Published online Jan 15, 2023. doi: 10.4251/wjgo.v15.i1.171
Peer-review started: October 1, 2022
First decision: October 20, 2022
Revised: October 26, 2022
Accepted: November 28, 2022
Article in press: November 28, 2022
Published online: January 15, 2023
Processing time: 100 Days and 18.6 Hours
Recently, many investigations have suggested that the rs3746444 T>C locus in the microRNA (miR)-499 gene may contribute to the occurrence of cancer. However, reports on the association between rs3746444 and hepatocellular carcinoma (HCC) are conflicting.
To further understand and explore the potential correlation between the single-nucleotide polymorphism of rs3746444 and the incidence of HCC.
In this meta-analysis, we obtained electronic literature by searching the PubMed, Embase and Chinese BioMedical Disc databases (through May 20, 2022). All eligible case-control, prospective cohort or nested case-control studies with sufficient data for calculating the odds ratios with their 95% confidence intervals were included.
Ultimately, a total of 17 independent studies were included. We identified that rs3746444 was associated with the development of HCC (C vs T: P = 0.019 and CC/CT vs TT: P = 0.016). In Asian individuals, rs3746444 was associated with susceptibility to HCC (C vs T: P = 0.013 and CC/CT vs TT: P = 0.016). In addition, this study identified that the miR-499 rs3746444 locus was associated with susceptibility to HCC in the normal/healthy control subgroup (C vs T: P = 0.034 and CC/CT vs TT: P = 0.024).
In summary, this meta-analysis highlights that rs3746444 in the miR-499 gene is involved in the occurrence of HCC, especially in Asian individuals.
Core Tip: Many investigations have suggested that the rs3746444 T>C locus in the microRNA (miR)-499 gene may contribute to the occurrence of cancer. However, reports on the association between rs3746444 and hepatocellular carcinoma (HCC) are conflicting. This meta-analysis highlights that rs3746444 in the miR-499 gene is involved in the occurrence of HCC, especially in Asian individuals.
- Citation: Jiang JK, Chen HS, Tang WF, Chen Y, Lin J. Rs3746444 T>C locus in miR-499 increases the susceptibility to hepatocellular carcinoma: A meta-analysis 14812 subjects. World J Gastrointest Oncol 2023; 15(1): 171-185
- URL: https://www.wjgnet.com/1948-5204/full/v15/i1/171.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i1.171
In 2020, liver cancer (LC) was the seventh most frequent malignancy, with 905677 new cases worldwide[1]. Accordingly, LC is ranked as the second leading cause of malignancy-related deaths, as it has resulted in death for 830180 individuals[1]. The incidence rates of LC and LC-related deaths remain higher in China than other parts of the world[2]. Hepatocellular carcinoma (HCC) is the predominant subtype of LC, accounting for approximately 75%-85% of primary LC cases[3]. Overall, the survival rate of HCC remains poor. To date, the etiology of HCC is not well established. Although it has been reported that chronic virus infection[4], type 2 diabetes[5,6], obesity[7,8], smoking[9,10], heavy alcohol intake[11-13] and aflatoxin-contaminated food stuffs[14] may contribute to the occurrence of HCC, other risk factors may also lead to the development of HCC, such as hereditary factors[15-18].
MicroRNAs (miRs) are noncoding RNAs of approximately 22 nucleotides in size. They may be implicated in the regulation of target genes and are involved in a number of cellular processes (e.g., growth, proliferation, differentiation, apoptosis, migration and invasion)[19-24]. Recently, several investigations have reported that the expression profiling of serum miRs could be used as a marker for hepatitis C virus-related cases of HCC[25]. Variants within miRs may alter target recognition, transcription, or posttranscriptional processing and then lead to malignant diseases[26]. Additionally, most of the established miRs may influence many target genes; single-nucleotide polymorphisms (SNPs) in miRs could affect the level of multifarious proteins. MiR-499 is located within chromosome 20q. MiR-499 is involved in infection and inflammatory diseases[27]. Rs3746444 T>C in miR-499 was identified to be correlated with the development of ankylosing spondylitis[28], arthritis susceptibility[29], and bronchial asthma[27].
Additionally, a number of investigations have suggested that the rs3746444 SNP in miR-499 may contribute to the occurrence of cancer. Liu et al[30] reported that miR-499-5p could promote the metastasis of colorectal cancer and might be used as a vital target for colorectal cancer therapy. Additionally, a previous study identified that in HepG2 cells, miR-499 could inhibit the level of the E26 transformation specific sequence 1, which is an important proto-oncogene in the development of HCC[31]. The miR-499 variant rs3746444 has been suggested to play an important role in the occurrence of various malignancies, such as adenocarcinoma of the esophagogastric junction[32], prostate cancer[33], cervical squamous cell carcinoma[34], oral squamous cell cancer[35], and lung cancer[36]. Recently, a number of studies have focused on the relationship between rs3746444 in miR-499 and HCC[36-40]; however, the obtained findings are conflicting. Several meta-analyses also reported controversial results. Some pooled analyses have suggested that the rs3746444 C allele could not confer a risk to HCC[41-44]. However, other publications have reported that the rs3746444 C allele may contribute to the occurrence of HCC[40,45-47]. These controversial findings may be due to the limited sample sizes included in these analyses. Recently, some case-control studies have been conducted to further explore this potential association[48-50]. An updated meta-analysis is needed to shed new light on the relationship between rs3746444 in miR-499 and HCC regarding all available publications. Therefore, this meta-analysis involved a large sample size to verify whether the miR-499 rs3746444 SNP could influence the occurrence of HCC. And these possible relationships might be beneficial to the prevention of liver carcinogenesis.
In this meta-analysis, we obtained electronic literature by searching the PubMed, Embase and Chinese BioMedical Disc (CBM) databases (through May 20, 2022). We used the following keywords: (SNP OR variant OR polymorphism) AND (neoplasm OR carcinoma OR tumor OR cancer) AND (hepatocellular OR liver) AND (microRNA499 OR miR499 OR microRNA-499 OR miR-499 OR rs3746444). The references included in the retrieved publications and relevant reviews, as well as published meta-analyses, were hand-searched to obtain more related data. Due to no restriction on language, a large amount of data was collected. We also cited high-quality articles in Reference Citation Analysis (https://www.referencecitationanalysis.com).
The inclusion criteria for the eligible literature were as follows: (1) Assessing the relationship of rs3746444 in miR-499 with HCC susceptibility; (2) Full-text study; (3) Designed as a case-control study, a prospective cohort or a nested case-control study; and (4) Sufficient data could be used to calculate the odds ratios (Ors) with their 95% confidence intervals (CIs). When a publication contained more than one investigation, it was treated as an independent case-control study. Accordingly, letters, reviews, comments, non-case-control studies, studies that violated Hardy-Weinberg equilibrium (HWE), literature without sufficient data and duplicated data were excluded.
Two authors (Jiang JK and Lin J) reviewed the eligible literature and extracted the data independently. The following information was collected: The first author, year of publication, mean age (years), sex (male, %), drinking status (%), smoking status (%), country/ethnicity, hepatitis B surface antigen (HBsAg) (positive, %), number of subjects, HWE, genotyping method and genotype data. In a case of a conflicting assessment, another author (Tang WF) took part in a discussion until a consensus opinion was obtained.
The results of this meta-analysis were assessed in four genetic models: A dominant model (CC/TC vs TT), recessive model (CC vs TT/TC), homozygote comparison (CC vs TT) and allelic model (C vs T). The correlation between rs3746444 in miR-499 and HCC susceptibility was determined by using Ors and the corresponding 95%CIs. The heterogeneity among the eligible studies was assessed by using the I2 test and Q test. For heterogeneity, the level of significance was P < 0.1 and/or I2≥ 50%. When it was significant, we used a random-effects model (DerSimonian and Laird) to assess the association between rs3746444 in miR-499 and HCC susceptibility[51,52]. Otherwise, we used a fixed-effects model (Mantel-Haenszel) to determine the potential association[53]. In this study, a Galbraith radial plot was used to confirm the source of the heterogeneity. Sensitivity analysis was performed to explore whether an individual investigation might significantly influence the assessment. We used Egger’s test and Begg’s funnel plots to measure the possible bias among the publications. For publication bias, the level of significance was P < 0.1. STATA 12.0 software (Stata Corp., College Station, Texas) was used to conduct statistical analysis. All P-values were measured with two-sided tests. By using Power-SampleSize software, the power value (α = 0.05) was also used to assess the stability of our study[54]. We used the Newcastle-Ottawa Quality Assessment Scale to assess the quality of eligible studies and defined scores ≥ 7 stars as high-quality studies[55].
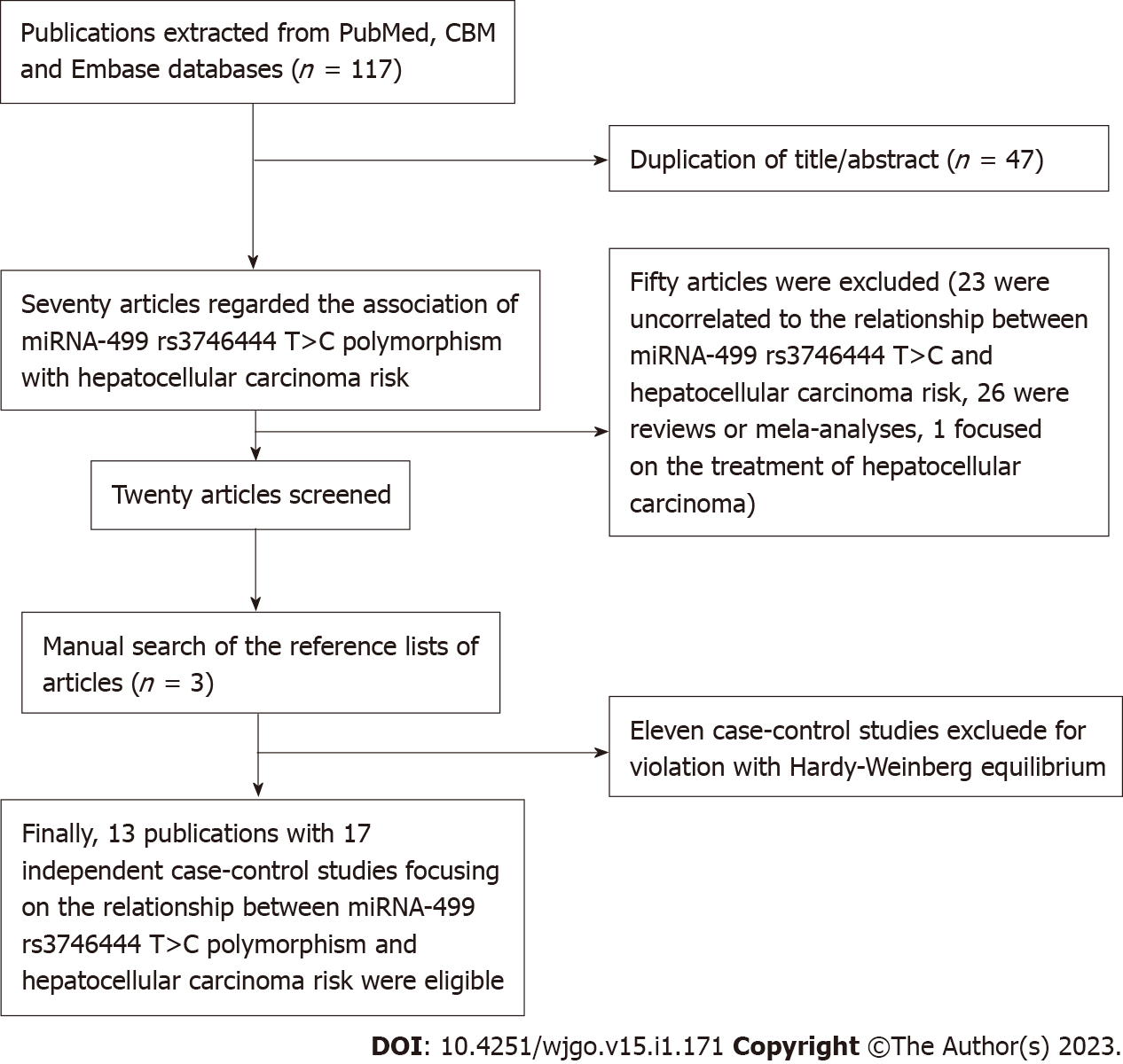
An electronic search of the CBM, PubMed and Embase databases obtained 117 publications. After the titles or abstracts were reviewed by two authors (Jiang JK and Lin J), 47 duplicates were removed. Fifty articles were excluded based on the inclusion criteria (Figure 1). Thus, 20 articles were reviewed in full text. Three publications were included after reading the references of eligible articles. However, 11 case-control studies were excluded for violating HWE. Finally, 13 publications with 17 independent case-control studies focusing on the relationship between the rs3746444 polymorphism and HCC risk were included[40,48,50,56-65].
These included studies were published between 2012 and 2020, and in the eligible case-control studies, the participant number ranged from 100 to 1507. Table 1 shows the included terms in the eligible studies. In summary, 7 case-control studies involving Caucasian individuals were found[50,56-58], and the others focused on Asian individuals[40,48,59-65]. The distributions of the rs3746444 genotypes and alleles in the miR-499 SNP and the results of the quality assessment are summarized in Tables 1 and 2.
| Ref. | Country | Ethnicity | Study design | Sex, male (%); case/control | Mean age (yr); case/control | Smoking (%); case/control | Drinking (%); case/control | HBsAg, positivee (%); case/control | Number cases/controls | Type of control | Case | Control | HWE | ||||
| TT | TC | CC | TT | TC | CC | ||||||||||||
| Zhang et al[59], 2016 | China | Asian | HB | 70.29/56.29 | 56.13/54.96 | 34.29/30.79 | 50.29/36.09 | NA | 175/302 | Normal or healthy control | 115 | 49 | 11 | 197 | 87 | 18 | 0.052 |
| Li et al[60], 2015 | China | Asian | PB | 75.56/75.56 | ≥ 55 yr, 55.26/≥55 yr, 53.38 | 36.47/31.58 | 47.37/36.47 | 41.35/12.03 | 266/250 | Normal or healthy control | 150 | 92 | 24 | 166 | 83 | 17 | 0.140 |
| Yan et al[61], 2015 | China | Asian | PB | 77.74/63.41 | ≥ 55 yr, 55.84/≥ 55 yr, 45.43 | 47.81/42.68 | 58.76/40.55 | 61.31/10.37 | 274/328 | Normal or healthy control | 147 | 98 | 29 | 188 | 112 | 28 | 0.060 |
| Qi et al[62], 2014 | China | Asian | PB | 83.8/83.8 | 50.7/49.6 | 38.9/NA | 27.4/NA | 83.2/0.0 | 314/406 | Normal or healthy control | 195 | 117 | 2 | 301 | 101 | 4 | 0.157 |
| Chu et al[63], 2014 | China | Asian | HB | 72.34/74.78 | < 45 yr, 5.05, 45-59 yr, 30.85, ≥ 60 yr, 63.83/< 45 yr, 7.12, 45-59 yr, 40.06, ≥ 60 yr, 52.82 | 42.55/33.23 | 36.17/40.36 | 42.55/13.23 | 188/337 | Normal or healthy control | 119 | 60 | 9 | 281 | 55 | 1 | 0.321 |
| Zhou et al[64], 2012 | China | Asian | PB | 82.8/NA | 52.1/NA | NA/NA | NA/NA | NA/NA | 186/483 | Normal or healthy control | 141 | 41 | 4 | 371 | 100 | 12 | 0.100 |
| Xiang et al[65], 2012 | China | Asian | HB | 82/39 | 48.55/47.02 | NA/NA | NA/NA | NA/NA | 100/100 | Hepatitis or virus related control | 36 | 40 | 24 | 52 | 35 | 13 | 0.081 |
| Xiang et al[65], 2012 | China | Asian | HB | 82/50 | 48.55/45.12 | NA/NA | NA/NA | NA/NA | 100/100 | Normal or healthy control | 36 | 40 | 24 | 54 | 36 | 10 | 0.284 |
| Kim et al[40], 2012 | Korea | Asian | PB | NA/NA | NA/NA | NA/NA | NA/NA | NA/NA | 159/201 | NA/NA | 109 | 47 | 3 | 120 | 74 | 7 | 0.278 |
| Zhang et al[48], 2020 | China | Asian | HB | 89.90/90.47 | 53.17/53.72 | 35.96/35.43 | 29.11/16.03 | 70.55/9.21 | 584/923 | Normal or healthy control | 409 | 154 | 12 | 669 | 230 | 22 | 0.673 |
| Toraih et al[50], 2016 | Egypt | Caucasian | HB | NA/NA | NA/NA | NA/NA | NA/NA | NA/NA | 60/150 | Normal or healthy control | 28 | 23 | 9 | 57 | 66 | 27 | 0.307 |
| Fteah et al[56], 2019, Abdel-Hamid et al[57], 2018 | Egypt | Caucasian | HB | 80.00/81.33 | 50.12/50.11 | 54.7/0.0 | NA/NA | NA/NA | 75/75 | Normal or healthy control | 41 | 32 | 2 | 31 | 30 | 14 | 0.175 |
| Egypt | Caucasian | HB | 78.0/70.0 | 55.8/54.4 | 34.0/34.0 | NA/NA | 6.0/0.0 | 50/50 | Normal or healthy control | 3 | 32 | 15 | 16 | 23 | 11 | 0.617 | |
| Al-Qahtani et al[58], 2017 | Saudi Arabia | Caucasian | HB | NA/68.4 | NA/40.29 | NA/NA | NA/NA | NA/100.00 | 145/585 | Hepatitis or virus related control | 48 | 70 | 27 | 219 | 273 | 93 | 0.607 |
| Al-Qahtani et al[58], 2017 | Saudi Arabia | Caucasian | HB | NA/79.7 | NA/36.33 | NA/NA | NA/NA | NA/100.00 | 145/222 | Hepatitis or virus related control | 48 | 70 | 27 | 87 | 100 | 35 | 0.486 |
| Al-Qahtani et al[58], 2017 | Saudi Arabia | Caucasian | HB | NA/94.25 | NA/37.49 | NA/NA | NA/NA | NA/0.0 | 145/400 | Normal or healthy control | 48 | 70 | 27 | 148 | 187 | 65 | 0.647 |
| Al-Qahtani et al[58], 2017 | Saudi Arabia | Caucasian | HB | NA/96.30 | NA/30.80 | NA/NA | NA/NA | NA/0.0 | 145/600 | Normal or healthy control | 48 | 70 | 27 | 216 | 291 | 93 | 0.758 |
| Ref. | Selection | Comparability of the cases and controls | Exposure | Total stars | |||||
| Adequate case definition | Representativeness of the cases | Selection of the controls | Definition of Controls | Ascertainment of exposure | Same ascertainment method for cases and controls | Non-response rate | |||
| Zhang et al[59], 2016 | ★ | ★ | - | ★ | ★★ | ★ | ★ | - | 7 |
| Li et al[60], 2015 | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | - | 8 |
| Yan et al[61], 2015 | ★ | ★ | ★ | ★ | ★ | ★ | ★ | - | 7 |
| Qi et al[62], 2014 | ★ | ★ | ★ | ★ | ★ | ★ | ★ | - | 7 |
| Chu et al[63], 2014 | ★ | ★ | - | ★ | ★★ | ★ | ★ | - | 7 |
| Zhou et al[64], 2012 | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | - | 8 |
| Xiang et al[65], 2012 | ★ | ★ | - | ★ | ★ | ★ | ★ | - | 6 |
| Xiang et al[65], 2012 | ★ | ★ | - | ★ | ★ | ★ | ★ | - | 6 |
| Kim et al[40], 2012 | ★ | ★ | ★ | ★ | - | ★ | ★ | - | 6 |
| Zhang et al[48], 2020 | ★ | ★ | - | ★ | ★★ | ★ | ★ | - | 7 |
| Toraih et al[50], 2016 | ★ | ★ | - | ★ | - | ★ | ★ | - | 5 |
| Fteah et al[56], 2019 | ★ | ★ | - | ★ | ★★ | ★ | ★ | - | 7 |
| Abdel-Hamid et al[57], 2018 | ★ | ★ | - | ★ | ★★ | ★ | ★ | - | 7 |
| Al-Qahtani et al[58], 2017 | ★ | ★ | - | ★ | - | ★ | ★ | - | 5 |
| Al-Qahtani et al[58], 2017 | ★ | ★ | - | ★ | - | ★ | ★ | - | 5 |
| Al-Qahtani et al[58], 2017 | ★ | ★ | - | ★ | - | ★ | ★ | - | 5 |
| Al-Qahtani et al[58], 2017 | ★ | ★ | - | ★ | - | ★ | ★ | - | 5 |
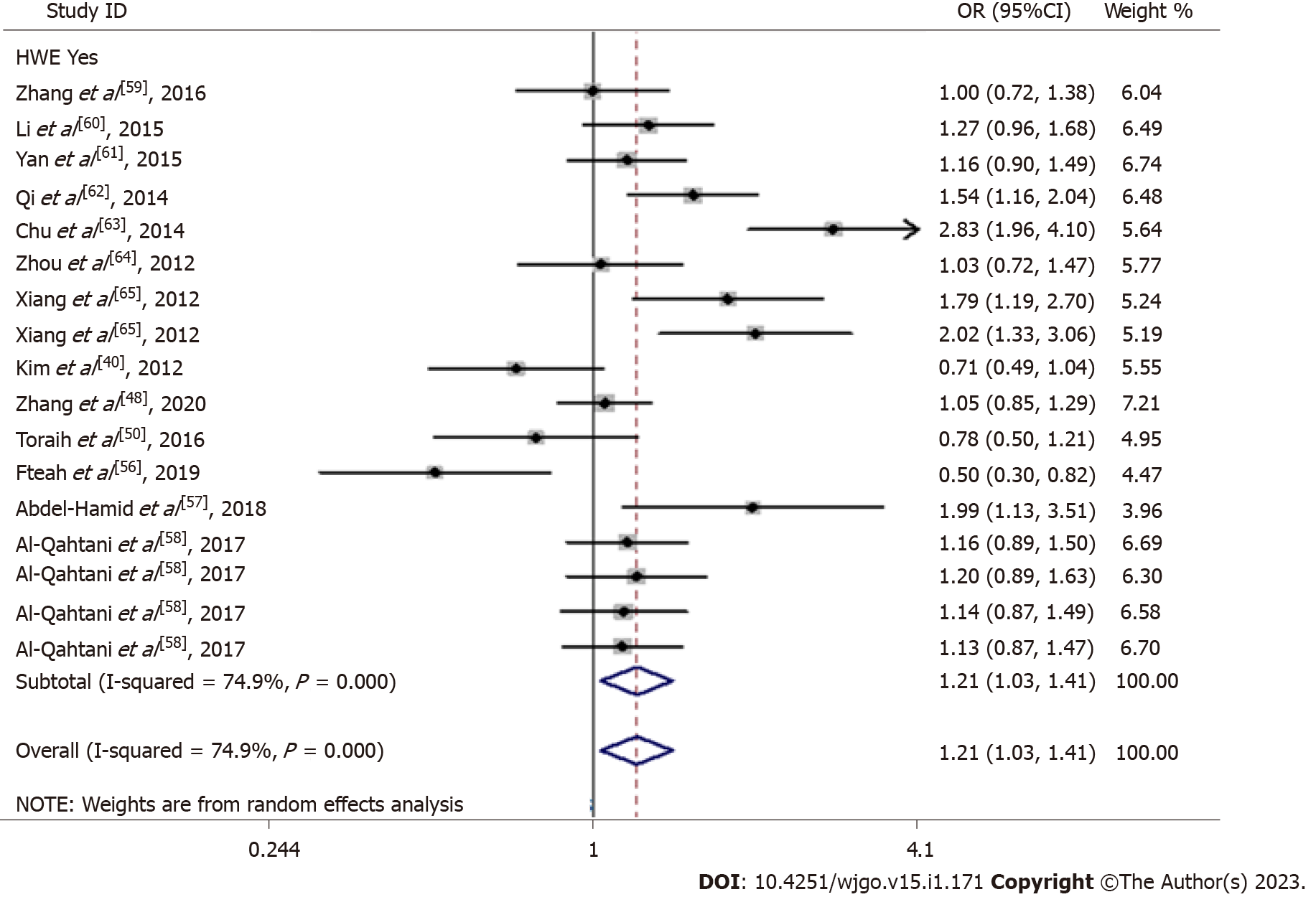
The main results are summarized in Table 3. When we combined the included case-control studies, we identified that rs3746444 in miR-499 was associated with the development of HCC (C vs T: P = 0.019 and CC/CT vs TT: P = 0.016, Figure 2). In a subgroup analysis by different races, rs3746444 in miR-499 was found to be associated with susceptibility to HCC in the Asian population (C vs T: P = 0.013 and CC/CT vs TT: P = 0.016). When we considered the source of disease, the miR-499 rs3746444 locus was identified to be associated with susceptibility to HCC in normal/healthy control individuals (C vs T: P = 0.034 and CC/CT vs TT: P = 0.024) and hepatitis/virus-related control individuals (C vs T: P = 0.007, CC vs TT: P = 0.014 and CC/CT vs TT: P = 0.018).
| Genetic comparison | Population | OR (95%CI) | P value | Test of heterogeneity | Model | Power value | |
| P value | I2 | ||||||
| C vs T | All | 1.21 (1.03-1.41) | 0.019 | < 0.001 | 74.9% | R | 1.000 |
| Ethnicity | |||||||
| Asians | 1.32 (1.06-1.64) | 0.013 | < 0.001 | 79.4% | R | 1.000 | |
| Caucasians | 1.06 (0.86-1.32) | 0.586 | 0.010 | 64.2% | R | - | |
| Study design | |||||||
| HB | 1.25 (1.01-1.54) | 0.039 | < 0.001 | 79.0% | R | 0.999 | |
| PB | 1.13 (0.90-1.42) | 0.285 | 0.027 | 63.6% | R | - | |
| Controls | |||||||
| Normal or healthy control | 1.22 (1.02-1.48) | 0.034 | < 0.001 | 77.2% | R | 0.998 | |
| Hepatitis or virus related control | 1.27 (1.07-1.52) | 0.007 | 0.192 | 39.4% | F | 0.785 | |
| NA | 0.71 (0.49-1.04) | 0.080 | - | - | - | - | |
| Nos quality assessment | |||||||
| ≥ 7.0 | 1.24 (0.97-1.58) | 0.088 | < 0.001 | 81.2% | R | ||
| < 7.0 | 1.17 (0.96-1.43) | 0.216 | 0.004 | 66.2% | R | ||
| CC vs TT | All | 1.33 (0.98-1.80) | 0.071 | 0.002 | 57.3% | R | - |
| Ethnicity | |||||||
| Asians | 1.48 (0.97-2.26) | 0.073 | 0.021 | 54.0% | R | - | |
| Caucasians | 1.16 (0.72-1.87) | 0.534 | 0.008 | 65.3% | R | - | |
| Study design | |||||||
| HB | 1.44 (0.97-2.16) | 0.074 | < 0.001 | 67.7% | R | - | |
| PB | 1.20 (0.83-1.73) | 0.344 | 0.551 | 0.0% | F | - | |
| Controls | |||||||
| Normal or healthy control | 1.31 (0.88-1.93) | 0.183 | 0.001 | 62.8% | R | - | |
| Hepatitis or virus related control | 1.56 (1.10-2.23) | 0.014 | 0.329 | 10.1% | F | 0.725 | |
| NA | 0.47 (0.12-1.87) | 0.285 | - | - | - | - | |
| Nos quality assessment | |||||||
| ≥ 7.0 | 1.26 (0.71-2.25) | 0.436 | 0.02 | 67.3% | R | ||
| < 7.0 | 1.41 (1.01-1.96) | 0.014 | 0.086 | 43.8% | R | 0.881 | |
| CC/CT vs TT | All | 1.26 (1.04-1.51) | 0.016 | < 0.001 | 70.0% | R | 0.999 |
| Ethnicity | |||||||
| Asians | 1.34 (1.06-1.71) | 0.016 | < 0.001 | 75.7% | R | 0.999 | |
| Caucasians | 1.12 (0.83-1.51) | 0.468 | 0.021 | 59.7% | R | - | |
| Study design | |||||||
| HB | 1.32 (1.03-1.70) | 0.031 | < 0.001 | 73.1% | R | 0.999 | |
| PB | 1.16 (0.87-1.54) | 0.309 | 0.014 | 67.9% | R | - | |
| Controls | |||||||
| Normal or healthy control | 1.29 (1.03-1.60) | 0.024 | < 0.001 | 72.5% | R | 0.999 | |
| Hepatitis or virus related control | 1.37 (1.06-1.77) | 0.018 | 0.397 | 0.0% | F | 0.697 | |
| NA | 0.68 (0.44-1.05) | 0.084 | - | - | - | - | |
| Nos quality assessment | |||||||
| ≥ 7.0 | 1.33 (1.01-1.76) | 0.044 | < 0.001 | 78.2% | R | 0.997 | |
| < 7.0 | 1.18 (0.92-1.51) | 0.191 | 0.026 | 56.1% | R | ||
| CC vs TT/CT | All | 1.21 (0.96-1.53) | 0.109 | 0.049 | 39.4% | R | - |
| Ethnicity | |||||||
| Asians | 1.37 (0.95-1.97) | 0.095 | 0.077 | 42.1% | R | - | |
| Caucasians | 1.09 (0.87-1.37) | 0.448 | 0.127 | 39.6% | F | - | |
| Study design | |||||||
| HB | 1.25 (0.93-1.70) | 0.145 | 0.014 | 53.7% | R | - | |
| PB | 1.15 (0.80-1.65) | 0.449 | 0.640 | 0.0% | F | - | |
| Controls | |||||||
| Normal or healthy control | 1.18 (0.87-1.60) | 0.284 | 0.029 | 47.5% | R | - | |
| Hepatitis or virus related control | 1.36 (0.99-1.87) | 0.061 | 0.421 | 0.0% | F | - | |
| NA | 0.53 (0.14-2.10) | 0.368 | - | - | - | - | |
| Nos quality assessment | |||||||
| ≥ 7.0 | 1.08 (0.68-1.71) | 0.744 | 0.027 | 53.7% | R | ||
| < 7.0 | 1.30 (1.05-1.60) | 0.017 | 0.284 | 18.5% | F | 0.734 | |
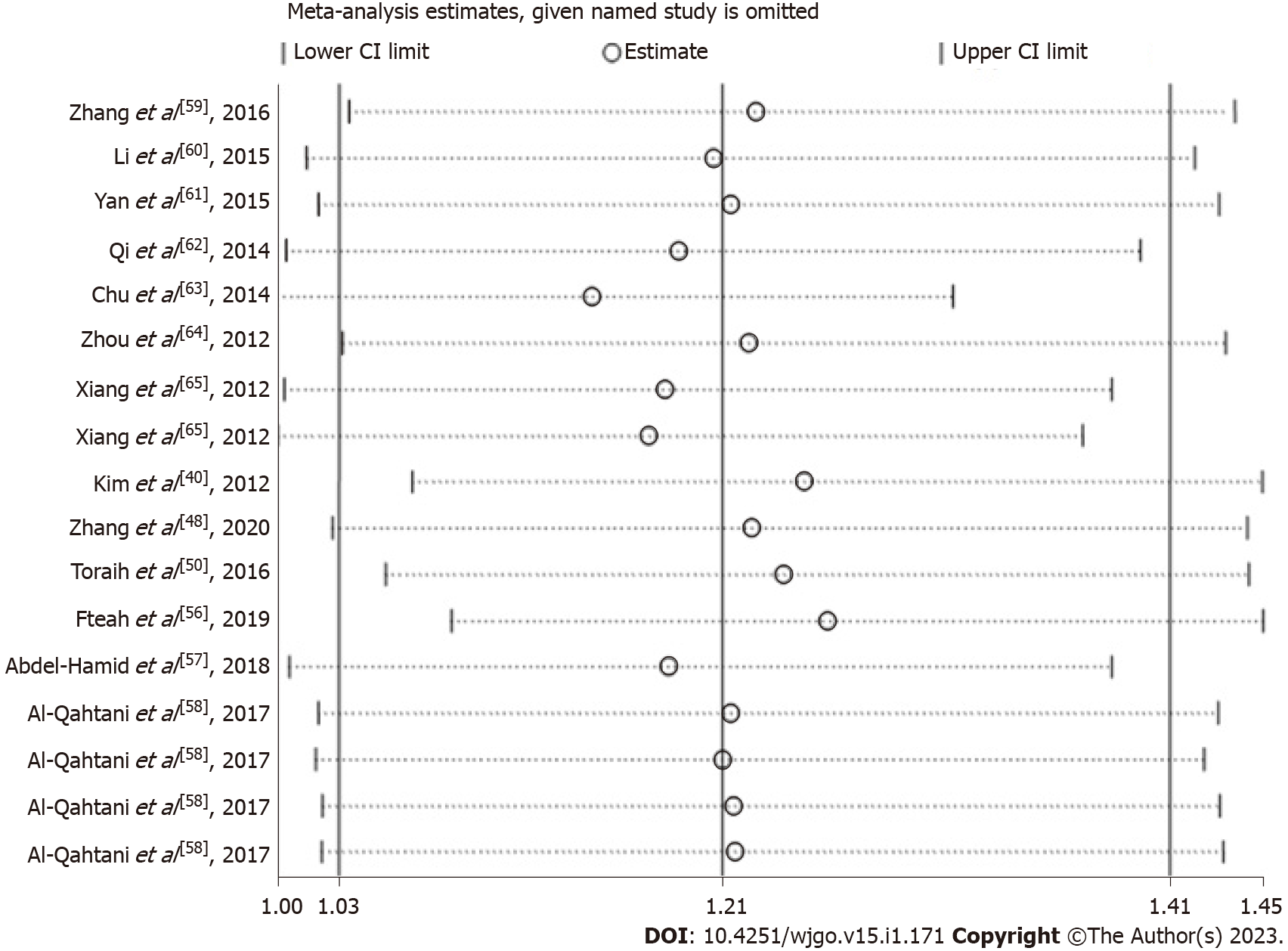
To confirm the stability of our findings, we conducted a sensitivity analysis in this meta-analysis. We deleted an individual study in turn and calculated the Ors and CIs of the remaining studies to determine the influence of each datum. The findings suggested that these evaluations could not be altered by any eligible study (Figure 3).
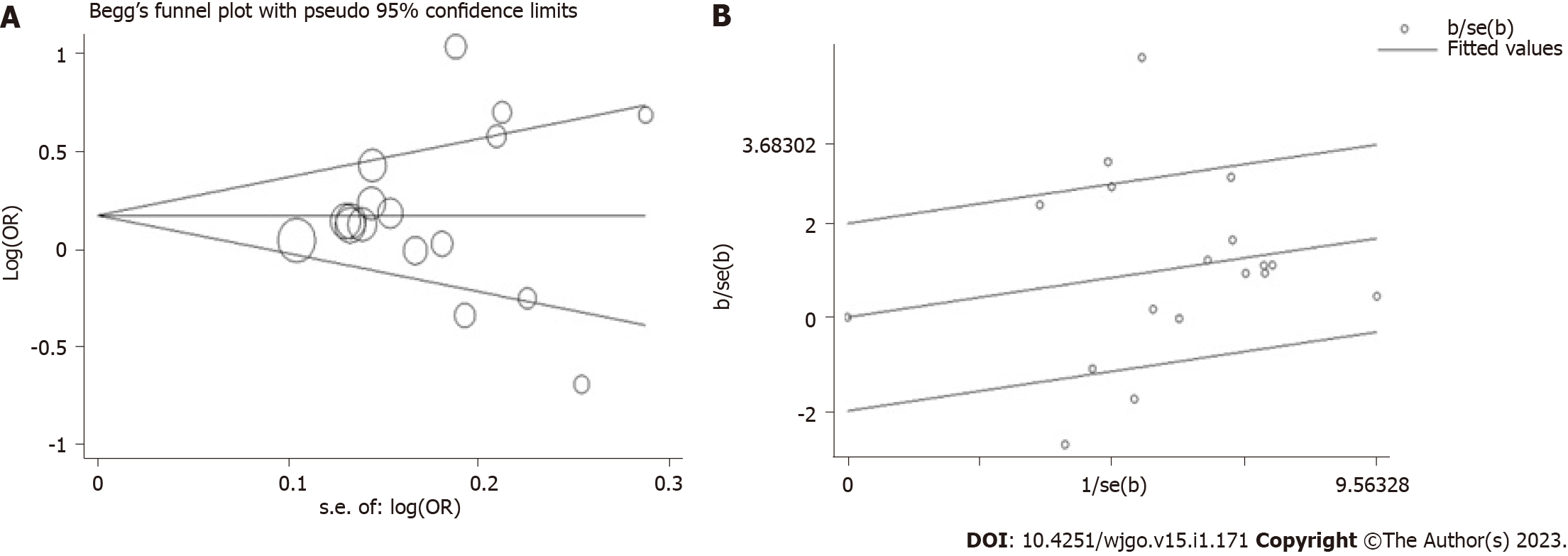
By using Begg’s and Egger’s tests, publication bias among the eligible studies was determined. There was no significant bias among the eligible studies (Figure 4A, P > 0.1, data not shown).
In this meta-analysis, significant heterogeneity was identified. We conducted stratified analyses to explore the source of heterogeneity. Newcastle-Ottawa Scale (Nos) was used to evaluate the literature quality. We found an association between hospital-based (HB) studies, high-quality studies (Nos ≥ 7.0), Asian individuals, and normal/healthy control subgroups and significant heterogeneity. The Galbraith radial plot test suggested that 4 outliers[40,56,63,65] might contribute to the significant heterogeneity (Figure 4B).
By using Power-Sample Size software, the power value (α = 0.05) was also used to assess the stability of our study. As summarized in Table 3, in the overall comparison, the power value was more than 0.8 in the allele and dominant genetic models. In the subgroup analyses, the power value was more than 0.8 in Asian individuals and the normal/healthy control subgroups in the allele genetic model and in Asian individuals and the normal/healthy control subgroups in the dominant genetic model.
Recently, rs3746444 in miR-499 and its importance to the occurrence of HCC have been extensively investigated. However, several meta-analyses reported controversial results, which might be due to the limited sample sizes included in these analyses. Recently, some case-control studies have been conducted to further explore this potential association in different populations. Thus, an updated meta-analysis should be conducted to shed new light on the relationship between rs3746444 in miR-499 and HCC. As summarized in Table 3, we identified that rs3746444 in miR-499 was associated with the development of HCC in the allele and the dominant genetic models (the value of power ≥ 0.8).
The merit of this updated meta-analysis was that the present pooled analysis included a larger sample size to verify whether the miR-499 rs3746444 SNP could influence the occurrence of HCC. In this study, we identified that the miR-499 rs3746444 SNP could confer a risk to HCC. Some meta-analyses have focused on the potential correlation between rs3746444 in miR-499 and the risk of HCC. A previous pooled analysis suggested that the rs3746444T allele in the miR-499 gene could not play a vital role in the tumorigenesis of HCC[42]. However, other meta-analyses reported that rs3746444 in miR-499 might confer susceptibility to HCC[40,45-47]. Additionally, some more recent case-control studies have been conducted to explore the potential association between rs3746444 in the miR-499 SNP and the risk of HCC[48-50]. The potential association was more controversial. Thus, we included 28 independent case-control studies with 5948 cases and 8864 controls and conducted an updated meta-analysis to focus on the relationship between rs3746444 in the miR-499 SNP and the risk of HCC. In this study, we identified that the miR-499 rs3746444 SNP could confer a risk to HCC.
Toraih et al[26] reported that in silico data analysis, the T to C substitution in the miR-499 rs3746444 SNP did not prominently affect the structure of the hairpin loop. Functional prediction revealed that different miR-499 rs3746444 alleles have different targets. The miR-499 rs3746444*C allele only has 58.2% of the gene targets of the rs3746444*T variant and generates 763 new gene targets. The miR-499 gene can target both alcohol dehydrogenase 1 beta polypeptide (ADH1B) and aldehyde dehydrogenase 1 family member A3 (ALDH1A3) genes. Pettinelli et al[66] suggested that hepatic ALDH1A3 was expressed at lower levels and was inversely correlated with the level of plasma retinol in nonalcoholic steatohepatitis cases, which may alter the risk for HCC. Recently, some studies have identified that the ADH1B gene may be involved in the development of HCC[67-69]. A previous study indicated that rs3746444 in miR-499 was correlated with susceptibility to ulcerative colitis and that the expression of miR-499 was decreased (5-fold) in ulcerative colitis cases with the rs3746444 TC genotype compared with those with the rs3746444 TT variant[70]. Taken together, these results indicate that the rs3746444 C allele in the miR-499 gene could decrease the expression of the miR-499 gene and alter the levels of the ADH1B and ALDH1A3 genes. Finally, this SNP could be implicated in the occurrence of HCC. However, the relationship between rs3746444 in miR-499 and HCC in different subgroups could not be well explained. In the future, more attention should be given to the potential mechanism by which hepatitis B virus infection acts in different ethnicities or statuses.
Since significant heterogeneity was found in this meta-analysis, subgroup analysis was performed to observe the major source of heterogeneity. The findings of the subgroup analysis indicated that the normal/healthy control, Asian and HB subgroups could greatly increase the heterogeneity. Additionally, the Galbraith radial plot identified 4 outliers[40,56,63,65], which could contribute to the major source of heterogeneity.
There are some limitations in this meta-analysis. First, the electronic literature was only searched in the PubMed, Embase and CBM databases, and bias might have occurred. Second, all investigations have been conducted in Caucasian and Asian populations; thus, our findings were only appropriate for these populations. Third, due to insufficient data (e.g., HBsAg, drinking, smoking, sex, age, body mass index and lifestyle) in this study, we did not consider these factors in the subgroup analysis. Fourth, due to the lack of environmental factors, we also did not take into account gene-environment interactions. Fifth, in this meta-analysis, significant heterogeneity was identified. Sixth, we did not pay close attention to the expression level of target genes, which could be controlled by rs3746444 T>C locus. Finally, in this study, we only included the relationship between rs3746444 in miR-499 and HCC risk. The potential role of other vital miR loci can’t be ignored.
In summary, this meta-analysis highlights that rs3746444 in miR-499 is involved in the occurrence of HCC, especially in Asian individuals. In the future, more investigations are needed to confirm our results.
This meta-analysis highlights that rs3746444 in microRNA (miR)-499 is involved in the occurrence of hepatocellular carcinoma (HCC), especially in Asian individuals. These possible relationships might be beneficial to the prevention of liver carcinogenesis. In the future, more investigations are needed to confirm.
Recently, a number of studies have focused on the relationship between rs3746444 in miR-499 and HCC. However, the obtained findings are conflicting.
In summary, this meta-analysis highlights that rs3746444 in miR-499 is involved in the occurrence of HCC, especially in Asian individuals. In the future, more investigations are needed to confirm our results.
This meta-analysis involved a large sample size to verify whether the miR-499 rs3746444 single-nucleotide polymorphism could influence the occurrence of HCC. These possible relationships might be beneficial to the prevention of liver carcinogenesis.
Reports on the association between rs3746444 and HCC are conflicting.
The results of this meta-analysis were assessed in four genetic models: A dominant model (CC/TC vs TT), recessive model (CC vs TT/TC), homozygote comparison (CC vs TT) and allelic model (C vs T). The correlation between rs3746444 in miR-499 and HCC susceptibility was determined by using odd ratios and the corresponding 95% confidence intervals. We used a random-effects model (DerSimonian and Laird) to assess the association between rs3746444 in miR-499 and HCC susceptibility. Otherwise, we used a fixed-effects model (Mantel-Haenszel) to determine the potential association. We used the Newcastle-Ottawa Quality Assessment Scale to assess the quality of eligible studies and defined scores ≥ 7 stars as high-quality studies.
Reports on the association between rs3746444 and HCC are conflicting. This meta-analysis highlights that rs3746444 in miR-499 is involved in the occurrence of HCC, especially in Asian individuals. These possible relationships might be beneficial to the prevention of liver carcinogenesis.
We wish to thank Dr. Hao Ding (Affiliated People’s Hospital of Jiangsu University, China) for technical support.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Keikha M, Iran; Skrypnik D, Poland S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64133] [Article Influence: 16033.3] [Reference Citation Analysis (174)] |
| 2. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13196] [Article Influence: 1466.2] [Reference Citation Analysis (3)] |
| 3. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55701] [Article Influence: 7957.3] [Reference Citation Analysis (132)] |
| 4. | Zhang CH, Xu GL, Jia WD, Li JS, Ma JL, Ge YS. Effects of interferon treatment on development and progression of hepatocellular carcinoma in patients with chronic virus infection: a meta-analysis of randomized controlled trials. Int J Cancer. 2011;129:1254-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 5. | Elemeery MN, Mohamed MA, Madkour MA, Shamseya MM, Issa NM, Badr AN, Ghareeb DA, Pan CH. MicroRNA signature in patients with hepatocellular carcinoma associated with type 2 diabetes. World J Gastroenterol. 2019;25:6322-6341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Michel M, Kalliga E, Labenz C, Straub BK, Wörns MA, Galle PR, Schattenberg JM. A young patient with type 2 diabetes associated non-alcoholic steatohepatitis, liver cirrhosis, and hepatocellular carcinoma. Z Gastroenterol. 2020;58:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 7. | Chong YC, Lim TE, Fu Y, Shin EM, Tergaonkar V, Han W. Indian Hedgehog links obesity to development of hepatocellular carcinoma. Oncogene. 2019;38:2206-2222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Chen Y, Zhang F, Zhao Y, He K, Zheng X, Pan Y, Shao D, Shang P, Yang Y, Zhang D, Xie Y, Yao X, Chen L, Li J, Zhang X. Obesity-associated miR-27a upregulation promotes hepatocellular carcinoma metastasis through suppressing SFRP1. Onco Targets Ther. 2018;11:3281-3292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 9. | Guarino M, Dufour JF. Smoking favours hepatocellular carcinoma. Ann Transl Med. 2019;7:S99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Li CL, Lin YK, Chen HA, Huang CY, Huang MT, Chang YJ. Smoking as an Independent Risk Factor for Hepatocellular Carcinoma Due to the α7-Nachr Modulating the JAK2/STAT3 Signaling Axis. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 11. | Mukaiya M, Nishi M, Miyake H, Hirata K. Chronic liver diseases for the risk of hepatocellular carcinoma: a case-control study in Japan. Etiologic association of alcohol consumption, cigarette smoking and the development of chronic liver diseases. Hepatogastroenterology. 1998;45:2328-2332. [PubMed] |
| 12. | Okada S, Ishii H, Nose H, Okusaka T, Kyogoku A, Yoshimori M, Shimada K, Yamamoto J, Kosuge T, Yamasaki S, Sakamoto M, Hirohashi S. Effect of heavy alcohol intake on long-term results after curative resection of hepatitis C virus-related hepatocellular carcinoma. Jpn J Cancer Res. 1996;87:867-873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Ioannou GN, Green P, Kerr KF, Berry K. Models estimating risk of hepatocellular carcinoma in patients with alcohol or NAFLD-related cirrhosis for risk stratification. J Hepatol. 2019;71:523-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 152] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 14. | Hifnawy MS, Mangoud AM, Eissa MH, Nor Edin E, Mostafa Y, Abouel-Magd Y, Sabee EI, Amin I, Ismail A, Morsy TA, Mahrous S, Afefy AF, el-Shorbagy E, el-Sadawy M, Ragab H, Hassan MI, el-Hady G, Saber M. The role of aflatoxin-contaminated food materials and HCV in developing hepatocellular carcinoma in Al-Sharkia Governorate, Egypt. J Egypt Soc Parasitol. 2004;34:479-488. [PubMed] |
| 15. | Yang J, Liu J, Chen Y, Tang W, Bo K, Sun Y, Chen J. Investigation of ICOS, CD28 and CD80 polymorphisms with the risk of hepatocellular carcinoma: a case-control study in eastern Chinese population. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Yang J, Liu J, Chen Y, Tang W, Liu C, Sun Y, Chen J. Association of CTLA-4 tagging polymorphisms and haplotypes with hepatocellular carcinoma risk: A case-control study. Medicine (Baltimore). 2019;98:e16266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Zhang S, Lin J, Jiang J, Chen Y, Tang W, Liu L. Association between methylenetetrahydrofolate reductase tagging polymorphisms and susceptibility of hepatocellular carcinoma: a case-control study. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Zhang S, Jiang J, Chen Z, Wang Y, Tang W, Chen Y, Liu L. Relationship of PPARG, PPARGC1A, and PPARGC1B polymorphisms with susceptibility to hepatocellular carcinoma in an eastern Chinese Han population. Onco Targets Ther. 2018;11:4651-4660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Wang L, Hu K, Chao Y, Wang X. MicroRNA-1296-5p suppresses the proliferation, migration, and invasion of human osteosarcoma cells by targeting NOTCH2. J Cell Biochem. 2020;121:2038-2046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 20. | Hu XH, Zhao ZX, Dai J, Geng DC, Xu YZ. MicroRNA-221 regulates osteosarcoma cell proliferation, apoptosis, migration, and invasion by targeting CDKN1B/p27. J Cell Biochem. 2019;120:4665-4674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Zhao X, Lu C, Chu W, Zhang Y, Zhang B, Zeng Q, Wang R, Li Z, Lv B, Liu J. microRNA-214 Governs Lung Cancer Growth and Metastasis by Targeting Carboxypeptidase-D. DNA Cell Biol. 2016;35:715-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Jian Y, Xu CH, Li YP, Tang B, Xie SH, Zeng EM. Down-regulated microRNA-30b-3p inhibits proliferation, invasion and migration of glioma cells via inactivation of the AKT signaling pathway by up-regulating RECK. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Wang MJ, Zhang H, Li J, Zhao HD. microRNA-98 inhibits the proliferation, invasion, migration and promotes apoptosis of breast cancer cells by binding to HMGA2. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 24. | Li J, Jin B, Wang T, Li W, Wang Z, Zhang H, Song Y, Li N. Serum microRNA expression profiling identifies serum biomarkers for HCV-related hepatocellular carcinoma. Cancer Biomark. 2019;26:501-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 25. | Chen LB, Zheng HK, Zhang L, An Z, Wang XP, Shan RT, Zhang WQ. A single nucleotide polymorphism located in microRNA-499a causes loss of function resulting in increased expression of osbpl1a and reduced serum HDL level. Oncol Rep. 2017;38:3515-3521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 26. | Toraih EA, Hussein MH, Al Ageeli E, Riad E, AbdAllah NB, Helal GM, Fawzy MS. Structure and functional impact of seed region variant in MIR-499 gene family in bronchial asthma. Respir Res. 2017;18:169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Xu HY, Wang ZY, Chen JF, Wang TY, Wang LL, Tang LL, Lin XY, Zhang CW, Chen BC. Association between ankylosing spondylitis and the miR-146a and miR-499 polymorphisms. PLoS One. 2015;10:e0122055. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Toraih EA, Ismail NM, Toraih AA, Hussein MH, Fawzy MS. Precursor miR-499a Variant but not miR-196a2 is Associated with Rheumatoid Arthritis Susceptibility in an Egyptian Population. Mol Diagn Ther. 2016;20:279-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 29. | Liu X, Zhang Z, Sun L, Chai N, Tang S, Jin J, Hu H, Nie Y, Wang X, Wu K, Jin H, Fan D. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis. 2011;32:1798-1805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 30. | Wei W, Hu Z, Fu H, Tie Y, Zhang H, Wu Y, Zheng X. MicroRNA-1 and microRNA-499 downregulate the expression of the ets1 proto-oncogene in HepG2 cells. Oncol Rep. 2012;28:701-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 31. | Tang W, Wang Y, Pan H, Qiu H, Chen S. Association of miRNA-499 rs3746444 A>G variants with adenocarcinoma of esophagogastric junction (AEG) risk and lymph node status. Onco Targets Ther. 2019;12:6245-6252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Nouri R, Ghorbian S. Association of single nucleotide polymorphism in hsa-miR-499 and hsa-miR-196a2 with the risk of prostate cancer. Int Urol Nephrol. 2019;51:811-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 33. | Srivastava S, Singh S, Fatima N, Mittal B, Srivastava AN. Pre-microRNA Gene Polymorphisms and Risk of Cervical Squamous Cell Carcinoma. J Clin Diagn Res. 2017;11:GC01-GC04. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 34. | Zhang E, Xu Z, Duan W, Huang S, Lu L. Association between polymorphisms in pre-miRNA genes and risk of oral squamous cell cancer in a Chinese population. PLoS One. 2017;12:e0176044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 35. | Li D, Zhu G, Di H, Li H, Liu X, Zhao M, Zhang Z, Yang Y. Associations between genetic variants located in mature microRNAs and risk of lung cancer. Oncotarget. 2016;7:41715-41724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Akkiz H, Bayram S, Bekar A, Akgöllü E, Üsküdar O. Genetic variation in the microRNA-499 gene and hepatocellular carcinoma risk in a Turkish population: lack of any association in a case-control study. Asian Pac J Cancer Prev. 2011;12:3107-3112. [PubMed] |
| 37. | Wang XH, Wang FR, Tang YF, Zou HZ, Zhao YQ. Association of miR-149C>T and miR-499A>G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genet Mol Res. 2014;13:5048-5054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 38. | Kou JT, Fan H, Han D, Li L, Li P, Zhu J, Ma J, Zhang ZH, He Q. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol Lett. 2014;8:1255-1260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 39. | Shan YF, Huang YH, Chen ZK, Huang KT, Zhou MT, Shi HQ, Song QT, Yu ZP, Deng AM, Zhang QY. miR-499A>G rs3746444 and miR-146aG>C expression and hepatocellular carcinoma risk in the Chinese population. Genet Mol Res. 2013;12:5365-5371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, Kim NK. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 41. | Zheng L, Zhuang C, Zhao J, Ming L. Functional miR-146a, miR-149, miR-196a2 and miR-499 polymorphisms and the susceptibility to hepatocellular carcinoma: An updated meta-analysis. Clin Res Hepatol Gastroenterol. 2017;41:664-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Yu JY, Hu F, Du W, Ma XL, Yuan K. Study of the association between five polymorphisms and risk of hepatocellular carcinoma: A meta-analysis. J Chin Med Assoc. 2017;80:191-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 43. | Ye LX, Fu CW, Jiang F, Cui YX, Meng W. [A meta-analysis of microRNA-149, microRNA-499 gene polymorphism and susceptibility to hepatocellular carcinoma]. Zhonghua Yu Fang Yi Xue Za Zhi. 2016;50:445-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 44. | Liu F, Lin H, Cheng Y, Yang L, Liu Y. rs3746444 polymorphism and susceptibility to hepatocellular carcinoma: evidence from published studies. Cell Biochem Biophys. 2014;70:1957-1961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 45. | Qiu D, Han F, Zhuang H. MiR-499 rs3746444 polymorphism and hepatocellular carcinoma risk: A meta-analysis. J Cancer Res Ther. 2018;14:S490-S493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 46. | Yu H, Wang Y, Wang S, Sun N. Association between miR-499 rs3746444 and the susceptibility of hepatocellular carcinoma. Cell Mol Biol (Noisy-le-grand). 2016;62:42-45. [PubMed] |
| 47. | Zhu SL, Zhong JH, Gong WF, Li H, Li LQ. Association of the miR-196a2 C>T and miR-499 A>G polymorphisms with hepatitis B virus-related hepatocellular carcinoma risk: an updated meta-analysis. Onco Targets Ther. 2016;9:2111-2119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 48. | Zhang S, Chen L, Wang Y, Tang W, Chen Y, Liu L. Investigation of the Association of miRNA-499, miRNA-146a, miRNA-196a2 Loci with Hepatocellular Carcinoma Risk: A Case-Control Study Involving 1507 Subjects. DNA Cell Biol. 2020;39:379-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 49. | Farokhizadeh Z, Dehbidi S, Geramizadeh B, Yaghobi R, Malekhosseini SA, Behmanesh M, Sanati MH, Afshari A, Moravej A, Karimi MH. Association of MicroRNA Polymorphisms With Hepatocellular Carcinoma in an Iranian Population. Ann Lab Med. 2019;39:58-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 50. | Toraih EA, Fawz MS, Elgazzaz MG, Hussein MH, Shehata RH, Daoud HG. Combined Genotype Analyses of Precursor miRNA196a2 and 499a Variants with Hepatic and Renal Cancer Susceptibility a Preliminary Study. Asian Pac J Cancer Prev. 2016;17:3369-3375. |
| 51. | Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39087] [Cited by in RCA: 46306] [Article Influence: 2104.8] [Reference Citation Analysis (3)] |
| 52. | DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26739] [Cited by in RCA: 30338] [Article Influence: 777.9] [Reference Citation Analysis (0)] |
| 53. | Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719-748. [PubMed] |
| 54. | Tang W, Qiu H, Ding H, Sun B, Wang L, Yin J, Gu H. Association between the STK15 F31I polymorphism and cancer susceptibility: a meta-analysis involving 43,626 subjects. PLoS One. 2013;8:e82790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 55. | Wang W, Shao Y, Tang S, Cheng X, Lian H, Qin C. Peroxisome proliferator-activated receptor-γ (PPARγ) Pro12Ala polymorphism and colorectal cancer (CRC) risk. Int J Clin Exp Med. 2015;8:4066-4072. [PubMed] |
| 56. | Fteah AM, Ahmed AI, Mosaad NA, Hassan MM, Mahmoud SH. Association of MicroRNA 196a and 499 Polymorphisms with Development of Cirrhosis and Hepatocellular Carcinoma Post-HCV Infection in Egyptian Patients. Asian Pac J Cancer Prev. 2019;20:3479-3485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 57. | Abdel-Hamid M, Elshaer S, Darwish A. Association of MicroRNA related single nucleotide polymorphisms 196A-2 and 499 with the risk of hepatocellular carcinoma in Egyptian patients. Meta Gene. 2018;16:139-142. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 58. | Al-Qahtani AA, Al-Anazi MR, Nazir N, Wani K, Abdo AA, Sanai FM, Khan MQ, Al-Ashgar HI, Albenmousa A, Al-Hamoudi WK, Alswat KA, Al-Ahdal MN. Association of single nucleotide polymorphisms in microRNAs with susceptibility to hepatitis B virus infection and HBV-related liver complications: A study in a Saudi Arabian population. J Viral Hepat. 2017;24:1132-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 59. | Zhang LH, Hao BB, Zhang CY, Dai XZ, Zhang F. Contributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinoma. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 60. | Li X, Li K, Wu Z. Association of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:9560-9566. [PubMed] |
| 61. | Yan P, Xia M, Gao F, Tang G, Zeng H, Yang S, Zhou H, Ding D, Gong L. Predictive role of miR-146a rs2910164 (C>G), miR-149 rs2292832 (T>C), miR-196a2 rs11614913 (T>C) and miR-499 rs3746444 (T>C) in the development of hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:15177-15183. [PubMed] |
| 62. | Qi JH, Wang J, Chen J, Shen F, Huang JT, Sen S, Zhou X, Liu SM. High-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patients. BMC Cancer. 2014;14:643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 63. | Chu YH, Hsieh MJ, Chiou HL, Liou YS, Yang CC, Yang SF, Kuo WH. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One. 2014;9:e89930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 64. | Zhou J, Lv R, Song X, Li D, Hu X, Ying B, Wei Y, Wang L. Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol. 2012;31:524-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 65. | Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. 2012;39:7019-7023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 66. | Pettinelli P, Arendt BM, Teterina A, McGilvray I, Comelli EM, Fung SK, Fischer SE, Allard JP. Altered hepatic genes related to retinol metabolism and plasma retinol in patients with non-alcoholic fatty liver disease. PLoS One. 2018;13:e0205747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 67. | Udali S, Guarini P, Ruzzenente A, Ferrarini A, Guglielmi A, Lotto V, Tononi P, Pattini P, Moruzzi S, Campagnaro T, Conci S, Olivieri O, Corrocher R, Delledonne M, Choi SW, Friso S. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics. 2015;7:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 68. | Liu J, Yang HI, Lee MH, Jen CL, Hu HH, Lu SN, Wang LY, You SL, Huang YT, Chen CJ. Alcohol Drinking Mediates the Association between Polymorphisms of ADH1B and ALDH2 and Hepatitis B-Related Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev. 2016;25:693-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 69. | Gaviria-Calle M, Duque-Jaramillo A, Aranzazu M, Di Filippo D, Montoya M, Roldán I, Palacio N, Jaramillo S, Restrepo JC, Hoyos S, Navas MC. Polymorphisms in alcohol dehydrogenase (ADH1) and cytochrome p450 2E1 (CYP2E1) genes in patients with cirrhosis and/or hepatocellular carcinoma. Biomedica. 2018;38:555-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 70. | Ranjha R, Meena NK, Singh A, Ahuja V, Paul J. Association of miR-196a-2 and miR-499 variants with ulcerative colitis and their correlation with expression of respective miRNAs. PLoS One. 2017;12:e0173447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |