Published online Sep 15, 2022. doi: 10.4251/wjgo.v14.i9.1874
Peer-review started: March 16, 2022
First decision: June 12, 2022
Revised: June 30, 2022
Accepted: July 31, 2022
Article in press: July 31, 2022
Published online: September 15, 2022
Processing time: 176 Days and 21.1 Hours
Twist is a repressor of E-cadherin transcription that induces epithelial-mesen
To investigate the prognostic and clinicopathological value of Twist expression in esophageal cancer.
Published literature in databases such as EMBASE, Web of Science, PubMed, China National Knowledge Infrastructure, Wanfang, and VIP databases was searched for eligible articles. Participants with esophageal cancer whose tumor tissues underwent immunohistochemistry to detect the expression of Twist were considered. Our meta-analysis was conducted using Stata version 12.0. The hazard ratio (HR) and relative ratio (RR) with their 95%CI were pooled. Heterogeneity was estimated by I2 statistics.
Eleven articles published between 2009 and 2021 fulfilled the selection criteria. The pooled HR for overall survival was 1.88 (95%CI: 1.32-2.69, I2 = 68.6%), and the pooled HR for disease-free survival/relapse-free survival/progression-free survival was 1.84 (95%CI: 1.12-3.02, I2 = 67.1%), suggesting that high Twist expression is associated with poor prognosis in esophageal cancer patients. In addition, overexpression of Twist was correlated with T stage (T3 + T4 vs T1 + T2, RR = 1.38, 95%CI: 1.14-1.67), lymph node metastasis (yes vs no, RR = 1.34, 95%CI: 1.11-1.60), distant metastasis (yes vs no, RR = 1.18, 95%CI: 1.02-1.35), tumor, node and metastasis (TNM) stage (III + IV vs I + II, RR = 1.35, 95%CI: 1.14-1.60), and clinical stage (III + IV vs I + II, RR = 1.58, 95%CI: 1.34-1.87). However, no correlation between Twist expression and age, gender, tumor location, differentiation, or venous invasion was observed.
High expression of Twist is associated with poor esophageal cancer prognosis. Moreover, Twist overexpression is correlated with T stage, lymph node metastasis, distant metastasis, TNM stage, and clinical stage, which indicates that Twist might accelerate esophageal cancer progression and metastasis.
Core Tip: Esophageal cancer is a leading cause of cancer mortality worldwide. Twist is a transcription factor involved in the process of epithelial-mesenchymal transition and esophageal cancer metastasis. However, the prognostic value of Twist expression in patients with esophageal cancer remains controversial. Therefore, we conducted a meta-analysis to investigate the prognostic and clinicopathological value of Twist expression in esophageal cancer in terms of overall survival, disease-free survival/relapse-free survival/progression-free survival, age, gender, tumor location, T stage, differentiation, lymph node metastasis, distant metastasis, tumor, node and metastasis stage, clinical-stage, and venous invasion.
- Citation: Song WP, Wang SY, Zhou SC, Wu DS, Xie JY, Liu TT, Wu XZ, Che GW. Prognostic and clinicopathological value of Twist expression in esophageal cancer: A meta-analysis. World J Gastrointest Oncol 2022; 14(9): 1874-1886
- URL: https://www.wjgnet.com/1948-5204/full/v14/i9/1874.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i9.1874
According to the latest global cancer burden report, there were an estimated 572000 new esophageal cancer cases and 509000 deaths in 2020, ranking seventh and fifth in morbidity and mortality, respectively[1]. Among esophageal cancers, 90% of the histological types are esophageal squamous cell carcinoma (ESCC)[1-3]. Although a slew of breakthroughs in terms of the diagnosis and treatment of esophageal cancer has been achieved[4], the 5-year survival rate of ESCC is only 15%–20%[5] due to invasion and distant metastasis. Therefore, there is an urgent need for the identification of new prognostic biomarkers to address the poor prognosis of esophageal cancer.
Epithelial-mesenchymal transition (EMT) describes a key developmental program in which epithelial cells change to motile mesenchymal cells[6]. Tumor cells can undergo EMT to promote local invasion[7], which is the first step of tumor metastasis[8]. Twist is reported to be a helix-loop-helix transcription factor that can directly bind to the promoter of E-cadherin, a tumor suppressor gene associated with EMT, and downregulate E-cadherin expression[9,10]. Thus, Twist can induce EMT and tumor metastasis. The prognostic value of Twist in esophageal cancer has been investigated in many studies[11-21] with controversial results. Some studies[12,13,15,17] have shown that Twist overexpression is closely related to the poor prognosis of esophageal cancer, while others show that it is unrelated[11,14,16,18-21]. Therefore, we performed a meta-analysis to combine relevant studies and clarify whether Twist could be a promising biomarker for predicting prognosis in esophageal cancer.
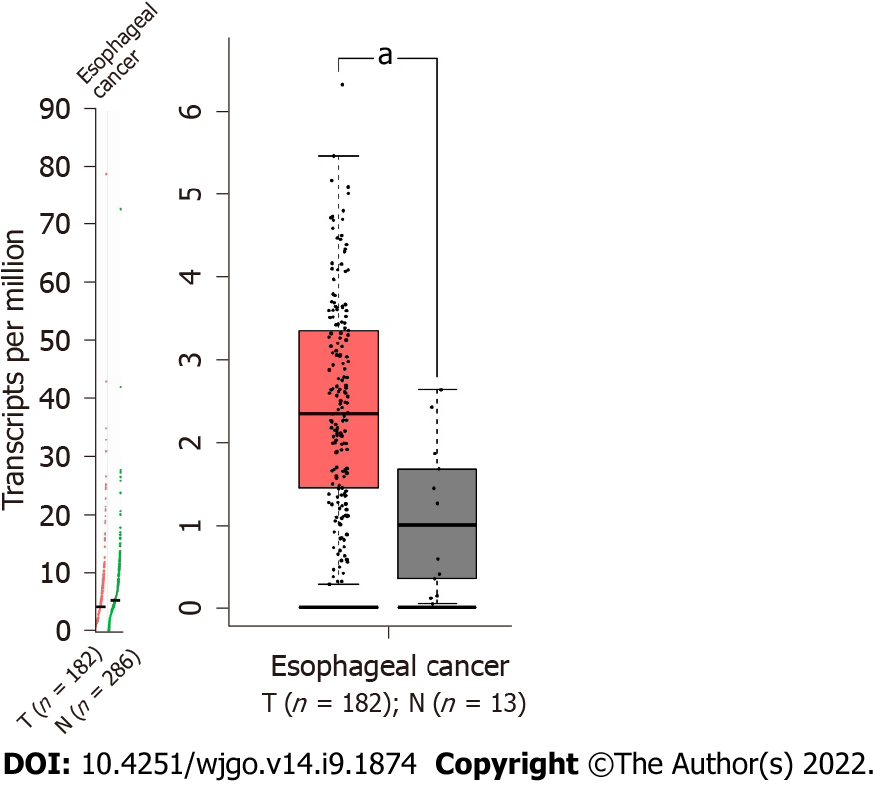
Gene expression profiling interactive analysis 2[22] (GEPIA2) is a valuable and efficient web server with which we can perform gene expression analysis based on the The Cancer Genome Atlas and the Genotype-Tissue Expression databases. We used GEPIA2 to analyze the expression of Twist in esophageal cancer tissues and normal tissue. Scatter diagrams and box plots were generated to assess the expression of Twist in esophageal cancer tissues and normal tissues.
A systematic literature search of the EMBASE, Web of Science, PubMed, China National Knowledge Infrastructure, Wanfang, and VIP databases was conducted to identify relevant studies up to December 28, 2021. The following keywords were variably combined: “Twist”, “esophageal”, “esophagus”, “tumor”, “cancer”, “carcinoma”, and “neoplasm”. Moreover, relevant meta-analysis articles, reviews, and references from the included studies were also screened.
The inclusion criteria in the present meta-analysis were as follows: (1) Twist expression was analyzed in human esophageal cancer tissues; (2) The hazard ratio (HR) with 95%CI was reported or available to be calculated indirectly; (3) Correlations between Twist expression and clinicopathologic characteristics were investigated; and (4) The reports were published in English or Chinese. The exclusion criteria were as follows: (1) Duplicate studies; (2) Reviews, animal experiments, case reports, and conference abstracts; and (3) The HR or 95%CI were unavailable.
Two of the authors (Wen-Peng Song and Su-Yan Wang) independently extracted the following data from each eligible study: the first author, year of publication, country, sample size, tumor location, positive proportion of Twist, tumor, node and metastasis (TNM) stage, clinical stage, venous invasion, detection method, cutoff value, antibodies against Twist, follow-up time, survival analysis, and HR estimates for positive or high expression of Twist vs negative or low expression of Twist, with their 95%CIs.
Two of the authors (Wen-Peng Song and Su-Yan Wang) independently assessed the quality of the included studies with the Newcastle–Ottawa scale (NOS) criteria. Included studies with NOS scores ≥ 6 were considered high-quality studies[23].
Our meta-analysis was conducted using Stata version 12.0 (StataCorp, College Station, Texas 77845 United States). We derived pooled HRs and their 95%CIs for all types of survival outcomes [overall survival (OS), disease-free survival (DFS), relapse-free survival (RFS), progression-free survival (PFS)]. Heterogeneity of the effect across the included studies was estimated by I2 statistics. We used a random-effects model if I2 > 50% and/or P < 0.10, which indicated the presence of significant heterogeneity. Otherwise, we used a fixed-effects model[24]. Moreover, we further investigated the correlations between Twist expression and clinicopathologic characteristics. These clinicopathologic characteristics included age, gender, tumor location (e.g., upper thorax, middle thorax, lower thorax), T stage, differentiation, lymph node metastasis, distant metastasis, TNM, clinical stage, and venous invasion. We performed sensitivity analyses to estimate the stability of the meta-analysis results. Publication bias was assessed with Egger’s test and Begg’s funnel plots[25,26]. P values less than 0.05 indicated the presence of significant publication bias[27]. In addition, we used the Reference Citation Analysis database (https://www.referencecitationanalysis.com/) to retrieve and supplement cutting-edge research results.
We used the GEPIA2 web server to detect the expression of Twist in esophageal cancer tissues and normal tissues. The expression of Twist was significantly higher in esophageal cancer tissues than in normal tissues (Figure 1). Therefore, we further explored the prognostic value of Twist overexpression in esophageal cancer by meta-analysis.
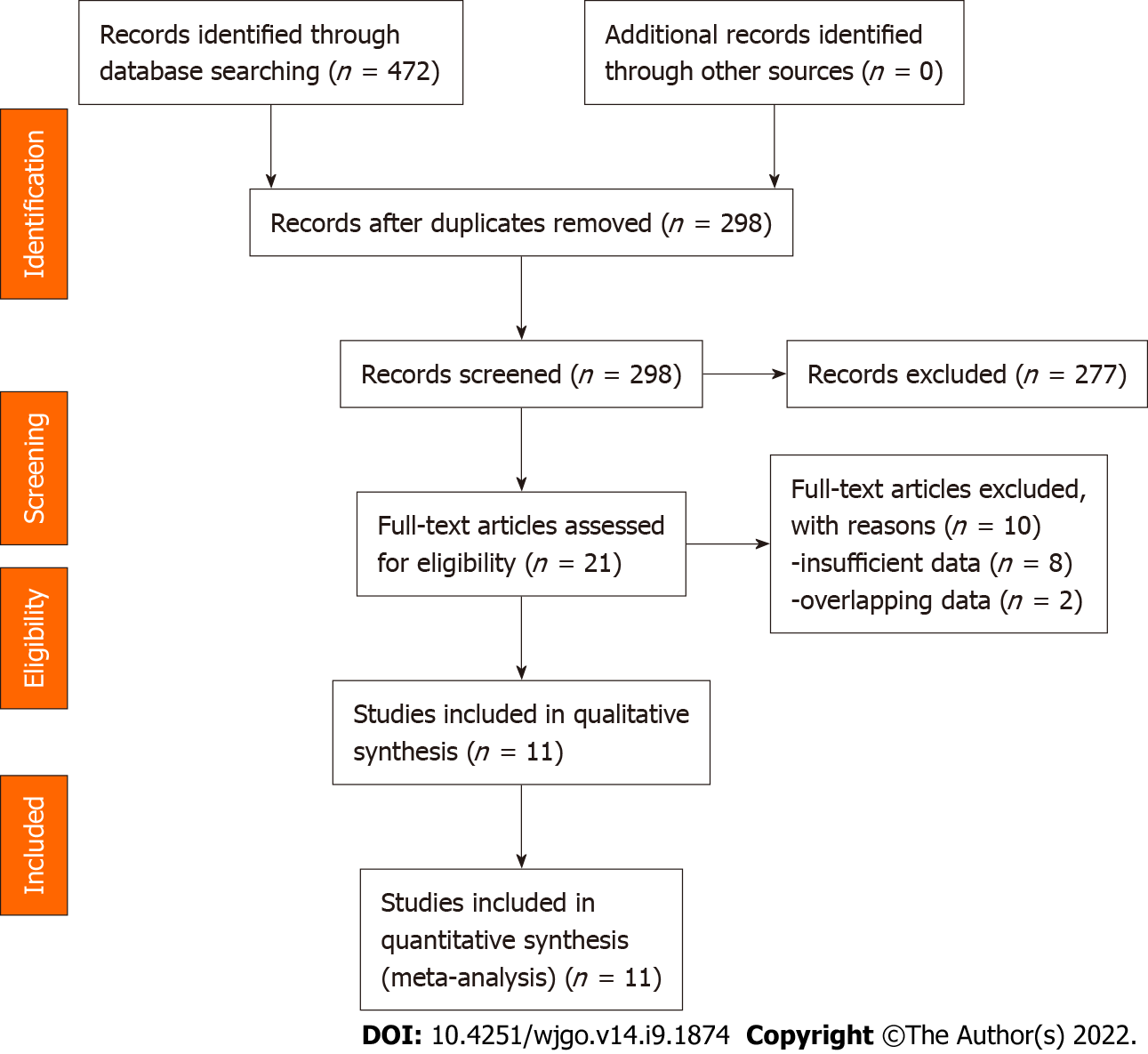
Figure 2 shows the flow diagram for the literature search and selection. We finally identified 11 eligible studies in this meta-analysis[11-21].
The baseline characteristics of the included studies are shown in Table 1. Among all eligible studies, six studies were published in English[11-14,17,18], while five were published in Chinese[15,16,19-21]. All included studies examined the expression of Twist in esophageal cancer tissue with immunohistochemistry (IHC). Two metrics for IHC staining were used in some studies[11,12,18-21]: The percentage of positively stained cells and the staining intensity. However, some studies[13-17] evaluated Twist expression using only one metric for IHC staining, which resulted in assessing the expression of Twist at various cutoff values. In addition, HRs were directly reported in some studies[11-14,17,20], while others[15,16,18,19,21] were indirectly calculated from survival curves.
| Ref. | Country | Sample size | TNM stage | Detection method | Antibody | Method of quantification | Cut-off value | Positive proportion (%) | Outcome | Source of HR | Follow-up time (mo) | NOS score |
| Sasaki et al[11], 2009 | Japan | 166 | I-IV | IHC | Anti-Twist (sc-15393, Santa Cruz) | Multiply percentage score and intensity score | Low: 0-5; High: 6-7 | 40.2 | OS | R | 24 (1-181) | 8 |
| Xie et al[12], 2009 | China | 112 | I-IV | IHC | Anti-Twist (sc-15393, Santa Cruz) | Multiply percentage score and intensity score | Negative: 0-3; Positive: 4-5+; 6-8++; ≥ 9+++ | 79.5 | OS | R | 35.8 (3.4-87) | 7 |
| Lee et al[13], 2012 | South Korea | 165 | I-IV | IHC/RT-PCR | Anti-Twist1 (ab50887, Abcam) | Intensity score | Negative: No expression; Positive: Weak, moderate, strong | 50.9 | OS/DFS | R/E | 115 (2-155) | 6 |
| Nakajima et al[14], 2012 | Japan | 54 | I-IVA | IHC | Anti-Twist (sc-15393, Santa Cruz) | Intensity score | Faint: 1; Moderate: 2; Strong: 3 | 37 | OS/RFS | R | NA | 7 |
| Sun et al[15], 2013 | China | 164 | I-III | IHC | Anti-Twist1 (ab50887, Abcam) | Percentage of stained cells | Negative: 0%-10%; Positive: > 10% | 34.1 | OS | E | 96-120 | 7 |
| Chen et al[16], 2016 | China | 50 | NR | IHC | Anti-Twist1 (Abcam) | Percentage of stained cells | NA | 50 | OS | E | > 60 | 7 |
| Yeo et al[17], 2017 | Korea | 169 | I-IV | IHC | Anti-Twist1 (Abcam) | Intensity score | Negative: 1; Positive: 2-3 | 89.9 | OS/DFS | R | NA | 7 |
| Xu et al[18], 2021 | China | 229 | I-IV | IHC | Anti-Twist1 (ab175430; Abcam) | Multiply percentage score and intensity score | Negative: 0-5; Positive: ≥ 6 | 59 | OS/PFS | E | NA | 6 |
| Du et al[19], 2021 | China | 72 | I-III | IHC | Anti-Twist (bs-2441R, Bioss) | Multiply percentage score and intensity score | Negative: 0-2; Positive: ≥ 3 | 61.1 | OS | E | 14-90 | 6 |
| Tang et al[20], 2021 | China | 40 | II-IV | IHC | Anti-Twist1 (ab50581, Abcam) | Multiply percentage score and intensity score | Negative: 0-2; Positive: ≥ 3 | 15 | OS | R | 17 (13.9-20.1) | 7 |
| Wang et al[21], 2021 | China | 72 | I-III | IHC | Anti-Twist1 (bs-2441R, Bioss) | Multiply percentage score and intensity score | Negative: 0-3; Positive: ≥ 4 | 61.1 | OS | E | 14-90 | 6 |
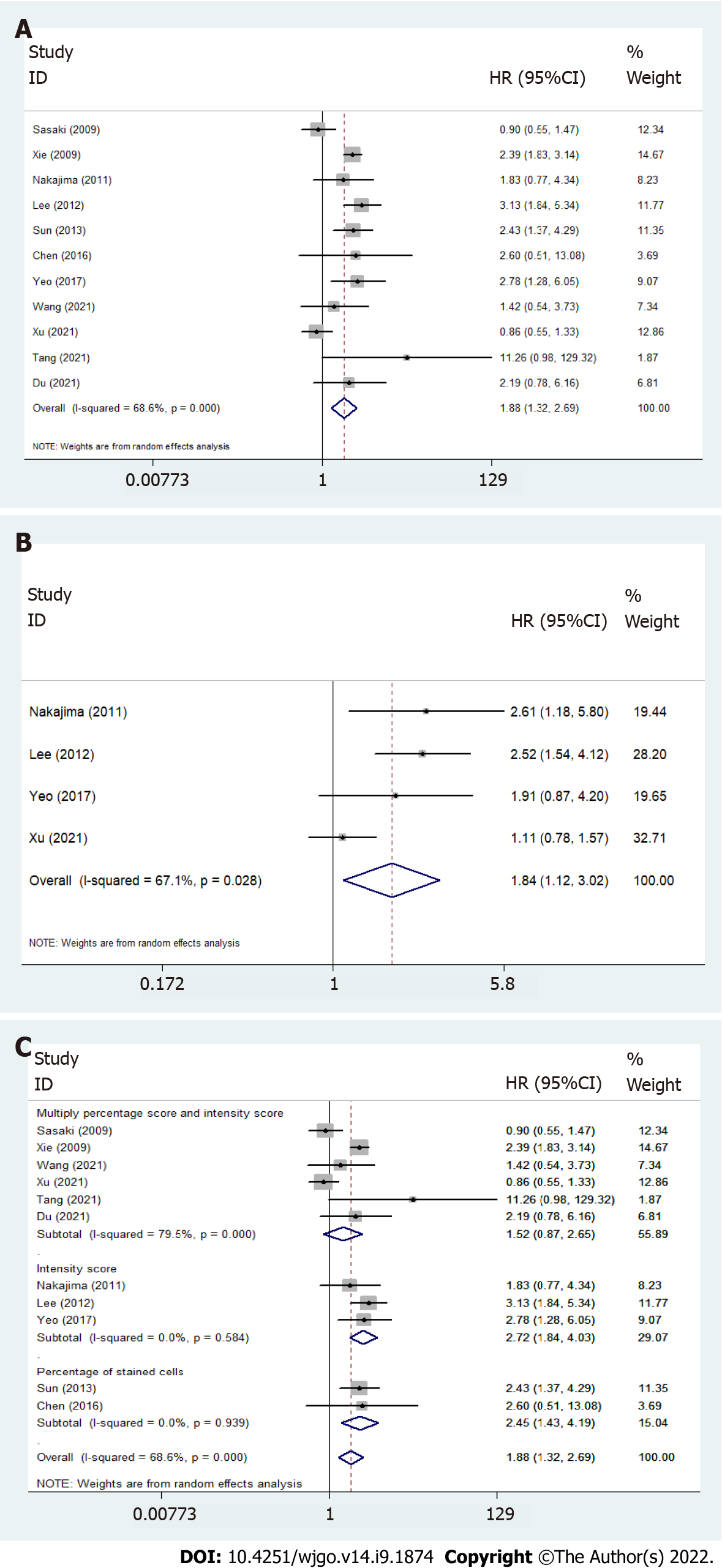
All included studies reported HRs of OS, and four reported DFS/RFS/PFS (Table 2, Figure 3). Both the pooled HR for OS (HR = 1.88, 95%CI: 1.32-2.69, I2 = 68.6%) and the pooled HR for DFS/RFS/PFS (HR = 1.84, 95%CI: 1.12-3.02, I2 = 67.1%) suggested that Twist overexpression was associated with poor prognosis in esophageal cancer patients. Heterogeneity was explored by subgroup analysis based on the detection method. Immunoreactivity scored by multiplying the percentage score and intensity score (pooled OS; HR = 1.517, 95%CI: 0.869-2.649, I2 = 79.5%) showed very high heterogeneity when compared with scoring by staining intensity (pooled OS; HR = 2.72, 95%CI: 1.84-4.03, I2 = 0%) or percentage of stained cells (pooled OS; HR = 2.45, 95%CI: 1.43-4.19, I2 = 0%) (Table 2 and Figure 3C).
| Meta-analysis | Endpoints | HR (95%CI) | Heterogeneity test (I2) | P value | Number of studies |
| TWIST (+) vs TWIST (−) | OS | 1.88 (1.32-2.69)a | 68.6% | 0.000 | 11 |
| DFS/RFS/PFS | 1.84 (1.12-3.02)a | 67.1% | 0.028 | 4 | |
| Method of quantification | Multiply percentage score and intensity score | 1.52 (0.87-2.65) | 79.5% | 0.319 | 6 |
| Intensity score | 2.72 (1.84-4.03)a | 0.00 | 0.062 | 9 | |
| Percentage of stained cells | 2.45 (1.43-4.19)a | 68.6% | 0.199 | 4 |
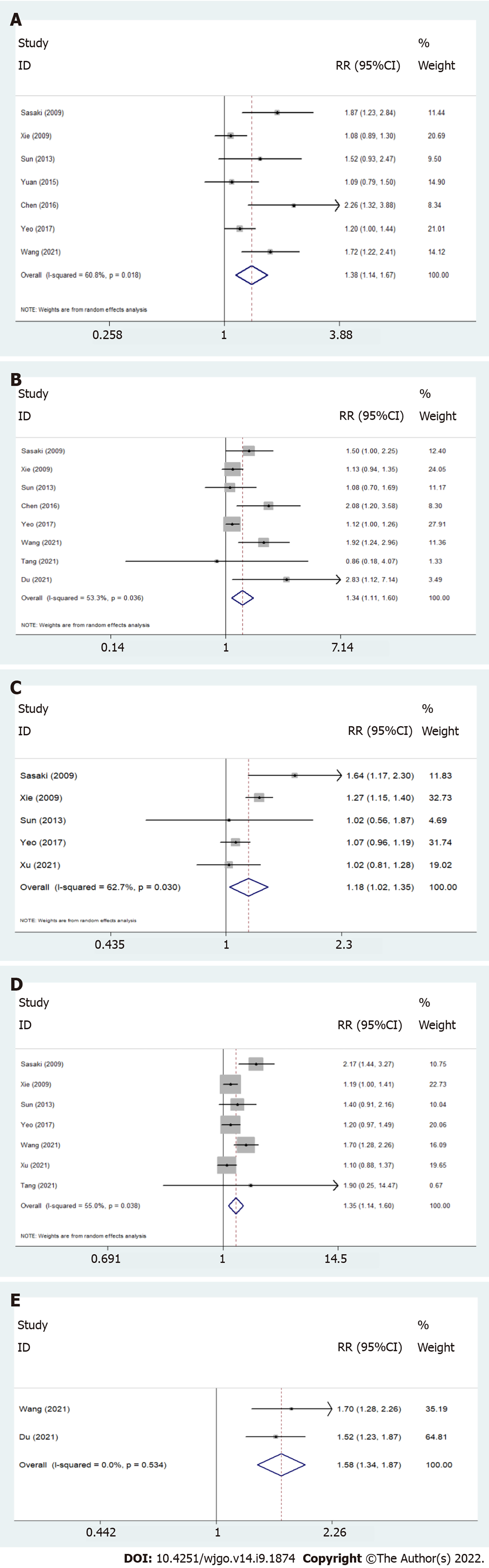
As shown in Table 3 and Figure 4, Twist overexpression was correlated with T stage (T3 + T4 vs T1 + T2, RR = 1.38, 95%CI: 1.14-1.67), lymph node metastasis (yes vs no, RR = 1.34, 95%CI: 1.11-1.60), distant metastasis (yes vs no, RR = 1.18, 95%CI: 1.02-1.35), TNM stage (III + IV vs I + II, RR = 1.35, 95%CI: 1.14-1.60), and clinical stage (III + IV vs I + II, RR = 1.58, 95%CI: 1.34-1.87), which indicated that Twist overexpression might accelerate esophageal progression and metastasis. However, no correlation between Twist expression and age, gender, tumor location, differentiation, or venous invasion was observed.
| Clinical features | RR (95%CI) | Heterogeneity test (I2) | P value | Number of studies |
| Age (≥ 60 vs < 60) | 1.07 (0.95-1.21) | 5.88 | 0.319 | 6 |
| Gender (male vs female) | 1.02 (0.89-1.18)a | 14.85 | 0.062 | 9 |
| Location (upper + middle vs lower) | 0.89 (0.80-1.00) | 4.66 | 0.199 | 4 |
| T stage (T3 + T4 vs T1 + T2) | 1.38 (1.14-1.67)a | 15.30 | 0.018 | 7 |
| Differentiation (high + moderate vs low) | 0.94 (0.81-1.09)a | 21.26 | 0.003 | 8 |
| Lymph node metastasis (yes vs no) | 1.34 (1.11- 1.60)a | 14.99 | 0.036 | 8 |
| Distant metastasis (yes vs no) | 1.18 (1.02-1.35)a | 10.74 | 0.030 | 5 |
| TNM stage (III + IV vs I + II) | 1.35 (1.14-1.60)a | 13.34 | 0.038 | 7 |
| Clinical stage (III + IV vs I + II) | 1.58 (1.34-1.87) | 0.39 | 0.534 | 2 |
| Venous invasion (yes vs no) | 1.46 (0.83-2.56)a | 4.49 | 0.034 | 2 |
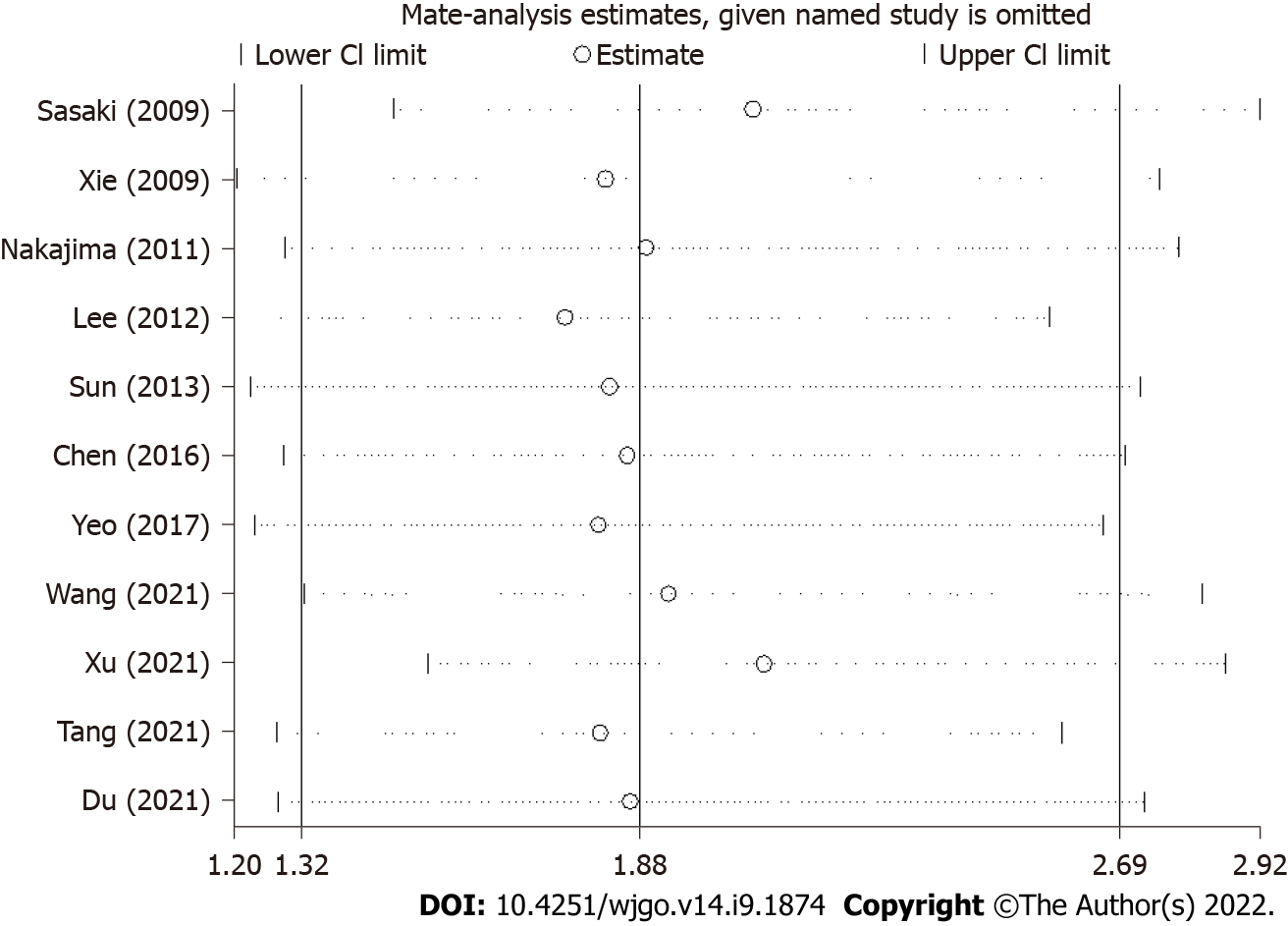
The sensitivity analyses for the association between Twist expression and esophageal cancer prognosis suggested that the results of this meta-analysis were stable and reliable (Figure 5).
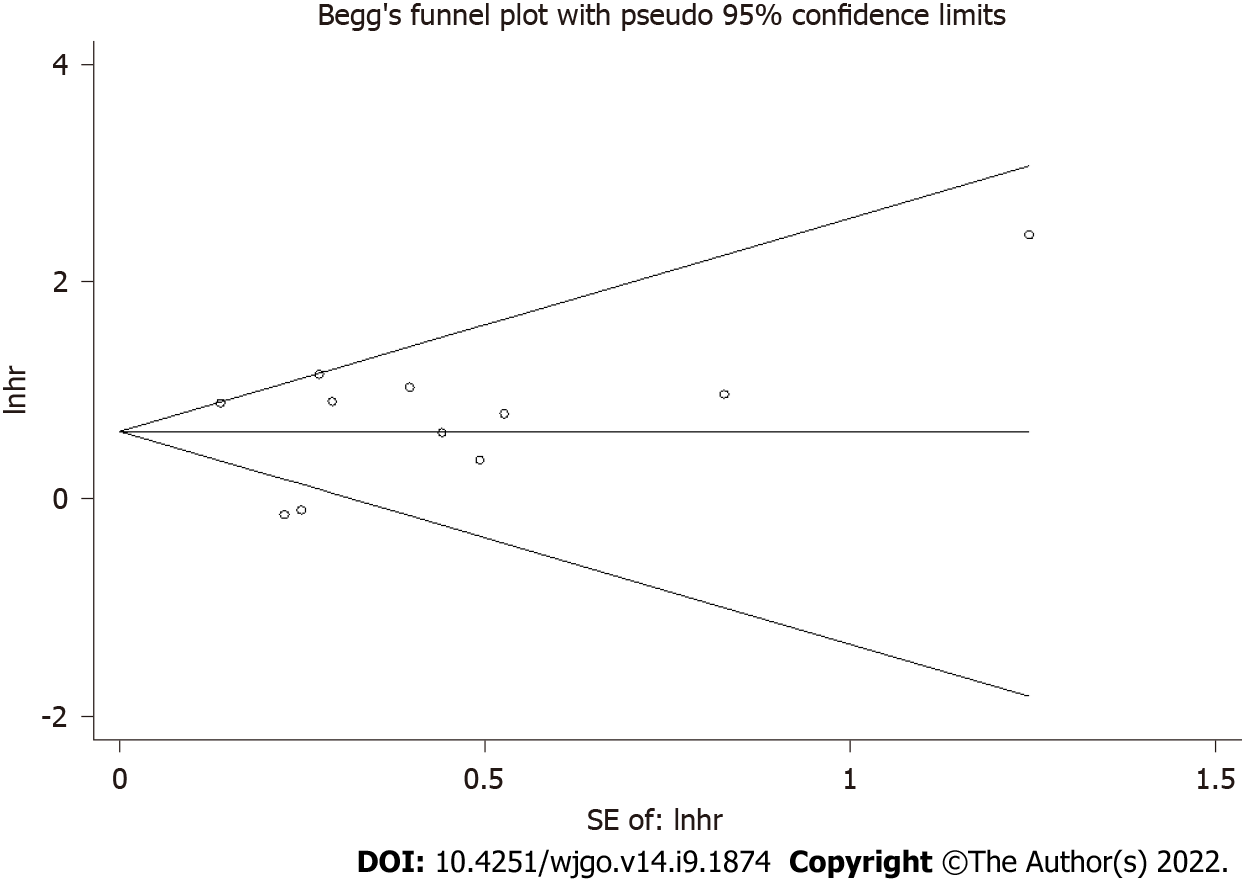
Publication bias was assessed, and the results showed symmetrical Begg’s funnel plots for OS with a P value of 0.78 (Figure 6), suggesting that no obvious publication bias existed.
This meta-analysis suggests that high expression of Twist is associated with poor prognosis in esophageal cancer. The subgroup analyses by the detection method of Twist expression imply that major heterogeneity is derived from evaluating Twist expression by different metrics for IHC staining. Several clinicopathological parameters, such as T stage, lymph node metastasis, distant metastasis, TNM stage, and clinical stage, were positively correlated with Twist expression. Some meta-analyses have investigated the relationship between Twist expression and prognosis in other cancers. For example, Zeng et al[28] investigated the prognostic value of Twist in lung cancer and found that high expression of Twist indicated a worse prognosis. Similarly, several meta-analyses revealed that Twist overexpression indicated poor prognosis in breast cancer[29], head and neck carcinoma[30], colorectal cancer[31], hepatocellular carcinoma, urinary cancer, and female reproductive cancer[32]. Our meta-analysis presents similar results and suggests that Twist might be a valuable prognostic biomarker in esophageal cancer.
The human Twist gene constitutes one intron and two exons localized on 7q21.2[33]. Twist is widely expressed in various cancers, such as lung cancer[34], breast cancer[35,36], esophageal cancer[37], and prostate cancer[38,39]. Twist not only plays an important role in mesodermal development but can also participate in the EMT of some epithelium-derived tumor cells. Twist could interact with the Mi2/NuRD chromatin remodeling and gene repression complex (MTA2, RbAp46, Mi2, and HDAC2)[40]. Twist recruits MTA2 to the E-cadherin promoter and reduces the level of acetylation in the promoter region, thereby inhibiting the expression of E-cadherin and promoting the invasive progression of ESCC[41]. Moreover, integrin-mediated adhesion to interstitial matrix proteins may differentially regulate nuclear/cytoplasmic translocation and DNA binding of Twist1, thereby activating the transcription of N-cadherin[38]. In malignant melanoma, increased N-cadherin expression following the loss of E-cadherin mRNA expression has been shown to play an important role in the regulation of cell migration, invasion, and survival[42].
Although all eligible studies used IHC to detect Twist expression, the type of primary antibody used, the degree of antibody dilution, and the quantification of the method were not the same. Second, immunohistochemical scores were classified into three categories in the included studies: scored by intensity, scored by the percentage of stained cells, and multiplied by the percentage score and intensity score, which may be the main sources of heterogeneity. The subgroup analysis found that immunoreactivity scored by multiplying the percentage score and intensity score showed very high heterogeneity (I2 = 79.5%), indicating that different scoring methods for IHC could contribute to potential publication bias. In addition, the scoring criteria and cutoff points for immunohistochemistry were subjective and not uniform in the included studies.
According to Sun et al[15], the positive expression of the Twist gene in ESCC stromal fibroblasts was associated with poor overall survival. Similarly, Yeo et al[17] found high Twist protein expression in cancer-associated fibroblasts of ESCC and concluded that Twist was an independent predictor of poor prognosis for OS. Therefore, more research is needed to explore the clinical significance of Twist expression in stromal fibroblasts. Nakajima et al[14] studied the expression of Twist in 54 patients who consecutively received 5-fluorouracil neoadjuvant chemotherapy followed by surgery. The results also showed that high Twist expression was positively associated with a worse esophageal cancer prognosis. In addition, Tang et al[20] detected tumor samples of 55 ESCC and 31 EAC obtained by endoscopy instead of surgery, while other included studies all detected Twist expression in tissues obtained from patients who underwent surgical treatment. Therefore, the conclusions of the studies discussed above are consistent with the results of our meta-analysis.
This study might have several limitations. First, only 11 studies including 1293 patients were included. Second, all of the patients were from Asian countries, and most were from China, which limited the application of our findings in other countries and regions. Third, the use of different anti-Twist antibodies in the included studies might cause heterogeneity in our meta-analysis. Hence, more evidence is urgently needed to assess the correlation between the expression of Twist and prognostic value in esophageal cancer patients.
Many aspects of Twist deserve further research. Except for the study of Tang et al[20], our meta-analysis only included ESCC patients who underwent surgery. We found few studies investigating the clinicopathological and prognostic significance of the Twist gene in other histological types of esophageal cancer. Furthermore, Lee et al[13] demonstrated that TWIST-positive circulating tumor cells (CTCs) were common in ESCC patients (75% of the total study population), and a proportion of TWIST (+) CTCs ≥ 0.5 was significantly associated with advanced histologic grade[43]. IHC staining is mostly used in studies on the clinical significance of TWIST in esophageal cancer, but this is not conducive to the application of Twist in the diagnosis and treatment of esophageal cancer. As a novel noninvasive biomarker for the diagnosis and prediction of tumor progression, CTCs are needed for more studies to evaluate the clinical prognostic value of TWIST (+) CTCs in esophageal cancer patients and overcome the challenges of standard CTC isolation and the diversity of CTC counting methods.
In summary, this meta-analysis suggests that Twist overexpression is associated with a poor esophageal cancer prognosis despite the limitations encountered by our study. Twist overexpression is correlated with T stage, lymph node metastasis, distant metastasis, TNM stage, and clinical stage, which indicates that Twist might accelerate esophageal cancer progression and metastasis. Furthermore, the sensitivity analyses implied that our meta-analysis yielded a stable and reliable estimate.
Twist can induce epithelial–mesenchymal transition (EMT) and cancer metastasis. However, the prognostic value of Twist expression in patients with esophageal cancer remains controversial.
To clarify whether Twist could be a promising biomarker for predicting prognosis in esophageal cancer.
To investigate the prognostic and clinicopathological value of Twist expression in esophageal cancer.
Published literature in several databases was searched for eligible articles. Participants with esophageal cancer whose tumor tissues underwent immunohistochemistry to detect the expression of Twist were considered when they met the inclusion criteria. The hazard ratio (HR) and relative ratio (RR) with their 95%CI were pooled. Heterogeneity was estimated by I2 statistics.
The pooled HR for overall survival was 1.88 (95%CI: 1.32-2.69, I2 = 68.6%), and the pooled HR for disease-free survival/relapse-free survival/progression-free survival was 1.84 (95%CI: 1.12-3.02, I2 = 67.1%). In addition, overexpression of Twist was correlated with T stage (T3 + T4 vs T1 + T2, RR = 1.38, 95%CI: 1.14-1.67), lymph node metastasis (yes vs no, RR = 1.34, 95%CI: 1.11-1.60), distant metastasis (yes vs no, RR = 1.18, 95%CI: 1.02-1.35), tumor, node and metastasis (TNM) stage (III + IV vs I + II, RR = 1.35, 95%CI: 1.14-1.60), and clinical stage (III + IV vs I + II, RR = 1.58, 95%CI: 1.34-1.87).
Twist overexpression indicates poor esophageal cancer prognosis. Moreover, Twist overexpression is correlated with T stage, lymph node metastasis, distant metastasis, TNM stage, and clinical stage, which indicates that Twist might accelerate esophageal cancer progression and metastasis.
Our meta-analysis suggests that Twist might be a valuable prognostic biomarker in esophageal cancer.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bairwa DBL, India; Casella C, Italy S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64631] [Article Influence: 16157.8] [Reference Citation Analysis (176)] |
| 2. | Zhang S, Sun K, Zheng R, Zeng H, Wang S, Chen R, Wei W, He J. Cancer incidence and mortality in China, 2015. J Natl Cancer Cent. 2021;1:2-11. [RCA] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 240] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 3. | Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390:2383-2396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 847] [Article Influence: 105.9] [Reference Citation Analysis (0)] |
| 4. | Hirano H, Kato K. Systemic treatment of advanced esophageal squamous cell carcinoma: chemotherapy, molecular-targeting therapy and immunotherapy. Jpn J Clin Oncol. 2019;49:412-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 5. | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11065] [Cited by in RCA: 12187] [Article Influence: 1523.4] [Reference Citation Analysis (3)] |
| 6. | Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori G, Dedhar S, Derynck R, Ford HL, Fuxe J, García de Herreros A, Goodall GJ, Hadjantonakis AK, Huang RYJ, Kalcheim C, Kalluri R, Kang Y, Khew-Goodall Y, Levine H, Liu J, Longmore GD, Mani SA, Massagué J, Mayor R, McClay D, Mostov KE, Newgreen DF, Nieto MA, Puisieux A, Runyan R, Savagner P, Stanger B, Stemmler MP, Takahashi Y, Takeichi M, Theveneau E, Thiery JP, Thompson EW, Weinberg RA, Williams ED, Xing J, Zhou BP, Sheng G; EMT International Association (TEMTIA). Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21:341-352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 624] [Cited by in RCA: 1387] [Article Influence: 277.4] [Reference Citation Analysis (0)] |
| 7. | Jung HY, Fattet L, Yang J. Molecular pathways: linking tumor microenvironment to epithelial-mesenchymal transition in metastasis. Clin Cancer Res. 2015;21:962-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 238] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 8. | Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192-2206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 805] [Cited by in RCA: 930] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 9. | Vesuna F, van Diest P, Chen JH, Raman V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun. 2008;367:235-241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 283] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 10. | Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2783] [Cited by in RCA: 2982] [Article Influence: 142.0] [Reference Citation Analysis (0)] |
| 11. | Sasaki K, Natsugoe S, Ishigami S, Matsumoto M, Okumura H, Setoyama T, Uchikado Y, Kita Y, Tamotsu K, Sakamoto A, Owaki T, Aikou T. Significance of Twist expression and its association with E-cadherin in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2009;28:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 12. | Xie F, Li K, Ouyang X. Twist, an independent prognostic marker for predicting distant metastasis and survival rates of esophageal squamous cell carcinoma patients. Clin Exp Metastasis. 2009;26:1025-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Lee KW, Kim JH, Han S, Sung CO, Do IG, Ko YH, Um SH, Kim SH. Twist1 is an independent prognostic factor of esophageal squamous cell carcinoma and associated with its epithelial-mesenchymal transition. Ann Surg Oncol. 2012;19:326-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 14. | Nakajima TE, Yoshida H, Okamoto N, Nagashima K, Taniguchi H, Yamada Y, Shimoda T, Masutomi K. Nucleostemin and TWIST as predictive markers for recurrence after neoadjuvant chemotherapy for esophageal carcinoma. Cancer Sci. 2012;103:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 15. | Sun FD, Cui Y, Zhang BL, Chu JN, Xuan YH. Expression of Twist1 in esophageal squamous cell carcinoma tissues and its clinical significance. Linchuang Yu Shiyan Bingli Xue Za Zhi. 2013;29:836-839. [DOI] [Full Text] |
| 16. | Chen HS, Wang P, Lu SH. Study on correlations between epithelial mesenchymal transition related proteins and clinicopathological characteristics and prognosis in primary esophageal squamous cell carcinoma. Jiaotong Yi Xue. 2016;30:214-217. |
| 17. | Yeo SY, Ha SY, Lee KW, Cui Y, Yang ZT, Xuan YH, Kim SH. Twist1 is highly expressed in cancer-associated fibroblasts of esophageal squamous cell carcinoma with a prognostic significance. Oncotarget. 2017;8:65265-65280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Xu S, Zhou Y, Biekemitoufu H, Wang H, Li C, Zhang W, Ma Y. Expression of Twist, Slug and Snail in esophageal squamous cell carcinoma and their prognostic significance. Oncol Lett. 2021;21:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Du QS, Hao XW, Zhang ZW. Correlations of Cofilin1 and Twist1 with clinicopathological features and prognosis of patients with esophageal cancer. Zhonghua Shiyong Zhenduan Yu Zhiliao Za Zhi. 2021;35:1115-1118. [DOI] [Full Text] |
| 20. | Tang T, Zhang H, Wang Y, Sun XM, Huang R, Wu H. Expression of SOX2 and Twistl in intermediate to advanced squamous esophageal carcinoma and their effect on the efficacy of radiotherapy and chemotherapy. Zhonghua Zhongliu Fangzhi Za Zhi. 2021;28:840-846. [DOI] [Full Text] |
| 21. | Wang J, Wu HF, Li Y, Hua CX. The expression of Twist and DAB2IP in esophageal squamous cell carcinoma and its clinical pathological characteristics,prognostic relationship. Zhongyi Linchuang Yanjiu. 2021;13:13-16. [DOI] [Full Text] |
| 22. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3074] [Article Influence: 512.3] [Reference Citation Analysis (0)] |
| 23. | Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8858] [Cited by in RCA: 12661] [Article Influence: 844.1] [Reference Citation Analysis (0)] |
| 24. | Barili F, Parolari A, Kappetein PA, Freemantle N. Statistical Primer: heterogeneity, random- or fixed-effects model analyses? Interact Cardiovasc Thorac Surg. 2018;27:317-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 188] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 25. | Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34245] [Cited by in RCA: 40541] [Article Influence: 1447.9] [Reference Citation Analysis (2)] |
| 26. | Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-1101. [PubMed] |
| 27. | Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1326] [Cited by in RCA: 1558] [Article Influence: 82.0] [Reference Citation Analysis (1)] |
| 28. | Zeng J, Zhan P, Wu G, Yang W, Liang W, Lv T, Song Y. Prognostic value of Twist in lung cancer: systematic review and meta-analysis. Transl Lung Cancer Res. 2015;4:236-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 29. | Qiao W, Jia Z, Liu H, Liu Q, Zhang T, Guo W, Li P, Deng M, Li S. Prognostic and clinicopathological value of Twist expression in breast cancer: A meta-analysis. PLoS One. 2017;12:e0186191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Zhuo X, Luo H, Chang A, Li D, Zhao H, Zhou Q. Is overexpression of TWIST, a transcriptional factor, a prognostic biomarker of head and neck carcinoma? Sci Rep. 2015;5:18073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Ahmadiankia N, Khosravi A. Significance of epithelial-to-mesenchymal transition inducing transcription factors in predicting distance metastasis and survival in patients with colorectal cancer: A systematic review and meta-analysis. J Res Med Sci. 2020;25:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (1)] |
| 32. | Zhang P, Hu P, Shen H, Yu J, Liu Q, Du J. Prognostic role of Twist or Snail in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest. 2014;44:1072-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 33. | Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 355] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 34. | Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS, Wu KJ. Prognostic significance of hypoxia-inducible factor-1alpha, TWIST1 and Snail expression in resectable non-small cell lung cancer. Thorax. 2009;64:1082-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 172] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 35. | Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 391] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 36. | Mehrotra J, Vali M, McVeigh M, Kominsky SL, Fackler MJ, Lahti-Domenici J, Polyak K, Sacchi N, Garrett-Mayer E, Argani P, Sukumar S. Very high frequency of hypermethylated genes in breast cancer metastasis to the bone, brain, and lung. Clin Cancer Res. 2004;10:3104-3109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 37. | Gong T, Xue Z, Tang S, Zheng X, Xu G, Gao L, Zhao G, Hong L, Tang G, Zhang H, Wang R, Jiang Y, Fan D. Nuclear expression of Twist promotes lymphatic metastasis in esophageal squamous cell carcinoma. Cancer Biol Ther. 2012;13:606-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Alexander NR, Tran NL, Rekapally H, Summers CE, Glackin C, Heimark RL. N-cadherin gene expression in prostate carcinoma is modulated by integrin-dependent nuclear translocation of Twist1. Cancer Res. 2006;66:3365-3369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 191] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 39. | Yuen HF, Chua CW, Chan YP, Wong YC, Wang X, Chan KW. Significance of TWIST and E-cadherin expression in the metastatic progression of prostatic cancer. Histopathology. 2007;50:648-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 40. | Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 41. | Dai SL, Wei SS, Zhang C, Li XY, Liu YP, Ma M, Lv HL, Zhang Z, Zhao LM, Shan BE. MTA2 promotes the metastasis of esophageal squamous cell carcinoma via EIF4E-Twist feedback loop. Cancer Sci. 2021;112:1060-1074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Na YR, Lee JS, Lee SJ, Seok SH. Interleukin-6-induced Twist and N-cadherin enhance melanoma cell metastasis. Melanoma Res. 2013;23:434-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 43. | Lee HJ, Kim GH, Park SJ, Kwon CH, Lee MW, Lee BE, Baek DH, I H. Clinical Significance of TWIST-Positive Circulating Tumor Cells in Patients with Esophageal Squamous Cell Carcinoma. Gut Liver. 2021;15:553-561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |