Published online Dec 15, 2023. doi: 10.4251/wjgo.v15.i12.2169
Peer-review started: July 29, 2023
First decision: September 23, 2023
Revised: October 1, 2023
Accepted: October 30, 2023
Article in press: October 30, 2023
Published online: December 15, 2023
Processing time: 138 Days and 0.7 Hours
Gastroesophageal reflux disease (GERD) affects approximately 13% of the global population. However, the pathogenesis of GERD has not been fully elucidated. The development of metabolomics as a branch of systems biology in recent years has opened up new avenues for the investigation of disease processes. As a po
To analyze of the relationship between 486 blood metabolites and GERD.
Two-sample MR analysis was used to assess the causal relationship between blood metabolites and GERD. A genome-wide association study (GWAS) of 486 metabolites was the exposure, and two different GWAS datasets of GERD were used as endpoints for the base analysis and replication and meta-analysis. Bon
In MR analysis, four blood metabolites are negatively correlated with GERD: Levulinate (4-oxovalerate), stearate (18:0), adrenate (22:4n6) and p-acetamidophenylglucuronide. However, we also found a positive correlation between four blood metabolites and GERD: Kynurenine, 1-linoleoylglycerophosphoethanolamine, butyrylcarnitine and guanosine. And bonferroni correction showed that butyrylcarnitine (odd ratio 1.10, 95% confidence interval: 1.05-1.16, P = 7.71 × 10-5) was the most reliable causal metabolite. In addition, one significant pathways, the “glycerophospholipid metabolism” pathway, can be involved in the pathogenesis of GERD.
Our study found through the integration of genomics and metabolomics that butyrylcarnitine may be a potential biomarker for GERD, which will help further elucidate the pathogenesis of GERD and better guide its treatment. At the same time, this also contributes to early screening and prevention of GERD. However, the results of this study require further confirmation from both basic and clinical real-world studies.
Core Tip: At present, there is no study on blood metabolomics of gastroesophageal reflux disease (GERD). This may be the first study combining metabolomics and genomics to explore the causal relationship between serum metabolites and GERD. We found that there was a significant correlation between eight metabolites and GERD, among which butyrylcarnitine was the most reliable pathogenic metabolite (odd ratio 1.10, 95% confidence interval: 1.05-1.16). Glycerophospholipid metabolism may be involved in the pathogenesis of GERD. The results provide a reference direction for the early screening, prevention and treatment of GERD and the design of future clinical research.
- Citation: Hu JY, Lv M, Zhang KL, Qiao XY, Wang YX, Wang FY. Evaluating the causal relationship between human blood metabolites and gastroesophageal reflux disease. World J Gastrointest Oncol 2023; 15(12): 2169-2184
- URL: https://www.wjgnet.com/1948-5204/full/v15/i12/2169.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i12.2169
Gastroesophageal reflux disease (GERD) refers to a disease in which gastric contents reflux into the esophagus, causing corresponding esophageal symptoms and/or complications. This condition affects approximately 13% of the global population[1]. GERD is not life-threatening, but it impairs patients' quality of life and increases the risk of other esophageal complications such as esophagitis, Barrett's esophagus (BE), and esophageal adenocarcinoma[2]. Previous epidemiological studies have identified several possible risk factors for GERD, including smoking, alcohol consumption and diabetes[3-5], which have played a role in the prevention of GERD. However, there are no studies on the blood metabolomics of GERD.
The development of metabolomics as a branch of systems biology in recent years has opened up new avenues for the investigation of disease processes. By identifying altered metabolites or metabolic pathways, metabolomics can spe
As a powerful statistical tool, Mendelian randomization (MR) is widely used to explore the causal relationship between exposure and outcome[13]. In particular, MR was able to circumvent the drawbacks of randomized controlled expe
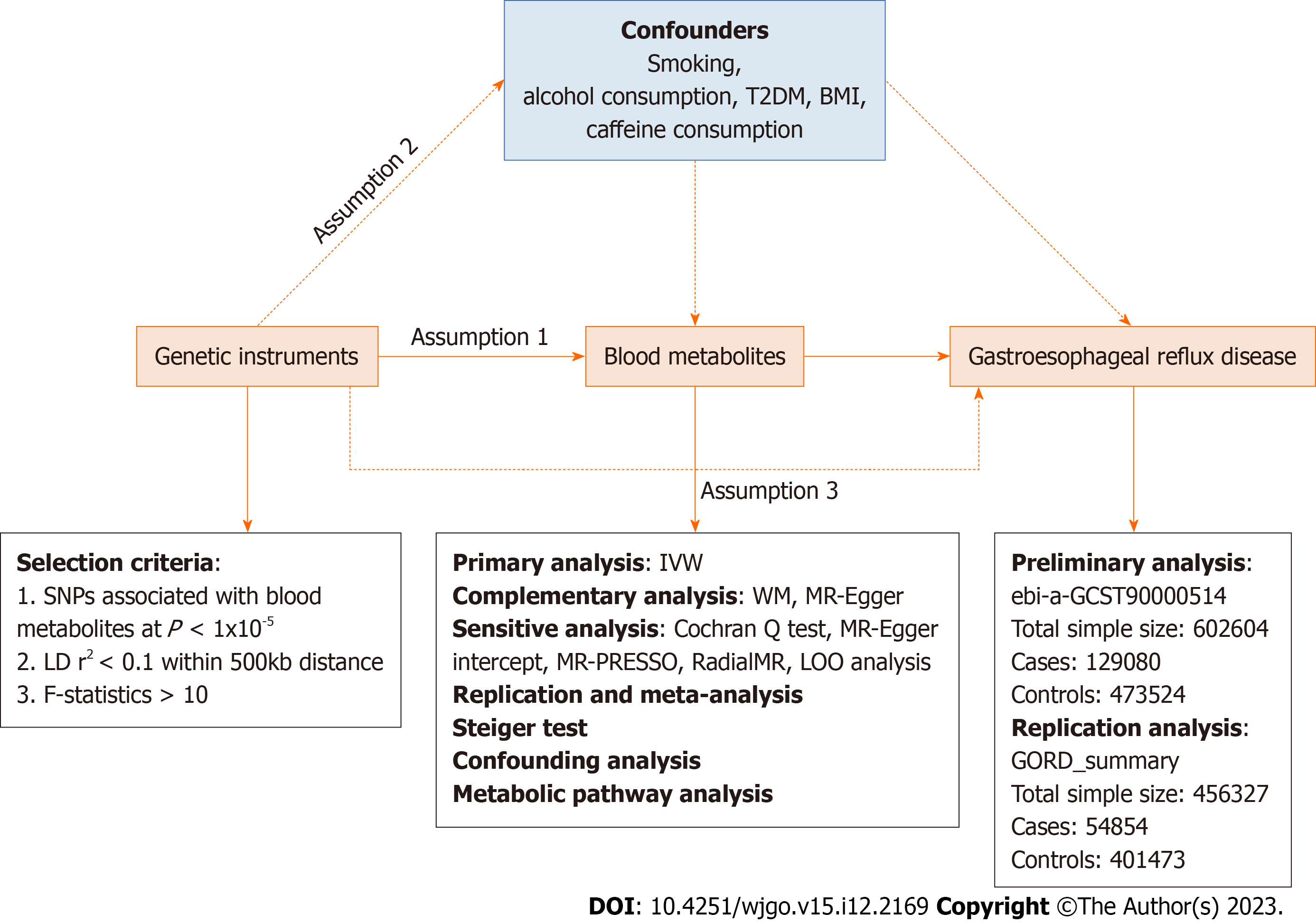
An effective MR study should follow three assumptions: (1) IVs are closely related to exposure factors; (2) IVs are not related to confounding factors; and (3) IVs are not related to outcomes and affect outcome only via exposures[16]. Two independent GWAS alliances give the genetic information of GERD, subjected to preliminary and replication analyses, followed by meta-analysis .The overview of the study is showed in Figure 1.
Genetic data for blood metabolites were obtained from the Metabolomics GWAS server (https://metabolomics.helmholtz-muenchen.de/gwas/). Shin et al[17] identified nearly 2.1 million SNPs of 486 metabolites related to human genetic variation through genome-wide association scanning and high-throughput metabolic analysis. Of the 486 metabolites, 107 are defined as unknown because their chemical properties are still unclear. Another 309 metabolites were chemically identified and assigned to eight broad metabolomes, including amino acid, carbohydrate, cofactors and vitamin, energy, lipid, nucleotide, peptide, and xenobiotic metabolism. The detailed names of 486 metabolites are shown in Supplementary Table 1, among which the chemical properties of the metabolites named X - are unknown.
Download GERD's GWAS summary data from IEU (https://gwas.mrcieu.ac.uk/). The GWAS directory login number is ebi-a-GCST9000514. Specifically, GWAS data containing 2320781 SNPs were obtained from a previous GERD-related GWAS study conducted by Ong et al[18] and colleagues with a total sample size of 602604 Europeans containing 129080 cases and 473524 controls. The above GWAS data were used for the preliminary analysis of GERD. To validate our results by conducting replication analysis and meta-analysis, we repeated the MR analysis using the GERD data (54854 GERD patients and 401473 healthy controls) published by Wu et al[19]. This data is publicly available on the website: https://cnsgenomics.com/content/data.
We performed a series of steps to select eligible genetic variants associated with metabolites. Given the small number of metabolite-related SNPs, we relaxed the significance threshold of P < 1 × 10-5 to select metabolite-related SNPs. We then clumped SNPs by removing linkage disequilibrium (R2 > 0.1 and within 500kb). This criterion has been widely applied in previous studies[20-22]. To satisfy hypothesis (3), we removed the SNPs associated with the results in the IVs (P < 1 × 10-5). We eliminate bias caused by weak IVs by calculating the R2 and F statistics for each SNP to measure statistical strength. SNPs with F < 10 were defined as weak genetic variants and were deleted. We further coordinated the SNPs of exposure and outcome, and removed the SNPs with palindromic effects and allele discordance (e.g. A/G vs A/C). Then, the final results were subjected to MR analysis.
The causal relationship between blood metabolites and GERD was mainly assessed based on the results of random-effect inverse variance weighted (IVW). IVW is based on the hypothesis that there is no horizontal pleiotropy for all SNPs and the results from the pooled analysis of Walden ratios for all genetic variants, under the premise that IVW provides the most accurate assessment of causal effects[23]. Therefore, we used IVW-based estimates to initially screen for blood metabolites that have a causal effect on GERD. To obtain more reliable results, we used two additional methods to further evaluate metabolites with significant estimates (IVW derived P < 0.05). The MR-Egger and weighted median (WM) methods are used as complementary analyses. These two methods can provide more robust estimates under the relaxed. WM assumes that at least half of the tools are valid[24] and MR-Egger provides horizontal pleiotropy and heterogeneity detection in the presence of horizontal pleiotropy for all SNPs[25]. When consistent with the InSide hypothesis (IVs intensity independent of direct effects), MR-Egger regression can provide unbiased estimates[26].
For the initially determined significant estimates (IVW P < 0.05), sensitivity analysis will be performed to assess any deviation from the MR hypothesis. Horizontal pleiotropy was observed when IVs affected the results through other pathways than exposure. Horizontal pleiotropy was assessed based on the Egger intercept. Cochran Q test was used to test for the presence of heterogeneity,. Heterogeneity was considered to exist when P < 0.05, I2 > 25%[27]. For data with significant associations, Radial MR was used to identify heterogeneous values, and MR analysis was repeated after eliminating heterogeneous SNPs to obtain more accurate results[28]. Finally, we used MR-PRESSO to check again for the presence of heterogeneous SNPs[29]. We used leave-one-out (LOO) analysis to ensure the robustness of the results. By discarding each SNP in turn and then performing MR analysis to assess whether the results are heavily influenced by a single SNP.
In conclusion, we rigorously screened blood metabolites with potential causal relationship with GERD by multiple criteria: (1) Significant P-values for preliminary analysis (IVW derived P < 0.05); (2) The direction and amplitude of the three MR methods were consistent; (3) There was no heterogeneity or level pleiotropy in Mr results; and (4) MR estimates are not significantly confounded by individual SNPs.
To fully assess the robustness of candidate metabolites identified based on the above criteria, we repeated the IVW analysis in another GERD cohort. In brief, the data from IEU with code ebi-a-GCST90000514 was used for the preliminary analysis and the data with the title GORD_summary was used for the replication analysis. Meta-analyses were based on a random-effects IVW model and performed on Review Manager 5.4 software.
We verified whether the observed causality was biased by reversal of causality using the Steiger test[30]. Using the Steiger test, we determined whether the included SNPs explained the variability of GERD better than the detected metabolites. When the combination of SNPs was found to contribute more to the genetic risk of GERD than metabolites (Steiger P > 0.05), it indicated that the direction of causal inference may be biased.
Although we evaluated the horizontal pleiotropy of Mr results through a series of sensitivity analyses to detect any SNPs that violated the MR hypothesis, there may also be a small number of residual confounding SNPs. Therefore, we examined the IVs of metabolites on the Phenoscanner V2 website (http://www.phenoscanner.medschl.cam.ac.uk/) to assess whether each SNP was associated with known risk factors for GERD, such as smoking, alcohol consumption, type 2 diabetes, and body mass index (BMI). If any SNP was observed to be associated with the above confounding factors (P < 1 × 10−5), then MR analysis was repeated after removing these SNPs to verify the reliability of the results.
To clarify the biological mechanisms underlying the effects of blood metabolites on GERD, we further performed metabolic pathway analysis using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/)[29] to explore the potential pathogenesis of GERD.
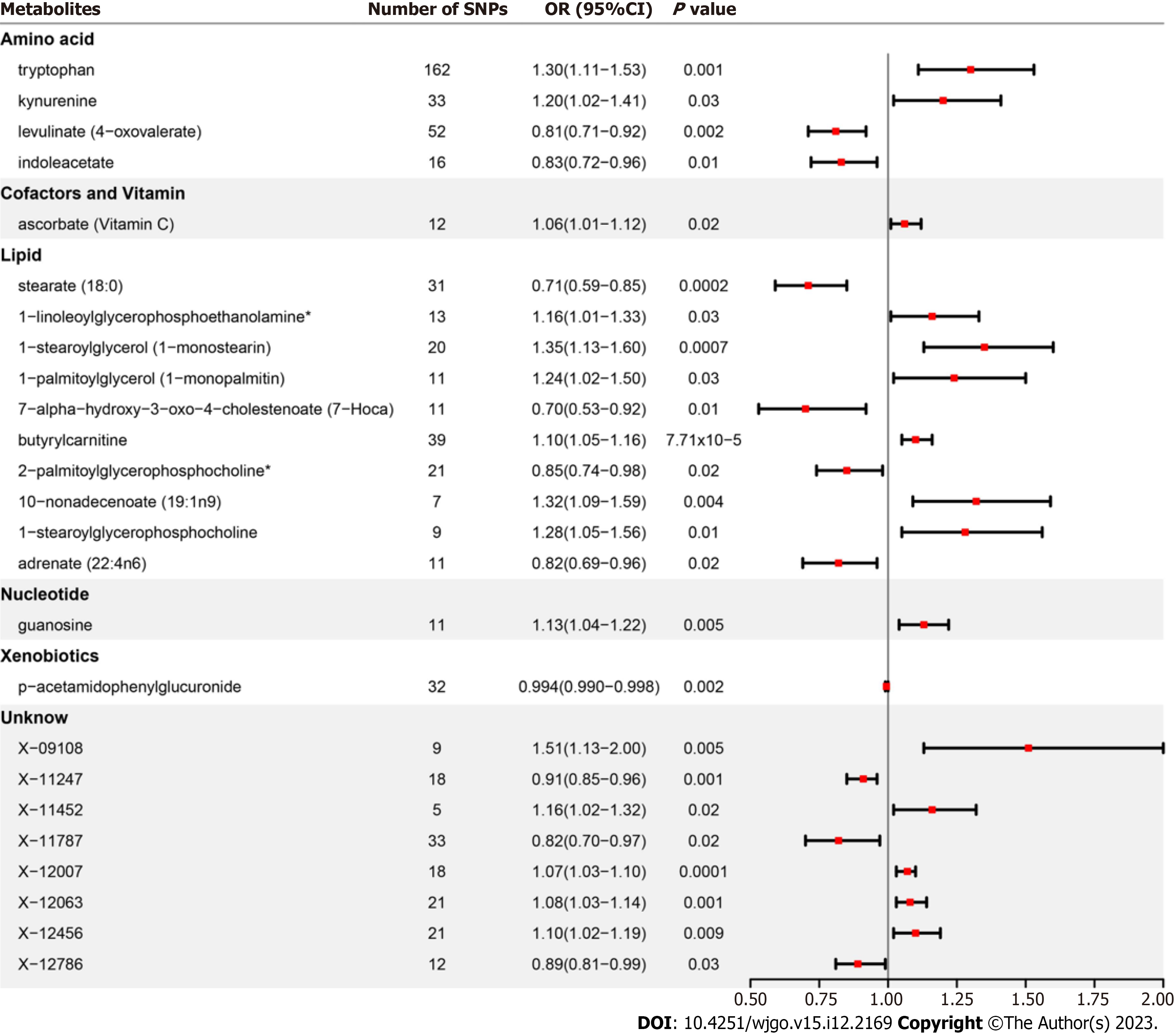
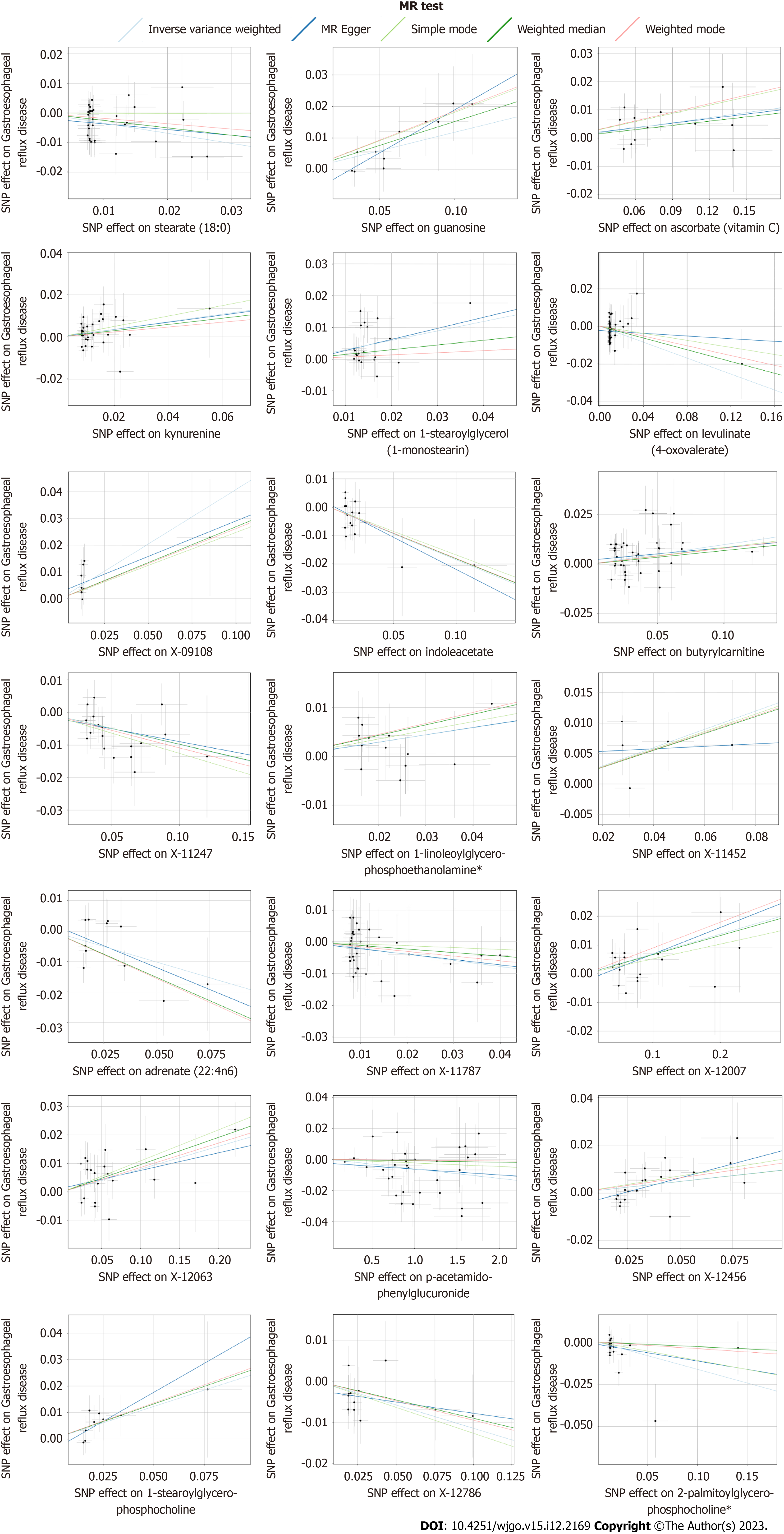
After strict control of the quality of IVs, SNPs of 25 metabolites were obtained. Filtered IVs contain 5 to 162 SNPs (X-11452 consists of 5 SNPs, while tryptophan sulfate consists of 162 SNPs). All metabolite-related SNPs had F-statistics greater than 10, which shows the strong power of the IVs. Supplementary Table 2 displays the specific IV data. Prior to MR analysis, radial MR locates and eliminates all outliers (Supplementary Table 3). IVW analysis initially identified 25 metabolites with a potential causal relationship with GERD, including 17 metabolites with known chemical identity and 8 metabolites with unknown chemical identity. These 17 known metabolites include amino acids, cofactors, vitamins, lipids, nucleotides, and xenobiotic metabolism factors (Figure 2). Among the 17 known metabolic traits, butyrylcarnitine was significantly associated with GERD after Bonferroni correction. Twenty-one metabolites that passed the strict screening requirements were used for the follow-up analysis by sensitivity analysis (Figure 3). In short, the MR estimates derived from WM and MR–Egger regression presented consistent directions and amplitudes, supporting the robustness of causality (Table 1). P values and I2 associated with Cochran Q indicated that no heterogeneity was found. In addition, the MR-Egger intercept term indicated a low risk of horizontal pleiotropy (Table 1). The LOO analysis did not find any high-impact SNPs biasing the pooled effect estimates (Supplementary Figure 1), and 21 metabolites that met the above criteria were included in the next study.
| Metabolites | n | MR analysis | Heterogeneity | Pleiotropy | ||||
| Method | OR (95%CI) | P value | Q | P value | Intercept | P value | ||
| Amino acid | ||||||||
| Tryptophan | 162 | ME | 0.72 (0.37-1.40) | 0.34 | 160 | 0.58 | 0.002 | 0.07 |
| WM | 1.42 (1.11-1.82) | 0.006 | ||||||
| Kynurenine | 33 | ME | 1.19 (0.78-1.81) | 0.44 | 31 | 0.88 | 0.002 | 0.96 |
| WM | 1.16 (0.90-1.48) | 0.24 | ||||||
| Levulinate (4-oxovalerate) | 52 | ME | 0.96 (0.73-1.27) | 0.80 | 50 | 0.94 | 0.002 | 0.15 |
| WM | 0.86 (0.67-1.09) | 0.20 | ||||||
| Indoleacetate | 16 | ME | 0.79 (0.60-1.05) | 0.13 | 14 | 0.74 | 0.003 | 0.72 |
| WM | 0.83 (0.67-1.04) | 0.11 | ||||||
| Cofactors and Vitamin | ||||||||
| Ascorbate (Vitamin C) | 12 | ME | 1.06 (0.89-1.25) | 0.54 | 10 | 0.48 | 0.006 | 0.94 |
| WM | 1.05 (0.98-1.13) | 0.17 | ||||||
| Lipid | ||||||||
| Stearate (18:0) | 31 | ME | 0.82 (0.51-1.32) | 0.43 | 29 | 0.49 | 0.003 | 0.51 |
| WM | 0.78 (0.60-1.01) | 0.06 | ||||||
| 1-linoleoylglycerophosphoethanolamine | 13 | ME | 1.16 (0.80-1.68) | 0.44 | 11 | 0.85 | 0.004 | 0.99 |
| WM | 1.24 (1.03-1.50) | 0.03 | ||||||
| 1-stearoylglycerol (1-monostearin) | 20 | ME | 1.42 (0.62-3.25) | 0.42 | 18 | 0.49 | 0.006 | 0.90 |
| WM | 1.16 (0.90-1.50) | 0.25 | ||||||
| 1-palmitoylglycerol (1-monopalmitin) | 11 | ME | 0.88 (0.48-1.60) | 0.68 | 9 | 0.89 | 0.005 | 0.27 |
| WM | 1.09 (0.83-1.43) | 0.54 | ||||||
| 7-alpha-hydroxy-3-oxo-4-cholestenoate (7-Hoca) | 11 | ME | 1.12 (0.52-2.41) | 0.78 | 9 | 0.36 | 0.006 | 0.23 |
| WM | 0.70 (0.48-1.01) | 0.06 | ||||||
| Butyrylcarnitine | 39 | ME | 1.06 (0.99-1.15) | 0.11 | 37 | 0.30 | 0.002 | 0.26 |
| WM | 1.07 (1.00-1.14) | 0.04 | ||||||
| 2-palmitoylglycerophosphocholine* | 21 | ME | 0.91 (0.72-1.14) | 0.41 | 19 | 0.76 | 0.002 | 0.50 |
| WM | 0.97 (0.79-1.20) | 0.80 | ||||||
| 10-nonadecenoate (19:1n9) | 7 | ME | 0.95 (0.60-1.50) | 0.83 | 5 | 0.52 | 0.006 | 0.19 |
| WM | 1.27 (1.00-1.61) | 0.05 | ||||||
| 1-stearoylglycerophosphocholine | 9 | ME | 1.55 (0.85-2.83) | 0.20 | 7 | 0.87 | 0.006 | 0.53 |
| WM | 1.30 (1.01-1.67) | 0.04 | ||||||
| Adrenate (22:4n6) | 11 | ME | 0.75 (0.50-1.12) | 0.19 | 9 | 0.20 | 0.005 | 0.66 |
| WM | 0.74 (0.60-0.91) | 0.004 | ||||||
| Nucleotide | ||||||||
| Guanosine | 11 | ME | 1.31 (1.06-1.64) | 0.04 | 9 | 0.99 | 0.005 | 0.16 |
| WM | 1.16 (1.05-1.29) | 0.005 | ||||||
| Xenobiotics | ||||||||
| p-acetamidophenylglucuronide | 32 | ME | 1.00 (0.99-1.00) | 0.41 | 30 | 0.25 | 0.004 | 0.60 |
| WM | 1.00 (0.99-1.00) | 0.75 | ||||||
| Unknow | ||||||||
| X-09108 | 9 | ME | 1.31 (0.72-2.37) | 0.41 | 7 | 0.71 | 0.005 | 0.61 |
| WM | 1.31 (0.89-1.93) | 0.17 | ||||||
| X-11247 | 18 | ME | 0.92 (0.78-1.10) | 0.37 | 16 | 0.68 | 0.004 | 0.82 |
| WM | 0.91 (0.83-0.99) | 0.03 | ||||||
| X-11452 | 5 | ME | 1.02 (0.67-1.56) | 0.93 | 3 | 0.45 | 0.008 | 0.58 |
| WM | 1.15 (0.97-1.36) | 0.10 | ||||||
| X-11787 | 33 | ME | 0.84 (0.64-1.12) | 0.25 | 31 | 0.94 | 0.002 | 0.84 |
| WM | 0.89 (0.72-1.11) | 0.31 | ||||||
| X-12007 | 18 | ME | 1.10 (1.03-1.17) | 0.01 | 16 | 0.68 | 0.003 | 0.39 |
| WM | 1.07 (1.01-1.13) | 0.02 | ||||||
| X-12063 | 21 | ME | 1.06 (0.98-1.16) | 0.16 | ||||
| WM | 1.10 (1.03-1.17) | 0.004 | 19 | 0.30 | 0.002 | 0.60 | ||
| X-12456 | 21 | ME | 1.26 (1.07-1.49) | 0.01 | 19 | 0.86 | 0.003 | 0.09 |
| WM | 1.10 (0.99-1.22) | 0.07 | ||||||
| X-12786 | 12 | ME | 0.95 (0.78-1.14) | 0.58 | 10 | 0.87 | 0.003 | 0.48 |
| WM | 0.91 (0.80-1.05) | 0.20 | ||||||
The meta-analysis further identified 14 metabolites (8 known and 6 unknown) that could affect GERD (Figure 4). In detail, levulinate (4-oxovalerate) [odd ratio (OR) 0.80, 95% confidence interval (CI): 0.72-0.89, P < 0.0001], stearate (18:0) (OR 0.77, 95%CI: 0.64-0.92, P = 0.004), adrenate (22:4n6) (OR 0.83, 95%CI: 0.74-0.94, P = 0.004), p-acetamidophenylglucuronide (OR 0.99, 95%CI: 0.99-1.00, P = 0.0002), X-11247 (OR 0.92, 95%CI: 0.88-0.96, P = 0.0006), X-12786 (OR 0.90, 95%CI: 0.82-0.98, P = 0.01) decreased risk of GERD, while kynurenine (OR 1.20, 95%CI: 1.07-1.35, P = 0.002), 1-linoleoylglycerophosphoethanolamine (OR 1.17, 95%CI: 1.05-1.31, P = 0.004), butyrylcarnitine (OR 1.09, 95%CI: 1.05-1.13, P < 0.0001), guanosine (OR 1.10, 95%CI: 1.03-1.18, P = 0.003), X-09108 (OR 1.40, 95%CI: 1.13-1.74, P = 0.002), X-11452 (OR 1.14, 95%CI: 1.02-1.29, P = 0.03), X-12063 (OR 1.07, 95%CI: 1.03-1.11, P = 0.0008), and X-12456 (OR 1.12, 95%CI: 1.06-1.19, P = 0.0001) increased susceptibility to GERD.
We further investigated genetic variants affecting metabolite levels and GERD. The 39 SNPs of IVs for butyrylcarnitine are shown in Table 2. Among them, rs4767937 showed a strong correlation with butyrylcarnitine (β = 0.1052; SE = 0.0035, P = 1.00 × 10-200). Notably, it had the strongest association with GERD (β = 0.0142; SE = 0.0048, P = 0.0032). The effect of this SNP on butyrylcarnitine and GERD suggests that the relevant genetic loci may provide valuable information for the biological mechanism of GERD, and butyrylcarnitine may be an important functional mediator of the biological processes affecting GERD.
| SNP | Gene | CHR | A1 | A2 | Butyrylcarnitine | GERD | ||||
| Beta | SE | P value | Beta | SE | P value | |||||
| rs10849832 | OASL | 12 | C | T | 0.0499 | 0.0067 | 1.06E-13 | 0.0106 | 0.0083 | 0.2040 |
| rs10849846 | P2RX7 | 12 | C | T | 0.0511 | 0.0097 | 1.34E-07 | 0.0079 | 0.0115 | 0.4916 |
| rs11065202 | CABP1 | 12 | C | T | -0.1297 | 0.004 | 1.00E-200 | -0.0087 | 0.0049 | 0.0751 |
| rs11065208 | MLEC | 12 | A | G | -0.0683 | 0.0106 | 1.17E-10 | -0.0075 | 0.0134 | 0.5745 |
| rs11065270 | SPPL3 | 12 | C | T | 0.0603 | 0.0069 | 1.65E-18 | 0.0198 | 0.0091 | 0.0298 |
| rs1109732 | RP11-210L7.1 | 12 | A | G | -0.0606 | 0.014 | 1.47E-05 | -0.0004 | 0.0187 | 0.9848 |
| rs1171617 | SLC16A9 | 10 | T | G | 0.0358 | 0.0047 | 1.61E-14 | 0.0049 | 0.0057 | 0.3849 |
| rs1186055 | P2RX7 | 12 | C | A | -0.0344 | 0.0043 | 1.30E-15 | -0.0042 | 0.0054 | 0.4358 |
| rs12025912 | KIF17 | 1 | C | T | 0.0228 | 0.0053 | 1.76E-05 | -0.0005 | 0.0069 | 0.9418 |
| rs12255141 | VTI1A | 10 | A | G | -0.0272 | 0.0062 | 1.15E-05 | -0.0093 | 0.0081 | 0.2478 |
| rs12257526 | GRID1 | 10 | C | T | 0.0248 | 0.0058 | 1.74E-05 | 0.0105 | 0.0074 | 0.1580 |
| rs12368199 | OASL | 12 | A | G | 0.1211 | 0.0052 | 1.67E-121 | 0.0062 | 0.0067 | 0.3564 |
| rs12562686 | RP11-410C4.4 | 1 | A | C | -0.036 | 0.0084 | 1.74E-05 | -0.0014 | 0.0103 | 0.8883 |
| rs1336584 | CTA-21C21.1 | 1 | C | T | -0.018 | 0.0042 | 1.91E-05 | -0.0079 | 0.0049 | 0.1064 |
| rs1469231 | NECAP1P1 | 7 | A | G | -0.0188 | 0.0043 | 1.00E-05 | -0.0098 | 0.0053 | 0.0653 |
| rs1557852 | RNA5SP192 | 5 | G | A | -0.0181 | 0.0042 | 1.53E-05 | -0.0067 | 0.0051 | 0.1904 |
| rs17050084 | AC007131.2 | 2 | C | T | -0.0413 | 0.0096 | 1.90E-05 | -0.0270 | 0.0123 | 0.0274 |
| rs17507671 | RNU4-1 | 12 | C | T | -0.0512 | 0.0096 | 1.03E-07 | 0.0036 | 0.0108 | 0.7423 |
| rs17686203 | WAC | 10 | C | T | -0.0518 | 0.0118 | 1.06E-05 | 0.0118 | 0.0142 | 0.4093 |
| rs1873745 | C8orf37-AS1 | 8 | A | G | -0.0195 | 0.0044 | 1.01E-05 | -0.0098 | 0.0059 | 0.0972 |
| rs1955919 | LINC01765 | 1 | A | G | -0.0467 | 0.0106 | 1.13E-05 | -0.0253 | 0.0123 | 0.0389 |
| rs1957910 | PRKCH | 14 | A | G | 0.0376 | 0.0088 | 1.96E-05 | -0.0046 | 0.0093 | 0.6255 |
| rs208294 | - | - | - | - | 0.0315 | 0.0036 | 1.08E-18 | 0.0058 | 0.0048 | 0.2293 |
| rs2631693 | FSIP1 | 15 | C | G | -0.0233 | 0.0053 | 1.14E-05 | -0.0085 | 0.0071 | 0.2302 |
| rs273914 | SLC22A4 | 5 | T | A | 0.0236 | 0.0041 | 1.17E-08 | 0.0016 | 0.0050 | 0.7519 |
| rs278136 | CIT | 12 | C | T | -0.0259 | 0.0043 | 1.96E-09 | 0.0040 | 0.0057 | 0.4776 |
| rs3767512 | CACNA1S | 1 | A | G | 0.0625 | 0.0146 | 1.91E-05 | 0.0252 | 0.0177 | 0.1544 |
| rs3817190 | CAMKK2 | 12 | A | T | 0.0249 | 0.0047 | 1.14E-07 | -0.0008 | 0.0049 | 0.8770 |
| rs4146382 | AC019050.1 | 2 | C | T | -0.0186 | 0.0043 | 1.25E-05 | -0.0001 | 0.0052 | 0.9912 |
| rs4766962 | COX6A1 | 12 | A | T | 0.0516 | 0.0043 | 1.70E-33 | 0.0078 | 0.0051 | 0.1238 |
| rs4767937 | SPPL3 | 12 | C | G | 0.1052 | 0.0035 | 1.00E-200 | 0.0142 | 0.0048 | 0.0032 |
| rs4870883 | FER1L6 | 8 | A | T | -0.0262 | 0.006 | 1.41E-05 | 0.0057 | 0.0053 | 0.2824 |
| rs4943508 | LINC01048 | 13 | C | T | 0.0153 | 0.0035 | 1.38E-05 | 0.0098 | 0.0048 | 0.0431 |
| rs646454 | SGO1-AS1 | 3 | T | C | 0.0257 | 0.0059 | 1.17E-05 | -0.0079 | 0.0074 | 0.2851 |
| rs6468765 | KB-1410C5.3 | 8 | C | T | 0.0156 | 0.0035 | 1.02E-05 | -0.0078 | 0.0049 | 0.1082 |
| rs6496996 | RN7SL599P | 15 | A | G | 0.0197 | 0.0045 | 1.05E-05 | 0.0035 | 0.0060 | 0.5583 |
| rs7295193 | TMEM117 | 12 | C | T | -0.0288 | 0.0067 | 1.58E-05 | 0.0115 | 0.0088 | 0.1884 |
| rs7303401 | HNF1A-AS1 | 12 | A | T | -0.0692 | 0.006 | 1.38E-30 | -0.0106 | 0.0077 | 0.1678 |
| rs7954772 | SLC38A4 | 12 | A | T | 0.0182 | 0.0042 | 1.22E-05 | -0.0009 | 0.0050 | 0.8617 |
| rs7979473 | HNF1A | 12 | G | A | 0.0184 | 0.0042 | 1.20E-05 | 0.0078 | 0.0050 | 0.1184 |
We performed the Steiger test to verify the direction of the effect from metabolites to GERD. The Steiger P value indicates that the identified causality is not biased by reverse causality. The results are shown in Supplementary Table 4.
We used Phenoscanner to examine all SNPs associated with the metabolites that were positive on initial screening (IVW < 0.05) (including glycine, N-acetylglycine, and 1-palmitoylglycerol (1 monopalmitin)) to test the validity of Hypothesis 2 (IVs are independent of confounders). The data were disregarded since the exclusion of glycine and N-acetylglycine was meaningless because rs715 was related to BMI and rs1260326 in 1-palmitoylglycerol (1-monopalmitin) was connected with alcohol use. Supplementary Table 5 displays the findings for the remaining 25 metabolites. In total, 14 SNPs were found to be associated with common GERD risk variables; however, even after eliminating these SNPs, the estimates were still significant. Two well-known metabolites, adrenate (22:4n6) and 1-linoylglycerophosphoethanolamine*, were unaffected by any confounding factors.
Unfortunately, we only found 1 metabolic pathway that may be involved in the etiology of GERD (Supplemen
In this study, we integrated two large-scale GWAS datasets to explore the causal effects of 486 blood metabolites on GERD through a rigorous MR design. Our study found 14 blood metabolites associated with GERD, among which butyrylcarnitine showed a significant positive correlation with GERD. This relationship is not affected by confounding factors such as smoking, alcohol consumption, BMI, and can be well replicated using samples from other data sources. In addition, we identified a metabolic pathway that may be involved in the biological mechanisms of GERD. This may be the first study to explore the causal relationship between serum metabolites and GERD by combining metabolomics and genomics. Given the unclear pathogenesis of GERD and the lack of blood metabolomics research related to GERD, this study is of great significance.
The onset of GERD mainly consists of two mechanisms: the invasion of reflux and the destruction of the anti-reflux barrier at the esophageal junction. Usually, the anti reflux defense mechanism of the esophagus is in balance with the erosive effect of reflux substances on the esophageal mucosa. When the person's defense mechanism decreases or the damaging effect increases, the balance is disrupted, which may lead to the occurrence of GERD[31]. At present, the exploration of the pathogenesis of GERD is still at the macro level, and multiple studies suggest that abnormal eso
A key clinical contribution of this study is the discovery of biomarkers. Our study supports a positive correlation between butyrylcarnitine and the risk of GERD from a causal perspective by combining genetics and metabolomics. Butyrylcarnitine belongs to the acyl carnitine group, which is composed of incomplete fatty acids β prooxidant compounds produced by oxidation. At present, there are no reports on the relationship between butyrylcarnitine and GERD, and we cannot accurately explain this relationship either. However, the carnitine shuttle pathway carries long-chain fatty acids from the cytoplasm to the mitochondria for later βoxidation, which necessitates acetyl-CoA and results in the esterification of L-carnitine to produce acyl carnitine derivatives[38]. The disturbance of the carnitine shuttle may lead to impaired mitochondrial function, which may reduce the ability of cells to process reactive oxygen species and increase the levels of inflammatory cytokines, leading to increased cell dysfunction and cell death[39]. This change may trigger GERD. On the other hand, weakened antioxidant capacity leads to a poor ability to prevent esophageal mucosal damage, which can also increase the severity of GERD[40,41]. Second, butyrylcarnitine is closely related to common GERD risk factors such as diabetes, obesity, anxiety and depression and other mental diseases. Studies have shown that butyrylcarnitine is involved in diet-induced insulin resistance, which in turn is related to the oxidation rate of fatty acids exceeding that of tricarboxylic acids and respiratory chains, leading to the accumulation of FAO intermediates such as acyl carnitine in mitochondria, and abnormal insulin signaling[42]. This indicates that butyryl carnitine plays an important role in the occurrence and development of diabetes. In addition, previous studies have found a positive correlation between butyrylcarnitine and obesity, showing similar results in both children and adults[43,44]. This result has been reported in individuals of Asian and European ancestry[45]. Obesity can lead to an increase in the number of brief relaxations of the lower esophageal sphincter, esophageal motility disorders, hiatal hernia, and elevated intra-abdominal pressure and is associated with complications such as BE and EA in GERD[46]. In addition, butyrylcarnitine has been found to be involved in the development of depression. Du et al[47] found that increasing neuronal differentiation is associated with symptoms of depression in later years, and the increase in neuronal differentiation is jointly regulated by an increase in butyrylcarnitine levels and a decrease in the levels of the glycerophospholipid PC35:1 (16:0/199:1). Zhao's study found cognitive improvement and decreased levels of butyrylcarnitine in schizophrenia patients treated with olanzapine[48]. These findings provide strong evidence for the involvement of butyrilcarnitine in the occurrence of mental illness. Therefore, we speculate that butyrylcarnitine may participate in the occurrence of GERD by increasing the risk factors for GERD. Finally, an increase in butyrylcarnitine is related to an increase in visceral fat content[49]. For example, research has found that compared to healthy controls, patients with steatosis and steatohepatitis have significantly higher levels of butyrylcarnitine[50]. Metabolically active visceral adipose tissue secretes adipokines and inflammatory cytokines, which may induce GERD and its complications. A recent MR study also confirmed a positive correlation between visceral adipose tissue accumulation and an increased risk of GERD[51]. In summary, butyrylcarnitine may participate in the occurrence of GERD through multiple pathways. Unfortunately, there is currently a lack of direct evidence linking butyrylcarnitine with GERD, including the pathogenesis of the latter. Our research for the first time discovered a relationship between genetics and metabolomics, which is also a key focus of our future research. Metabolomics has a noninvasive advantage compared to gastroscopy and pathological tissue biopsy, as it can determine the material basis for the occurrence and development of GERD and further speculate on the metabolic pathways involved. This research method combining microdetection and macroanalysis has strong technical support and extensive practical significance.
Genetic factors played a central role in our study of the relationship between metabolites and GERD, and SNP rs4767937 (corresponding to the sppl3 gene) was most significantly associated with butyrylcarnitine and GERD. SPPL3 is widely expressed in the human gastrointestinal tract, most notably in the esophagus[52]. Its main function is the in
The only metabolic pathway identified in our study was that of glycerolipid metabolism, which involves 1-lino
This MR analysis has several advantages. First, this is the most systematic and complete study to date on exploring the causal relationship between blood metabolites and GERD. Second, our results are convincing. The three MR estimates are highly consistent in direction and sensitivity analysis. The strict MR analysis allows us to avoid the rigorous MR analyses allow us to avoid the pitfalls of previous studies, such as reverse causality and confounding disturbances, and ensure the robustness of our results. Third, the reliability of the results was further verified by replication analysis and meta-analysis of additional GWAS data. Fourth, our study offers fresh insights into the molecular pathways underlying the patho
There are also some limitations to this study. First, given the small number of metabolite-related SNPs, our MR analysis set a slightly relaxed threshold. However, the F statistic of all SNPs associated with metabolites was greater than 10, indicating that IVS have a strong power. Furthermore, the Steiger test results' consistent causal direction support lends credence to our lenient threshold choice. Second, for the MR analysis, we solely used GWAS data from people with European ancestry in order to reduce the impact of ethnic differences. Therefore, it merits further investigation and validation to determine whether our findings hold true for other populations. Third, our study did not perform subgroup analysis on GERD. Because the existing dataset does not distinguish between GERD subtypes, which could be further subdivided when the data are more complete, there may be differences between subtypes. Moreover, although MR analysis provides valuable insights into the etiology, our findings should be rigorously confirmed by randomized controlled trials and basic research before clinical application.
In summary, this MR study revealed that eight known blood metabolites are causally associated with GERD, with butyrylcarnitine showing a significant association signal after Bonferroni correction. Our study also highlights the extent to which genetic factors (such as SPPL3) contribute to changes in metabolic levels and the development of GERD. Glycerolipid metabolism has also been found to be possibly related to the biological processes behind GERD. Although further validation of experimental data is needed, the discovery of these serum metabolites provides valuable insights into the early screening, prevention and treatment of GERD and the design of future clinical studies. This combined genomic and metabolomic MR analysis also provides a reference direction for exploring the etiology and pathogenesis of GERD. Future research should also include genetic and metabolomic data related to GERD related diseases. For example, non erosive reflux disease, reflux esophagitis, BE, and hiatal hernia. At the same time, it is necessary to compare the genetic and metabolomic differences among various diseases, which will help clarify the relationship between diseases and better explain GERD.
Gastroesophageal reflux disease (GERD) affects approximately 13% of the global population. However, the pathogenesis of GERD has not been fully elucidated. The development of metabolomics as a branch of systems biology in recent years has opened up new avenues for the investigation of disease processes. As a powerful statistical tool, Mendelian randomization (MR) is widely used to explore the causal relationship between exposure and outcome.
At present, there is still a significant lack of blood metabolomics research on GERD.
We used MR analysis to thoroughly investigate the causal relationships between 486 blood metabolites and GERD using data from a genome-wide association study (GWAS). Additionally, we identified the metabolic pathways that cause GERD. In addition to advancing our understanding of the pathophysiological mechanisms underlying GERD, the integration of metabolomics and genomics offers fresh perspectives on the early detection and management of the disease.
Two-sample MR analysis was used to assess the causal relationship between blood metabolites and GERD. A GWAS of 486 metabolites was the exposure, and two different GWAS datasets of GERD were used as endpoints for the base analysis and replication and meta-analysis. Using the MR Steiger filtration method to detect whether there is a reverse causal relationship between metabolites and GERD. In addition, metabolic pathway analysis was conducted using the online database based MetaboAnalyst 5.0 software.
The results of this study indicated significant associations between eight metabolites, levulinate (4-oxovalerate) [odd ratio (OR) 0.80, 95% confidence interval (CI): 0.72-0.89, P < 0.0001], stearate (18:0) (OR 0.77, 95%CI: 0.64-0.92, P = 0.004), adrenate (22:4n6) (OR 0.83, 95%CI: 0.74-0.94, P = 0.004), p-acetamidophenylglucuronide (OR 0.99, 95%CI: 0.99-1.00, P = 0.0002), kynurenine (OR 1.20, 95%CI: 1.07-1.35, P = 0.002), 1-linoleoylglycerophosphoethanolamine (OR 1.17, 95%CI: 1.05-1.31, P = 0.004), butyrylcarnitine (OR 1.09, 95%CI: 1.05-1.13, P < 0.0001), and guanosine (OR 1.10, 95%CI: 1.03-1.18, P = 0.003), and GERD. Bonferroni correction showed that butyrylcarnitine (OR 1.10, 95%CI: 1.05-1.16, P = 7.71 × 10-5) was the most reliable causal metabolite. Glycerophospholipid metabolism may be involved in the pathogenesis of GERD.
Through the integration of genomics and metabolomics, we found that butyrylcarnitine may be a potential biomarker for GERD.
The relationship between GERD and butyrilcarnitine needs further confirmation from basic and clinical real-world studies. Future research should also include genetic and metabolomic data related to GERD related diseases. For example, non erosive reflux disease, reflux esophagitis, BE, and hiatal hernia. At the same time, it is necessary to compare the genetic and metabolomic differences among various diseases, which will help clarify the relationship between diseases and better explain GERD.
We first appreciate the original GWAS data provided by So-Youn Shin et al and Jue-Sheng Ong et al and Yeda Wu et al and the GWAS Catalog database.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Caboclo JLF, Brazil; Yücel O, Turkey S-Editor: Qu XL L-Editor: A P-Editor: Xu ZH
| 1. | Richter JE, Rubenstein JH. Presentation and Epidemiology of Gastroesophageal Reflux Disease. Gastroenterology. 2018;154:267-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 363] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 2. | Mehta RS, Staller K, Chan AT. Review of Gastroesophageal Reflux Disease. JAMA. 2021;325:1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 3. | Mehta RS, Nguyen LH, Ma W, Staller K, Song M, Chan AT. Association of Diet and Lifestyle With the Risk of Gastroesophageal Reflux Disease Symptoms in US Women. JAMA Intern Med. 2021;181:552-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 4. | Ness-Jensen E, Hveem K, El-Serag H, Lagergren J. Lifestyle Intervention in Gastroesophageal Reflux Disease. Clin Gastroenterol Hepatol. 2016;14:175-82.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 182] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 5. | Yuan S, Larsson SC. Adiposity, diabetes, lifestyle factors and risk of gastroesophageal reflux disease: a Mendelian randomization study. Eur J Epidemiol. 2022;37:747-754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 56] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 6. | Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1116] [Cited by in RCA: 1811] [Article Influence: 201.2] [Reference Citation Analysis (0)] |
| 7. | Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert Rev Mol Diagn. 2008;8:617-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 495] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 8. | Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 519] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 9. | O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 771] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 10. | Bajaj JS, Reddy KR, O'Leary JG, Vargas HE, Lai JC, Kamath PS, Tandon P, Wong F, Subramanian RM, Thuluvath P, Fagan A, White MB, Gavis EA, Sehrawat T, de la Rosa Rodriguez R, Thacker LR, Sikaroodi M, Garcia-Tsao G, Gillevet PM. Serum Levels of Metabolites Produced by Intestinal Microbes and Lipid Moieties Independently Associated With Acute-on-Chronic Liver Failure and Death in Patients With Cirrhosis. Gastroenterology. 2020;159:1715-1730.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 11. | Di'Narzo AF, Houten SM, Kosoy R, Huang R, Vaz FM, Hou R, Wei G, Wang W, Comella PH, Dodatko T, Rogatsky E, Stojmirovic A, Brodmerkel C, Perrigoue J, Hart A, Curran M, Friedman JR, Zhu J, Agrawal M, Cho J, Ungaro R, Dubinsky MC, Sands BE, Suárez-Fariñas M, Schadt EE, Colombel JF, Kasarskis A, Hao K, Argmann C. Integrative Analysis of the Inflammatory Bowel Disease Serum Metabolome Improves Our Understanding of Genetic Etiology and Points to Novel Putative Therapeutic Targets. Gastroenterology. 2022;162:828-843.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 12. | Li Liu, Sheng Xie, Li-jian Liu. Plasma metabolomics study and clinical observation in patients with gastroesophageal reflux disease. Journal of Guangxi University (Natural Science Edition). 2022;47:254-261. [DOI] [Full Text] |
| 13. | Zuccolo L, Holmes MV. Commentary: Mendelian randomization-inspired causal inference in the absence of genetic data. Int J Epidemiol. 2017;46:962-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Richmond RC, Davey Smith G. Mendelian Randomization: Concepts and Scope. Cold Spring Harb Perspect Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 437] [Article Influence: 145.7] [Reference Citation Analysis (0)] |
| 15. | Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1330] [Cited by in RCA: 3787] [Article Influence: 315.6] [Reference Citation Analysis (1)] |
| 16. | Boef AG, Dekkers OM, le Cessie S. Mendelian randomization studies: a review of the approaches used and the quality of reporting. Int J Epidemiol. 2015;44:496-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 377] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 17. | Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E; Multiple Tissue Human Expression Resource (MuTHER) Consortium, Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmüller G, Spector TD, Soranzo N. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1097] [Cited by in RCA: 1100] [Article Influence: 100.0] [Reference Citation Analysis (1)] |
| 18. | Ong JS, An J, Han X, Law MH, Nandakumar P; 23andMe Research team; Esophageal cancer consortium, Schumacher J, Gockel I, Bohmer A, Jankowski J, Palles C, Olsen CM, Neale RE, Fitzgerald R, Thrift AP, Vaughan TL, Buas MF, Hinds DA, Gharahkhani P, Kendall BJ, MacGregor S. Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett's oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut. 2022;71:1053-1061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 120] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 19. | Wu Y, Murray GK, Byrne EM, Sidorenko J, Visscher PM, Wray NR. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat Commun. 2021;12:1146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 20. | Choi KW, Chen CY, Stein MB, Klimentidis YC, Wang MJ, Koenen KC, Smoller JW; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Assessment of Bidirectional Relationships Between Physical Activity and Depression Among Adults: A 2-Sample Mendelian Randomization Study. JAMA Psychiatry. 2019;76:399-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 467] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 21. | Yang J, Yan B, Zhao B, Fan Y, He X, Yang L, Ma Q, Zheng J, Wang W, Bai L, Zhu F, Ma X. Assessing the Causal Effects of Human Serum Metabolites on 5 Major Psychiatric Disorders. Schizophr Bull. 2020;46:804-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 22. | Cai J, Li X, Wu S, Tian Y, Zhang Y, Wei Z, Jin Z, Chen X, Chen WX. Assessing the causal association between human blood metabolites and the risk of epilepsy. J Transl Med. 2022;20:437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 87] [Reference Citation Analysis (0)] |
| 23. | Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178:1177-1184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 419] [Cited by in RCA: 920] [Article Influence: 76.7] [Reference Citation Analysis (0)] |
| 24. | Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4015] [Cited by in RCA: 5701] [Article Influence: 633.4] [Reference Citation Analysis (0)] |
| 25. | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2275] [Cited by in RCA: 6220] [Article Influence: 622.0] [Reference Citation Analysis (0)] |
| 26. | Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1644] [Cited by in RCA: 2866] [Article Influence: 358.3] [Reference Citation Analysis (0)] |
| 27. | Greco M FD, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34:2926-2940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 977] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 28. | Bowden J, Spiller W, Del Greco M F, Sheehan N, Thompson J, Minelli C, Davey Smith G. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47:1264-1278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 507] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 29. | Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2495] [Cited by in RCA: 5344] [Article Influence: 763.4] [Reference Citation Analysis (0)] |
| 30. | Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 411] [Cited by in RCA: 1408] [Article Influence: 176.0] [Reference Citation Analysis (0)] |
| 31. | Savarino V, Marabotto E, Zentilin P, Furnari M, Bodini G, De Maria C, Tolone S, De Bortoli N, Frazzoni M, Savarino E. Pathophysiology, diagnosis, and pharmacological treatment of gastro-esophageal reflux disease. Expert Rev Clin Pharmacol. 2020;13:437-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 32. | Schlottmann F, Andolfi C, Herbella FA, Rebecchi F, Allaix ME, Patti MG. GERD: Presence and Size of Hiatal Hernia Influence Clinical Presentation, Esophageal Function, Reflux Profile, and Degree of Mucosal Injury. Am Surg. 2018;84:978-982. [PubMed] |
| 33. | Lin S, Li H, Fang X. Esophageal Motor Dysfunctions in Gastroesophageal Reflux Disease and Therapeutic Perspectives. J Neurogastroenterol Motil. 2019;25:499-507. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 34. | Savarino E, Bredenoord AJ, Fox M, Pandolfino JE, Roman S, Gyawali CP; International Working Group for Disorders of Gastrointestinal Motility and Function. Expert consensus document: Advances in the physiological assessment and diagnosis of GERD. Nat Rev Gastroenterol Hepatol. 2017;14:665-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 35. | Zheng Z, Shang Y, Wang N, Liu X, Xin C, Yan X, Zhai Y, Yin J, Zhang J, Zhang Z. Current Advancement on the Dynamic Mechanism of Gastroesophageal Reflux Disease. Int J Biol Sci. 2021;17:4154-4164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 36. | Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A. The blood exposome and its role in discovering causes of disease. Environ Health Perspect. 2014;122:769-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 260] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 37. | Buas MF, Gu H, Djukovic D, Zhu J, Onstad L, Reid BJ, Raftery D, Vaughan TL. Candidate serum metabolite biomarkers for differentiating gastroesophageal reflux disease, Barrett's esophagus, and high-grade dysplasia/esophageal adenocarcinoma. Metabolomics. 2017;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Sharma S, Black SM. CARNITINE HOMEOSTASIS, MITOCHONDRIAL FUNCTION, AND CARDIOVASCULAR DISEASE. Drug Discov Today Dis Mech. 2009;6:e31-e39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 113] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 39. | Mitchell SL, Uppal K, Williamson SM, Liu K, Burgess LG, Tran V, Umfress AC, Jarrell KL, Cooke Bailey JN, Agarwal A, Pericak-Vance M, Haines JL, Scott WK, Jones DP, Brantley MA Jr. The Carnitine Shuttle Pathway is Altered in Patients With Neovascular Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci. 2018;59:4978-4985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 40. | Cheng HH, Chang CS, Wang HJ, Wang WC. Interleukin-1beta and -10 polymorphisms influence erosive reflux esophagitis and gastritis in Taiwanese patients. J Gastroenterol Hepatol. 2010;25:1443-1451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 41. | Rieder F, Biancani P, Harnett K, Yerian L, Falk GW. Inflammatory mediators in gastroesophageal reflux disease: impact on esophageal motility, fibrosis, and carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2010;298:G571-G581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 42. | Schooneman MG, Vaz FM, Houten SM, Soeters MR. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 2013;62:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 437] [Cited by in RCA: 487] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 43. | Butte NF, Liu Y, Zakeri IF, Mohney RP, Mehta N, Voruganti VS, Göring H, Cole SA, Comuzzie AG. Global metabolomic profiling targeting childhood obesity in the Hispanic population. Am J Clin Nutr. 2015;102:256-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 170] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 44. | Park S, Sadanala KC, Kim EK. A Metabolomic Approach to Understanding the Metabolic Link between Obesity and Diabetes. Mol Cells. 2015;38:587-596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 124] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 45. | Moore SC, Matthews CE, Sampson JN, Stolzenberg-Solomon RZ, Zheng W, Cai Q, Tan YT, Chow WH, Ji BT, Liu DK, Xiao Q, Boca SM, Leitzmann MF, Yang G, Xiang YB, Sinha R, Shu XO, Cross AJ. Human metabolic correlates of body mass index. Metabolomics. 2014;10:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 153] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 46. | Chang P, Friedenberg F. Obesity and GERD. Gastroenterol Clin North Am. 2014;43:161-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 47. | Du Preez A, Lefèvre-Arbogast S, González-Domínguez R, Houghton V, de Lucia C, Low DY, Helmer C, Féart C, Delcourt C, Proust-Lima C, Pallàs M, Sánchez-Pla A, Urpi-Sardà M, Ruigrok SR, Altendorfer B, Aigner L, Lucassen PJ, Korosi A, Manach C, Andres-Lacueva C, Samieri C, Thuret S. Impaired hippocampal neurogenesis in vitro is modulated by dietary-related endogenous factors and associated with depression in a longitudinal ageing cohort study. Mol Psychiatry. 2022;27:3425-3440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 48. | Zhao L, Liu H, Wang W, Wang Y, Xiu M, Li S. Carnitine metabolites and cognitive improvement in patients with schizophrenia treated with olanzapine: a prospective longitudinal study. Front Pharmacol. 2023;14:1255501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 49. | Pallister T, Jackson MA, Martin TC, Glastonbury CA, Jennings A, Beaumont M, Mohney RP, Small KS, MacGregor A, Steves CJ, Cassidy A, Spector TD, Menni C, Valdes AM. Untangling the relationship between diet and visceral fat mass through blood metabolomics and gut microbiome profiling. Int J Obes (Lond). 2017;41:1106-1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 50. | Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404-413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 428] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 51. | Zhao X, Ding R, Su C, Yue R. Sleep traits, fat accumulation, and glycemic traits in relation to gastroesophageal reflux disease: A Mendelian randomization study. Front Nutr. 2023;10:1106769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 52. | Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlén M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1959] [Cited by in RCA: 2744] [Article Influence: 228.7] [Reference Citation Analysis (0)] |
| 53. | Voss M, Künzel U, Higel F, Kuhn PH, Colombo A, Fukumori A, Haug-Kröper M, Klier B, Grammer G, Seidl A, Schröder B, Obst R, Steiner H, Lichtenthaler SF, Haass C, Fluhrer R. Shedding of glycan-modifying enzymes by signal peptide peptidase-like 3 (SPPL3) regulates cellular N-glycosylation. EMBO J. 2014;33:2890-2905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 54. | Kuhn PH, Voss M, Haug-Kröper M, Schröder B, Schepers U, Bräse S, Haass C, Lichtenthaler SF, Fluhrer R. Secretome analysis identifies novel signal Peptide peptidase-like 3 (Sppl3) substrates and reveals a role of Sppl3 in multiple Golgi glycosylation pathways. Mol Cell Proteomics. 2015;14:1584-1598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 55. | Calzada E, Onguka O, Claypool SM. Phosphatidylethanolamine Metabolism in Health and Disease. Int Rev Cell Mol Biol. 2016;321:29-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 330] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 56. | Reichel M, Hönig S, Liebisch G, Lüth A, Kleuser B, Gulbins E, Schmitz G, Kornhuber J. Alterations of plasma glycerophospholipid and sphingolipid species in male alcohol-dependent patients. Biochim Biophys Acta. 2015;1851:1501-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 57. | Xiao G, He Q, Liu L, Zhang T, Zhou M, Li X, Chen Y, Qin C. Causality of genetically determined metabolites on anxiety disorders: a two-sample Mendelian randomization study. J Transl Med. 2022;20:475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 82] [Reference Citation Analysis (0)] |
| 58. | Delomenède M, Buchet R, Mebarek S. Lansoprazole is an uncompetitive inhibitor of tissue-nonspecific alkaline phosphatase. Acta Biochim Pol. 2009;56:301-305. [PubMed] |
| 59. | Li M, Ding L, Hu YL, Qin LL, Wu Y, Liu W, Wu LL, Liu TH. Herbal formula LLKL ameliorates hyperglycaemia, modulates the gut microbiota and regulates the gut-liver axis in Zucker diabetic fatty rats. J Cell Mol Med. 2021;25:367-382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |