Published online Jul 15, 2022. doi: 10.4251/wjgo.v14.i7.1265
Peer-review started: January 25, 2022
First decision: May 9, 2022
Revised: May 18, 2022
Accepted: June 26, 2022
Article in press: June 26, 2022
Published online: July 15, 2022
Processing time: 168 Days and 22.7 Hours
Liver fibrosis and hepatocellular carcinoma (HCC) are common adverse consequences of chronic liver injury. The interaction of various risk factors may cause them to happen. Identification of specific biomarkers is of great significance for understanding the occurrence, development mechanisms, and determining the novel tools for diagnosis and treatment of both liver fibrosis and HCC.
To identify liver fibrosis-related core genes, we analyzed the differential expression pattern of core genes in liver fibrosis and HCC.
Gene expression profiles of three datasets, GSE14323, GSE36411, and GSE89377, obtained from the Gene Expression Omnibus (GEO) database, were analyzed, and differentially expressed genes (DEGs) between patients with liver cirrhosis and healthy controls were identified by screening via R software packages and online tool for Venn diagrams. The WebGestalt online tool was used to identify DEGs enriched in biological processes, molecular functions, cellular components, and Kyoto Encyclopedia of Genes and Genomes pathways. The protein–protein interactions of DEGs were visualized using Cytoscape with STRING. Next, the expression pattern of core genes was analyzed using Western blot and immunohistochemistry in a carbon tetrachloride (CCl4)-induced liver cirrhosis mouse model and in patient liver samples. Finally, Kaplan-Meier curves were constructed using the Kaplan-Meier plotter online server.
Forty-five DEGs (43 upregulated and 2 downregulated genes) associated with liver cirrhosis were identified from three GEO datasets. Ten hub genes were identified, which were upregulated in liver cirrhosis. Western blot and immunohistochemical analyses of the three core genes, decorin (DCN), dermatopontin (DPT), and SRY-box transcription factor 9 (SOX9), revealed that they were highly expressed in the CCl4-induced liver cirrhosis mouse model. The expression levels of DCN and SOX 9 were positively correlated with the degree of fibrosis, and SOX 9 level in HCC patients was significantly higher than that in fibrosis patients. However, high expression of DPT was observed only in patients with liver fibrosis, and its expression in HCC was low. The gene expression profiling interactive analysis server (GEPIA) showed that SOX9 was significantly upregulated whereas DCN and DPT were significantly downregulated in patients with HCC. In addition, the Kaplan-Meier curves showed that HCC patients with higher SOX9 expression had significantly lower 5-year survival rate, while patients with higher expression of DCN or DPT had significantly higher 5-year survival rates.
The expression levels of DCN, DPT, and SOX9 were positively correlated with the degree of liver fibrosis but showed different correlations with the 5-year survival rates of HCC patients.
Core Tip: GSE14323, GSE36411, and GSE89377 are available from the Gene Expression Omnibus database. Forty-five differentially expressed genes and 10 hub genes were identified between cirrhotic and healthy livers. quantitative polymerase chain reaction, Western blot, and immunohistochemical analyses showed that decorin (DCN), dermatopontin (DPT), and SRY-box transcription factor 9 (SOX9) were highly expressed in the CCl4-induced cirrhotic mouse model. The expression level of SOX9 was also significantly increased in HCC patients,and was associated with the fibrosis stage.. However, overexpression of DPT was only observed in patients with liver fibrosis. The Kaplan-Meier curves showed that HCC patients with higher SOX9 expression had significantly lower 5-year survival rate, while patients with higher expression of DCN or DPT had higher 5-year survival rates.
- Citation: Li Y, Yuan SL, Yin JY, Yang K, Zhou XG, Xie W, Wang Q. Differences of core genes in liver fibrosis and hepatocellular carcinoma: Evidence from integrated bioinformatics and immunohistochemical analysis. World J Gastrointest Oncol 2022; 14(7): 1265-1280
- URL: https://www.wjgnet.com/1948-5204/full/v14/i7/1265.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i7.1265
Chronic liver infection, including chronic hepatitis B (CHB), chronic hepatitis C, alcoholic liver disease, and non-alcoholic fatty liver disease can all result in liver fibrosis. Primary liver cancer is the seventh most common cancer worldwide[1]. Hepatocellular carcinoma (HCC), which is the dominant type of liver cancer, accounts for approximately 75% of all liver cancers worldwide[2] and is the second most fatal disease in China[3]. Liver cirrhosis is an advanced stage of liver fibrosis and is characterized by limited regeneration capacity and serious complications[4]. Most HCCs develop in the background of chronic liver injury, hepatic inflammation, and liver fibrosis. Unfortunately, to date, there are still no effective treatment strategies for liver cirrhosis, and the limited number of specific biomarkers for HCC related to fibrosis further compounds the problem of its diagnosis and treatment[5,6].
The pathogenesis of liver fibrosis and HCC is complex as the interaction of many factors may lead to their occurrence. In recent years, with the optimization of gene sequencing platforms, several differentially expressed genes (DEGs) have been identified using bioinformatics analysis[7,8]. To date, there is a huge collection of data stored in the Gene Expression Omnibus (GEO) database of gene expression that can be explored to find the relevant DEGs for a diseased condition. Chan et al[9] identified DEGs between cirrhotic and non-cirrhotic livers using microarray gene analysis. Many human genes may show differential expression patterns and functions with the onset of fibrosis and/or HCC. However, the study by Chan et al[9] was limited by the small sample size, which only included 24 patients with cirrhosis and 16 patients without cirrhosis. The results obtained solely from either bioinformatics or experimental approach may not elucidate relevant DEGs. Hence, integrating bioinformatics methods with experimental techniques may help us to better understand the underlying mechanisms behind fibrosis/HCC pathogenesis.
In this study, we analyzed three databases from GEO, R software packages and online tools to identify DEGs, including upregulated and downregulated genes between liver fibrosis and HCC. The molecular function, cellular component, biological process, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of DEGs were then assessed. We also constructed a protein-protein interaction (PPI) network using Cytoscape for further analysis. Using these methods, hub genes were identified and subjected to KEGG pathway enrichment analysis. Finally, real-time quantitative polymerase chain reaction (qPCR), Western blotting, and immunohistochemistry of the liver tissue samples from mouse model and patients were carried out to identify novel biomarkers of liver fibrosis. Our study identified fibrosis-related core genes and compared their phenotypic differences between liver fibrosis and HCC.
Liver tissue samples were collected from 5 healthy controls, 40 patients with CHB (n = 28) and CHB-associated HCC (n = 12) at the Beijing Ditan Hospital, Capital Medical University, Beijing, China. The diagnosis of CHB was based on the “Guidelines on prevention and treatment of chronic hepatitis B in China”[10]. Chronic HBV infection is defined as the persistence of HBsAg in blood serum for at least 6 mo. Patients who were diagnosed for Hepatitis C viral infection, drug-induced liver disease, non-alcoholic liver disease, alcoholic liver disease, autoimmune liver disease, cholestatic liver disease, or hereditary metabolic liver disease were excluded. All samples were analyzed by a clinician and two independent pathologists with no prior knowledge of demographic and clinical data. The degree of liver inflammation and fibrosis were scored according to the METAVIR system[11], and liver samples were divided into five groups: normal control, fibrosis grade 0 (S0), fibrosis grade 1-2 (S1-2), fibrosis grade 3-4 (S3-4), and HCC group. Meanwhile, we collected the clinical data, including sex, age, HBeAg, HBV DNA, alanine transaminase, aspartate transaminase, total bilirubin, albumin, cholinesterase and alpha-fetoprotein. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, as approved by the Ethics Committee of Beijing Ditan Hospital.
Six-week-old male C57BL/6 mice were purchased from Vital River Laboratory Animal Technology Co., Ltd, Beijing, China. All mice were housed in a specific pathogen free laboratory animal house (Institute of Zoology of Beijing, Chinese Academy of Sciences, China) at 24 ℃ with a 12 h light/dark cycle. All animal studies were approved by the Institutional Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences.
Twelve male C57BL/6 mice were randomly divided into two groups: control group and carbon tetrachloride (CCl4)-treated group. To induce liver cirrhosis, CCl4 (0.5 μL/g) mixed with corn oil was intraperitoneally injected into the mice three times per week for 12 wk. The control group was injected with an equal volume of corn oil. CCl4 was purchased from Sigma-Aldrich (St. Louis, MO, United States).
For hematoxylin and eosin (HE) staining, Masson’s trichrome staining, and immunohistochemical analysis, liver tissues collected from mouse model or patients were fixed with 4% paraformaldehyde solution and embedded in paraffin. For histological analysis, 5 μm thick sections were stained and observed under 10 × or 20 × objective lens. Masson’s trichrome kit (G1281, Solarbio, Beijing, China) was used according to the manufacturer’s instructions.
For immunohistochemical staining, 5% bovine serum albumin in 0.1% TritonX-100 tris-buffered saline was used as the blocking solution. The samples were incubated overnight at 4 ℃ with anti-decorin (DCN) (ab277636, Abcam), anti-dermatopontin (DPT) (10537-1-AP, Proteintech), and anti-SRY-box transcription factor 9 (SOX9) (ab185966; Abcam) antibodies. After incubation with a peroxidase-conjugated secondary antibody, the signal was visualized using a diaminobenzidine peroxidase substrate kit. The collagen area or positive area of immunohistochemical staining was quantified using ImageJ 1.52a software.
Total RNA from liver tissues was isolated using TRIzolTM reagent (Thermo Fisher Scientific, MA, United States). Isolated RNA was reverse-transcribed into complementary DNA (cDNA) using a high-capacity cDNA reverse transcription kit (Promega, WI, United States). The relative expression of genes was detected by real-time fluorescence qPCR system (Light Cycler 480, Roche, Sweden) with SYBR green master mix (Promega, WI, United States). The primer sequences used in this study were listed in Supplementary Table 1. Statistical significance between the control and CCl4-treated groups was defined at P < 0.05.
Total protein from liver tissues was extracted using radioimmunoprecipitation assay buffer. The protein concentration of the samples was measured by bicinchoninic acid assay. The same concentration of protein was loaded to 10% sodium dodecyl sulfate polyacrylamide gel and then transferred to a polyvinylidene fluoride membrane. The membranes were incubated overnight at 4℃ in anti-α-SMA (ab5694, Abcam), anti-DCN (ab277636, Abcam), anti-DPT (10537-1-AP, Proteintech), and anti-SOX9 (ab185966, Abcam) antibodies. All signals were visualized by density scanning (Image Quant TL7.0; GE Healthcare Biosciences, Uppsala, Sweden). The intensity of the bands was analyzed using ImageJ 1.52a software.
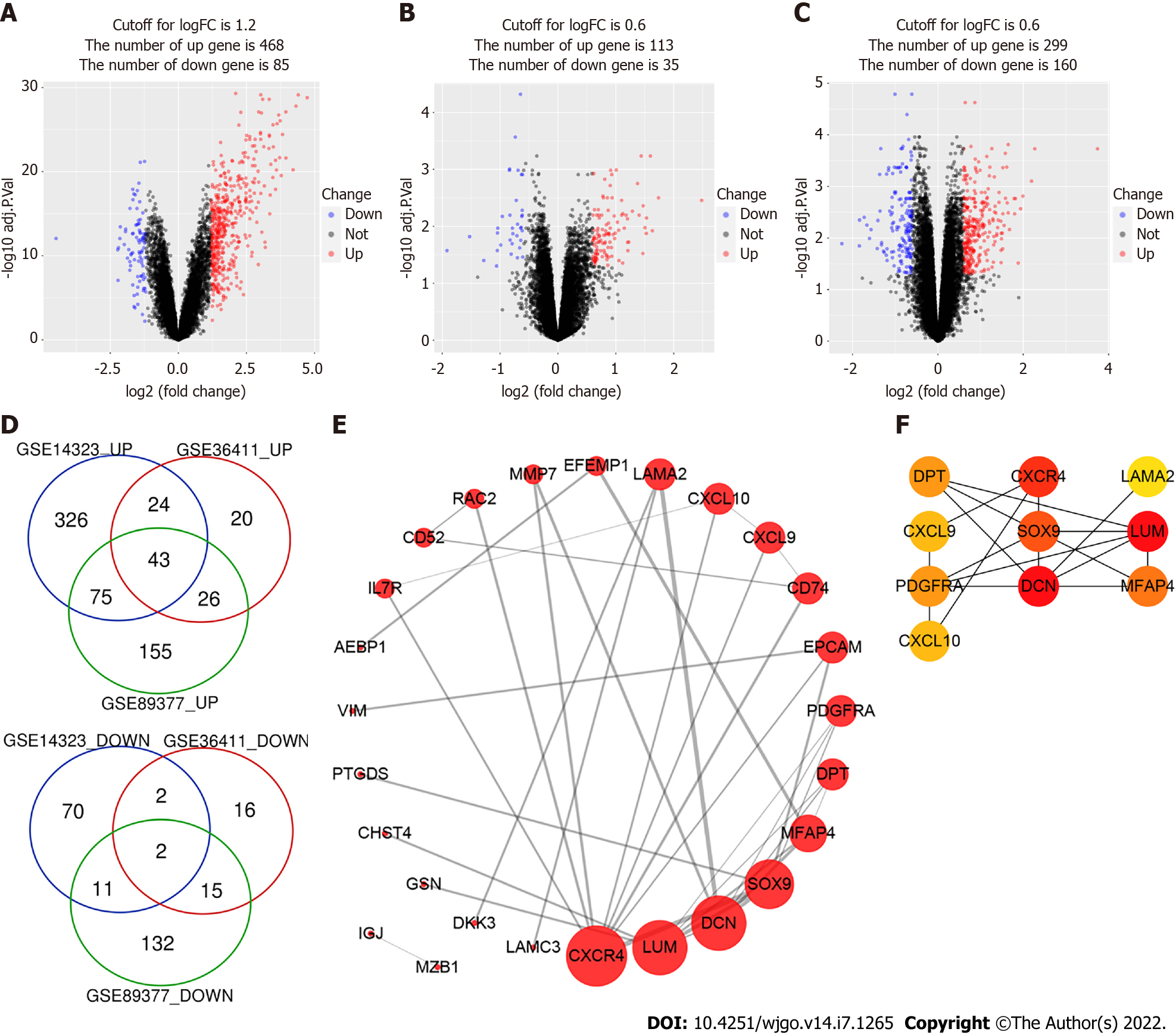
The RNA expression data of human cirrhotic and healthy livers were collected from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The GSE14323, GSE36411, and GSE89377 datasets contained gene expression data collected from 41, 21, and 12 cirrhotic and 19, 21, and 13 healthy liver tissues, respectively. The GEOquery R software package was used to download the GEO data and platform information. Then, the gene ID conversion was performed, and the maximum value of genes with the same name was selected. Ggplot2 package was used to plot the boxplot and density of the expression levels for each sample. Ggfortify package was used to perform the principal component analysis (PCA). DEGs between the cirrhosis and healthy liver tissue groups were identified using limma package by limiting the value of adjustment: P-value (adjust. P < 0.05) and the absolute value of logFC (|logFC| > 1.2 or 0.6). A volcano map was generated using the ggplot2 package, and Venn diagram online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to draw a Venn map. The DEGs were verified using the ONCOMINE server (https://www.Oncomine.org/resource/), which is an online available microarray database[12].
WebGestalt online tool (http://www.webgestalt.org) was used to identify DEGs enriched in biological processes, molecular functions, cellular component-related pathways, and KEGG pathways. The P-value of less than 0.05 was considered statistically significant.
STRING online database (https://string-db.org/; version 11.5) was used to build the PPI network[13]. The DEGs were submitted to the STRING database to construct the PPI network. Cytoscape (version 3.7.2) was used to draw the PPI network of DEGs, and the cytoHubba plugin was used to identify hub genes[14].
Gene expression profiling interactive analysis (GEPIA) online server (http://gepia.cancer-pku.cn/) was used to analyze the RNA sequencing expression data of tumors and healthy samples from the cancer genome atlas and genotype tissue expression projects[15]. We used this server to check whether the identified hub genes were differentially expressed in HCC tissues. Overall 5-year survival rates according to gene expression were obtained using the Kaplan-Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq)[16].
GraphPad Prism 6.0 (GraphPad Software Inc. La Jolla, CA, United States) was used for statistical analysis. Data were presented as mean ± SE or SD (for normally distributed data) or median with interquartile range (for non-normally distributed data). Statistically significant differences were determined using a two-tailed Student’s t-test or analysis of variance (ANOVA). Statistical significance was set and marked as aP < 0.05, bP < 0.01, cP < 0.001, and dP < 0.0001. Replicates are indicated in the figure legends, and (n) represents the number of experimental replicates.
We analyzed three gene expression datasets, GSE14323, GSE36411, and GSE89377, which included data from 74 cirrhotic and 53 healthy liver tissue samples in total. The analysis of processed sample data showed that the gene expression levels in different samples were primarily the same (Supple
| DEGs | Genes | ||
| Upregulated | C-X-C chemokine receptor type 4 (CXCR4) | SH3 domain-containing YSC84-like protein 1 (SH3YL1) | Laminins containing the α2 (LAMA2) |
| Lumican (LUM) | DNA-binding protein inhibitor ID-3 (ID3) | Microfibril-associated glycoprotein 4 (MFAP4) | |
| Prostaglandin-H2 D-isomerase (PTGDS) | Aldo-keto reductase family 1 member B10 (AKR1B10) | Marginal zone B- and B1-Cell-specific protein (MZB1) | |
| Dickkopf-related protein 3 (DKK3) | Ras-related protein Rac2 (RAC2) | Suppressor of lin-12-like protein 3 (SEL1L3) | |
| Dermatopontin (DPT) | Annexin A13 (ANXA13) | Defensin Beta 1 (DEFB1) | |
| H-2 class II histocompatibility antigen gamma chain (CD74) | CAMPATH-1 antigen (CD52) | Protein unc-93 homolog A (UNC93A) | |
| FXYD domain-containing ion transport regulator 2 (FXYD2) | Adipocyte enhancer-binding protein 1 (AEBP1) | Interleukin-7 receptor subunit alpha (IL7R) | |
| C-X-C motif chemokine 9 (CXCL9) | C-X-C motif chemokine 10 (CXCL10) | Ribonuclease pancreatic (RNASE1) | |
| SRY-Box transcription factor 9 (SOX9) | Gelsolin (GSN) | Carbohydrate sulfotransferase 4 (CHST4) | |
| Vimentin (VIM) | Galectin-3-binding protein (LGALS3BP) | Platelet-derived growth factor receptor alpha (PDGFRA) | |
| Lectin, galactoside-binding soluble 4 (LGALS4) | Laminin subunit gamma-3 (LAMC3) | Claudin-10 (CLDN10) | |
| Joining chain of multimeric IgA and IgM (JCHAIN) | Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 (ENPP2) | Cholesterol 25-hydroxylase (CH25H) | |
| Apolipoprotein L3 (APOL3) | Decorin (DCN) | Complement component C7 (C7) | |
| Epithelial cell adhesion molecule (EPCAM) | Keratin type I cytoskeletal 23 (KRT23) | EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) | |
| Matrix metallopeptidase 7 (MMP7) | |||
| Downregulated | Small conductance calcium-activated potassium channel protein 2 (KCNN2) | Cytochrome P450 2C19 (CYP2C19) | |
To understand the function of identified DEGs, we carried out Gene Ontology (GO) and KEGG pathways analyses using Webgestalt online server. The GO analysis results were listed according to P-values (Table 2), which showed that DEGs were significantly enriched in extracellular matrix organization (GO: 0030198), extracellular matrix structural constituent (GO: 0005201), collagen-containing extracellular matrix (GO: 0062023), extracellular matrix (GO: 0031012), cell adhesion (GO: 0007155), biological adhesion (GO: 0022610), and taxis (GO: 0042330). KEGG pathway analysis showed that the DEGs involved in the chemokine signaling pathway, focal adhesion, regulation of actin cytoskeleton, leukocyte transendothelial migration, pathways in cancer, and cytokine–cytokine receptor interaction, and arachidonic acid metabolism were highly enriched in patients with liver cirrhosis (Table 3).
| Gene set | Description | Count | P value |
| GO: 0030198 | Extracellular matrix organization | 11 | 3.38E-10 |
| GO: 0043062 | Extracellular structure organization | 11 | 1.56E-9 |
| GO: 0005201 | Extracellular matrix structural constituent | 7 | 7.48E-8 |
| GO: 0062023 | Collagen-containing extracellular matrix | 9 | 1.53E-7 |
| GO: 0031012 | Extracellular matrix | 10 | 1.78E-7 |
| GO: 0007155 | Cell adhesion | 15 | 2.78E-7 |
| GO: 0022610 | Biological adhesion | 15 | 3.00E-7 |
| GO: 0006935 | Chemotaxis | 10 | 0.000001284 |
| GO: 0042330 | Taxis | 10 | 0.000001323 |
| GO: 0005198 | Structural molecule activity | 11 | 0.000001624 |
| Gene set | Description | P value | Genes |
| hsa04062 | Chemokine signaling pathway | 0.00 | CXCR4, CXCL9, CXCL10, RAC2 |
| hsa04510 | Focal adhesion | 0.01 | LAMA2, LAMC3, PDGFRA, RAC2 |
| hsa04810 | Regulation of actin cytoskeleton | 0.01 | CXCR4, GSN, PDGFRA, RAC2 |
| hsa04670 | Leukocyte transendothelial migration | 0.01 | CLDN10, CXCR4, RAC2 |
| hsa05200 | Pathways in cancer | 0.01 | CXCR4, IL7R, LAMA2, LAMC3, PDGFRA, RAC2 |
| hsa05416 | Viral myocarditis | 0.02 | LAMA2 |
| hsa04060 | Cytokine-cytokine receptor interaction | 0.02 | CXCL9, CXCL10, CXCR4, IL7R |
| hsa00590 | Arachidonic acid metabolism | 0.02 | CYP2C19, PTGDS |
| hsa04976 | Bile secretion | 0.03 | FXYD2, KCNN2 |
| hsa04024 | cAMP signaling pathway | 0.02 | FXYD2, RAC2, SOX9 |
PPI network analysis aids in studying the molecular mechanisms of the disease pathogenesis. Using String v11 and Cytoscape software, we constructed a PPI network with 26 nodes and 36 edges (Figure 1E). These genes were upregulated in liver cirrhosis. The top 10 hub genes were identified using the CytoHubba plugin of Cytoscape, which included C-X-C motif chemokine ligand 9 (CXCL9), CXCL10, C-X-C motif chemokine receptor 4 (CXCR4), DCN, DPT, laminin subunit alpha 2 (LAMA2), lumican (LUM), microfibril associated protein 4 (MFAP4), platelet-derived growth factor receptor alpha (PDGFRA), and SOX9 (Figure 1F).
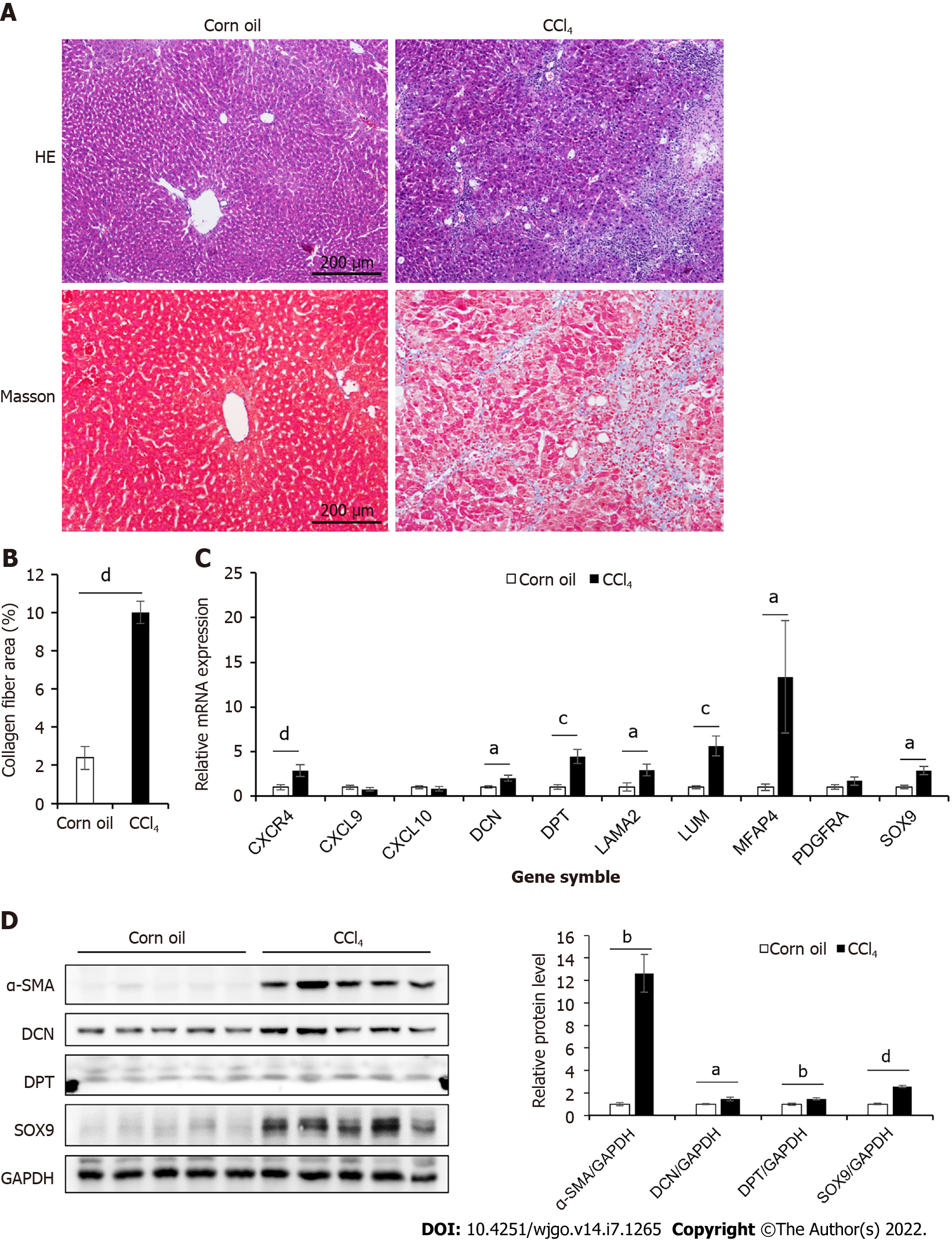
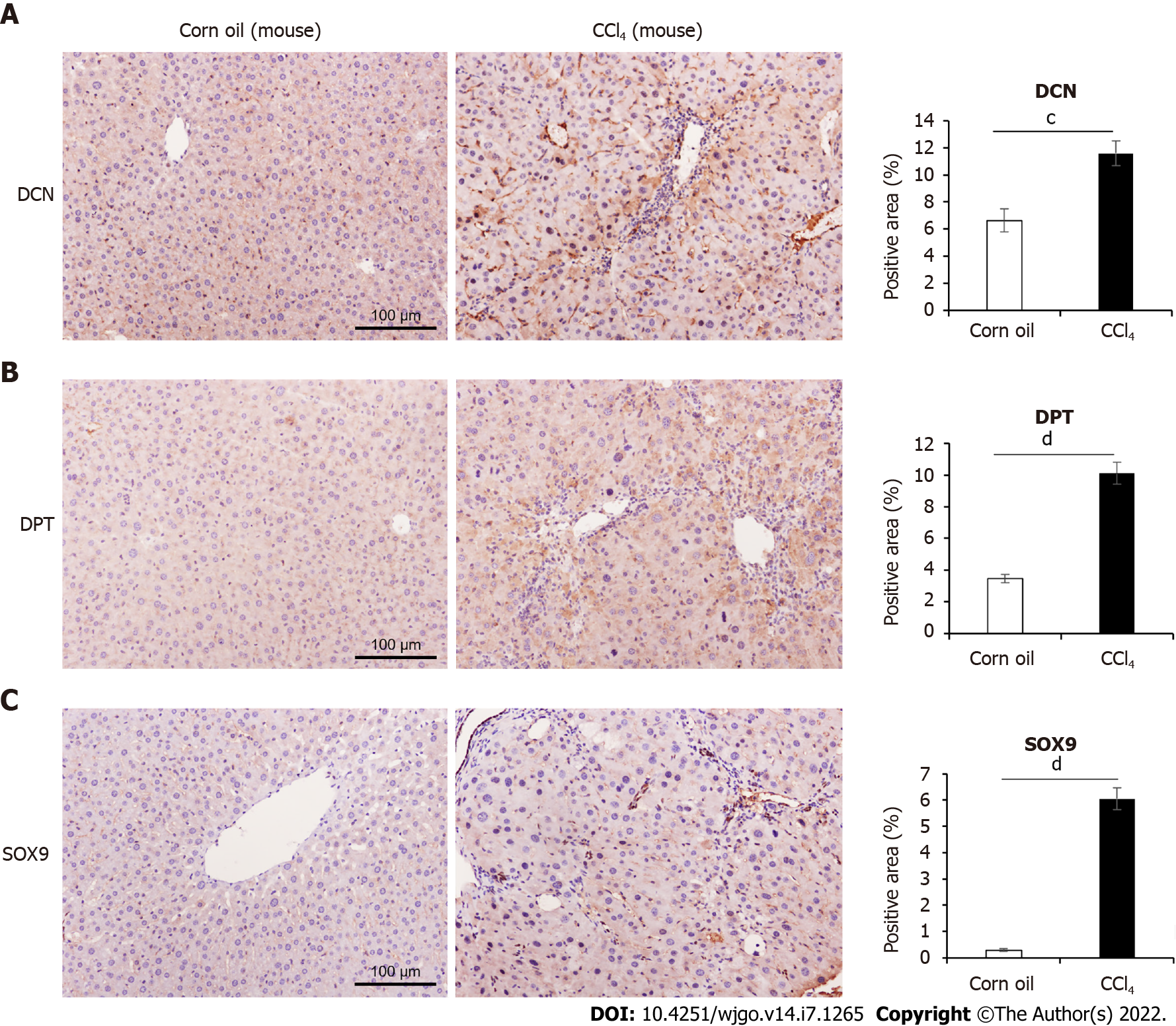
To study the role of the 10 hub genes in liver cirrhosis, we generated a CCl4-induced liver cirrhosis mouse model (Figure 2A–B). The relative mRNA levels of these genes were shown in Figure 2C. Compared to those in the control group, seven genes (CXCR4, DCN, DPT, LAMA2, LUM, MFAP4, and SOX9) were significantly upregulated in the liver cirrhosis mice, while the expression levels of PDGFRA,CXCL9, and CXCL10 were not significantly different between the two groups. For further validation, we performed Western blotting and immunohistochemical analysis (Figure 2D, Figure 3). These results confirmed that the protein levels of DCN, DPT, and SOX9 were significantly upregulated in the liver tissue of cirrhotic mice.
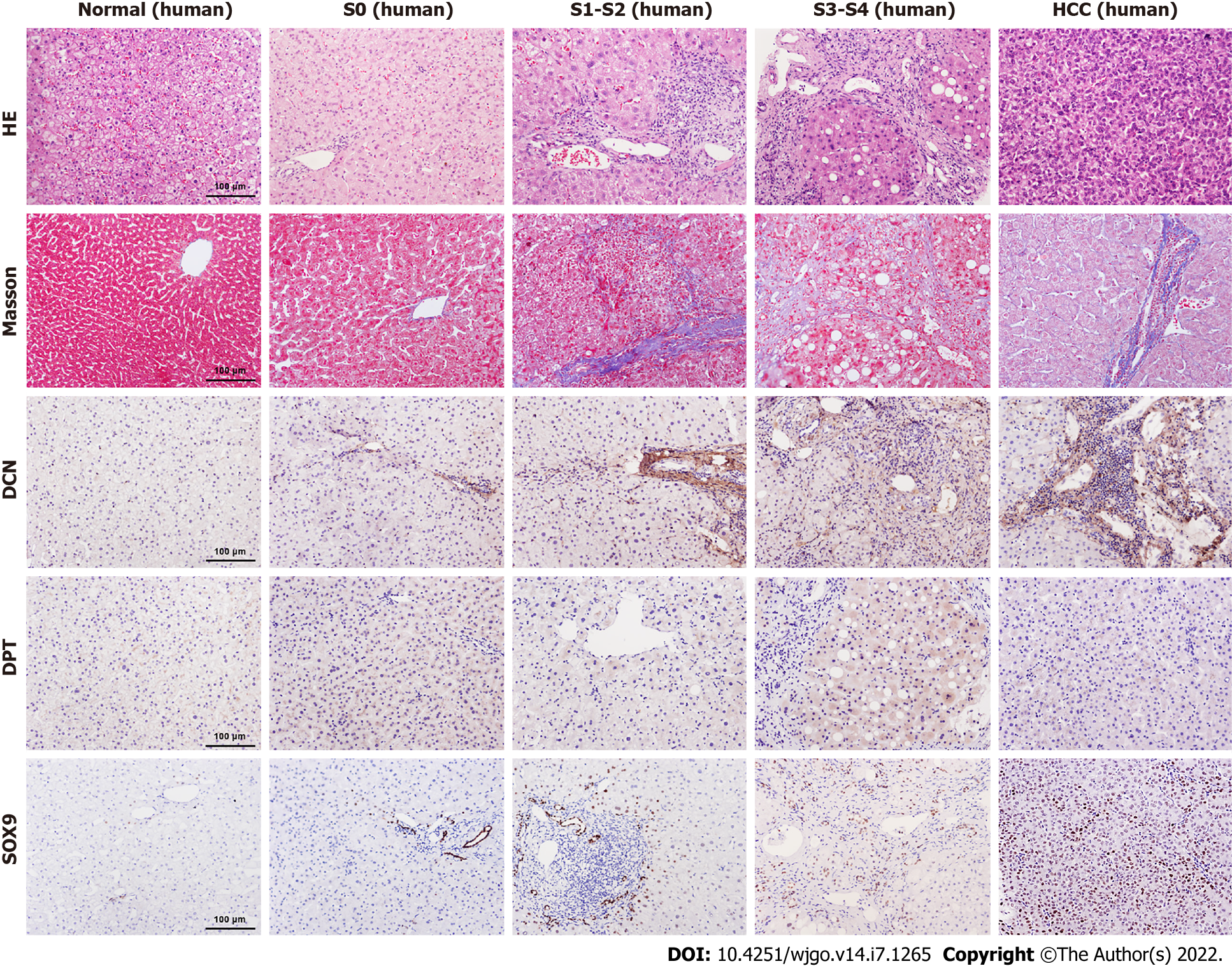
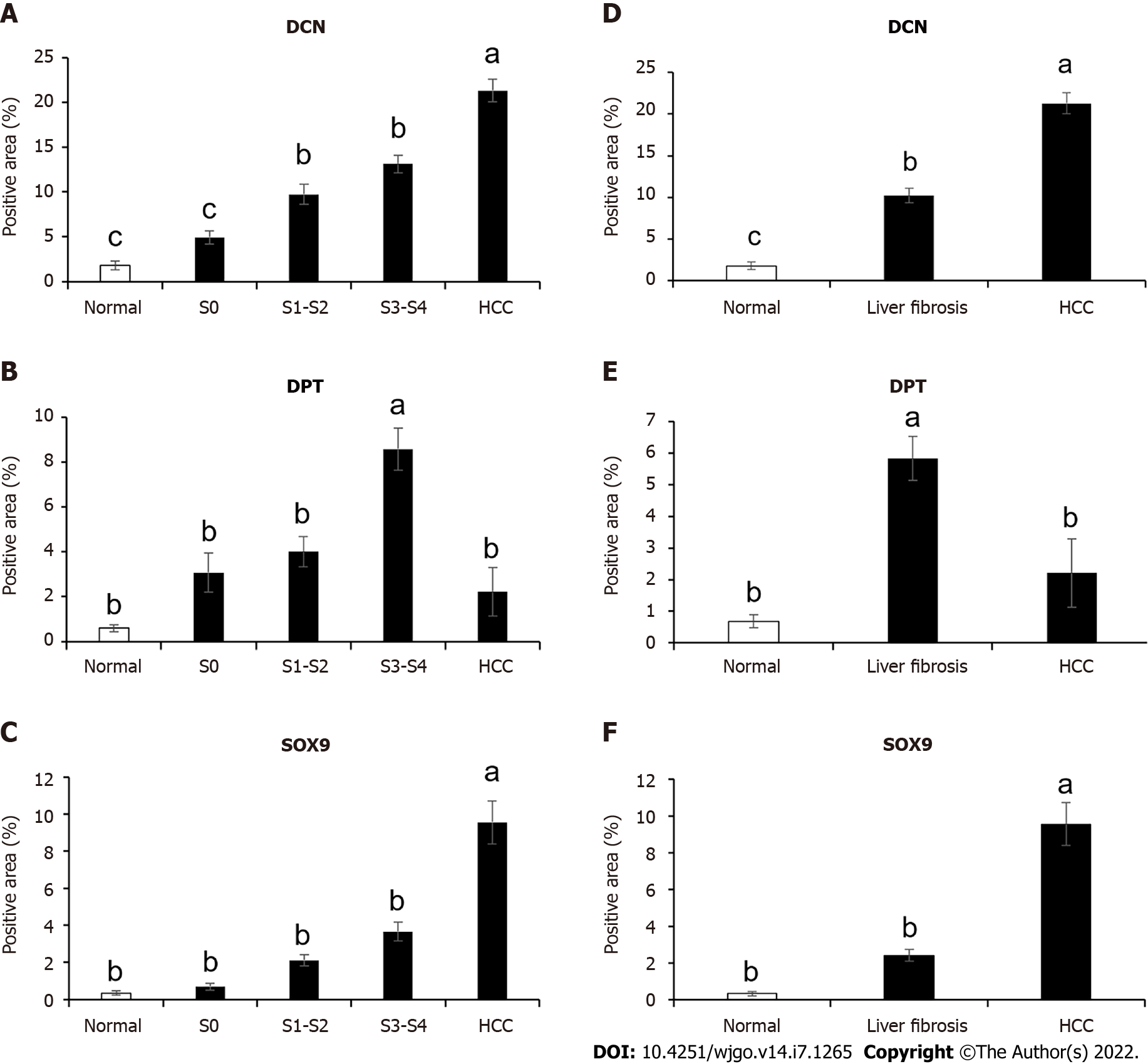
To further explore the relationship between DCN, DPT, and SOX9 protein expression and progression of liver cirrhosis, we collected liver biopsy tissue samples from 5 healthy controls, 28 patients with CHB, and 12 patients with CHB-associated HCC. All liver tissues were divided into 5 groups (normal, S0, S1-2, S3-4, and HCC) according to METAVIR system. The clinical profile of the patients enrolled in the study is summarized in Table 4. The results showed that males were the majority in S3-4 and HCC groups. The age, TBil, ALB, and CHE values in all groups and the AFP value in non-HCC group were in normal distribution, and the median AFP value in the HCC group was higher than the upper limit of normal value. In patients with CHB, HBV DNA was detected as positive, and most ALT and AST levels were elevated, which was consistent with the inflammatory activity of the liver. Most patients with HCC were detected negative for HBV DNA, which is related to antiviral treatment. Liver samples from patients with fibrosis showed increased collagen deposition, inflammatory cell infiltration, or atypical cells viewed with HE and Masson’s trichrome staining (Figure 4).
| Control (n = 5) | CHB-S0 (n = 4) | CHB-S1-2 (n = 13) | CHB-S3-4 (n = 11) | HCC (n = 12) | |
| Sex (M/F), n | 3/2 | 2/2 | 6/7 | 8/3 | 11/1 |
| Age, yr, mean ± SD | 41.8 ± 9.5 | 33.0 ± 10.3 | 38.2 ± 8.1 | 40.3 ± 6.1 | 52.8 ± 10.5 |
| HBeAg(+), n | 0 | 2 | 9 | 7 | 3 |
| HBV DNA, logIU/mL, median with IQR | - | 5.5 (2.4, 8.5) | 5.0 (2.5, 7.0) | 4.7 (2.0, 6.6) | 0 (0, 3.0) |
| ALT, U/L, median with IQR | 19.2 (16.8, 28.7) | 56.6 (45.0, 75.8) | 39.3 (17.9, 66.3) | 51.3 (29.6, 70.0) | 54.7 (24.1, 105.8) |
| AST, U/L, median with IQR | 20.7 (19.4, 22.5) | 36.3 (27.2, 44.6) | 27.3 (20.9, 39.9) | 31.8 (23.8, 49.7) | 54.8 (27.9, 111.6) |
| TBil, μmol/L, mean ± SD | 18.9 ± 14.1 | 14.0 ± 4.3 | 12.8 ± 4.9 | 12.9 ± 4.8 | 16.8 ± 14.6 |
| ALB, g/L, mean ± SD | 45.9 ± 4.8 | 47.8 ± 4.3 | 46.5 ± 3.9 | 44.4 ± 5.0 | 41.3 ± 5.3 |
| CHE, IU/L, mean ± SD | 6191.3 ± 1908.0 | 11514.5 ± 3416.1 | 8887.5 ± 1964.8 | 7708.5 ± 2064.4 | 6538.7 ± 7065.7 |
| AFP, ng/ml, mean ± SD or median with IQR | 2.5 ± 1.4 | 2.0 ± 0.8 | 4.7 ± 6.1 | 11.3 ± 22.1 | 37.9 (9.3, 388.9) |
Immunohistochemical results showed that the expression levels of DCN and SOX9 increased with the aggravation of liver fibrosis (Figure 4, Figure 5A, Figure 5C) and were significantly higher in the HCC group than those in healthy controls and fibrotic groups (Figure 4, Figure 5D, and Figure 5F). Further, compared with healthy controls, DPT expression was significantly increased in patients with liver fibrosis, particularly in the S3-S4 group but extremely reduced in patients with HCC (Figure 5B, Figure 5E). In addition, we found that DCN was mostly expressed in the portal vein region, which was highly consistent with the distribution of collagen fibers, while SOX9 and DPT were mostly expressed in hepatocytes and several other types of cells (Figure 4).
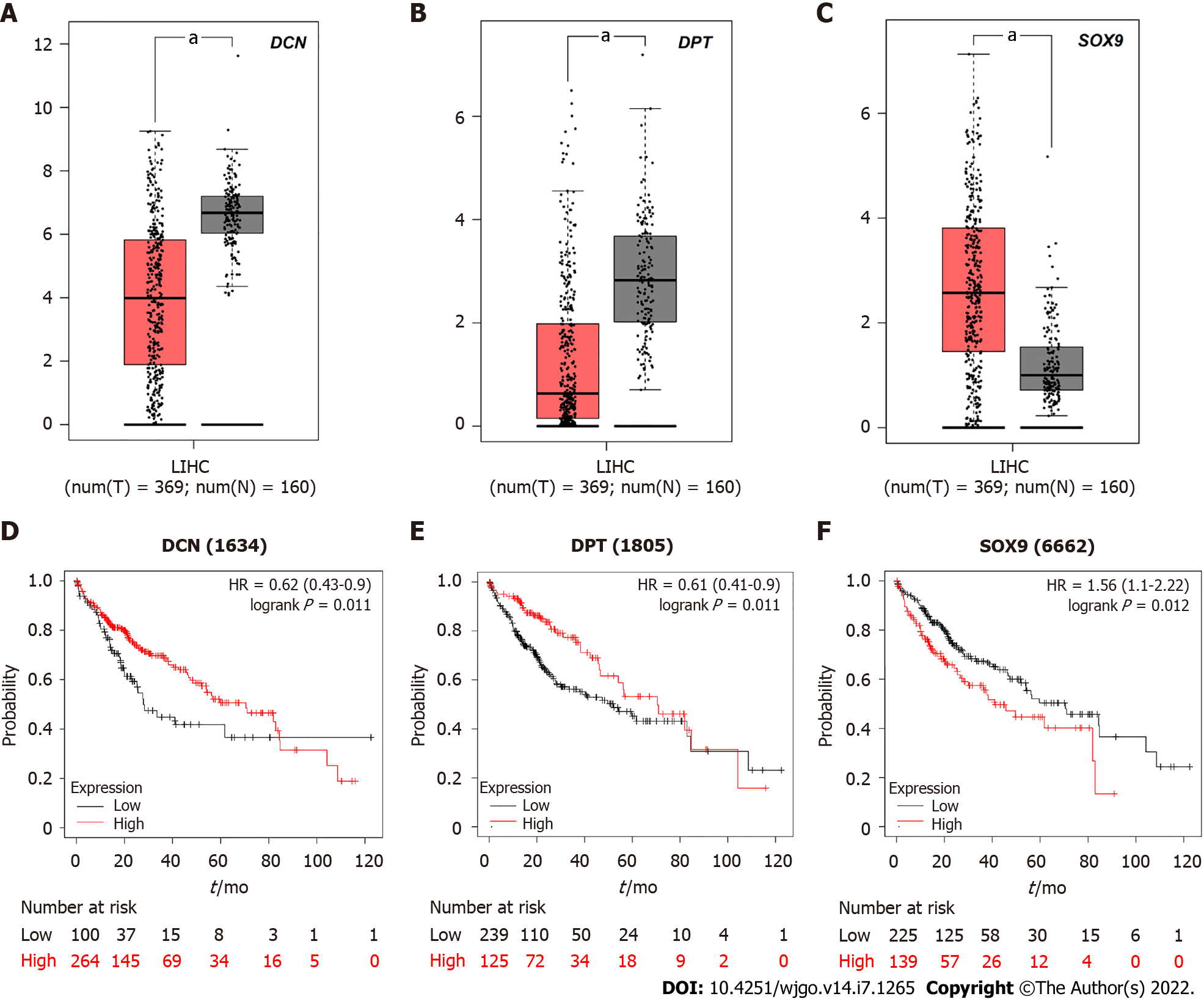
Due to the differential expression pattern and distribution, we decided to analyze the correlation between DCN, DPT, and SOX9 expression levels and the survival rate of patients with HCC. The GEPIA server was used to detect the mRNA expression levels in tissues of patients 369 liver hepatocellular carcinoma (LIHC) and 160 normal tissues (Figure 6A-C). The results showed that SOX9 was significantly upregulated whereas DCN and DPT were significantly downregulated in patients with LIHC.
Kaplan-Meier curves, depicting samples from these 369 patients with HCC, showed that the 5-year survival rate of patients with high expression of SOX 9 was significantly reduced, while that of patients with high expression of DCN or DPT was increased (Figure 6D-F).
Fibrosis is a common pathological symptom of severe liver damage caused by various chronic liver diseases. The most common primary liver cancer, HCC, occurs most often in people inflicted with chronic liver diseases[17]. Understanding the molecular mechanisms underlying cirrhosis can help in the development of effective treatments. Although bioinformatics tools can be used to study the relationship between gene function, liver fibrosis, and HCC, it is necessary to comprehensively analyze gene expression levels and distribution in the context of disease pathology via in vivo experiments.
GEO is an international public resource bank for high-throughput microarray and next-generation sequencing of functional genome datasets submitted by research groups. In this study, we identified 45 DEGs from GSE14323, GSE36411, and GSE89377 datasets in the GEO database for an in-depth analysis of their biological function. Most of these DEGs have been associated with liver diseases. CXCL9, ENPP2, CH25H, KRT23, IL7R, APOL3, and GSN are involved in HCV or HBV infection[18-23]. AEBP1, C7, and LUM are overexpressed in non-alcoholic steatohepatitis (NASH)[24-26]. CHST4, DEFB1, EFEMP1, MMP7, and SOX9 are associated with cholestasis[27-31]. AKR1B10, CLDN10, DKK3, EPCAM, LGALS4, and ITM2A are upregulated in patients with HCC[32-38]. The combined data-mining of three datasets from different sources yielded 45 DEGs, further indicating that the GEO database is indeed a useful resource for understanding the mechanism of liver diseases and using the GEO database can increase the efficiency of published resources.
Next, for detailed characterization of hub gene functions, we selected three representative genes (DCN, DPT, and SOX9), since their expression was consistently found to be associated with liver cirrhosis. We established a CCl4-induced mice model and verified their mRNA and protein levels using qPCR, Western blotting, and immunohistochemistry to confirm the expression and main patho
The DCN gene encodes a member of the small leucine-rich proteoglycan family of proteins, which can act as a tumor repressor in a variety of cancers[39]. DCN is a regulator of matrix assembly and not only targets transforming growth factor-beta 1 (TGF-β1) but is also involved in the maturation of collagen fibrils[40,41]. The enhanced deposition of DCN reflects the stimulatory effect of overproduction of TGF-β1[41]. Dudás et al[42] indicated that high amounts of TGF-β1 colocalize with DCN within the fibrotic areas of the liver using a cohort of liver pathologies, including chronic hepatitis, fibrosis, and cirrhosis, which is consistent with our results. However, contrary to our results, Shang et al[43] found DCN mRNA expression to be downregulated and not upregulated in patients with HCC via gene expression profile analyses. Although GEPIA analysis showed that the mRNA expression level of DCN in ILHC was lower than that normal tissues, the heterogeneity within each group, especially the ILHC group, was very different. In addition, our results were based on immunohistochemical analysis, different from the expression profile of the whole liver tissue used by Shang et al[43]. From the distribution of immunohistochemical sections, we concluded that almost all the increase in DCN expression was localized in the collagen-intensive area of the portal region and not in the hepatic lobule, which indicates that the upregulated DCN significantly represents an increase in matrix assembly. These may partly explain why the increased DCN is not associated with the lower 5-year survival rate of HCC patients, and why the expression pattern of DCN expression analyzed by GEPIA server was different from our study. Considering these inconsistent results, the expression, location and function of DCN deserves further study.
DPT is a downstream target of the vitamin D receptor. Fu et al[44] reported that mRNA expression of DPT was significantly downregulated in HCC, while its protein was weakly expressed in tumorous tissues compared to that in non-tumorous tissues. However, Lefebvre et al[45] suggested that DPT is upregulated in active NASH and fibrosis, and it is necessary for collagen deposition in profibrotic conditions. Our results also confirmed that the expression of DPT increased with the aggravation of liver fibrosis. Interestingly, a previous study showed that DPT interacts with DCN, which influences collagen fibrillogenesis and increases TGF-β1 signaling[46]. Our study identified these two molecules in a combined screening, suggesting that both DPT and DCN play an important role in the occurrence and development of liver fibrosis and HCC. However, the interactions between them needs to be further studied.
During tumorigenesis, SOX9 is upregulated in various tumors and plays an essential role in tumor progression as an oncogene[29], which regulates cellular proliferation, senescence, and self-renewal and is highly expressed in liver cancer stem cells[47]. In addition, SOX9 was the earliest marker expressed by biliary precursors[48]. It has been confirmed as a transcription factor that regulates bile duct development and contributes to liver regeneration and fibrosis[47]. In this study, we confirmed that SOX9 was positively correlated with the degree of fibrosis, and the high expression of SOX9 indicated a decline in the 5-year survival rate of patients with HCC, which is consistent with the results of other studies. In addition, our results showed that SOX9 was enriched and expressed in bile duct cells in mouse and human fibrotic livers, and the expression levels of SOX9 in hepatocytes were also increased significantly in patients with HCC. This observation can be explained by the fact that SOX9 mediates the transdifferentiation of hepatocytes into bile duct epithelial cells[47]. However, the detailed molecular mechanism of SOX9 overexpression in hepatocytes requires further elucidation.
Our current study has some limitations. First, we only analyzed the transcriptome, and many studies have shown that epigenetic modifications and non-coding RNAs also play an important role in the progression of liver diseases[33,49]. Secondly, the sample size in terms of number of patients was small, and immunohistochemistry was a semi-quantitative method. In addition, although our study had identified the signal transduction pathway involved in liver cirrhosis and HCC, it lacked in-depth analysis on the mechanism of action of these molecules, which needed to be further studied using in vivo studies or knock-out mice.
We screened GEO databases and obtained 45 DEGs and 10 hub genes (particularly DCN, DPT, and SOX9) in cirrhotic liver tissues. Upregulated expression of DCN, DPT, and SOX9 was all positively correlated with the degree of fibrosis, but there may be differences between their correlation with the 5-year survival rate of HCC patients.
Liver fibrosis and hepatocellular carcinoma (HCC) are common adverse consequences of chronic liver injury. Establishing more effective biomarkers is important for understanding the pathogenesis, occurrence, development mechanisms of liver fibrosis and HCC, as well as to identify new diagnostic and therapeutic tools.
Bioinformatics has screened out many differentially expressed genes related to liver fibrosis; however, it is unknown whether these genes are different in animal and human liver fibrosis tissues, especially among the different fibrotic degrees. Therefore, we should carefully analyze the research results of bioinformatics.
To identify liver fibrosis-related core genes, we observed and compared the differential expression pattern of core genes in patients with liver fibrosis and HCC.
In this study, we analyzed the expression pattern of hub genes of fibrosis and HCC. Bioinformatics analyses, quantitative polymerase chain reaction, Western blot, and immunohistochemistry of liver tissues from mouse model and patients were performed to identify novel biomarkers of liver fibrosis and HCC.
Ten hub genes (CXCL9, CXCL10, CXCR4, DCN, DPT, LAMA2, LUM, MFAP4, PDGFRA, and SOX9) associated with cirrhosis were screened from GSE14323, GSE36411, and GSE89377 datasets. DCN, DPT, and SOX9 were highly expressed in the CCl4-induced mouse model of liver cirrhosis and fibrotic patient liver samples, and their expression levels were associated with the degree of fibrosis. In patients with HCC, SOX9 was upregulated, while DCN and DPT were downregulated. However, the 5-year survival rate of HCC patients with high SOX 9 expression was significantly reduced, which is different from DPT or DCN.
We screened and identified 10 hub genes related to fibrosis. The expression levels of DCN, DPT, and SOX were positively correlated with the degree of liver fibrosis but showed different correlations with the survival rate of patients with HCC.
The integrated approach of bioinformatics and molecular biology is more efficient to research multi-factorial diseases, such as liver fibrosis and liver cancer. Future studies on the differences on DCN, DPT, and SOX9 expression may help in the better understanding of the mechanisms involved in the development of liver fibrosis and HCC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Beijing Medical Association Hepatology Branch.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mukthinuthalapati VVPK, United States; Papadopoulos VP, Greece; Silva LD, Brazil S-Editor: Gong ZM L-Editor: A P-Editor: Li X
| 1. | Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 386] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 2. | Harris PS, Hansen RM, Gray ME, Massoud OI, McGuire BM, Shoreibah MG. Hepatocellular carcinoma surveillance: An evidence-based approach. World J Gastroenterol. 2019;25:1550-1559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 156] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (4)] |
| 3. | Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N, Zhao Y. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res. 2020;10:2993-3036. [PubMed] |
| 4. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13161] [Article Influence: 1462.3] [Reference Citation Analysis (3)] |
| 5. | Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20:7312-7324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 335] [Cited by in RCA: 399] [Article Influence: 36.3] [Reference Citation Analysis (10)] |
| 6. | Thomson MJ, Lok AS, Tapper EB. Optimizing medication management for patients with cirrhosis: Evidence-based strategies and their outcomes. Liver Int. 2018;38:1882-1890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Huang X, Li Y, Guo X, Zhu Z, Kong X, Yu F, Wang Q. Identification of differentially expressed genes and signaling pathways in chronic obstructive pulmonary disease via bioinformatic analysis. FEBS Open Bio. 2019;9:1880-1899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Jiang CH, Yuan X, Li JF, Xie YF, Zhang AZ, Wang XL, Yang L, Liu CX, Liang WH, Pang LJ, Zou H, Cui XB, Shen XH, Qi Y, Jiang JF, Gu WY, Li F, Hu JM. Bioinformatics-based screening of key genes for transformation of liver cirrhosis to hepatocellular carcinoma. J Transl Med. 2020;18:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Chan KM, Wu TH, Wu TJ, Chou HS, Yu MC, Lee WC. Bioinformatics microarray analysis and identification of gene expression profiles associated with cirrhotic liver. Kaohsiung J Med Sci. 2016;32:165-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Chinese Society of Infectious Diseases; Chinese Medical Association; Chinese Society of Hepatology, Chinese Medical Association. [The guidelines of prevention and treatment for chronic hepatitis B (2019 version)]. Zhonghua Gan Zang Bing Za Zhi. 2019;27:938-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 109] [Reference Citation Analysis (0)] |
| 11. | Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2860] [Cited by in RCA: 3068] [Article Influence: 105.8] [Reference Citation Analysis (0)] |
| 12. | Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1471] [Cited by in RCA: 1661] [Article Influence: 92.3] [Reference Citation Analysis (0)] |
| 13. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11519] [Article Influence: 1919.8] [Reference Citation Analysis (1)] |
| 14. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 32904] [Article Influence: 1566.9] [Reference Citation Analysis (0)] |
| 15. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 6966] [Article Influence: 870.8] [Reference Citation Analysis (0)] |
| 16. | Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 335] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 17. | Müller M, Bird TG, Nault JC. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol. 2020;72:990-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 18. | Marrocco C, Rinalducci S, Mohamadkhani A, D'Amici GM, Zolla L. Plasma gelsolin protein: a candidate biomarker for hepatitis B-associated liver cirrhosis identified by proteomic approach. Blood Transfus. 2010;8 Suppl 3:s105-s112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 19. | Guzmán-Fulgencio M, Berenguer J, Jiménez-Sousa MA, Pineda-Tenor D, Aldámiz-Echevarria T, García-Broncano P, Carrero A, García-Álvarez M, Tejerina F, Diez C, Vazquez-Morón S, Resino S. Association between IL7R polymorphisms and severe liver disease in HIV/HCV coinfected patients: a cross-sectional study. J Transl Med. 2015;13:206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Xiang Y, Tang JJ, Tao W, Cao X, Song BL, Zhong J. Identification of Cholesterol 25-Hydroxylase as a Novel Host Restriction Factor and a Part of the Primary Innate Immune Responses against Hepatitis C Virus Infection. J Virol. 2015;89:6805-6816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 21. | Shrivastava S, Meissner EG, Funk E, Poonia S, Shokeen V, Thakur A, Poonia B, Sarin SK, Trehanpati N, Kottilil S. Elevated hepatic lipid and interferon stimulated gene expression in HCV GT3 patients relative to non-alcoholic steatohepatitis. Hepatol Int. 2016;10:937-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Farquhar MJ, Humphreys IS, Rudge SA, Wilson GK, Bhattacharya B, Ciaccia M, Hu K, Zhang Q, Mailly L, Reynolds GM, Ashcroft M, Balfe P, Baumert TF, Roessler S, Wakelam MJO, McKeating JA. Autotaxin-lysophosphatidic acid receptor signalling regulates hepatitis C virus replication. J Hepatol. 2017;66:919-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 23. | Kinast V, Leber SL, Brown RJP, Vieyres G, Behrendt P, Eßbach C, Strnad P, Vondran FWR, Cornberg M, Wex C, Pietschmann T, Haybaeck J, Todt D, Steinmann E. Identification of Keratin 23 as a Hepatitis C Virus-Induced Host Factor in the Human Liver. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 24. | Gerhard GS, Hanson A, Wilhelmsen D, Piras IS, Still CD, Chu X, Petrick AT, DiStefano JK. AEBP1 expression increases with severity of fibrosis in NASH and is regulated by glucose, palmitate, and miR-372-3p. PLoS One. 2019;14:e0219764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Wolff G, Taranko AE, Meln I, Weinmann J, Sijmonsma T, Lerch S, Heide D, Billeter AT, Tews D, Krunic D, Fischer-Posovszky P, Müller-Stich BP, Herzig S, Grimm D, Heikenwälder M, Kao WW, Vegiopoulos A. Diet-dependent function of the extracellular matrix proteoglycan Lumican in obesity and glucose homeostasis. Mol Metab. 2019;19:97-106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Hou W, Janech MG, Sobolesky PM, Bland AM, Samsuddin S, Alazawi W, Syn WK. Proteomic screening of plasma identifies potential noninvasive biomarkers associated with significant/advanced fibrosis in patients with nonalcoholic fatty liver disease. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Huang CC, Chuang JH, Chou MH, Wu CL, Chen CM, Wang CC, Chen YS, Chen CL, Tai MH. Matrilysin (MMP-7) is a major matrix metalloproteinase upregulated in biliary atresia-associated liver fibrosis. Mod Pathol. 2005;18:941-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Jinawath N, Chamgramol Y, Furukawa Y, Obama K, Tsunoda T, Sripa B, Pairojkul C, Nakamura Y. Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma. Hepatology. 2006;44:1025-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 29. | Yovchev MI, Xue Y, Shafritz DA, Locker J, Oertel M. Repopulation of the fibrotic/cirrhotic rat liver by transplanted hepatic stem/progenitor cells and mature hepatocytes. Hepatology. 2014;59:284-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 30. | Lou Y, Tian GY, Song Y, Liu YL, Chen YD, Shi JP, Yang J. Characterization of transcriptional modules related to fibrosing-NAFLD progression. Sci Rep. 2017;7:4748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 31. | Klag T, Thomas M, Ehmann D, Courth L, Mailänder-Sanchez D, Weiss TS, Dayoub R, Abshagen K, Vollmar B, Thasler WE, Stange EF, Berg CP, Malek NP, Zanger UM, Wehkamp J. β-Defensin 1 Is Prominent in the Liver and Induced During Cholestasis by Bilirubin and Bile Acids via Farnesoid X Receptor and Constitutive Androstane Receptor. Front Immunol. 2018;9:1735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Cheung ST, Leung KL, Ip YC, Chen X, Fong DY, Ng IO, Fan ST, So S. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res. 2005;11:551-556. [PubMed] |
| 33. | Ding Z, Qian YB, Zhu LX, Xiong QR. Promoter methylation and mRNA expression of DKK-3 and WIF-1 in hepatocellular carcinoma. World J Gastroenterol. 2009;15:2595-2601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Huang GW, Ding X, Chen SL, Zeng L. Expression of claudin 10 protein in hepatocellular carcinoma: impact on survival. J Cancer Res Clin Oncol. 2011;137:1213-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 35. | Bacigalupo ML, Manzi M, Rabinovich GA, Troncoso MF. Hierarchical and selective roles of galectins in hepatocarcinogenesis, liver fibrosis and inflammation of hepatocellular carcinoma. World J Gastroenterol. 2013;19:8831-8849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 54] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 36. | Ye X, Li C, Zu X, Lin M, Liu Q, Liu J, Xu G, Chen Z, Xu Y, Liu L, Luo D, Cao Z, Shi G, Feng Z, Deng H, Liao Q, Cai C, Liao DF, Wang J, Jin J, Cao D. A Large-Scale Multicenter Study Validates Aldo-Keto Reductase Family 1 Member B10 as a Prevalent Serum Marker for Detection of Hepatocellular Carcinoma. Hepatology. 2019;69:2489-2501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 37. | Zhang Z, Li J, He T, Ouyang Y, Huang Y, Liu Q, Wang P, Ding J. The competitive endogenous RNA regulatory network reveals potential prognostic biomarkers for overall survival in hepatocellular carcinoma. Cancer Sci. 2019;110:2905-2923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Zhang Z, Wen H, Weng J, Feng L, Liu H, Hu X, Zeng F. Silencing of EPCAM suppresses hepatic fibrosis and hepatic stellate cell proliferation in mice with alcoholic hepatitis via the PI3K/Akt/mTOR signaling pathway. Cell Cycle. 2019;18:2239-2254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Zhang W, Ge Y, Cheng Q, Zhang Q, Fang L, Zheng J. Decorin is a pivotal effector in the extracellular matrix and tumour microenvironment. Oncotarget. 2018;9:5480-5491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 135] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 40. | Zhang G, Ezura Y, Chervoneva I, Robinson PS, Beason DP, Carine ET, Soslowsky LJ, Iozzo RV, Birk DE. Decorin regulates assembly of collagen fibrils and acquisition of biomechanical properties during tendon development. J Cell Biochem. 2006;98:1436-1449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 315] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 41. | Baghy K, Iozzo RV, Kovalszky I. Decorin-TGFβ axis in hepatic fibrosis and cirrhosis. J Histochem Cytochem. 2012;60:262-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 42. | Dudás J, Kovalszky I, Gallai M, Nagy JO, Schaff Z, Knittel T, Mehde M, Neubauer K, Szalay F, Ramadori G. Expression of decorin, transforming growth factor-beta 1, tissue inhibitor metalloproteinase 1 and 2, and type IV collagenases in chronic hepatitis. Am J Clin Pathol. 2001;115:725-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 43. | Shang YK, Li F, Zhang Y, Liu ZK, Wang ZL, Bian H, Chen ZN. Systems analysis of key genes and pathways in the progression of hepatocellular carcinoma. Medicine (Baltimore). 2018;97:e10892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 44. | Fu Y, Feng MX, Yu J, Ma MZ, Liu XJ, Li J, Yang XM, Wang YH, Zhang YL, Ao JP, Xue F, Qin W, Gu J, Xia Q, Zhang ZG. DNA methylation-mediated silencing of matricellular protein dermatopontin promotes hepatocellular carcinoma metastasis by α3β1 integrin-Rho GTPase signaling. Oncotarget. 2014;5:6701-6715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 45. | Lefebvre P, Lalloyer F, Baugé E, Pawlak M, Gheeraert C, Dehondt H, Vanhoutte J, Woitrain E, Hennuyer N, Mazuy C, Bobowski-Gérard M, Zummo FP, Derudas B, Driessen A, Hubens G, Vonghia L, Kwanten WJ, Michielsen P, Vanwolleghem T, Eeckhoute J, Verrijken A, Van Gaal L, Francque S, Staels B. Interspecies NASH disease activity whole-genome profiling identifies a fibrogenic role of PPARα-regulated dermatopontin. JCI Insight. 2017;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 46. | Okamoto O, Fujiwara S, Abe M, Sato Y. Dermatopontin interacts with transforming growth factor beta and enhances its biological activity. Biochem J. 1999;337 (Pt 3):537-541. [PubMed] |
| 47. | Suda H, Yoshii D, Yamamura K, Yokouchi Y, Inomata Y. New insight into reactive ductular cells of biliary atresia provided by pathological assessment of SOX9. Pediatr Surg Int. 2014;30:481-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 707] [Cited by in RCA: 649] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 49. | Yang JJ, Tao H, Deng ZY, Lu C, Li J. Non-coding RNA-mediated epigenetic regulation of liver fibrosis. Metabolism. 2015;64:1386-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |