Published online Apr 15, 2022. doi: 10.4251/wjgo.v14.i4.794
Peer-review started: December 17, 2021
First decision: January 27, 2022
Revised: February 2, 2022
Accepted: March 17, 2022
Article in press: March 17, 2022
Published online: April 15, 2022
Processing time: 119 Days and 1.5 Hours
Esophageal cancer (EC) is a malignant cancer that still has a poor prognosis, although its prognosis has been improving with the development of multidisciplinary treatment modalities such as surgery, chemotherapy and radiotherapy. Therefore, identifying specific molecular markers that can be served as biomarkers for the prognosis and treatment response of EC is highly desirable to aid in the personalization and improvement of the precision of medical treatment. Sirtuins are a family of nicotinamide adenine dinucleotide (NAD+)-dependent proteins consisting of seven members (SIRT1-7). These proteins have been reported to be involved in the regulation of a variety of biological functions including apoptosis, metabolism, stress response, senescence, differentiation and cell cycle progression. Given the variety of functions of sirtuins, they are speculated to be associated in some manner with cancer progression. However, while the role of sirtuins in cancer progression has been investigated over the past few years, their precise role remains difficult to characterize, as they have both cancer-promoting and cancer-suppressing properties, depending on the type of cancer. These conflicting characteristics make research into the nature of sirtuins all the more fascinating. However, the role of sirtuins in EC remains unclear due to the limited number of reports concerning sirtuins in EC. We herein review the current findings and future prospects of sirtuins in EC.
Core Tip: Although there have been several reports on the function of sirtuins in cancer progression, the biological roles and clinical implications of sirtuins in esophageal cancer (EC) remain controversial. This is the first review to focus on sirtuins in the field of EC. In this review, we will briefly summarize the role of sirtuins in cancer and discuss the current findings and future prospects of sirtuins in EC.
- Citation: Otsuka R, Hayano K, Matsubara H. Role of sirtuins in esophageal cancer: Current status and future prospects. World J Gastrointest Oncol 2022; 14(4): 794-807
- URL: https://www.wjgnet.com/1948-5204/full/v14/i4/794.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i4.794
Esophageal cancer (EC) is the seventh leading cause of morbidity and the sixth leading cause of mortality worldwide[1], and the prognosis of advanced EC patients is extremely poor[2]. Therefore, identifying specific molecular markers that can be used as prognostic markers or therapeutic targets for EC is highly desirable to aid in the personalization and improvement of the precision of medical treatment, which can prevent side effects and extra expenses, thereby leading to a more effective multidisciplinary treatment.
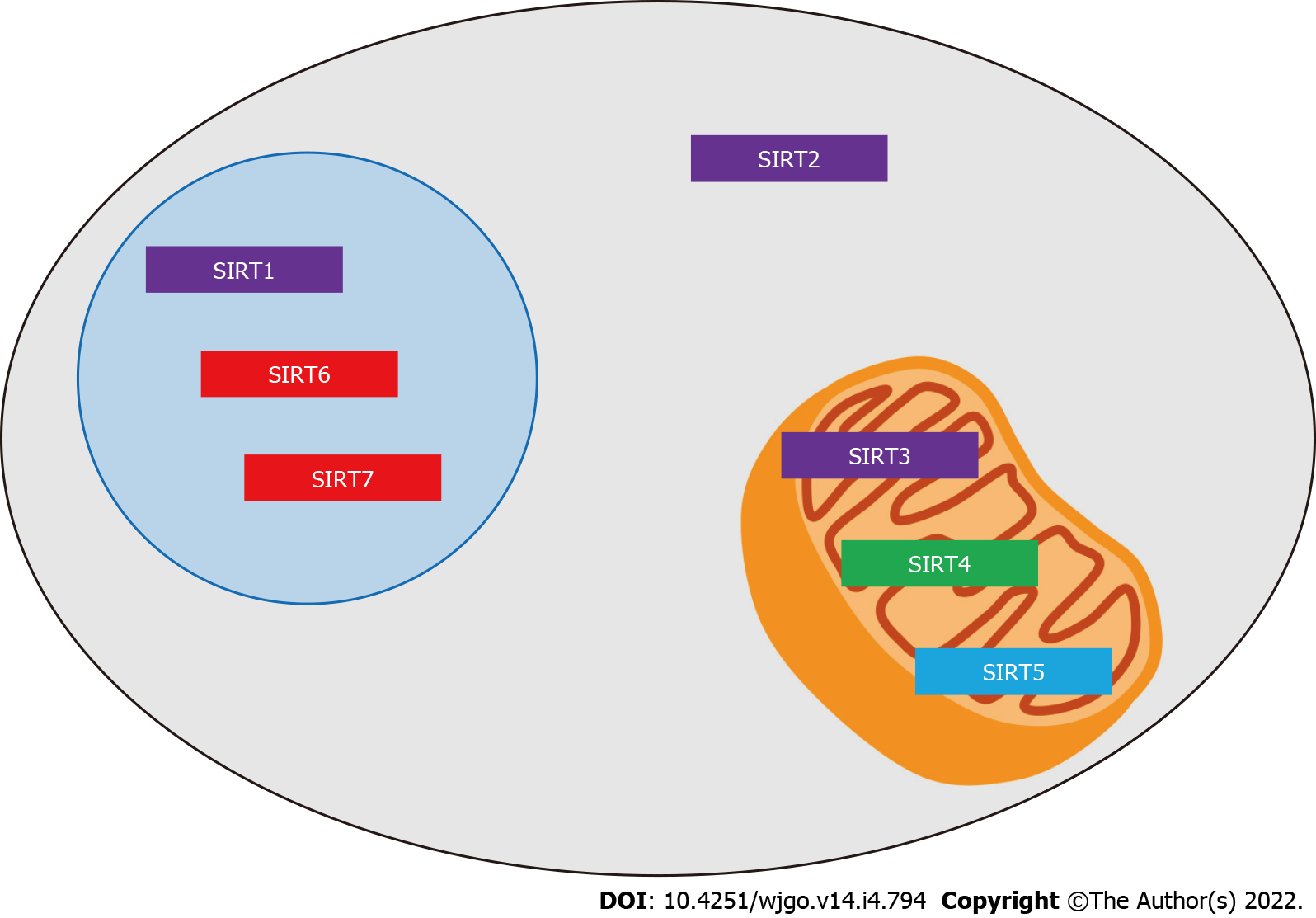
Sirtuins are a highly conserved family of proteins that exist in a wide range of prokaryotic and eukaryotic organisms, and their functional activity is dependent on the cofactor nicotinamide adenine dinucleotide (NAD+)[3]. The mammalian sirtuin family is a homolog of the yeast silent information regulator 2 (Sir2) protein and consists of seven members: SIRT1-7. Sirtuins are distinguished by their subcellular localization: SIRT1, SIRT6 and SIRT7 are mainly found in the nucleus; SIRT3, SIRT4 and SIRT5 are mainly located in the mitochondria; and SIRT2 is mainly found in the cytoplasm. Furthermore, the SIRT family proteins have conserved domains in their core catalytic activities, with SIRT1, SIRT2 and SIRT3 designated as class I; SIRT4 designated as class II; SIRT5 designated as class III; and SIRT6 and SIRT7 designated as class IV (Figure 1)[4]. Sirtuins are involved in the regulation of various biological functions, such as apoptosis, metabolism, stress response, aging, differentiation and cell cycle progression[5]. However, while the role of sirtuins in cancer progression has been investigated over the past few years, their precise role remains difficult to characterize, as they have both cancer-promoting and cancer-suppressing properties, depending on the type of cancer[6]. These conflicting characteristics make research into the nature of sirtuins all the more fascinating.
In EC, the clinical impact of sirtuins remains unclear due to the limited number of reports concerning sirtuins in EC. Therefore, this is the first review to focus on sirtuins in the field of EC. In this review, we will briefly summarize the role of sirtuins in cancer and discuss the current findings and future prospects of sirtuins in EC.
PubMed was searched to identify studies on sirtuins and cancer from inception until January 2022. The following search terms were applied: “Sirtuin” or “Silent mating type information regulation 2 homolog” or “SIRT” and “carcinoma” or “cancer”. The reference lists of all related articles were screened for other potentially relevant studies.
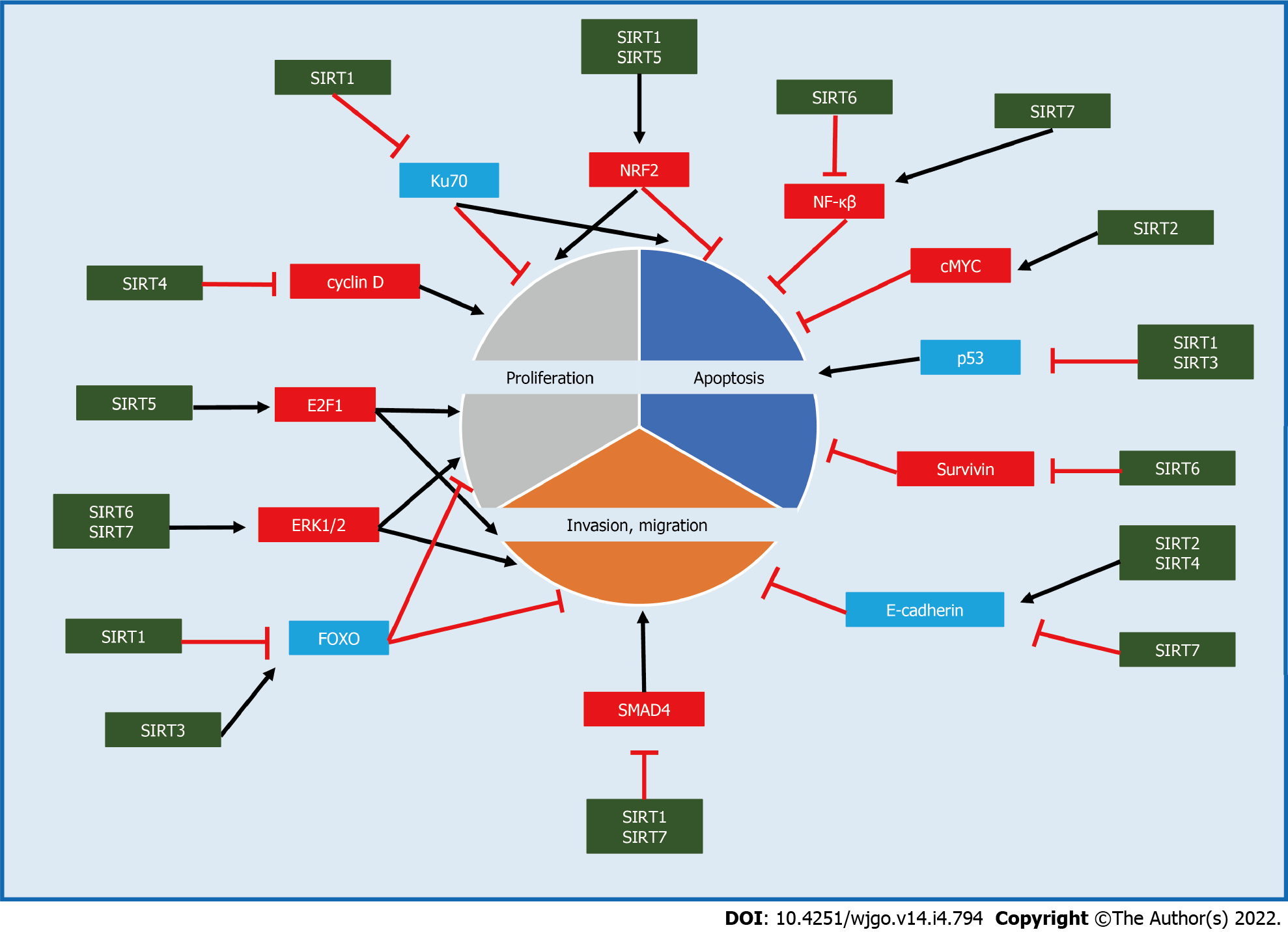
The role of SIRT1 in cancer progression is contradictory. This is because SIRT1 can both promote and inhibit tumorigenesis (Table 1 and Figure 2)[7].
| Role | Effect | Involved pathway or mechanism | |
| SIRT1 | Promotor | Promote proliferation | p53, FOXO family member, E2F1, p73, RB, Ku70, SFRP1, SFRP2, GATA4, GATA5, MLH1 |
| Inhibit apoptosis | p53, NF-κβ, FOXO3, Ku70, AKT, MAPK, NRF2 | ||
| Induce EMT, promote migration and metastasis | ZEB1 | ||
| Suppressor | Inhibit tumor formation and proliferation | β-catenin | |
| Induce apoptosis | survivin | ||
| Inhibit EMT | SMAD4, TGF-β signaling on MMP7 | ||
| SIRT2 | Promotor | Promote proliferation | Mediating immune evasion, altering the alkaline environment |
| Inhibit apoptosis | cMYC | ||
| Promote invasion and migration | Stimulating mitochondrial metabolism, mediating EMT | ||
| Suppressor | Inhibit proliferation | Inhibiting fibroblast activity and tumor angiogenesis | |
| Inhibit invasion and migration | MMP9, E-cadherin | ||
| SIRT3 | Promotor | Inhibit apoptosis and promote proliferation | p53, SHMT2, IDH2 |
| Promote invasion and metastasis | Reprogramming fatty acid metabolism | ||
| Suppressor | Induce cell arrest and apoptosis | Bcl-2, p53, HIF1α, PDC, SOD2, GOT2 | |
| Inhibit EMT and migration | FOXO3A, Wnt / β-catenin pathway | ||
| Inhibit tumorigenesis | PDH | ||
| SIRT4 | Promotor | Promote proliferation | PTEN |
| Suppressor | Inhibit glutamine metabolism and proliferation | mTORC1 pathway | |
| Inhibit EMT, invasion and migration | E-cadherin | ||
| Induce G1 cell cycle arrest | Cyclin D, cyclin E | ||
| SIRT5 | Promotor | Promote proliferation | GLUD1, SHMT2, NRF2, PKM2, SUN2 |
| Inhibit mitochondrial apoptosis | Cyt c | ||
| Promote autophagy | AMPK–mTOR pathway | ||
| Promote invasion and migration | E2F1 | ||
| Promote resistance to chemotherapy | SDHA | ||
| Suppressor | Inhibit carcinoma development | ACOX1 | |
| Inhibit proliferation | SOD1 | ||
| Represent protective mechanism for tumor cells | Inhibiting ammonia-induced autophagy | ||
| SIRT6 | Promotor | Promote proliferation and inhibit apoptosis | ERK1/2 pathway, AKT |
| Promote invasion and migration | ERK1/2/MMP9 signaling | ||
| Contribute to cancer development and progression | Regulating autophagy | ||
| Suppressor | Inhibit proliferation | PCBP2, ERK1/2 | |
| Inhibit Warburg effect | HIF-1α | ||
| Induce apoptosis | NF-κβ, Bax, survivin | ||
| Inhibit proliferation, invasion and migration | PKM2 | ||
| SIRT7 | Promotor | Promote proliferation | ERK1/2, H3K18ac |
| Inhibit apoptosis | miR34a, NF-κβ family subunits, mTOR/IGF2 pathway | ||
| Promote invasion | Vimentin, fibronectin, E-cadherin, β-catenin | ||
| Suppressor | Inhibit proliferation and invasion | SMAD4 | |
| Inhibit EMT | TGF-β signaling |
Several mechanisms that are responsible for the tumor-promoting nature of SIRT1 have been uncovered as follows: (1) SIRT1 contributes to cell proliferation by epigenetically suppressing the expression and activity of many tumor suppressor genes and proteins with DNA damage repair functions such as protein 53 (p53)[8], forkhead class O transcription factor (FOXO) family members[9], E2F transcription factor 1 (E2F1)[10], protein 73 (p73)[11], retinoblastoma protein (RB)[12], Ku70[13], secreted Frizzled-related protein 1(SFRP1), SFRP2, GATA4, GATA5 and mutL homolog 1 (MLH1)[14]; (2) SIRT1 acts as a regulator of apoptosis by deacetylating key apoptosis-related proteins and cell signaling molecules such as p53, nuclear factor kappa B subunit 1 (NF-κB), FOXO3, Ku70, protein kinase B (AKT), mitogen-activated protein kinase (MAPK), and nuclear factor erythroid 2-related factor 2 (NRF2), in response to DNA damage and oxidative stress[6]; and (3) SIRT1 induces epithelial-mesenchymal transition (EMT) and promotes cell migration and metastasis by cooperating with EMT transcription factors such as zinc finger E-box binding homeobox 1 (ZEB1) in prostate cancer[15]. It has also been reported that the high expression of SIRT1 is associated with an advanced stage and poor prognosis in certain types of cancer such as gastric cancer[16], lung adenocarcinoma[17] and colorectal cancer[18].
However, SIRT1 has also been reported to function as a tumor suppressor through the following mechanisms: (1) SIRT1 inhibits tumor formation and proliferation by deacetylating catenin beta (β-catenin) in colon cancer[19]; (2) SIRT1 induces apoptosis in breast cancer 1 (BRCA1)-related breast cancer by suppressing survivin, an inhibitor of apoptosis[20]; and (3) SIRT1 suppresses EMT in cancer by deacetylating SMAD family member 4 (SMAD4) and inhibiting the effect of transforming growth factor beta (TGF-β) signaling on matrix metallopeptidase 7 (MMP7), a target gene of SMAD4[21].
In EC, SIRT1 has been reported as a tumor-promoting factor (Table 2). Suppression of SIRT1 inhibits cell proliferation, cell migration and EMT in esophageal squamous cell carcinoma (ESCC) cell line[22,23]. SIRT1 has been suggested to be useful as a biomarker in EC as follows: (1) It has been reported that SIRT1 expression is associated with a poor prognosis in both ESCC and esophageal adenocarcinoma (EAC)[23-28]; (2) SIRT1 has also been demonstrated to be related to chemotherapy and chemoradiotherapy resistance in several ESCC studies[29-32]; and (3) SIRT1 has been described to be a useful biomarker for high-grade dysplasia and cancer of Barrett's esophagus[33]. Furthermore, we conducted a meta-analysis of these articles and demonstrated that a high expression of SIRT1 was correlated with a poor overall survival (OS), deeper tumors and a more advanced TNM stage in patients with ESCC[34]. In addition, recent studies have reported the potential utility of SIRT1 as a therapeutic target in EC. Liu et al[35] reported that rapamycin suppressed cell viability, migration, invasion and the phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathways in EC by negatively regulating SIRT1. Jiang et al[36] reported that sirtinol inhibited cell viability in EAC in a dose-dependent manner, affected proliferation in the long term and potentially suppressed resistant and recurrent tumors under hypoxic conditions. Taken together, these reports suggest that SIRT1 inhibition may play an important role in the therapeutic field of EC.
| Type | Role | Effect | Ref. | |
| SIRT1 | ESCC | Promotor | Suppression of SIRT1 inhibits cell proliferation, cell migration, and EMT in ESCC cell line | [22,23] |
| SIRT1 expression is associated with poor prognosis | [23-27,34] | |||
| SIRT1 enhances chemotherapy and chemoradiotherapy resistance | [29-32] | |||
| Rapamycin suppresses cell viability, migration, invasion by negatively regulating SIRT1 | [35] | |||
| EAC | SIRT1 is associated with poor overall survival | [28] | ||
| SIRT1 is a useful biomarker for high-grade dysplasia and cancer of Barrett's esophagus | [33] | |||
| Sirtinol, SIRT1 inhibitor, inhibits cell viability, affects proliferation in the long term, and potentially suppresses resistant and recurrent tumors under hypoxic conditions | [36] | |||
| SIRT2 | ESCC | Promotor | SIRT2 expression was associated with tumor invasion, lymph node metastasis, advanced clinical stage, poor progression-free survival, and overall survival | [47] |
| EAC | Suppressor | Dysregulation of SIRT2 is associated with poor prognosis | [48,49] | |
| SIRT3 | ESCC | Promotor | Serum SIRT3 levels are higher in ESCC patients compared to those in the control subjects | [63] |
| SIRT3 induces the proliferation inhibition and apoptosis | [64] | |||
| High SIRT3 expression is associated with poor survival outcome | [65,66] | |||
| EAC | No report | |||
| SIRT4 | ESCC | Suppressor | SIRT4 rescues the promoting effect of miR-424-5p on ESCC cell proliferation and migration | [76] |
| Low SIRT4 expression is associated with a high distant recurrence rate and poor prognosis, and in vitro, knockdown of SIRT4 promotes cell proliferation and migration | [77] | |||
| EAC | No report | |||
| SIRT5 | ESCC | No report | ||
| EAC | ||||
| SIRT6 | ESCC | Promotor | SIRT 6 is overexpressed in ESCC tissues and that it also promotes cell proliferation and induces the expression of Bcl2, an important anti-apoptotic factor, and autophagy in ESCC cells | [102] |
| EAC | No report | |||
| SIRT7 | ESCC | No report | ||
| EAC |
Similar to SIRT1, SIRT2 has been reported to have both tumor-promoting and tumor-suppressing effects depending on the cancer type (Table 1 and Figure 2).
SIRT2 has been reported to promote cell proliferation in hepatocellular carcinoma (HCC), pancreatic cancer and neuroblastoma[37,38]. SIRT2 also promotes cell growth by interacting with the tumor microenvironment, such as mediating immune evasion and altering the alkaline environment[39]. In cholangiocarcinoma, SIRT2 inhibits apoptosis via the peroxidation reaction through metabolic reprogramming by activating cMYC[40]. In addition, SIRT2 promotes invasion and migration in HCC by stimulating mitochondrial metabolism and mediating EMT[41,42].
Conversely, SIRT2 has been reported as a tumor suppressor that inhibits the growth of tumor cells through interaction with the tumor microenvironment, such as by inhibiting fibroblast activity and tumor angiogenesis[39]. In addition, the increased expression of matrix metalloproteinase 9 (MMP9) and decreased expression of cadherin 1 (E-cadherin) were shown to promote cell migration and invasion in SIRT2-deficient mouse embryonic fibroblasts[43]. In addition, a low expression of SIRT2 is reportedly associated with a poor prognosis in prostate cancer[44], cervical cancer[45] and colorectal cancer[46].
In EC, Yan et al[47] reported that SIRT2 expression was associated with tumor invasion, lymph node metastasis, advanced clinical stage, a poor progression-free survival and the OS in ESCC patients (Table 2). In contrast, SIRT2 has been reported to be a tumor suppressor in EAC. Ong et al[48] revealed that dysregulation of SIRT2 significantly increased the hazard ratio of death. Peters et al[49] also demonstrated that the dysregulation of SIRT2 was significantly associated with a poor prognosis in esophageal and junctional adenocarcinoma.
Whether SIRT3, a major mitochondrial deacetylase, functions as a tumor promoter or suppressor remains controversial (Table 1 and Figure 2).
SIRT3 regulates deacetylation to a variety of substrates, including p53, serine hydroxymethyltransferase 2 (SHMT2) and isocitrate dehydrogenase 2 (IDH2), preventing apoptosis and promoting cell proliferation[50-52]. In addition, Li et al[53] showed that SIRT3 promotes infiltration and metastasis of cervical cancer cells by reprogramming fatty acid metabolism.
In contrast, many studies have suggested the role of SIRT3 as a tumor suppressor. It has been reported that SIRT3 induces tumor-suppressing effects such as cell arrest and apoptosis by controlling Bcl-2, p53, hypoxia inducible factor 1 subunit alpha (HIF1α), pyruvate dehydrogenase complex (PDC), superoxide dismutase 2 (SOD2) and glutamic-oxaloacetic transaminase 2 (GOT2)[54-59]. Regarding metastasis, Li et al[60] revealed that SIRT3 promoted FOXO3A expression by weakening the Wnt/β-catenin pathway thereby inhibiting EMT and prostate cancer cell migration. Furthermore, Ozden et al[61] reported that activation of pyruvate dehydrogenase (PDH) by SIRT3 increased oxidative phosphorylation and reactive oxygen species production and reduced glycolysis which contributed to reduced tumorigenesis in cancer cells.
The relationship between SIRT3 expression and the clinical prognosis reportedly differs depending on the type of cancer and no clear consensus has yet been reached[62].
Regarding EC, several reports showed SIRT3 was a tumor promotor in ESCC (Table 2). Cobanoğlu et al[63] reported that serum SIRT3 Levels were significantly higher in ESCC patients than in the control subjects. Yang et al[64] showed that downregulation of SIRT3 induced the proliferation inhibition and apoptosis in ESCC cells. In addition, two articles demonstrated that a high SIRT3 expression was significantly associated with a poor survival outcome[65,66]. There have been no reports yet on the relationship between EAC and SIRT3.
SIRT4 has been reported primarily as a tumor suppressor (Table 1 and Figure 2). Wang et al[67] revealed that SIRT4 was downregulated in 30 cancers according to an analysis using data from The Cancer Genome Atlas (TCGA) database. SIRT4 is an important component of the DNA damage response pathway that inhibits glutamine metabolism, arrests the cell cycle and suppresses tumors. When SIRT4 is deficient, glutamine-dependent proliferation and stress-induced genomic instability increase resulting in a tumorigenic phenotype[68]. Csibi et al[69] also reported that the mammalian target of rapamycin complex 1 (mTORC1) pathway inhibited SIRT4 and stimulated glutamine metabolism and cell proliferation. In addition, SIRT4 has been reported to enhance E-cadherin and inhibit EMT, thereby decreasing migration and the invasion ability in gastric and colorectal cancer cells[70,71]. Furthermore, Hu et al[72] showed that overexpression of SIRT4 induced G1 cell cycle arrest through the inhibition of the phosphorylated extracellular signal-regulated kinases cyclin D and cyclin E. In addition, several studies have revealed that a low SIRT4 expression was significantly correlated with a poor prognosis in patients with various cancers[73].
In contrast, a small number of studies have reported the function of SIRT4 as a tumor-promoting factor (Table 1). Jeong et al[74] demonstrated that the overexpression of SIRT4 protected cancer cells from DNA damage or endoplasmic reticulum stress, and conversely, the loss of SIRT4 sensitized cells after drug treatment. Furthermore, when cells are starved of nutrients, SIRT4 cooperates with insulin-degrading enzymes to degrade phosphatase and tensin homolog (PTEN), a tumor-suppressing factor, and promote the survival of cancer cells[75].
In EC, SIRT4 has been reported as a tumor suppressor (Table 2). Cui et al[76] revealed that SRT4 was negatively regulated by miR-424-5p, and overexpression of SIRT4 strongly rescued the promoting effect of miR-424-5p on ESCC cell proliferation and migration capacity. In addition, Nakahara et al[77] reported that a low SIRT4 expression was significantly associated with a high distant recurrence rate and poor prognosis, and in vitro, knockdown of SIRT4 promoted glutamine dehydrogenase activity and stimulated cell proliferation and migration.
As with other sirtuins, the role of SIRT5 in cancer is highly controversial with some reports emphasizing the cancer-promoting function of SIRT5. (Table 1 and Figure 2). SIRT5 functionally activates glutamate dehydrogenase 1 (GLUD1), an important regulator of intracellular glutaminolysis, and is involved in cell proliferation[78]. In addition, Yang reported that SIRT5 mediated the desuccinylation of SHMT2 and enhanced its activity, which in turn promotes serine metabolism in tumor cells thereby promoting cancer cell growth[79]. Furthermore, studies have shown that SIRT3 promotes cell proliferation by targeting NRF2, pyruvate kinase M2 (PKM2), and Sad1 and UNC84 domain containing 2 (SUN2)[80-82]. Regarding apoptosis, SIRT5 has been reported to deacetylate cytochrome C (Cyt c) and induce mitochondrial apoptosis[83]. Gu et al[84] demonstrated that SIRT5 enhances autophagy and exerts tumor-promoting functions in gastric cancer cells. Moreover, SIRT5 promotes cancer cell invasion and migration by targeting E2F1[85]. Du et al[86] revealed that SIRT5 demalonylated and inactivated succinate dehydrogenase complex flavoprotein subunit A (SDHA) and accumulated its metabolite succinate resulting in resistance to chemotherapy.
In contrast, SIRT5 has also been reported to have tumor-suppressive effects (Table 1). Chen et al[87] revealed that SIRT5-mediated desuccinylation inhibited the activity of acyl-CoA oxidase 1 (ACOX1) and played an important role in the suppression of oxidative stress, protection of the liver and inhibition of HCC development. SIRT5 has been suggested to have a tumor-suppressor function via desuccinylation of superoxide dismutase 1 (SOD1)[88]. Furthermore, Polletta et al[89] demonstrated that SIRT5 inhibited ammonia-induced autophagy which is regarded as a protective mechanism for tumor cells. Therefore, activation of SIRT5 is thought to reduce the survival of tumor cells in response to stresses, such as chemotherapy, hypoxia and nutrient starvation.
The relationship between the SIRT5 expression and clinical prognosis has also been reported to vary by cancer type[78,90].
In EC, there have been no reports on the role of SIRT5, and there is much room for further investigation of the association between SIRT5 and EC.
SIRT6, like other sirtuins, functions as a double-edged sword in cancer (Table 1 and Figure 2). SIRT6 inhibits tumor growth by targeting poly(rC) binding protein 2 (PCBP2) and extracellular signal-regulated kinases 1/2 (ERK1/2)[91,92]. SIRT6 represses HIF-1α and regulates the expression of multiple glycolytic genes[93]. This indicates that SIRT6 plays a role in tumor suppression by inhibiting the Warburg effect. In addition, SIRT6 induces apoptosis in cancer cells by acting on NF-κB, BCL2 associated X (Bax) and survivin[94,95]. Bhardwaj et al[96] found that SIRT6 inhibited the oncogenic activity of PKM2, which has a non-metabolic nuclear carcinogenic function, resulting in a reduced cell proliferation, migration ability and invasiveness. One meta-analysis revealed that a high SIRT6 expression was associated with a longer OS in gastrointestinal cancers and a favorable TNM stage[97].
However, the role of SIRT6 as a tumor-promoting factor has also been reported. SIRT6 enhances HCC cell proliferation and inhibits apoptosis through the regulation of the ERK1/2 pathway[98]. In addition, Zhou et al[99] revealed that SIRT6 inhibited the acetylation of AKT and promoted its activation thereby preventing apoptosis and inducing cell proliferation. Bai et al[100] reported that the overexpression of SIRT6 in non-small-cell lung cancer cell lines promoted migration and invasion via ERK1/2/MMP9 signaling. SIRT6 has also been reported to positively regulate autophagy in melanoma cells and to exhibit tumor-promoting effects[101].
In EC, SIRT6 has been reported as a tumor-promoting factor (Table 2). Huang et al[102] demonstrated that SIRT6 was markedly overexpressed in ESCC tissues and that it also promoted cell proliferation and induced the expression of Bcl2, an important anti-apoptotic factor and autophagy in ESCC cells.
Like other sirtuins, SIRT7 has also been reported to have both tumor-promoting and tumor-suppressing roles (Table 1 and Figure 2). SIRT7 promotes cell proliferation by regulating ERK1/2 and histone H3 Lysine 18 acetylation (H3K18ac)[103,104]. In addition, SIRT7 induces apoptosis via miR34a, NF-κB family subunits and the mTOR/insulin like growth factor 2 (IGF2) pathway[105-107]. SIRT7 also influences the metastasis of cancer cells. Yu et al[103] showed that cells overexpressing SIRT7 had elevated levels of vimentin and fibronectin, which are markers of mesenchymal lineage, and decreased levels of E-cadherin and β-catenin, which are markers of epithelial lineage indicating enhanced invasion of colon cancer cells.
The role of SIRT7 as a tumor suppressor has been reported to include inhibition of growth and metastasis. Li et al[108] demonstrated that SIRT7 inhibited cell proliferation and invasion by deacetylating SMAD4 in oral squamous cell carcinoma. In addition, Tang et al[109] also revealed that loss of SIRT7 activated TGF-β signaling and promoted EMT.
Reports concerning the relationship between the SIRT7 expression and the prognosis are conflicting, with some citing a good prognosis while the others describe a poor prognosis[110].
In EC, there are no reports investigating the role of SIRT7, and whether it acts as a tumor-promoting factor or a tumor-suppressing factor remains unclear.
As mentioned above, sirtuins have been investigated in a variety of cancer types and play a dichotomous role depending on the situation. This trend is also true in the field of EC. SIRT1, SIRT2, SIRT3 and SIRT6 have been reported as tumor-promoting factors in ESCC, along with SIRT1 in EAC, while SIRT4 and SIRT2 have been reported as tumor suppressors in ESCC and EAC, respectively. SIRT5 and SIRT7 are interesting targets of study since their roles in both ESCC and EAC have not yet been reported.
One of the future points to be explored concerning sirtuins in EC is expected to be their utility as biomarkers. In most previous studies, the degree of sirtuin expression was assessed by immunohistochemistry. However, the cut-off values for sirtuin expression differed among studies, and this heterogeneity in assessment methods may have led to conflicting results among cancer types. Therefore, more accurate and less-invasive evaluations are anticipated in the future. Serum SIRT3 Levels have been reported to be a potentially useful biomarker, not only in EC[63] but also in lung cancer[111], suggesting that serum sirtuin levels merit exploration as a minimally invasive biomarker. Furthermore, in recent years, a wide variety of public databases, such as TCGA, have been used for analyses[67]. This is expected to make it possible to obtain more comprehensive and standardizable information in the future.
Since sirtuin enzymes play an important role in the regulation of various cellular events, there is strong interest in pursuing sirtuins as therapeutic targets. Although many reports related to the development of sirtuin inhibitors/activators have been found in electronic searches, only a very limited number of small-molecule compounds, such as reveratol and Ex-527, have been subjected to clinical trials[112]. In the field of EC, the effects of the SIRT1 inhibitors rapamycin and sirtinol have been reported in vitro and in vivo[35,36]. However, SIRT1, like other sirtuins, has been suggested to promote or inhibit cancer in a context-dependent manner so many comprehensive studies are needed to determine its clinical application. Although not yet presented, other sirtuin-targeted agents still have great therapeutic potential and advances in this area will contribute to the development of EC treatment.
In recent years, the breakthrough of immunotherapy has been considered an important topic in EC[113]. The involvement of sirtuins in immunity has been widely studied since the early discovery that SIRT1 regulates NF-κB, a transcription factor known to control inflammation and immune cell proliferation[114]. There have been no reports on the role of sirtuins in immunotherapy of esophageal cancer, although some reports have appeared in other cancer types. Zhang et al[115] showed that pharmacological inhibition of SIRT2 increased natural killer cell infiltration into the tumor and suppressed tumor growth in allograft melanoma. Furthermore, Xiang et al[116] demonstrated that SIRT7 suppressed myocyte enhancer factor 2D acetylation and programmed death ligand 1 expression and promoted HCC cell proliferation. Thus, the role of sirtuins in anti-tumor immunity in EC is an issue that deserves further attention and research.
In summary, sirtuins may be a key target for EC treatment in the future. However, much research is still needed to determine the clinical application as many aspects remain unresolved. We hope that this review will contribute to the development of this field.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Kalayarasan R, India; Yang X, China; Zhuge YZ, China S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Gong ZM
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64442] [Article Influence: 16110.5] [Reference Citation Analysis (176)] |
| 2. | Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1956] [Cited by in RCA: 1960] [Article Influence: 163.3] [Reference Citation Analysis (5)] |
| 3. | Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1402] [Cited by in RCA: 1372] [Article Influence: 76.2] [Reference Citation Analysis (0)] |
| 4. | Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1039] [Cited by in RCA: 1092] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 5. | Jaiswal A, Xudong Z, Zhenyu J, Saretzki G. Mitochondrial sirtuins in stem cells and cancer. FEBS J. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Zhao E, Hou J, Ke X, Abbas MN, Kausar S, Zhang L, Cui H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 7. | Li K, Luo J. The role of SIRT1 in tumorigenesis. N Am J Med Sci (Boston). 2011;4:104-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 8. | Sasca D, Hähnel PS, Szybinski J, Khawaja K, Kriege O, Pante SV, Bullinger L, Strand S, Strand D, Theobald M, Kindler T. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014;124:121-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 9. | Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1039] [Cited by in RCA: 1086] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 10. | Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 349] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 11. | Dai JM, Wang ZY, Sun DC, Lin RX, Wang SQ. SIRT1 interacts with p73 and suppresses p73-dependent transcriptional activity. J Cell Physiol. 2007;210:161-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 108] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J. 2007;407:451-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 104] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 13. | Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1456] [Cited by in RCA: 1503] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 14. | Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 306] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 15. | Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, Dai Y. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31:4619-4629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 262] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 16. | Noguchi A, Kikuchi K, Zheng H, Takahashi H, Miyagi Y, Aoki I, Takano Y. SIRT1 expression is associated with a poor prognosis, whereas DBC1 is associated with favorable outcomes in gastric cancer. Cancer Med. 2014;3:1553-1561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 17. | Li C, Wang L, Zheng L, Zhan X, Xu B, Jiang J, Wu C. SIRT1 expression is associated with poor prognosis of lung adenocarcinoma. Onco Targets Ther. 2015;8:977-984. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 18. | Zu G, Ji A, Zhou T, Che N. Clinicopathological significance of SIRT1 expression in colorectal cancer: A systematic review and meta analysis. Int J Surg. 2016;26:32-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 473] [Cited by in RCA: 468] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 20. | Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, Lee MH, Xiao C, Vassilopoulos A, Chen W, Gardner K, Man YG, Hung MC, Finkel T, Deng CX. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 281] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 21. | Simic P, Williams EO, Bell EL, Gong JJ, Bonkowski M, Guarente L. SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Rep. 2013;3:1175-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 187] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 22. | Qin T, Liu W, Huo J, Li L, Zhang X, Shi X, Zhou J, Wang C. SIRT1 expression regulates the transformation of resistant esophageal cancer cells via the epithelial-mesenchymal transition. Biomed Pharmacother. 2018;103:308-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Ma MC, Chiu TJ, Lu HI, Huang WT, Lo CM, Tien WY, Lan YC, Chen YY, Chen CH, Li SH. SIRT1 overexpression is an independent prognosticator for patients with esophageal squamous cell carcinoma. J Cardiothorac Surg. 2018;13:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 24. | Chen GQ, Tian H, Yue WM, Li L, Li SH, Qi L, Gao C, Si LB, Lu M, Feng F. SIRT1 expression is associated with lymphangiogenesis, lymphovascular invasion and prognosis in pN0 esophageal squamous cell carcinoma. Cell Biosci. 2014;4:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | He Z, Yi J, Jin L, Pan B, Chen L, Song H. Overexpression of Sirtuin-1 is associated with poor clinical outcome in esophageal squamous cell carcinoma. Tumour Biol. 2016;37:7139-7148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 26. | Han F, Zhang S, Liang J, Qiu W. Clinicopathological and predictive significance of SIRT1 and peroxisome proliferator-activated receptor gamma in esophageal squamous cell carcinoma: The correlation with EGFR and Survivin. Pathol Res Pract. 2018;214:686-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Yan L, Zhao Q, Liu L, Jin N, Wang S, Zhan X. Expression of SIRT1 and survivin correlates with poor prognosis in esophageal squamous cell carcinoma. Medicine (Baltimore). 2020;99:e21645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Zhu L, Dong L, Feng M, Yang F, Jiang W, Huang Z, Liu F, Wang L, Wang G, Li Q. Profiles of autophagy-related genes in esophageal adenocarcinoma. BMC Cancer. 2020;20:943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Cao B, Shi Q, Wang W. Higher expression of SIRT1 induced resistance of esophageal squamous cell carcinoma cells to cisplatin. J Thorac Dis. 2015;7:711-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 30. | Chen Y, Xie T, Ye Z, Wang F, Long D, Jiang M, Fang J, Lin Q, Li K, Wang Z, Fu Z. ADC correlation with Sirtuin1 to assess early chemoradiotherapy response of locally advanced esophageal carcinoma patients. Radiat Oncol. 2019;14:192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Kuo IY, Huang YL, Lin CY, Lin CH, Chang WL, Lai WW, Wang YC. SOX17 overexpression sensitizes chemoradiation response in esophageal cancer by transcriptional down-regulation of DNA repair and damage response genes. J Biomed Sci. 2019;26:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Ye Z, Xie T, Yan F, Wang L, Fang J, Wang Z, Hu F, Wang F, Fu Z. MiR-34a reverses radiation resistance on ECA-109 cells by inhibiting PI3K/AKT/mTOR signal pathway through downregulating the expression of SIRT1. Int J Radiat Biol. 2021;97:452-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Zhang S, Wang XI. SIRT1 is a useful biomarker for high-grade dysplasia and carcinoma in Barrett's esophagus. Ann Clin Lab Sci. 2013;43:373-377. [PubMed] |
| 34. | Otsuka R, Sakata H, Murakami K, Kano M, Endo S, Toyozumi T, Matsumoto Y, Suito H, Takahashi M, Sekino N, Hirasawa S, Kinoshita K, Sasaki T, Matsubara H. SIRT1 Expression Is a Promising Prognostic Biomarker in Esophageal Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Canner Diagn Progn. 2022;In press. |
| 35. | Liu T, Liang X, Sun Y, Yang S. Rapamycin suppresses the PI3K/AKT/mTOR signaling pathway by targeting SIRT1 in esophageal cancer. Exp Ther Med. 2021;22:1190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 36. | Jiang H, Patil K, Vashi A, Wang Y, Strickland E, Pai SB. Cellular molecular and proteomic profiling deciphers the SIRT1 controlled cell death pathways in esophageal adenocarcinoma cells. Cancer Treat Res Commun. 2021;26:100271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Xie HJ, Jung KH, Nam SW. Overexpression of SIRT2 contributes tumor cell growth in hepatocellular carcinomas. Mol Cell Toxicol. 2011;7:367-374. [RCA] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 38. | Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, Koach J, Tee AE, Haber M, Norris MD, Toon C, Rooman I, Xue C, Cheung BB, Kumar S, Marshall GM, Biankin AV, Liu T. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013;20:503-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 39. | Chen G, Huang P, Hu C. The role of SIRT2 in cancer: A novel therapeutic target. Int J Cancer. 2020;147:3297-3304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 40. | Xu L, Wang L, Zhou L, Dorfman RG, Pan Y, Tang D, Wang Y, Yin Y, Jiang C, Zou X, Wu J, Zhang M. The SIRT2/cMYC Pathway Inhibits Peroxidation-Related Apoptosis In Cholangiocarcinoma Through Metabolic Reprogramming. Neoplasia. 2019;21:429-441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 41. | Chen J, Chan AW, To KF, Chen W, Zhang Z, Ren J, Song C, Cheung YS, Lai PB, Cheng SH, Ng MH, Huang A, Ko BC. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology. 2013;57:2287-2298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 42. | Huang S, Zhao Z, Tang D, Zhou Q, Li Y, Zhou L, Yin Y, Wang Y, Pan Y, Dorfman RG, Ling T, Zhang M. Downregulation of SIRT2 Inhibits Invasion of Hepatocellular Carcinoma by Inhibiting Energy Metabolism. Transl Oncol. 2017;10:917-927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 43. | Nguyen P, Lee S, Lorang-Leins D, Trepel J, Smart DK. SIRT2 interacts with β-catenin to inhibit Wnt signaling output in response to radiation-induced stress. Mol Cancer Res. 2014;12:1244-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 44. | Damodaran S, Damaschke N, Gawdzik J, Yang B, Shi C, Allen GO, Huang W, Denu J, Jarrard D. Dysregulation of Sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer. 2017;17:874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 45. | Yang LP, Feng HQ, Ma JC, Wu H, Liu CR, Hou JD. SIRT2 expression exhibits potential to serve as a biomarker for disease surveillance and prognosis in the management of cervical cancer patients. Medicine (Baltimore). 2020;99:e18668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 46. | Du F, Li Z, Zhang G, Shaoyan S, Geng D, Tao Z, Qiu K, Liu S, Zhou Y, Zhang Y, Gu J, Wang G, Li L, Wu W. SIRT2, a direct target of miR-212-5p, suppresses the proliferation and metastasis of colorectal cancer cells. J Cell Mol Med. 2020;24:9985-9998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 47. | Yan L, Zhan X, Jia Z, Liu L, Jin N. Sirtuin 2 (Sirt2) Expression Predicts Lymph Node Metastasis and Poor Overall Survival of Patients with Esophageal Squamous Cell Carcinoma. Clin Lab. 2018;64:669-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Ong CA, Shapiro J, Nason KS, Davison JM, Liu X, Ross-Innes C, O'Donovan M, Dinjens WN, Biermann K, Shannon N, Worster S, Schulz LK, Luketich JD, Wijnhoven BP, Hardwick RH, Fitzgerald RC. Three-gene immunohistochemical panel adds to clinical staging algorithms to predict prognosis for patients with esophageal adenocarcinoma. J Clin Oncol. 2013;31:1576-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 49. | Peters CJ, Rees JR, Hardwick RH, Hardwick JS, Vowler SL, Ong CA, Zhang C, Save V, O'Donovan M, Rassl D, Alderson D, Caldas C, Fitzgerald RC; Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Study Group. A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology. 2010;139:1995-2004.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 121] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 50. | Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One. 2010;5:e10486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 51. | Wei Z, Song J, Wang G, Cui X, Zheng J, Tang Y, Chen X, Li J, Cui L, Liu CY, Yu W. Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nat Commun. 2018;9:4468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 52. | Bergaggio E, Riganti C, Garaffo G, Vitale N, Mereu E, Bandini C, Pellegrino E, Pullano V, Omedè P, Todoerti K, Cascione L, Audrito V, Riccio A, Rossi A, Bertoni F, Deaglio S, Neri A, Palumbo A, Piva R. IDH2 inhibition enhances proteasome inhibitor responsiveness in hematological malignancies. Blood. 2019;133:156-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 53. | Xu LX, Hao LJ, Ma JQ, Liu JK, Hasim A. SIRT3 promotes the invasion and metastasis of cervical cancer cells by regulating fatty acid synthase. Mol Cell Biochem. 2020;464:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 54. | Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669-2677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 117] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 55. | Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19:416-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 675] [Cited by in RCA: 649] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 56. | Xiao K, Jiang J, Wang W, Cao S, Zhu L, Zeng H, Ouyang R, Zhou R, Chen P. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep. 2013;30:1323-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 57. | Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, Mohammadi M, Britton LM, Garcia BA, Alečković M, Kang Y, Kaluz S, Devi N, Van Meir EG, Hitosugi T, Seo JH, Lonial S, Gaddh M, Arellano M, Khoury HJ, Khuri FR, Boggon TJ, Kang S, Chen J. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell. 2014;53:534-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 235] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 58. | Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, Zhao S, Yang Y, Ling ZQ, Guan KL, Xiong Y, Ye D. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 2015;34:1110-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 157] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 59. | Yu W, Denu RA, Krautkramer KA, Grindle KM, Yang DT, Asimakopoulos F, Hematti P, Denu JM. Loss of SIRT3 Provides Growth Advantage for B Cell Malignancies. J Biol Chem. 2016;291:3268-3279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 60. | Li R, Quan Y, Xia W. SIRT3 inhibits prostate cancer metastasis through regulation of FOXO3A by suppressing Wnt/β-catenin pathway. Exp Cell Res. 2018;364:143-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 61. | Ozden O, Park SH, Wagner BA, Song HY, Zhu Y, Vassilopoulos A, Jung B, Buettner GR, Gius D. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic Biol Med. 2014;76:163-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 151] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 62. | Torrens-Mas M, Oliver J, Roca P, Sastre-Serra J. SIRT3: Oncogene and Tumor Suppressor in Cancer. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 63. | Cobanoğlu U, Dülger C, Kemik O, Celik S, Sayir F. A novel screening test for esophageal squamous cell carcinoma: sirtuin-3. Eur Rev Med Pharmacol Sci. 2017;21:5399-5401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 64. | Yang M, Yang C, Pei Y. Effects of downregulation of SIRT3 expression on proliferation and apoptosis in esophageal squamous cell carcinoma EC9706 cells and its molecular mechanisms. Biomed Mater Eng. 2014;24:3883-3890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 65. | Zhao Y, Yang H, Wang X, Zhang R, Wang C, Guo Z. Sirtuin-3 (SIRT3) expression is associated with overall survival in esophageal cancer. Ann Diagn Pathol. 2013;17:483-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 66. | Yan SM, Han X, Han PJ, Chen HM, Huang LY, Li Y. SIRT3 is a novel prognostic biomarker for esophageal squamous cell carcinoma. Med Oncol. 2014;31:103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 67. | Wang YS, Du L, Liang X, Meng P, Bi L, Wang YL, Wang C, Tang B. Sirtuin 4 Depletion Promotes Hepatocellular Carcinoma Tumorigenesis Through Regulating Adenosine-Monophosphate-Activated Protein Kinase Alpha/Mammalian Target of Rapamycin Axis in Mice. Hepatology. 2019;69:1614-1631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 68. | Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, Xu X, Li C, Wang RH, Lee J, Csibi A, Cerione R, Blenis J, Clish CB, Kimmelman A, Deng CX, Haigis MC. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 348] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 69. | Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2021;184:2256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 70. | Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, Nishimura J, Hata T, Takemasa I, Mizushima T, Doki Y, Mori M, Ishii H. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer. 2015;113:492-499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 123] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 71. | Sun H, Huang D, Liu G, Jian F, Zhu J, Zhang L. SIRT4 acts as a tumor suppressor in gastric cancer by inhibiting cell proliferation, migration, and invasion. Onco Targets Ther. 2018;11:3959-3968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 72. | Hu Y, Lin J, Lin Y, Chen X, Zhu G, Huang G. Overexpression of SIRT4 inhibits the proliferation of gastric cancer cells through cell cycle arrest. Oncol Lett. 2019;17:2171-2176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 73. | Wang C, Liu Y, Zhu Y, Kong C. Functions of mammalian SIRT4 in cellular metabolism and research progress in human cancer. Oncol Lett. 2020;20:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 74. | Jeong SM, Hwang S, Seong RH. SIRT4 regulates cancer cell survival and growth after stress. Biochem Biophys Res Commun. 2016;470:251-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 75. | Liu M, Wang Z, Ren M, Yang X, Liu B, Qi H, Yu M, Song S, Chen S, Liu L, Zhang Y, Zou J, Zhu WG, Yin Y, Luo J. SIRT4 regulates PTEN stability through IDE in response to cellular stresses. FASEB J. 2019;33:5535-5547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 76. | Cui Y, Yang J, Bai Y, Zhang Y, Yao Y, Zheng T, Liu C, Wu F. miR-424-5p regulates cell proliferation and migration of esophageal squamous cell carcinoma by targeting SIRT4. J Cancer. 2020;11:6337-6347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 77. | Nakahara Y, Yamasaki M, Sawada G, Miyazaki Y, Makino T, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mimori K, Mori M, Doki Y. Downregulation of SIRT4 Expression Is Associated with Poor Prognosis in Esophageal Squamous Cell Carcinoma. Oncology. 2016;90:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 78. | Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang JL, Zou TH, Sun DF, Gao QY, Chen YX, Fang JY. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat Commun. 2018;9:545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 79. | Yang X, Wang Z, Li X, Liu B, Liu M, Liu L, Chen S, Ren M, Wang Y, Yu M, Wang B, Zou J, Zhu WG, Yin Y, Gu W, Luo J. SHMT2 Desuccinylation by SIRT5 Drives Cancer Cell Proliferation. Cancer Res. 2018;78:372-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 163] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 80. | Lu W, Zuo Y, Feng Y, Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol. 2014;35:10699-10705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 81. | Lv XB, Liu L, Cheng C, Yu B, Xiong L, Hu K, Tang J, Zeng L, Sang Y. SUN2 exerts tumor suppressor functions by suppressing the Warburg effect in lung cancer. Sci Rep. 2015;5:17940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 82. | Xiangyun Y, Xiaomin N, Linping G, Yunhua X, Ziming L, Yongfeng Y, Zhiwei C, Shun L. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget. 2017;8:6984-6993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 83. | Zhang R, Wang C, Tian Y, Yao Y, Mao J, Wang H, Li Z, Xu Y, Ye M, Wang L. SIRT5 Promotes Hepatocellular Carcinoma Progression by Regulating Mitochondrial Apoptosis. J Cancer. 2019;10:3871-3882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 84. | Gu W, Qian Q, Xu Y, Xu X, Zhang L, He S, Li D. SIRT5 regulates autophagy and apoptosis in gastric cancer cells. J Int Med Res. 2021;49:300060520986355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 85. | Chang L, Xi L, Liu Y, Liu R, Wu Z, Jian Z. SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol Med Rep. 2018;17:342-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 86. | Du Z, Liu X, Chen T, Gao W, Wu Z, Hu Z, Wei D, Gao C, Li Q. Targeting a Sirt5-Positive Subpopulation Overcomes Multidrug Resistance in Wild-Type Kras Colorectal Carcinomas. Cell Rep. 2018;22:2677-2689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 87. | Chen XF, Tian MX, Sun RQ, Zhang ML, Zhou LS, Jin L, Chen LL, Zhou WJ, Duan KL, Chen YJ, Gao C, Cheng ZL, Wang F, Zhang JY, Sun YP, Yu HX, Zhao YZ, Yang Y, Liu WR, Shi YH, Xiong Y, Guan KL, Ye D. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 204] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 88. | Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, Jin W, Huang HH, Chen X. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun. 2013;441:191-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 214] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 89. | Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T, Schutkowski M, Pellegrini L, Sansone L, Villanova L, Runci A, Pucci B, Morgante E, Fini M, Mai A, Russo MA, Tafani M. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;11:253-270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 248] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 90. | Chen X, Xu Z, Zeng S, Wang X, Liu W, Qian L, Wei J, Yang X, Shen Q, Gong Z, Yan Y. SIRT5 downregulation is associated with poor prognosis in glioblastoma. Cancer Biomark. 2019;24:449-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 91. | Chen X, Hao B, Liu Y, Dai D, Han G, Li Y, Wu X, Zhou X, Yue Z, Wang L, Cao Y, Liu J. The histone deacetylase SIRT6 suppresses the expression of the RNA-binding protein PCBP2 in glioma. Biochem Biophys Res Commun. 2014;446:364-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 92. | Zhang ZG, Qin CY. Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signalregulated kinase signaling pathway. Mol Med Rep. 2014;9:882-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 93. | Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 847] [Cited by in RCA: 795] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 94. | Fukuda T, Wada-Hiraike O, Oda K, Tanikawa M, Makii C, Inaba K, Miyasaka A, Miyamoto Y, Yano T, Maeda D, Sasaki T, Kawana K, Fukayama M, Osuga Y, Fujii T. Putative tumor suppression function of SIRT6 in endometrial cancer. FEBS Lett. 2015;589:2274-2281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 95. | Ouyang L, Yi L, Li J, Yi S, Li S, Liu P, Yang X. SIRT6 overexpression induces apoptosis of nasopharyngeal carcinoma by inhibiting NF-κB signaling. Onco Targets Ther. 2018;11:7613-7624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 96. | Bhardwaj A, Das S. SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc Natl Acad Sci U S A. 2016;113:E538-E547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 121] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 97. | Shi L, Wang Y, Oppong TB, Fu X, Yang H. Prognostic role of SIRT6 in gastrointestinal cancers: a meta-analysis. Open Med (Wars). 2020;15:358-365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 98. | Zhang C, Yu Y, Huang Q, Tang K. SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Mol Med Rep. 2019;20:1575-1582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 99. | Zhou HZ, Zeng HQ, Yuan D, Ren JH, Cheng ST, Yu HB, Ren F, Wang Q, Qin YP, Huang AL, Chen J. NQO1 potentiates apoptosis evasion and upregulates XIAP via inhibiting proteasome-mediated degradation SIRT6 in hepatocellular carcinoma. Cell Commun Signal. 2019;17:168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 100. | Bai L, Lin G, Sun L, Liu Y, Huang X, Cao C, Guo Y, Xie C. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget. 2016;7:40377-40386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 101. | Garcia-Peterson LM, Ndiaye MA, Singh CK, Chhabra G, Huang W, Ahmad N. SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes Cancer. 2017;8:701-712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 102. | Huang N, Liu Z, Zhu J, Cui Z, Li Y, Yu Y, Sun F, Pan Q, Yang Q. Sirtuin 6 plays an oncogenic role and induces cell autophagy in esophageal cancer cells. Tumour Biol. 2017;39:1010428317708532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 103. | Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X, Zhou Y, Wang H, Pan C, Huang W. Overexpression of sirt7 exhibits oncogenic property and serves as a prognostic factor in colorectal cancer. Clin Cancer Res. 2014;20:3434-3445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 104. | Wei W, Jing ZX, Ke Z, Yi P. Sirtuin 7 plays an oncogenic role in human osteosarcoma via downregulating CDC4 expression. Am J Cancer Res. 2017;7:1788-1803. [PubMed] |
| 105. | Wang HL, Lu RQ, Xie SH, Zheng H, Wen XM, Gao X, Guo L. SIRT7 Exhibits Oncogenic Potential in Human Ovarian Cancer Cells. Asian Pac J Cancer Prev. 2015;16:3573-3577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 106. | Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, Hu Y, Cai T. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5:9787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 107. | Yu W, Cui X, Wan Z, Yu Y, Liu X, Jin L. Silencing forkhead box M1 promotes apoptosis and autophagy through SIRT7/mTOR/IGF2 pathway in gastric cancer cells. J Cell Biochem. 2018;119:9090-9098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 108. | Li W, Zhu D, Qin S. SIRT7 suppresses the epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis by promoting SMAD4 deacetylation. J Exp Clin Cancer Res. 2018;37:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 109. | Tang X, Shi L, Xie N, Liu Z, Qian M, Meng F, Xu Q, Zhou M, Cao X, Zhu WG, Liu B. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat Commun. 2017;8:318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 167] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 110. | Wu D, Li Y, Zhu KS, Wang H, Zhu WG. Advances in Cellular Characterization of the Sirtuin Isoform, SIRT7. Front Endocrinol (Lausanne). 2018;9:652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 111. | Tao F, Gu C, Li N, Ying Y, Feng Y, Ni D, Zhang Q, Xiao Q. SIRT3 acts as a novel biomarker for the diagnosis of lung cancer: A retrospective study. Medicine (Baltimore). 2021;100:e26580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 112. | Curry AM, White DS, Donu D, Cen Y. Human Sirtuin Regulators: The "Success" Stories. Front Physiol. 2021;12:752117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 113. | Puhr HC, Preusser M, Ilhan-Mutlu A. Immunotherapy for Esophageal Cancers: What Is Practice Changing in 2021? Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 114. | Warren JL, MacIver NJ. Regulation of Adaptive Immune Cells by Sirtuins. Front Endocrinol (Lausanne). 2019;10:466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 115. | Zhang M, Acklin S, Gillenwater J, Du W, Patra M, Yu H, Xu B, Yu J, Xia F. SIRT2 promotes murine melanoma progression through natural killer cell inhibition. Sci Rep. 2021;11:12988. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 116. | Xiang J, Zhang N, Sun H, Su L, Zhang C, Xu H, Feng J, Wang M, Chen J, Liu L, Shan J, Shen J, Yang Z, Wang G, Zhou H, Prieto J, Ávila MA, Liu C, Qian C. Disruption of SIRT7 Increases the Efficacy of Checkpoint Inhibitor via MEF2D Regulation of Programmed Cell Death 1 Ligand 1 in Hepatocellular Carcinoma Cells. Gastroenterology. 2020;158:664-678.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |