Published online Jan 15, 2023. doi: 10.4251/wjgo.v15.i1.76
Peer-review started: September 28, 2022
First decision: October 21, 2022
Revised: November 1, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: January 15, 2023
Processing time: 103 Days and 22 Hours
As reported, multiple circular RNAs (circRNAs) interfere with colorectal cancer (CRC) progression. Here, circRNA_0001658 (circ_0001658) is focused on studying how it works in CRC.
Clarify the expression pattern, biological function, and underlying mechanism of circ_0001658 of CRC tumorigenesis.
In CRC-related chip data retrieved using the database named Gene Expression Omnibus, different expressions of circRNAs between CRC and normal tissue samples were identified. Quantitative Real-time PCR and Western blot ensured the analysis on circ_0001658, microRNA-590-5P (miR-590-5p), and methyltransferase-like 3 (METTL3) mRNA expressions in tissues and cells. Cell counting kit-8 and flow cytometry were used to detect cell proliferation, apoptosis and migration. The targeting relations between circ_0001658, miR-590-5p, and METTL3 mRNA 3'-untranslated region were under the verification of bioin
Circ_0001658 and METTL3 mRNA was elevated in CRC tissues and cells, whereas miR-590-5p was decreased. Circ_0001658 overexpression promoted the proliferation of HT29 cells, inhibited apoptosis, and accelerated the cell cycle. In SW480 cells, knocking down circ_0001658 had the opposite effect. Circ_0001658 could specifically bind to miR-590-5p and negatively modulate its expressions; METTL3 is a miR-590-5p target that can be positively regulated by circ 0001658. Circ 0001658 was inversely associated with miR-590-5p expression while positively with METTL3 expressions.
Circ_0001658 regulates the miR-590-5p/METTL 3-axis to increase CRC cell growth and decrease apoptosis.
Core Tip: As we know, the progression of colorectal cancer (CRC) is significantly influenced by circular RNAs (circRNAs). This study focused on circRNA_0001658 (circ_0001658) and delved into how it works in CRC. The results confirmed that circ_0001658 and methyltransferase-like 3 (METTL3) mRNA expression in CRC tissues and cells were increased, while miR-590-5p was decreased. Circ_0001658 was inversely associated with miR-590-5p expression while positively with METTL3 expressions. In a word, circ_0001658 accelerates the progression of CRC through miR-590-5p/METTL3 regulatory axis.
- Citation: Lu Y, Wang XM, Li ZS, Wu AJ, Cheng WX. Hsa_circ_0001658 accelerates the progression of colorectal cancer through miR-590-5p/METTL3 regulatory axis. World J Gastrointest Oncol 2023; 15(1): 76-89
- URL: https://www.wjgnet.com/1948-5204/full/v15/i1/76.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i1.76
Colorectal Cancer (CRC) is a kind of prevalent gastrointestinal cancer worldwide, with the incidence rate of the third worldwide and the tumor-related mortality of the fourth[1,2]. For CRC diagnosis, the current standard of treatments for CRC includes surgical procedures, chemotherapy, and radiotherapy. Although these treatments progressed continuously[3], the high rates of metastasis and recurrence in patients with CRC have resulted in a 5-year relative survival rate of 65% in sufferers at stage I-III CRC and 12% for patients at stage IV[4,5]. Consequently, it is essential to discover and create effective biomarkers and individualized treatment.
Circular RNA (circRNA) is a category of non-coding RNA (ncRNA) with closed-loop structure, absent of 5'to 3 'polarity and polyadenylate tail[6,7]. Considering this special structure, circRNA is endowed with high stability, and can mediate tumor progression through a variety of mechanisms; circRNA interacts with RNA-binding protein, acts as splicing and transcription regulators, and sponges microRNA (miRNA)[8]. For example, circ_0084927 boosts the progression of cervical cancer via adsorbing miR-634 and up-regulating tumor protein D52 (TPD52) expression[9]. Recent studies have found that circRNA_0001658 (circ_0001658) features prominently in the progression of tumors through being a biomarker for the diagnostic and prognostic purposes[10,11]. For example, circ_0001658 has a high level of expression in osteosarcoma, and overexpression of this circRNA promotes the proliferation and metastasis[10]. Circ_0001658 is up-regulated in non-small cell lung cancer (NSCLC), and depleting circ_0001658 can inhibit the activity of NSCLC cells and expedite apoptosis[11]. How circ_0001658 interferes with the progression of CRC deserves our furtherance.
As reported, miR-590-5p expression is low in CRC tissues and cells and is associated with adverse clinical and pathological indicators in patients; miR-590-5p overexpression inhibits the growth and migration of CRC cells[12,13]. In addition, overexpression of methyltransferase-like 3 (METTL3) boosts CRC cell multiplication, migration and restrains apoptosis[14]. In this study, the bioinformatics analysis showed that circ_0001658 targeted miR-590-5p and miR-590-5p directly targeted METL3. However, for the CRC, the function of the circ_0001658/miR-590-5p/mETL3 axis is inconclusive.
42 CRC tissue samples and their normal tissue were all selected via the surgically removed tumor tissues and the corresponding normal tissues in PKUCare Luzhong Hospital. The samples were stored in liquid nitrogen within 30 min after isolation and then in the refrigerator for subsequent RNA extraction. All the patients were not treated with radiotherapy, chemotherapy, and other related treatments before surgery. This research was authorized by the Hospital Ethics Committee Hospital and conducted in compliance with the Declaration of Helsinki and the standards of the Hospital Ethics Committee.
The CircRNA chip dataset (GSE172229) was downloaded from Gene Expression Omnibus (GEO). Subsequently, the GEO2R online analysis tool retrieved the associated raw data. With the Excel tool, circRNAs with P < 0.05 and > log2 (fold change) > 1 were filtered out from each data set.
The American Type Culture Collection (Rockville, MD, USA) possessed the CRC cell line (HT29, SW480, LoVo, and DLD-1) and the normal colonic epithelial cell line (FHC).
All cells were placed in Roswell Park Memorial Institute-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 ℃ in 5% CO2 with 95% relative humidity.
Overexpressing circ_0001658 plasmid (circ-OE), empty plasmid (NC), small interfering RNA (siRNA) targeting circ_0001658 (si-circ-1, si-circ-2), siRNA negative control (si-NC), miR-590-5p mimics, and miR-590-5p inhibitors and their control (mimics NC and inhibitors NC) were provided by RiboBio (Guangzhou, China). CRC cell transfection was conducted by Lipo-fectaminTM 3000 (Invitrogen, Carlsbad, CA, USA) as instructions. The effectiveness was below the detection threshold of quantitative real-time polymerase chain reaction (qRT-PCR).
TRIzol (Invitrogen) was used to extract total RNA from tissues and cell lines, which was then reverse-transcribed into cDNA using the PrimeScript RT kit (TaKaRa, Dalian, China). The PCR reaction was then conducted via the Miscript Sybr Green PCR system (Qiagen, GMBH, Hillen, Germany) by the PCR machine named Rotorgene 3000 Series (Corbett Research, Sydney, Australia). Ultimately, the quantitative analysis of miRNA and mRNA was tackled with the Rotor Gene software, with U6 and GAPDH as standardized internal references. The relative expression of miR-590-5p and METTL3 mRNA were calculated by 2−ΔΔCT. The prim sequences: circ_0001658-F: 5’- CTCTCCTGTTGGCTCTCCTG-3’, circ_0001658-R: 3’- CCACCTAGGAGGAACTGACAA-5’; miR-590-5p-F: 5’- AGAAGGCTGGGGCTCATTTG-3’; miR-590-5p-R: 3’- AGGGGCCATCCACAGTCTTTC-5’; METTL3-F: 5’- CTATCTCCTGGCACTCGCAAGA-3’; METTL3-R: 5’- -GCTTGAACCGTGCAACCACATC-3’; GAPDH-F: 5’-GGGAAACTGTGGCGTGAT-3’; GAPDH-R: 5’-GAGTGGGTGTCGCTGTTGA-3’; U6-F: 5’-CTCGCTTCGGGCAGCACA-3’; U6-R:5’-AACGCTCTCACGAATTTGCGT-3’. With GAPDH or U6 as internal references.
Total RNA (2 μg) was incubated for 30 min at under the degrees Celsius of 37 with or without 3 U/μg of RNase R (Epicentre; Illumina, Inc, Madison, WI, USA). Afterward, circ_0001658 and GAPDH mRNA expression were under examination by qRT-PCR.
The cell countering kit-8 (CCK-8) kit (Invitrogen, Shanghai, China) was utilized to determine the viability of cells. The transfected SW480 and HT29 cells were inoculated into a 96-well plate at a density of 3000 cells per well at 37 ℃ for 48 h and then mixed with 10 μL CCK-8 reagent (Invitrogen) for 4 h. The absorbance optical density at 450 nm was below the microplate reader's value.
Cell cycle distributions were determined using by flow cytometer for fluorescence-activated cell sorting flow cytometer (BD Bioscience, San Jose, CA, USA). The cells were harvested 48 h after transfection and trypsinized, subsequently fixed overnight at 4 ℃ in 70% ethanol, and then stained with 50 μg /mL propidium iodide (BD Bioscience) in darkness at ambient temperature for 30 min. Cell cycle distributions were under the analysis of the FACS Calibur system and ModFit 3.0 software. The Annexin-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Southern Biotechnology, Birmingham, AL, USA) was used to detect apoptosis. Three triplicate wells of HT29 and SW480 cells transfected for 48 h were derived and washed twice using pre-cooled PBS. Following the addition of 5 μL of Annexin V-FITC and 5 L of PI working solution, the cells were suspended in 500 μL of binding buffer. After mixing, the cells were allowed to rest at room temperature for 15 min in a dark environment. The green (Annexin V-FITC) and red (PI) fluorescence were probed by flow cytometry. Ultimately, the apoptotic rate was tackled by FlowJo software.
The cytoplasm and nuclei were isolated from HT29 and SW480 cells by RNA Isolation Kit (Thermo Fisher Scientific, Shanghai, China). RNA was then separated from the cytoplasm and nucleus, and the expression of circ_0001658 was under the determination of qRT-PCR. The cytoplasmic and nuclear controls, namely GAPDH and U6, respectively.
Circinteractome software and TargetScan database predicted the binding sites between circ_0001658 and miR-590-5p as well as those between miR-590-5p and METTL3'--untranslated region (UTR), respectively. These critical regions were amplified by PCR and introduced into the plasmid vector pGL3-Promoter (Promega, Madison, WI, USA) and wild-type (WT) and mutant type (MUT) circ_0001658 and METTL3 dual luciferase reporter gene vectors (circ_0001658-WT, METTL3-WT) were constructed. Site-directed mutagenesis was used to generate the mutant circ_0001658 and METTL3 dual luciferase reporter gene vectors (circ_0001658-MUT and METTL3-MUT, respectively). The corresponding vectors were then co-transfected with miR-590-5p or miR-NC into SW480 and HT29 cells. The luciferase intensity was under the exploration of a dual luciferase reporter gene assay system (Promega).
The cells were lysed in radioimmunoprecipitation assay lysis buffer (Pierce, Rockford, IL, USA), and the total protein was extracted, with concentrations assessed by a bicinchoninic acid protein assay kit (Pierce). The protein was denatured by boiling with Loading Buffer and applied to Sodium Dodecyl Sulfone-Polyacrylamide gel with 6% concentration gel and 10% separation gel. Gel electrophoresis voltage was adjusted at 80–120 V, while wet transport and film transfer volta ge were controlled at 100 mV for 45–70 min. Proteins were transferred to a polyvinylidene fluoride (Pierce) membrane by an electroporator. The membranes were followingly blocked under the 5% skimmed milk for 1 h at ambient temperature and incubated overnight as described with anti-METTL3 antibodies (1: 1000, ab195352, Abcam, Cambridge, MA, USA) and internal reference GAPDH antibodies (1: 1000, ab9485, Abcam) at 4 ℃. Next day, the membranes were cleaned thrice for five minutes at a time, and incubated with tris buffered saline tween and secondary antibodies (1:1000, ab205718, Abcam) over 2 h at room temperature and then washed again, and the blots were developed by chemiluminescent substrate, with the grayscale under the analysis of gel imaging analysis system.
Data processing was tackled using software named SPSS 22.0 and GraphPad Prism 8.0. Measurement data were utilized to represent as mean ± SD. One-way analysis of variance was adopted for mean comparison among multiple groups, and Student’s t test for that between two groups. The correlation among the expressions of circ_0001658, miR-590-5p, and METTL3 mRNA in CRC tissues was under the examination of Pearson correlation analysis. P <0.05 signifies a statistically significant distinction.
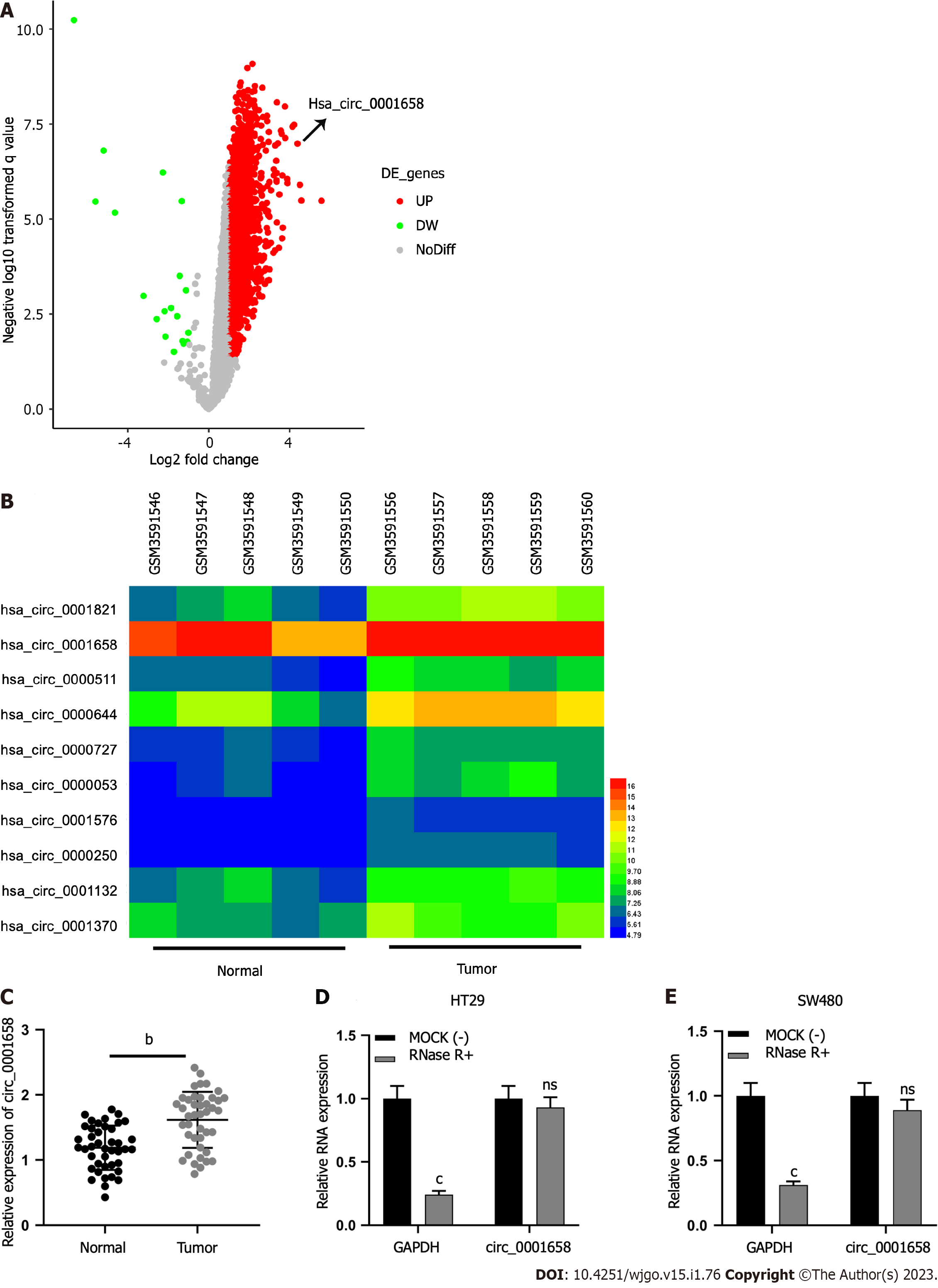
Microarray data set GES172229 available from the GEO database displayed significantly up-regulated or down-regulated circRNAs as per the screening criterion of │log2FC│> 1 and P < 0.05 (Figure 1A). Circ_0001658 was greatly raised in CRC tissues as opposed to controls (Figure 1B). qRT-PCR uncovered that in 42 pairs of tumors and precancerous tissues, a significant increase in circ_0001658 Levels in CRC tissues was observed (Figure 1C). RNase-R treatment has witnessed circ_0001658’s resistance to RNase-R as against GAPDH (Figure 1D).
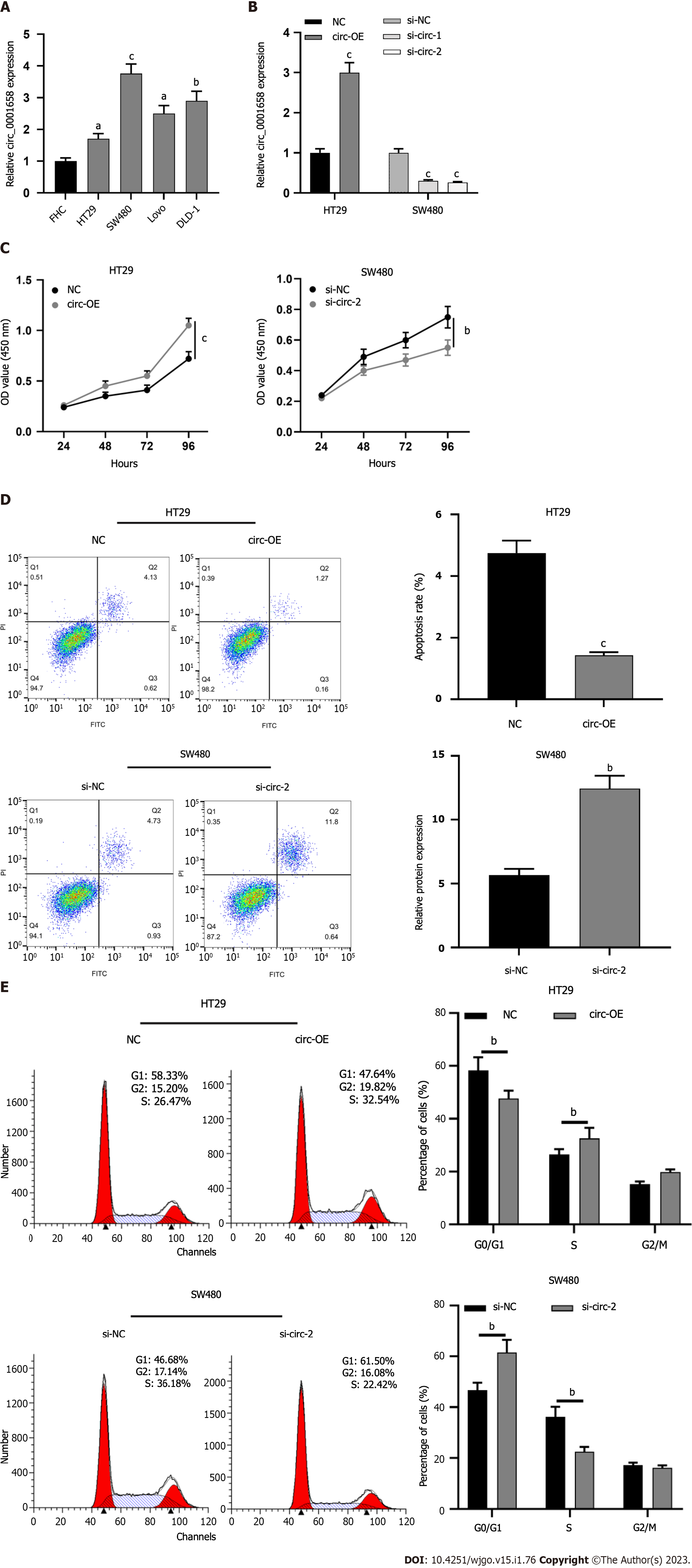
Circ_0001658 expression was significantly up-regulated in CRC cell lines (HT29, SW480, LoVo, and DLD-1) as opposed to human normal colorectal mucosal cell FHC (Figure 2A). Of the four CRC cells, expressions of circ_0001658 were the lowest in HT29 cells, while those of SW480 were the highest. Therefore, we transfected HT29 cells with the circ_0001658 overexpression plasmid and NC, respectively, and transfected SW480 cells with si-circ-1, si-circ-2, and si-NC, respectively. qRT-PCR (Figure 2B) demonstrated the effectiveness of the transfections. CCK-8 assay focused that as opposed to the control, as can be seen, circ_0001658 overexpression greatly promoted the viability of HT29 cells, while the knockdown worked oppositely on SW480 cells (Figure 2C). Flow cytometry demonstrated that circ 0001658 overexpression dramatically prevented HT29 cell death and accelerated cell cycle progression, while depleting circ_0001658 functioned oppositely on SW480 cells (Figure 2D-E).
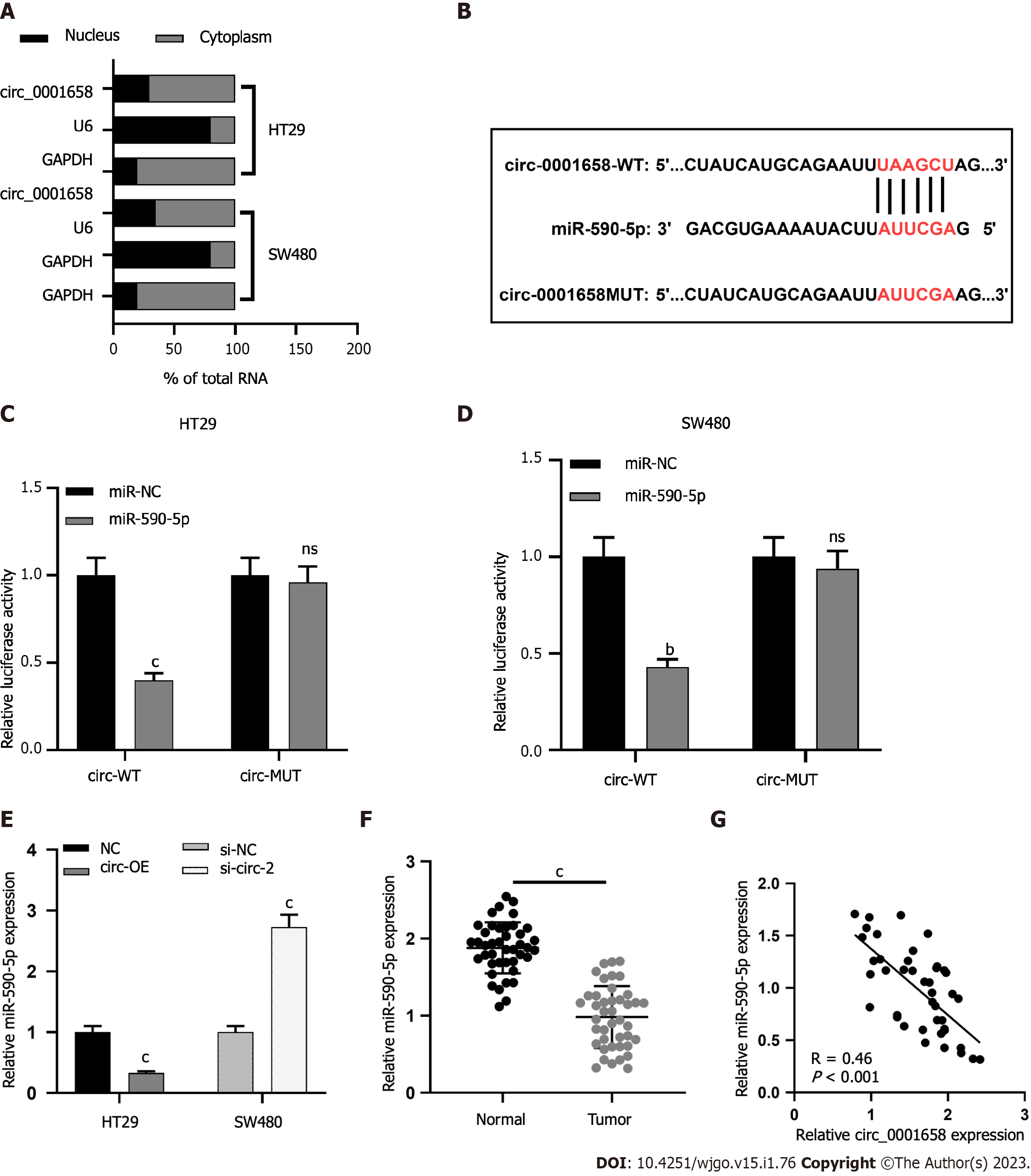
To study the subcellular distribution of circ 0001658 in SW480 and HT29 cells, a nuclear mass separation experiment was conducted. The findings uncovered that circ_0001658 was mainly present in the cytoplasm as compared with U6 and GAPDH (Figure 3A). Bioinformatics prediction displayed a binding site between miR-590-5p and circ_0001658 (Figure 3B). The dual luciferase reporter gene experiment highlighted that, as compared with miR-NC, overexpression of miR-590-5p inhibited the activity of circ_0001658 WT in SW480 and HT29 cells. However, the activity of circ 0001658 MUT was not dramatically impacted (Figure 3C-D). qRT-PCR demonstrated that circ_0001658 overexpression in SW480 cells suppressed miR-590-5p expression considerably, whereas circ_0001658 knockdown in HT29 cells caused an increase in miR-590-5p expression (Figure 3E). Levels of miR-590-5p in CRC tissues were significantly reduced when as to adjacent non-neoplastic tissues (Figure 3F). In addition, there was a negative association between the expressions of circ_0001658 and miR-590-5p in CRC tissues (Figure 3G).
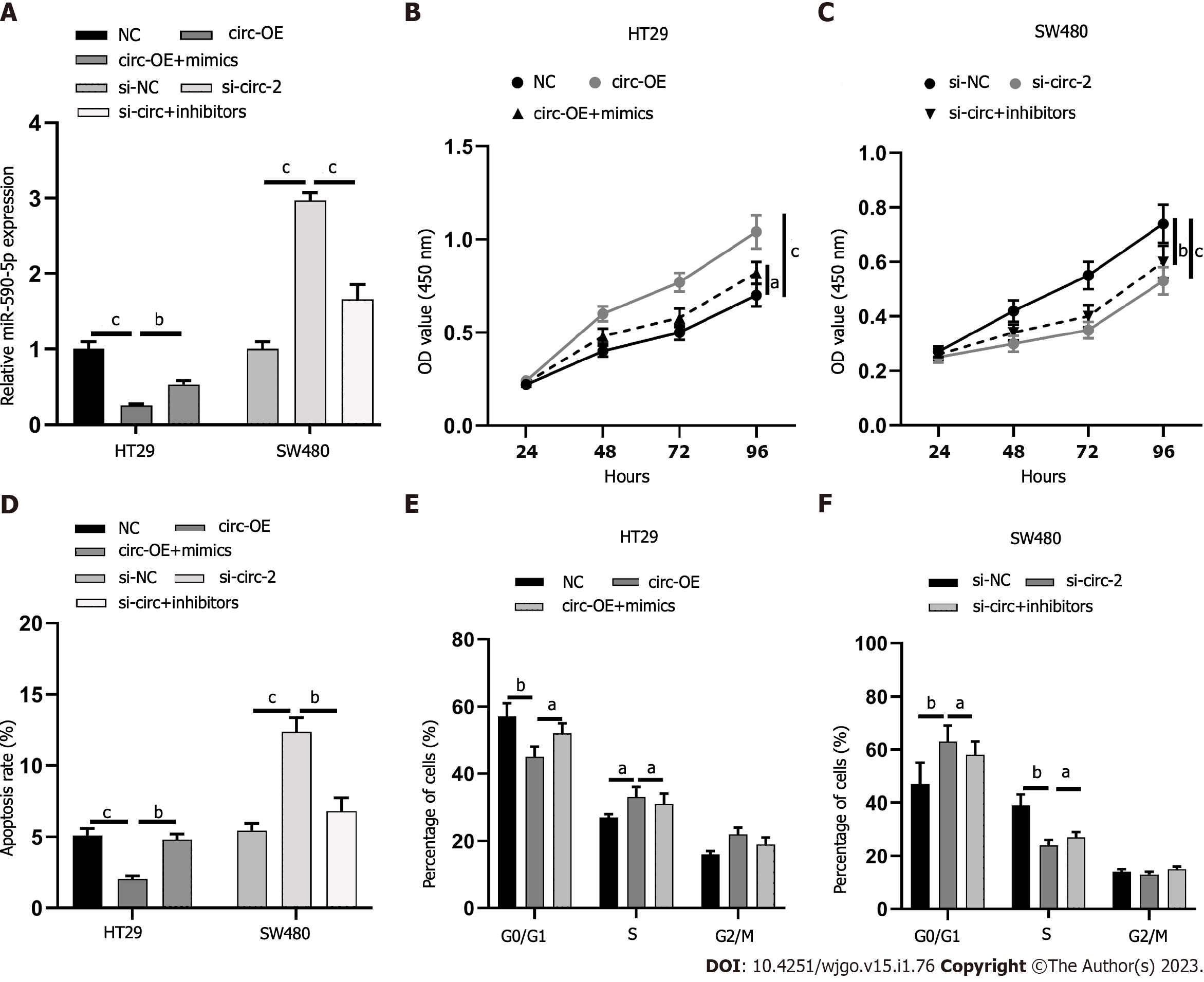
The proliferation effects of circ_0001658 and miR-590-5p, apoptosis, and cycle of CRC cells were the next focus. Circ-OE+mimics and si-circ+inhibitors were co-transfected into HT29, and SW480 cells, respectively, with qRT-PCR, which verified it a success (Figure 4A). miR-590-5p overexpression significantly restrained SW480 cell proliferation, accelerated apoptosis, and arrested cell cycle as compared to circ_0001658 transfection alone, as demonstrated by CCK-8 assay and flow cytometry (Figure 4B-F). Inhibiting miR-590-5p significantly accelerated cell proliferation, impeded apoptosis, and accelerated cell cycle when as opposed to transfection of si-circ alone (Figure 4B-F).
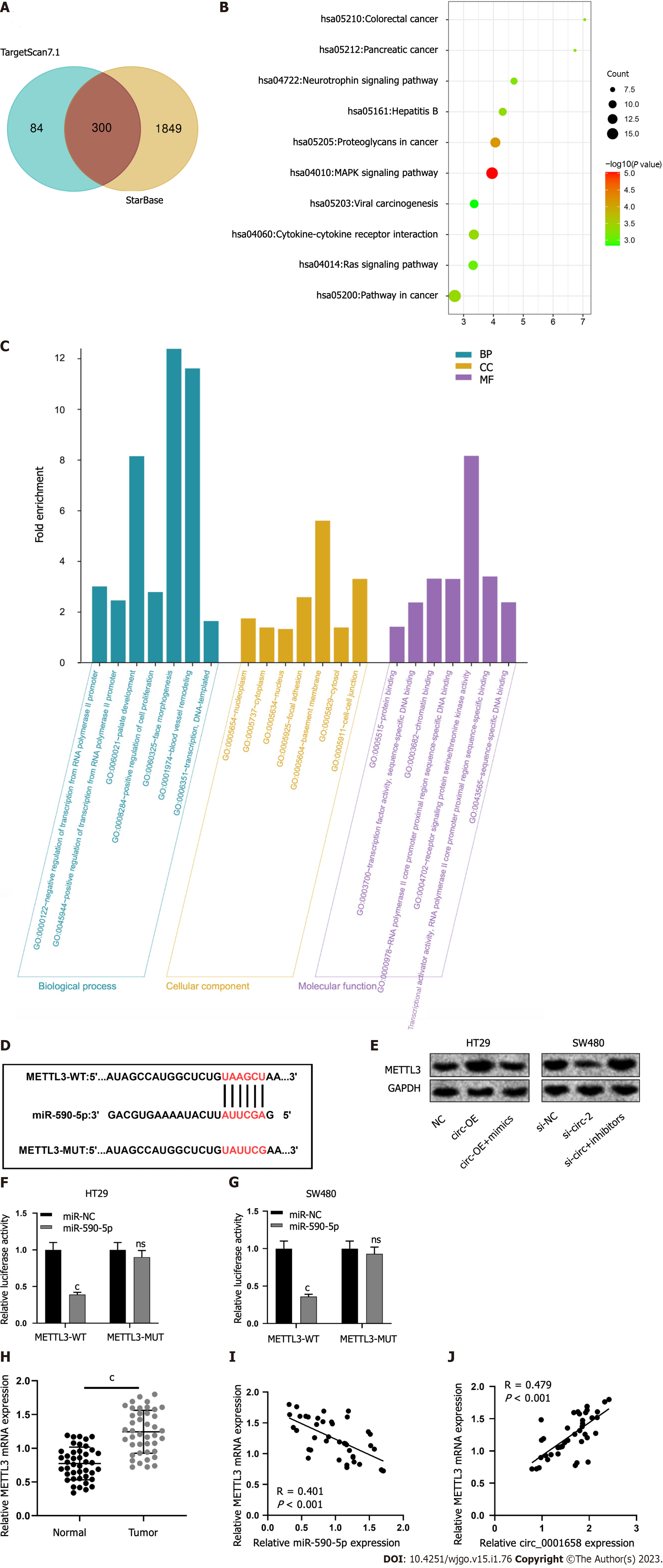
To determine the downstream mechanism of action of miR-590-5p, we screened candidate miR-590-5p targets through the StarBase and TartgetScan7.1 databases, and the results show that miR-590-5p had 300 candidate targets (Figure 5A). Then, the above targets were tackled with the Database for Annotation, Visualization and Integrated Discovery database for enrichment research by the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO). KEGG analysis showed that the above target genes MAPK and Ras signal pathways were significantly enriched (Figure 5B). GO analysis showed significant enrichment of the above target genes in biological processes, cellular components, and molecular functions (Figure 5C). Among them, METTL3, related to CRC progression, is one of the candidate targets for miR-590-5p, and the binding sequence of the two is shown in Figure 5D. Western blot results uncovered that circ_0001658 overexpression promoted TTL 3 protein expression, while miR-590-5p overexpression weakened this effect; knockdown of circ_0001658 inhibited METTL3 protein expression, and downregulation of miR-590-5p reversed this effect (Figure 5E). Dual luciferase reporter gene assay depicted that the overexpression of miR-590-5p in HT29 and SW480 cells reduced the luciferase activity of METTL3-WT substantially (Figure 5F-G). METTL3 mRNA and protein levels were demonstrably increased in CRC tissues as against normal tissues (Figure 5H). The correlation analysis indicated a negative relation among the mRNA expressions of miR-590-5p and TTL 3 in CRC tissues (Figure 5I) and a positive relation among circ_0001658 and TTL 3 mRNA expression in CRC tissues (Figure 5J).
More and more reports have pointed out that circRNA interferes with different biological processes. CircRNA, as a cancer-promoter or tumor deterrent, participates in the formation, spread, and incidence of cancers[15,16]. Reportedly, circRNAs have important biological functions in gastric cancer, liver cancer, and CRC[17-20]. Moreover, circRNAs are also widely involved in the onset and development of CRC. For instance, circ_001680 expression is upregulated in CRC. Circ_001680 overexpression boosts cancer stem cell growth in CRC-like populations and enhances the resistance of tumor cells to irinotecan by upregulating the expression of the Bmi1 polycomb ring finger oncogene[20]. Circ-Erbin activates Hypoxia-Induced Factor-1α by upregulating Eukaryotic translation initiation factor 4E binding protein one expression, promoting the proliferation and migration of CRC cells as well as the growth of xenografts in CRC cells in vivo[21]. There are also a large number of reports regarding the role of circ_0001658 in tumors. For example, circ_0001658 is raised in gastric cancer tissues, and circ_0001658 may interfere with the development of gastric cancer by regulating the miR-375/PAX6 axis[22]. Another study demonstrates that circ 0001658 is significantly expressed in osteosarcoma and is related to poor clinical pathology; Additional studies have shown circ_0001658 accelerates the multiplication and metastasis of osteosarcoma cells via regulating the miR-382-5P/y box-binding protein one axis[10]. Circ_0001658 is upregulated in NSCLC and significantly correlated with increased tumor-node-metastasis staging and decreased degree of differentiation, and it stimulated NSCLC cell growth and inhibited apoptosis through the regulating miR-409-3p/TWIST1 axis[11]. In this study, we determined that circ_0001658 is significant in CRC tissues and cells. Additional research revealed that overexpression of circ_0001658 greatly increased CRC cell proliferation and prevented apoptosis; depleting circ_0001658 exerted opposite effects. Collectively, circ_0001658 plays a pro-cancer role in CRC.
MiRNA is a form of non-coding, single-stranded RNA with a length of between 22 and 25 nucleotides. It regulates gene expression post-transcriptionally by inducing mRNA cleavage or inhibiting mRNA translation and participates in a number of crucial biological processes, including cell development, proliferation, differentiation, and apoptosis[23]. More and more studies have indicated that miRNAs can impact the progression of various cancers, including CRC, by targeting multiple target genes[24,25]. For example, miR-590-5p is inhibited in CRC and ultimately inhibits CRC lung metastasis and CRC angiogenesis in nude mice by specifically regulating nuclear factor 90, which represses the expression of vascular endothelial growth factor[12]. As reported, miR-590-5p is inhibited in CRC, and this miRNA inhibits the growth, migration, and aggressiveness of CRC cells by targeting the recombination signal binding protein for immunoglobulin kappa J region[25]. MiR-590-5p directly targets Yes1 associated transcriptional regulators and inhibits the tumorigenesis of CRC cells in vitro and in vivo[26]. Here we have confirmed the repressed miR-590-5p in CRC. In addition, circ_0001658 was an upstream target of miR-590-5p. By targeting miR-590-5p, Circ_0001658 has the potential to promote CRC cell survival and inhibit apoptosis.
Compared to DNA methylation and histone modification, m6A-RNA methylation is an epigenetic alteration. It is a biological process that is dynamic and reversible, regulated by methyltransferase, dimethyl transferase, and related proteins, which exert different biological effects on mRNA, including mRNA cleavage, nucleation, and degradation, affecting mRNA stability and translation efficiency[27,28]. METTL3, a key component of the m6A methyltransferase complex, affects the malignant phenotype of the tumor by regulating the m6A modification[29]. As reported, METTL3 shows a pro-cancer effect in CRC. For example, METTL3 is significantly expressed in CRC and has been related to a patient’s poor prognosis. Further studies have shown that METTL3 promotes SOX2 expression in CRC cells through m6A-IGF2BP2-dependent mechanism and accelerates cell self-renewal in vitro, thus promoting CRC occurrence and metastasis[30]. By targeting the M6A site in the yippee like 5 transcript region, METTL3 inhibits the expression of yippee like 5 in an m6A-YTHDF2-dependent manner, thus promoting the growth and metastasis of CRC tumors[31]. METTL3 increases the stability of PTTG3P and upregulates its expression through the m6A-IGF2BP2 mechanism, thus boosting the proliferation of CRC cells[32]. Here we discovered that METTL3, as a target gene of miR-590-5p, was positively pertinent to circ_0001658 and negative to miR-590-5p. In addition, we found that circ_0001658 promoted the malignant phenotype of CRC cells by adsorbing miR-590-5p and upregulating METTL3 expressions.
Notably, the present approach has certain limitations. First, the present study only performed in vitro experiments, and in the following research, in vivo models are employed to further validate the biological function of circ_0001658. Secondly, as is well known, a circRNA may have multiple target miRNAs, and a miRNA may have multiple target genes. That implies that circ_0001658 may participate in CRC progression via other mechanisms, which remains to be investigated in the following research. Last but not least, to further estimate the prognostic value of circ_0001658, a larger cohort of patients should be enrolled.
On all accounts, circ_0001658 is increased in the tissues and cells of patients with CRC. circ_0001658 promotes cell proliferation, accelerates the cell cycle, and depresses apoptosis of CRC by regulating the miR-590-5p/METTL3 axis. Collectively, circ_0001658 is anticipated to become a novel therapeutic direction and target for CRC.
According to reports, circular RNAs (circRNAs) have a major role in cancer biology. Some circRNAs have been reported to function as oncogenes or tumor suppressors in colorectal cancer (CRC).
To further clarify the function of circRNAs for the development of CRC.
This paper aims to clarify the expression pattern, biological function, and underlying mechanism of circ_0001658 of CRC tumorigenesis.
A series of in vitro experiments were performed. CircRNA expression profile using the GEO database was analyzed, and circRNAs with differential expression in CRC and normal tissue samples were detected. Quantitative real-time polymerase chain reaction and western blot were performed for the analysis of the expression of circ_0001658, miR-590-5p, and methyltransferase-like 3 (METTL3) mRNA expression levels in tissues and cells. Using Cell counting kit-8 and flow cytometry, cell proliferation, apoptosis, and the cell cycle were observed and studied. The targeting relations between circ_0001658, miR-590-5p, and METTL3 mRNA 3'UTR were under the verification of bioinformatics prediction and dual luciferase reporter gene assay.
circ_0001658 is significantly expressed in CRC tissues and cell lines. It enhances cancer cells' malignant biological activities, including proliferation, resistance to apoptosis, and cell cycle progression, via repressing miR-590-5p and up-regulating METTL3.
circ_0001658 is an oncogenic circRNA in CRC, and it works as an endogenous RNA that competes with miR-590-5p and METTL3.
circ_0001658 may have the potential to give and employ a therapeutic target and diagnostic biomarker for CRC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kim SY, South Korea; Nogueira-Costa G, Portugal S-Editor: Liu GL L-Editor: A P-Editor: Zhang XD
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55806] [Article Influence: 7972.3] [Reference Citation Analysis (132)] |
| 2. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 4893] [Article Influence: 699.0] [Reference Citation Analysis (1)] |
| 3. | Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2417] [Cited by in RCA: 3072] [Article Influence: 512.0] [Reference Citation Analysis (0)] |
| 4. | Yu IS, Cheung WY. Metastatic Colorectal Cancer in the Era of Personalized Medicine: A More Tailored Approach to Systemic Therapy. Can J Gastroenterol Hepatol. 2018;2018:9450754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Effendi-Ys R. Cancer Stem Cells and Molecular Biology Test in Colorectal Cancer: Therapeutic Implications. Acta Med Indones. 2017;49:351-359. [PubMed] |
| 6. | Yuan X, Yuan Y, He Z, Li D, Zeng B, Ni Q, Yang M, Yang D. The Regulatory Functions of Circular RNAs in Digestive System Cancers. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 7. | Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 1013] [Article Influence: 202.6] [Reference Citation Analysis (0)] |
| 8. | Yin Y, Long J, He Q, Li Y, Liao Y, He P, Zhu W. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10:5015-5021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 181] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 9. | Shi P, Zhang X, Lou C, Xue Y, Guo R, Chen S. Hsa_circ_0084927 Regulates Cervical Cancer Advancement via Regulation of the miR-634/TPD52 Axis. Cancer Manag Res. 2020;12:9435-9448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Wang L, Wang P, Su X, Zhao B. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochem Funct. 2020;38:77-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 11. | Yu X, Fu X, Zhang X, Bai C, Wang Y. Circ_0001658 regulates gefitinib resistance of non-small cell lung cancer through miR-409-3p/TWIST1 axis. Anticancer Drugs. 2022;33:158-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Zhou Q, Zhu Y, Wei X, Zhou J, Chang L, Sui H, Han Y, Piao D, Sha R, Bai Y. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axis. Cell Death Dis. 2016;7:e2413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 13. | Yu M, Luo Y, Cong Z, Mu Y, Qiu Y, Zhong M. MicroRNA-590-5p Inhibits Intestinal Inflammation by Targeting YAP. J Crohns Colitis. 2018;12:993-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 14. | Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 288] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 15. | Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869-881.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1086] [Cited by in RCA: 1183] [Article Influence: 197.2] [Reference Citation Analysis (0)] |
| 16. | Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 833] [Cited by in RCA: 1159] [Article Influence: 144.9] [Reference Citation Analysis (0)] |
| 17. | Wang N, Lu K, Qu H, Wang H, Chen Y, Shan T, Ge X, Wei Y, Zhou P, Xia J. CircRBM33 regulates IL-6 to promote gastric cancer progression through targeting miR-149. Biomed Pharmacother. 2020;125:109876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 18. | Li C, Tian Y, Liang Y, Li Q. Circ_0008035 contributes to cell proliferation and inhibits apoptosis and ferroptosis in gastric cancer via miR-599/EIF4A1 axis. Cancer Cell Int. 2020;20:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 19. | Luo X, Liu Y, Li H, Cheng T, Wu J, Chen L, Ju L, Cai W, Bian Z. Hsa_circ_0013290 Acts as Cancer-Promoting Gene in Hepatocellular Carcinoma. Cancer Control. 2021;28:10732748211055681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X, Chen L, Yang M, Peng K, Zhang Z, Liu T, Ye Y, Jiao H, Wang S, Zhou W, Ding Y, Li T. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 21. | Liu YT, Chuang YC, Lo YS, Lin CC, Hsi YT, Hsieh MJ, Chen MK. Asiatic Acid, Extracted from Centella asiatica and Induces Apoptosis Pathway through the Phosphorylation p38 Mitogen-Activated Protein Kinase in Cisplatin-Resistant Nasopharyngeal Carcinoma Cells. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Jiang F, Shen XB. miRNA and mRNA expression profiles in gastric cancer patients and the relationship with circRNA. Neoplasma. 2019;66:879-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' Action through miRNA Editing. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 438] [Cited by in RCA: 601] [Article Influence: 100.2] [Reference Citation Analysis (0)] |
| 24. | Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 535] [Cited by in RCA: 769] [Article Influence: 109.9] [Reference Citation Analysis (0)] |
| 25. | Chen GQ, Liao ZM, Liu J, Li F, Huang D, Zhou YD. LncRNA FTX Promotes Colorectal Cancer Cells Migration and Invasion by miRNA-590-5p/RBPJ Axis. Biochem Genet. 2021;59:560-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 26. | Ou C, Sun Z, Li X, Ren W, Qin Z, Zhang X, Yuan W, Wang J, Yu W, Zhang S, Peng Q, Yan Q, Xiong W, Li G, Ma J. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 27. | Liu S, Zhuo L, Wang J, Zhang Q, Li Q, Li G, Yan L, Jin T, Pan T, Sui X, Lv Q, Xie T. METTL3 plays multiple functions in biological processes. Am J Cancer Res. 2020;10:1631-1646. [PubMed] |
| 28. | Zheng W, Dong X, Zhao Y, Wang S, Jiang H, Zhang M, Zheng X, Gu M. Multiple Functions and Mechanisms Underlying the Role of METTL3 in Human Cancers. Front Oncol. 2019;9:1403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 29. | Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 396] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 30. | Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, Chen D, Li B, Kang TB, Xie D, Lin D, Ju HQ, Xu RH. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 302] [Cited by in RCA: 581] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 31. | Zhou D, Tang W, Xu Y, Xu B, Fu S, Wang Y, Chen F, Chen Y, Han Y, Wang G. METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Mol Oncol. 2021;15:2172-2184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 32. | Zheng Y, Wang Y, Liu Y, Xie L, Ge J, Yu G, Zhao G. N6-Methyladenosine Modification of PTTG3P Contributes to Colorectal Cancer Proliferation via YAP1. Front Oncol. 2021;11:669731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |