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 104. | Hou W, Xie Y, Song X, Sun X, Lotze MT, Zeh HJ 3rd, Kang R, Tang D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1171] [Cited by in RCA: 1658] [Article Influence: 184.2] [Reference Citation Analysis (0)] |
| 105. | Gryzik M, Asperti M, Denardo A, Arosio P, Poli M. NCOA4-mediated ferritinophagy promotes ferroptosis induced by erastin, but not by RSL3 in HeLa cells. Biochim Biophys Acta Mol Cell Res. 2021;1868:118913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 108] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 106. | Fang Y, Chen X, Tan Q, Zhou H, Xu J, Gu Q. Inhibiting Ferroptosis through Disrupting the NCOA4-FTH1 Interaction: A New Mechanism of Action. ACS Cent Sci. 2021;7:980-989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 313] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 107. | Gu C, Chang W, Wu J, Yao Y, Liu G, Yuan Y, Quan W, Sun Z, Shang A, Li D. NCOA4: An Immunomodulation-Related Prognostic Biomarker in Colon Adenocarcinoma and Pan-Cancer. J Oncol. 2022;2022:5242437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 108. | Hu Q, Wei W, Wu D, Huang F, Li M, Li W, Yin J, Peng Y, Lu Y, Zhao Q, Liu L. Blockade of GCH1/BH4 Axis Activates Ferritinophagy to Mitigate the Resistance of Colorectal Cancer to Erastin-Induced Ferroptosis. Front Cell Dev Biol. 2022;10:810327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 126] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 109. | Hasan M, Reddy SM, Das NK. Ferritinophagy is not required for colon cancer cell growth. Cell Biol Int. 2020;44:2307-2314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 110. | Li B, Wei S, Yang L, Peng X, Ma Y, Wu B, Fan Q, Yang S, Li X, Jin H, Tang S, Huang M, Li H, Liu J. CISD2 Promotes Resistance to Sorafenib-Induced Ferroptosis by Regulating Autophagy in Hepatocellular Carcinoma. Front Oncol. 2021;11:657723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 111. | Li J, Yuan J, Li Y, Wang J, Xie Q, Ma R, Ren M, Lu D, Xu Z. d-Borneol enhances cisplatin sensitivity via autophagy dependent EMT signaling and NCOA4-mediated ferritinophagy. Phytomedicine. 2022;106:154411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 112. | Zhang Z, Yao Z, Wang L, Ding H, Shao J, Chen A, Zhang F, Zheng S. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14:2083-2103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 362] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 113. | Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, Xie Y, Klionsky DJ, Kroemer G, Lotze MT, Zeh HJ, Kang R, Tang D. AMPK-Mediated BECN1 Phosphorylation Promotes Ferroptosis by Directly Blocking System X(c)(-) Activity. Curr Biol. 2018;28:2388-2399.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 564] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 114. | Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun (Lond). 2018;38:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 567] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 115. | Baird L, Swift S, Llères D, Dinkova-Kostova AT. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol Adv. 2014;32:1133-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 116. | Lu MC, Ji JA, Jiang ZY, You QD. The Keap1-Nrf2-ARE Pathway As a Potential Preventive and Therapeutic Target: An Update. Med Res Rev. 2016;36:924-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 595] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 117. | Yamamoto M, Kensler TW, Motohashi H. The KEAP1-NRF2 System: a Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol Rev. 2018;98:1169-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 696] [Cited by in RCA: 1317] [Article Influence: 188.1] [Reference Citation Analysis (1)] |
| 118. | Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1546] [Cited by in RCA: 1914] [Article Influence: 127.6] [Reference Citation Analysis (0)] |
| 119. | Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561-13566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 397] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 120. | Jiang T, Harder B, Rojo de la Vega M, Wong PK, Chapman E, Zhang DD. p62 Links autophagy and Nrf2 signaling. Free Radic Biol Med. 2015;88:199-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 500] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 121. | Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 1492] [Article Influence: 165.8] [Reference Citation Analysis (0)] |
| 122. | Chen J, Zhang J, Chen T, Bao S, Li J, Wei H, Hu X, Liang Y, Liu F, Yan S. Xiaojianzhong decoction attenuates gastric mucosal injury by activating the p62/Keap1/Nrf2 signaling pathway to inhibit ferroptosis. Biomed Pharmacother. 2022;155:113631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 123. | Wu Y, Pi D, Chen Y, Zuo Q, Zhou S, Ouyang M. Ginsenoside Rh4 Inhibits Colorectal Cancer Cell Proliferation by Inducing Ferroptosis via Autophagy Activation. Evid Based Complement Alternat Med. 2022;2022:6177553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 124. | Celesia A, Morana O, Fiore T, Pellerito C, D'Anneo A, Lauricella M, Carlisi D, De Blasio A, Calvaruso G, Giuliano M, Emanuele S. ROS-Dependent ER Stress and Autophagy Mediate the Anti-Tumor Effects of Tributyltin (IV) Ferulate in Colon Cancer Cells. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 125. | Jarc E, Petan T. Lipid Droplets and the Management of Cellular Stress. Yale J Biol Med. 2019;92:435-452. [PubMed] |
| 126. | Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, McNiven MA. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61:1896-1907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 244] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 127. | Bai Y, Meng L, Han L, Jia Y, Zhao Y, Gao H, Kang R, Wang X, Tang D, Dai E. Lipid storage and lipophagy regulates ferroptosis. Biochem Biophys Res Commun. 2019;508:997-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 341] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 128. | Jin L, Zhu LY, Pan YL, Fu HQ, Zhang J. Prothymosin α promotes colorectal carcinoma chemoresistance through inducing lipid droplet accumulation. Mitochondrion. 2021;59:123-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 129. | Bejarano E, Cuervo AM. Chaperone-mediated autophagy. Proc Am Thorac Soc. 2010;7:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 130] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 130. | Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 944] [Article Influence: 157.3] [Reference Citation Analysis (0)] |
| 131. | Wu Z, Geng Y, Lu X, Shi Y, Wu G, Zhang M, Shan B, Pan H, Yuan J. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci U S A. 2019;116:2996-3005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 435] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 132. | Yu S, Li Z, Zhang Q, Wang R, Zhao Z, Ding W, Wang F, Sun C, Tang J, Wang X, Zhang H, Huang R, Wu Q, Jiang J, Zhao X. GPX4 degradation via chaperone-mediated autophagy contributes to antimony-triggered neuronal ferroptosis. Ecotoxicol Environ Saf. 2022;234:113413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 133. | Liu C, Sun W, Zhu T, Shi S, Zhang J, Wang J, Gao F, Ou Q, Jin C, Li J, Xu JY, Tian H, Xu GT, Lu L. Glia maturation factor-β induces ferroptosis by impairing chaperone-mediated autophagic degradation of ACSL4 in early diabetic retinopathy. Redox Biol. 2022;52:102292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 94] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 134. | Zhang S, Hu B, You Y, Yang Z, Liu L, Tang H, Bao W, Guan Y, Shen X. Sorting nexin 10 acts as a tumor suppressor in tumorigenesis and progression of colorectal cancer through regulating chaperone mediated autophagy degradation of p21(Cip1/WAF1). Cancer Lett. 2018;419:116-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 135. | Chen T, Yuan Z, Lei Z, Duan J, Xue J, Lu T, Yan G, Zhang L, Liu Y, Li Q, Zhang Y. Hippocalcin-Like 1 blunts liver lipid metabolism to suppress tumorigenesis via directly targeting RUVBL1-mTOR signaling. Theranostics. 2022;12:7450-7464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 136. | Chen X, Song X, Li J, Zhang R, Yu C, Zhou Z, Liu J, Liao S, Klionsky DJ, Kroemer G, Tang D, Kang R. Identification of HPCAL1 as a specific autophagy receptor involved in ferroptosis. Autophagy. 2023;19:54-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 74] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 137. | Chen F, Cai X, Kang R, Liu J, Tang D. Autophagy-Dependent Ferroptosis in Cancer. Antioxid Redox Signal. 2023;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 54] [Reference Citation Analysis (0)] |
| 138. | Shan X, Li S, Sun B, Chen Q, Sun J, He Z, Luo C. Ferroptosis-driven nanotherapeutics for cancer treatment. J Control Release. 2020;319:322-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 146] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 139. | Zhou D, Yao Y, Zong L, Zhou G, Feng M, Chen J, Liu G, Chen G, Sun K, Yao H, Liu Y, Shi X, Zhang W, Shi B, Tai Q, Wu G, Sun L, Hu W, Zhu X, He S. TBK1 Facilitates GLUT1-Dependent Glucose Consumption by suppressing mTORC1 Signaling in Colorectal Cancer Progression. Int J Biol Sci. 2022;18:3374-3389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 140. | Jiffry J, Thavornwatanayong T, Rao D, Fogel EJ, Saytoo D, Nahata R, Guzik H, Chaudhary I, Augustine T, Goel S, Maitra R. Oncolytic Reovirus (pelareorep) Induces Autophagy in KRAS-mutated Colorectal Cancer. Clin Cancer Res. 2021;27:865-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 141. | Chen Y, Chen Y, Zhang J, Cao P, Su W, Deng Y, Zhan N, Fu X, Huang Y, Dong W. Fusobacterium nucleatum Promotes Metastasis in Colorectal Cancer by Activating Autophagy Signaling via the Upregulation of CARD3 Expression. Theranostics. 2020;10:323-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 142. | Chen XL, Liu P, Zhu WL, Lou LG. DCZ5248, a novel dual inhibitor of Hsp90 and autophagy, exerts antitumor activity against colon cancer. Acta Pharmacol Sin. 2021;42:132-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 143. | Kim J, Choi S, Kim JO, Kim KK. Autophagy-mediated upregulation of cytoplasmic claudin 1 stimulates the degradation of SQSTM1/p62 under starvation. Biochem Biophys Res Commun. 2018;496:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 144. | Xia Y, Liu S, Li C, Ai Z, Shen W, Ren W, Yang X. Discovery of a novel ferroptosis inducer-talaroconvolutin A-killing colorectal cancer cells in vitro and in vivo. Cell Death Dis. 2020;11:988. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 145. | Liu MY, Li HM, Wang XY, Xia R, Li X, Ma YJ, Wang M, Zhang HS. TIGAR drives colorectal cancer ferroptosis resistance through ROS/AMPK/SCD1 pathway. Free Radic Biol Med. 2022;182:219-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 74] [Article Influence: 24.7] [Reference Citation Analysis (0)] |