Published online Mar 15, 2022. doi: 10.4251/wjgo.v14.i3.664
Peer-review started: July 27, 2021
First decision: October 3, 2021
Revised: November 10, 2021
Accepted: February 10, 2022
Article in press: February 10, 2022
Published online: March 15, 2022
Processing time: 225 Days and 21.4 Hours
O6-methylguanine-DNA methyltransferase (MGMT) is a suicide enzyme that repairs the mispairing base O6-methyl-guanine induced by environmental and experimental carcinogens. It can transfer the alkyl group to a cysteine residue in its active site and became inactive. The chemical carcinogen N-nitroso compounds (NOCs) can directly bind to the DNA and induce the O6-methylguanine adducts, which is an important cause of gene mutation and tumorigenesis. However, the underlying regulatory mechanism of MGMT involved in NOCs-induced tumorigenesis, especially in the initiation phase, remains largely unclear.
To investigate the molecular regulatory mechanism of MGMT in NOCs-induced gastric cell malignant transformation and tumorigenesis.
We established a gastric epithelial cell malignant transformation model induced by N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) or N-methyl-N-nitroso-urea (MNU) treat
We observed a constant increase in MGMT mRNA and protein expression in gastric epithelial cell malignant transformation induced by MNNG or MNU treatment. Moreover, we found a reduction of MGMT gene promoter methylation level by methylation-specific PCR and bisulfite sequencing PCR in MNNG/MNU-treated cells. Inhibition of the MGMT expression by O6-benzylguanine promoted the MNNG/MNU-induced malignant phenotypes. Overexpression of MGMT partially reversed the cell malignant transformation process induced by MNNG/MNU. Clinical gastric tissue analysis showed that MGMT was upregulated in the precancerous lesions and metaplasia tissues, but downregulated in the gastric cancer tissues.
Our finding indicated that MGMT upregulation is induced via its DNA promoter hypomethy
Core Tip: This study revealed a molecular regulatory mechanism of O6-methylguanine-DNA methyltransferase (MGMT) gene upregulation in the early stage of tumor development, and improved the under
- Citation: Chen YX, He LL, Xiang XP, Shen J, Qi HY. O6-methylguanine DNA methyltransferase is upregulated in malignant transformation of gastric epithelial cells via its gene promoter DNA hypomethylation. World J Gastrointest Oncol 2022; 14(3): 664-677
- URL: https://www.wjgnet.com/1948-5204/full/v14/i3/664.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i3.664
Gastric cancer (GC) is currently the fifth most frequently diagnosed and the third leading cause of cancer death worldwide with a high prevalence in many Asia countries, particularly in China, Japan, and South Korea[1,2]. Previous studies have reported that epigenetic alterations are widely recognized to be involved in the initiation and progression of gastric tumorigenesis[3,4]. DNA methylation is a common significant epigenetic modification and plays an important role in the development and prognosis of GC[5-8].
O6-methylguanine-DNA methyltransferase (MGMT) is a suicide enzyme that efficiently removes alkylating lesions at the O6 position of guanine induced by DNA alkylating agents[9]. Following the transfer of the alkyl group to itself, MGMT becomes inactive and it is ubiquitinated and targeted for proteasomal degradation. MGMT is frequently regulated by epigenetic silencing mediated Fits gene promoter DNA methylation in gliomas[10,11]. The abnormal modifications of histone and aberrant expression of transcriptional activators and repressors, also contribute to the regulation of MGMT expression in different tumors[11]. O6-methylguanine is a potent mutagenic lesion that leads to base mismatching and double-strand breaks, promoting gene mutagenesis and tumor initiation. MGMT can restore this type of DNA damage and play an important role in maintaining genomic stability[12]. Inhibiting MGMT function can induce G:C to A:T mutation of the onco-suppressors p53 and PTEN to promote human carcinogenesis[13]. In the TCGA database, the probability of point mutation of p53 and PTEN was higher in MGMT promoter methylated tissues than in non-methylated tissues of glioma. In colon cancer, lung cancer, and GC, the reduction of MGMT expression induced by DNA methylation in its promoter regions was also observed[14-17]. Yet, MGMT expression can be increased by che
The monofunctional alkylating agent N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) and N-methyl-N-nitroso-urea (MNU) are widely accepted model chemical carcinogens for studying the mechanisms of mutagenesis and carcinogenesis induced by N-nitroso compounds (NOCs). They generate adducts with DNA and protein, such as O6-methylguanine, which lead to point mutations, chromosomal aberrations, initiation and promotion of various cancer, specially increasing the risk of gastrointestinal cancers[19,20]. Our previously studies showed that MNNG and MNU treatments can stimulate multiple cellular responses, including epigenetic events[21,22]. We revealed a dysregulation of histone modifications and DNA methylation, which contributed to numerous cancer-related gene expression changes promoting cell malignant transformation upon NOCs treatment[21,22]. These findings prompted us to speculate that the abnormal epigenetic regulation could be the critical molecular mechanism of chemical carcinogens-induced gastric carcinogenesis.
In the present study, we investigated the epigenetic changes of MGMT in NOCs-induced human gastric cell malignant transformation. And we demonstrated that DNA methylation level of MGMT promoter was strongly decreased, which resulted the inhibition of MGMT expression, contributing the malignant phenotypes during the cell malignant transformation process.
A total of 93 clinical gastric tissue samples collected by endoscopic biopsy at the Second Affiliated Hospital of Zhejiang University were used in this study, including 25 cases of gastritis, 18 cases of gastric metaplasia (used as precancerous lesion), 50 pairs of GC and adjacent normal tissues (early stage, 19 cases; advanced stage, 31 cases). The study was approved by the ethics committee of Zhejiang University School of Medicine (No. 2017026). The tissue samples were formalin-fixed and paraffin-embedded for immunohistochemistry or used for mRNA isolation to detect gene expression.
The human gastric normal epithelium cell line GES-1 (Cell Bank of the Chinese Academy of Science, Xiangya, China) was cultured in DMEM (Gibco, Grand Island, NY, United States) supplemented with 10% fetal bovine serum (FBS; Gibco), streptomycin (100 g/mL), and penicillin (100 U/mL) at 37 °C in an atmosphere containing 5% CO2. And GC cell lines, including AGS, MKN45, SGC7901, KATOIII, and NCI-N87 (Cell Bank of the Chinese Academy of Science, Shanghai, China), were cultured in DMEM or PRIM-1640 supplemented with 10% FBS, streptomycin, and penicillin. The authenticity of cell lines used in this study had been verified by short tandem repeat profiling.
Cells were exposed to MNNG or MNU as described in a previous study to establish the cell transformation model[22]. Briefly, cells were exposed to MNU (TRC, Toronto, Canada) or MNNG (Sigma, St. Louis, MO, United States) for 2 h in serum free medium. Then, the medium was removed and cells were recovered in fresh medium at 37 °C. MNU and MNNG exposure was repeated once a week for 4 wk. After 4 wk of treatment and 4 wk of restoration, characteristics related to malignant phenotype were measured.
Cells were plated in 6-well plates and allowed to reach 90% confluence. The monolayer was scratched with a 10-mL sterile pipette tip. Images of the scratches were taken using an inverted microscope at × 10 magnification at 0, 24, and 48 h of incubation. ImageJ software was used to analyze the percentage of wound closure.
Cells (1.2 × 104 cells/well) were seeded in 24-well plates and cultured for 24, 48, 72, 96, and 120 h. Cells were digested by trypsin every 24 h and then re-suspended in fresh medium and counted.
For soft agar assay, cells (1000 cells/well) were suspended in a culture medium containing 0.4% agarose (A9045-5G) (Sigma) and seeded onto a base layer of 0.7% agar bed in 12-well plates. The culture medium was changed every 3 d. After 2 wk, colonies were stained with crystal violet and photo
For colony formation assay, cells (1000 cells/well) were seeded in 6-well plates and cultured. The culture medium was changed every 3 d. After 2 wk, colonies were stained with crystal violet and photographed. Colonies ≥ 0.05 mm in diameter were counted.
The MGMT promoter sequence (-954/+24) was amplified from the extracted genomic DNA and cloned into pGL3-promoter vector (Promega, Madison, WI, United States). After seeding MNNG/MNU-transformed cells for 24 h, the cells were co-transfected with 0.5 μg of pGL3-MGMT-promoter and 0.02 μg of pRL-SV40 renilla luciferase reporter plasmid using X-treme GENE HP (Roche, Basel, Switzerland). Dual-Luciferase Reporter Assay System was used for testing relative luciferase activity after transfection for 24 h (Promega).
Total DNA (5 × 106 cells) was isolated from the MNNG/MNU-transformed cells with the Qiagen DNA Isolation Kit. Then, bisulfite conversion was performed with 500 ng of genomic DNA using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, United States). The converted DNA was eluted in 100 mL of nuclease-free water. Methylation specific polymerase chain reaction (PCR) (MSP) analysis was performed in a 25-μL reaction system that consisted of 50 ng of sodium bisulfite-treated DNA, 12.5 μL of 2 × Master Mix (Qiagen, Germany), ddH2O, and 3 μL of isometric mixture of MGMT gene methylated and un-methylated primers. MGMT methylated and un-methylated primers used are: Forward 5’-TTTCGACGTTCGTAGGTTTTCGC-3’ and reverse, 5’-GCACTCTTCCGAAAACGAAACG-3’; forward, 5’-TTTGTGTTTTGATGTTTGTAGGTTTTTGT-3’ and reverse, 5’-ACTCCACACTCTTCCAAAAAC AAAACA-3’. Bisulfite genomic sequence (BSP) analysis was performed by Xiangyin Biological Corporation. Bisulfite treatments of the genomic DNA samples were carried out with the Qiagen EpiTect kit according to the manufacturer’s instructions, followed by the PCR amplification procedure (30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s; 72 °C for 30 min; and held at 4 °C) using KAPA2G Fast Multiplex Mix and KAPA 2G Robust HS. The PCR products were identified by electrophoresis and gel-purified with the Gel and PCR Clean-up System (Promega). The purified PCR products were inserted into PMD-18T Vector and sequenced by Sanger sequencing.
For chromatin immunoprecipitation (ChIP) assay, the malignant transformed cells were cross-linked with 1% formaldehyde for 10 min at 37 °C. Then, cells were isolated and lysed for preparation of sheared chromatin. The cell lysates were sonicated for 1 min and repeated ten times at 1-min intervals. After centrifugation at 13000 g at 4 °C for 30 min, the cell lysates were diluted with IP buffer and incubated with anti-DNMT1, anti-H3K9Me3, and anti-H3K4Me2 antibodies (CST, Massachusetts, United States) overnight, respectively. For collecting the bound DNA, the coated beads were added in the samples and incubated for 4 h at 4 °C. The beads were collected, washed, and eluted with elution buffer. Then, the bound DNA was extracted with a DNA extraction kit (Qiagen) for quantification by qPCR. Primers used for detecting the binding sites in MGMT promoter are: Forward, 5’-GCCCCTAGAACGCTTTGC-3’ and reverse, 5’-CAACACCTGGGAGGCACTT-3’.
Immunohistochemistry was performed using an Envision Detection System (DAKO, Carpinteria, CA, United States) according to the manufacturer's instructions. Mouse monoclonal anti-human MGMT antibody (dilution, 1:150) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, United States) and used for immunohistochemistry. The staining results were assessed and confirmed by two independent investigators blinded to the clinical data.
Total RNA was extracted from cell lines and tissue samples with TRIzol reagent (Invitrogen). For gene expression, mRNA was reverse transcribed using a Prime-Script RT reagent Kit (TaKaRa). qPCR was carried out with SYBR Premix Ex Taq (TaKaRa). Experiments were performed in triplicate and values were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2−ΔΔCt method for gene expression analysis. The primers used for MGMT and GAPDH amplification are: Forward, 5’-AACGCTGCCCTTGCTCTATT-3’ and reverse, 5’-AGCTTTCTAGTGTGGACGGC-3’ for MGMT; forward, 5’-ATGGGGAAGGTGAAGGTCGGAGT-3’ and reverse, 5’-TGACAAGCTTCCCGTTCTCA GCC-3’ for GAPDH.
Cells were lysed with RIPA buffer, and the total protein was quantified by Bradford assay. Cell lysates (50 mg) were separated on a 10% SDS-PAGE gel and then transferred onto a nitrocellulose membrane (Whatman, Maidstone, United Kingdom). The membrane was blocked with 5% skim milk solution for 2 h and incubated overnight with diluted primary antibody at 4 °C. Then, the membrane was incubated with IRDye 800- or IRDye 680-conjugated secondary antibody (LI-COR Biosciences, Lincoln, NE, United States) and detected with an Odyssey infrared imaging system. Mouse monoclonal anti-human MGMT antibody (dilution, 1:1000) and mouse monoclonal anti-human GADPH antibody (dilution, 1:2000) were purchased from Santa Cruz Biotechnology.
Balb/c nude mice (4 wk) were purchased from Shanghai Slac Laboratory Animal Co. LTD. Thirty-six Balb/c nude mice were randomly divided into three groups: Control group, MNNG-induced subclone injected group, and MNU-induced subclone injected group. The mice were subcutaneously injected with 1 × 106 MNNG/MNU-transformed cells (100 μL). Three days after injection, the long diameter (a) and short diameter (b) of the tumors were measured, after which the volume (V) was calculated using the formula V = 1/2 × a × b2. Mice were sacrificed, and the tumor tissues were obtained and weighed. The animal experiments were conducted in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals. The Committee on the Use of Animals of Zhejiang University, China approved the study protocol of our experiments.
The MGMT protein coding sequences was subcloned into PCDNA3.1 vector. The transformed cells were transfected with the MGMT overexpression plasmid and PCDNA3.1 empty vector (EV), respectively. Then, the proliferative activity of cells was analyzed by colony formation and soft agar assays.
SiRNAs targeting human MGMT (GenePharma, Shanghai, China) were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. A siRNA negative control (siRNA NC) was also used.
The two-tailed Student's t test and one-way analysis of variance were used for statistical analyses. The data are expressed as the mean ± SD from three separate experiments. P ≤ 0.05 was considered statistically significant.
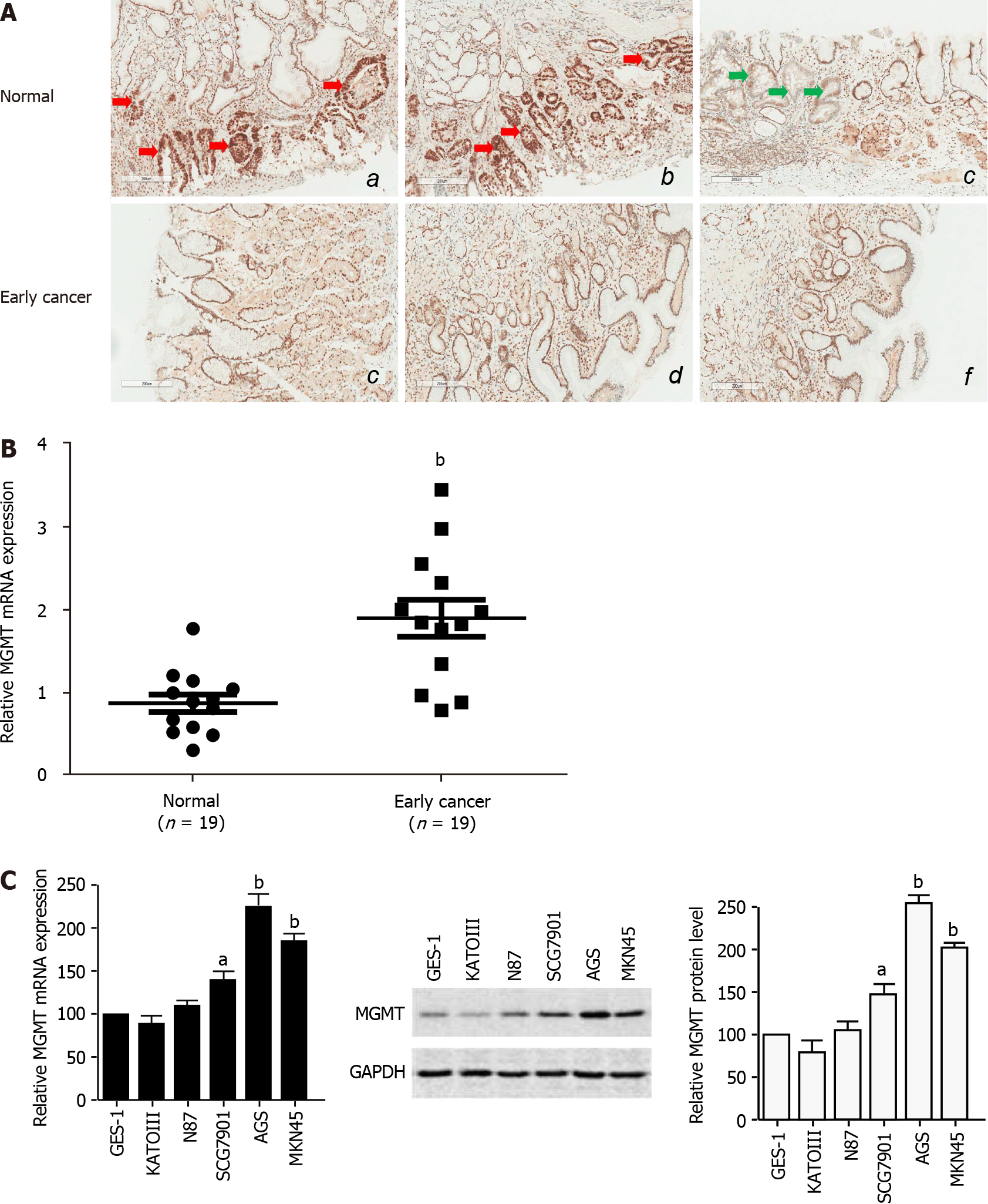
To study the role of MGMT in GC development, especially in the early events and tumor initiation, we detected the expression of MGMT in 19 clinical early stage GC tissues. The immunohistochemistry analysis showed that MGMT expression was increased in early stage cancer tissues compared with the normal tissue, though there was an individual difference (Figure 1A). qPCR analysis of endoscopic biopsy samples confirmed the upregulation of MGMT mRNA expression in early stage cancer tissues compared with adjacent normal tissues (Figure 1B). Moreover, MGMT expression was also enhanced in the GC cell lines at both the mRNA and protein levels (Figure 1C). Collectively, these results suggest that MGMT expression is upregulated in early stage GC.
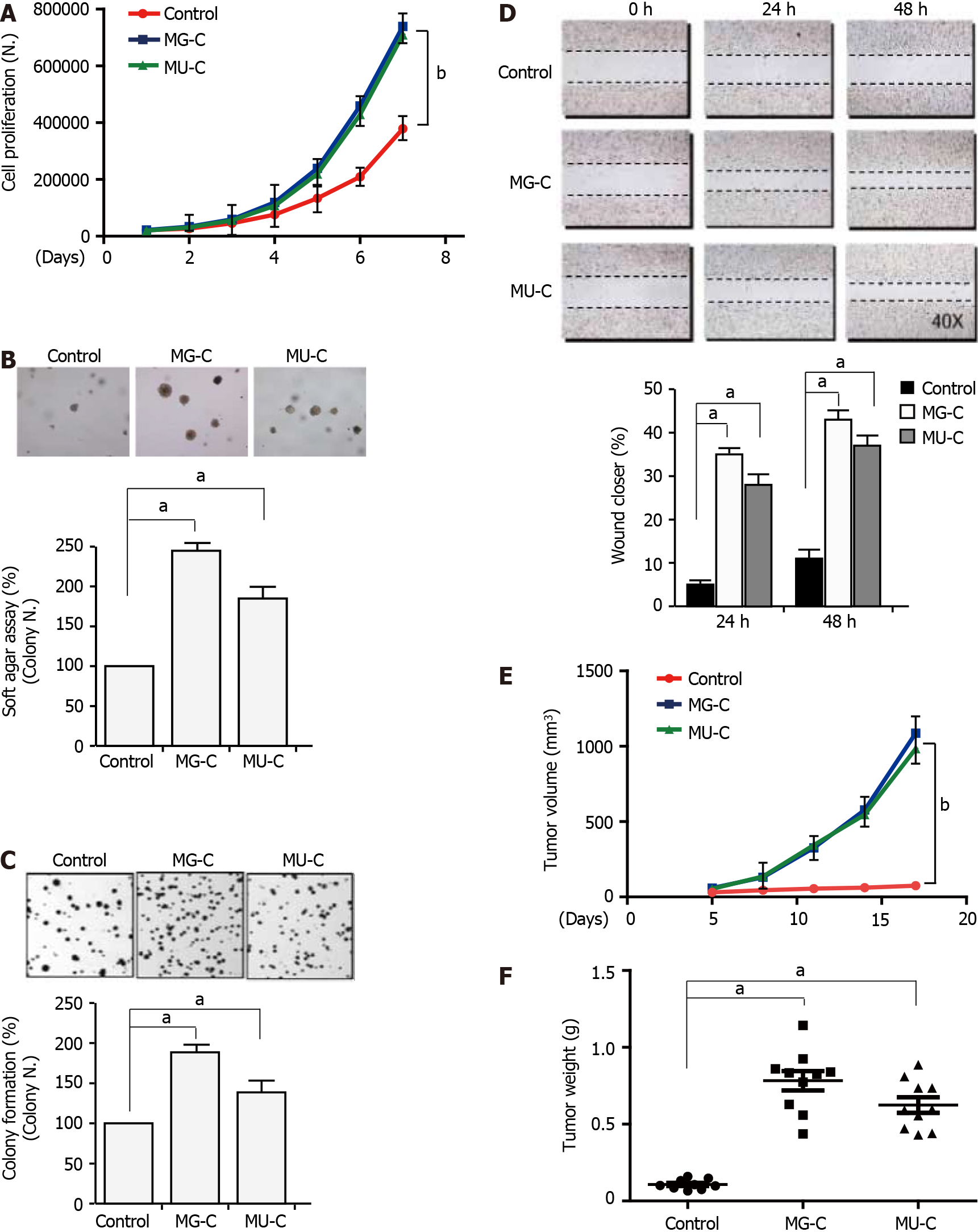
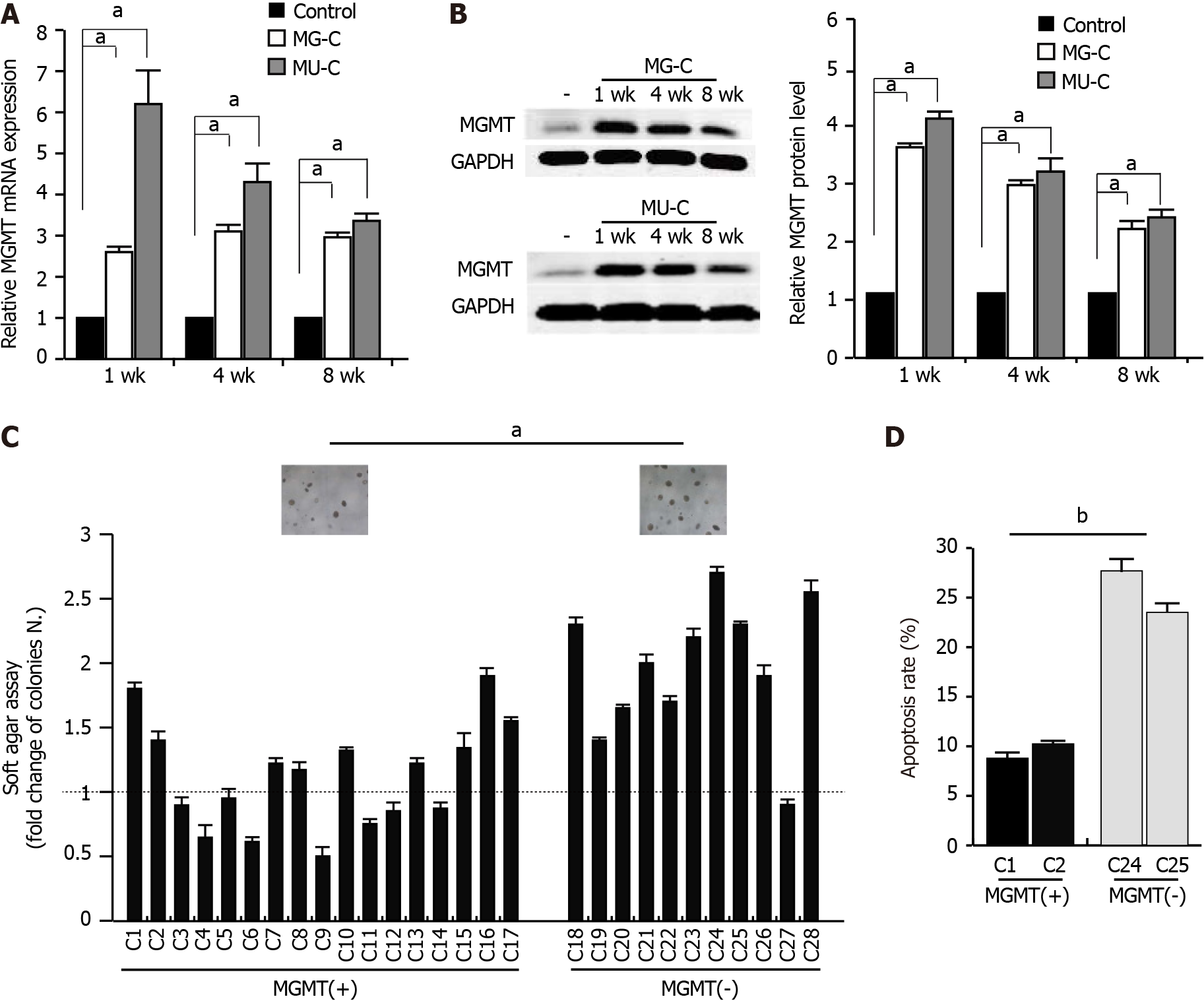
To investigate the molecular mechanism of MGMT upregulation, we established a gastric epithelial cell (GES-1) malignant transformation model following MNNG and MNU exposure. MNNG/MNU-treated cells showed an increase of cell proliferation, anchorage-independent growth capability, and colony formation ability, as demonstrated by cell proliferative assay, soft agar assay, and colony formation assays, respectively (Figure 2A-C). We also observed that the cell migration was enhanced upon MNNG/MNU treatment by wound healing assay (Figure 2D). Xenograft assay showed that MNNG/ MNU-induced transformed cells demonstrated increased tumor growth (Figure 2E and F), further confirming the malignant phenotypes of NOCs-induced transformed GES-1 cells. Then, we detected the MGMT expression in MNNG/MNU-transformed cells. MGMT expression was persistently increased during the malignant transformation process (Figure 3A and B). But the extent of MGMT upregulation was decreased after removal of MNNG/MNU exposure for 12 wk (data not shown), suggesting that MGMT expression demonstrated a dynamic change in MNNG/MNU-induced cell malignant trans
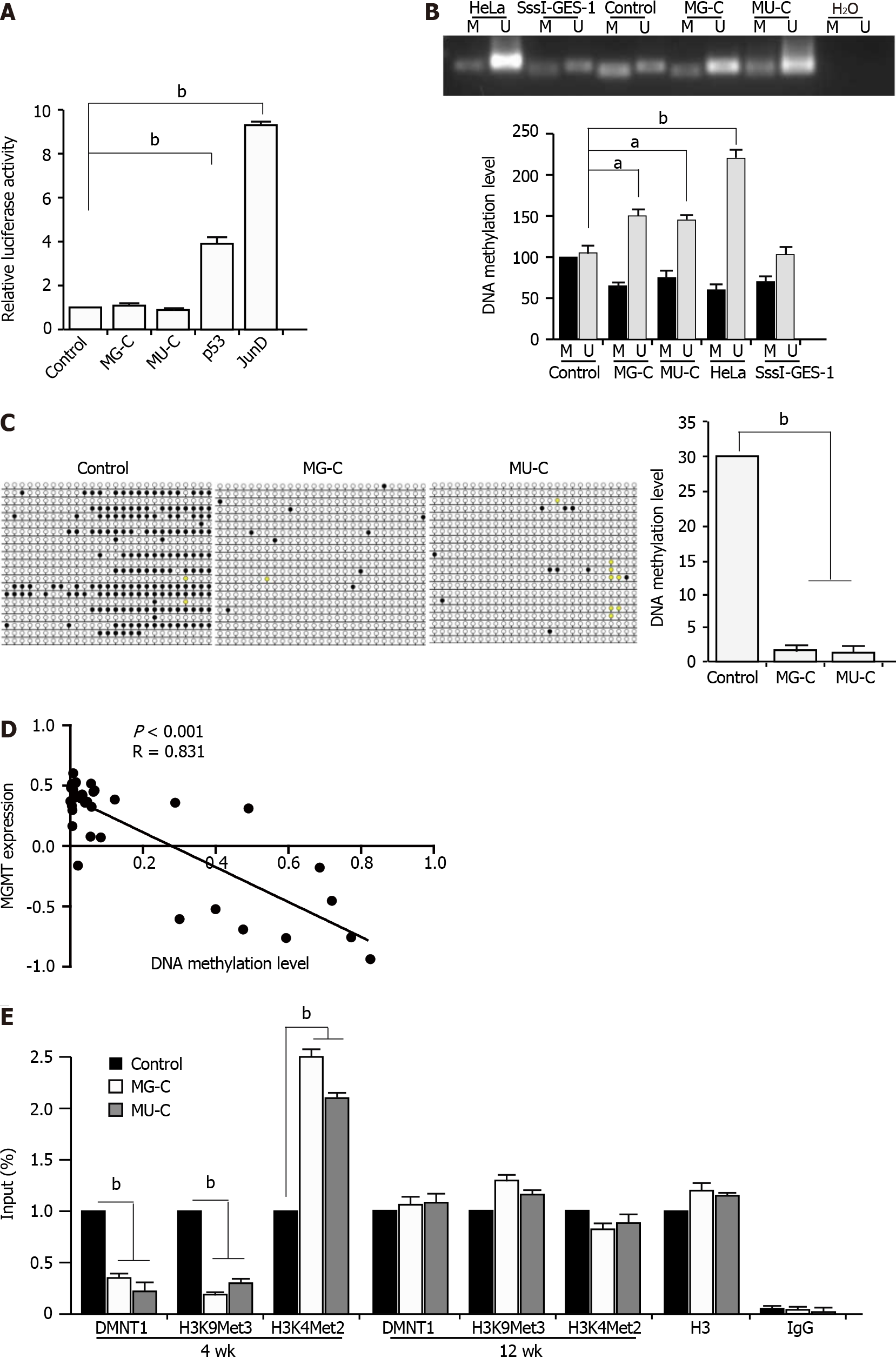
To further investigate the regulatory mechanism underlying the MGMT upregulation upon MNNG/MNU treatment, we constructed a MGMT gene promoter luciferase reporter. We did not find an increase of the MGMT gene promoter in MNNG/MNU-induced cells by dual-luciferase reporter assay. We used p53 and JunD as the positive controls, since it was reported that they are the transcriptional activators of the MGMT promoter (Figure 4A). It is known that MGMT expression was closely related with its promoter DNA methylation in different cancers. We preformed MSP to detect the DNA methylation level in the MGMT promoter, which showed that in MNNG/MNU-transformed subcolones, unmethylated DNA was accumulated, indicating a reduction of DNA methylation level in the MGMT gene promoter (Figure 4B). Since it is known that HeLa cells exhibit high expression of MGMT with a low DNA methylation level in the promoter region, they were used as a positive control. M. SssI (CpG methyltransferase) treated cell was used as a negative control. As shown by BSP analysis, MNNG/MNU-transformed subcolones showed few DNA methylation sites compared with the controls, confirming the reduction of DNA methylation level in the MGMT gene promoter (Figure 4C). Furthermore, we used 5-aza, a DNMT specific inhibitor, to treat CES-1 cells. After 48 and 72 h, MGMT expression was increased upon 5-aza treatment (Supplementary Figure 1). Moreover, based on the CCLE database, we found that MGMT expression was negatively related with DNA methylation levels (Figure 4D), indicating that the DNA methylation level is involved in the upregulation of MGMT.
Next, we preformed ChIP-PCR with anti-DNMT1 and anti-H3K9Met3 and anti-H3K4Met2 (against specific methylation sites) antibodies. H3K9Met3 was known as a transcriptional inhibition signal and H3K4Met2 was reported as a transcriptional activation signal. The results showed that the DMNT1 recruitment was significantly decreased to the promoter region of MGMT. We also detected the reduction of H3K9Met3 and the augment of H3K4Met2 located in the promoter region of MGMT, as well as a reduction of DNMT1 binding to the MGMT promoter (Figure 4E). The results suggested that the upregulation of MGMT expression was dependent on the DNA hypomethylation in its promoter. Interestingly, after removal of NOCs exposure for 12 wk, the binding of DNMT1 to the MGMT gene promoter returned to the baseline level. The same changes of H3K9Met3 and H3K4Met2 levels in the MGMT gene promoter were also observed. These data suggest that the binding of DNMT1 to the MGMT gene promoter changes dynamically, which could help us to understand the dynamic changes of MGMT expression regulated by DNA methylation in NOCs-induced malignant transformation.
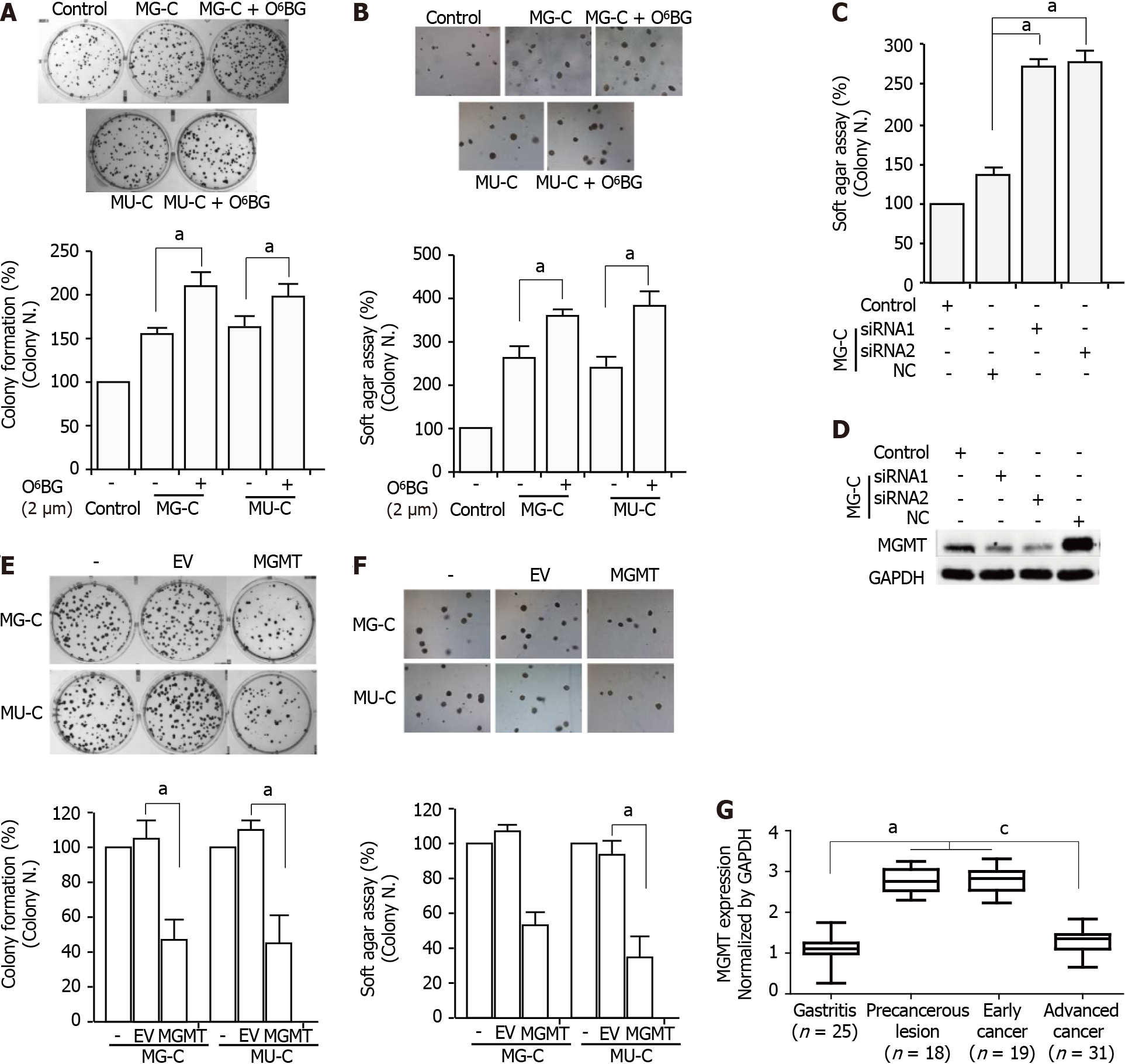
To evaluate the role of MGMT in NOCs-induced cell malignant transformation, we used O6-benzylguanine (O6-BG), a specific inhibitor of MGMT, to inhibit the activity of MGMT. After treatment with O6-BG at different concentrations, we found a reduction of MGMT expression (Supplementary Figure 1). MTT assay showed that treatment with O6-BG at low doses did not induce a decrease of cell viability (Supplementary Figure 1). Therefore, we used 2 μM O6-BG in the subsequent experiments. O6-BG exposure increased the proliferative activity and anchorage-independent growth capability of MNNG/MNU-induced cells (Figure 5A and B). Knock-down of MGMT in MNNG/MNU-transformed cells also resulted in the increase of cell reproductive activity (Figure 5C and D). Moreover, overexpressed MGMT resulted in the decrease of cell proliferative activity in MNNG/MNU-transformed cells (Figure 5E and F). In addition, MGMT was upregulated in precancerous lesions (gastric metaplasia) and early stage GC compared with non-cancerous lesions (gastritis), but the MGMT level was reduced in the advanced GC tissues compared with the precancerous lesion and early tumor tissues (Figure 5G), indicating a protective role of MGMT in GC progression.
By causing DNA damages and activating downstream pathways that promote cancer initiation and development, NOCs can directly induce cell malignant transformation, thus contributing to gastric carcinogenesis[23,24]. The formation of DNA adducts induced by NOCs has been studied in different studies[19,20,25,26]. The present study focused on early events and the molecular mechanisms of MGMT gene dysregulation in cell malignant transformation and gastric tumorigenesis following MNNG/MNU exposure. Our data showed persistent upregulation of MGMT expression in gastric epithelial cell malignant transformation induced by NOCs. The reduction of MGMT gene promoter DNA methylation level was responsible for the increase of MGMT expression in MNNG/MNU-treated cells. Inhibited MGMT expression promoted the MNNG/MNU-induced malignant phenotype, while overexpression of MGMT partially reversed the cell malignant transformation phenotype, suggesting that stable MGMT upregulation induced by its promoter DNA hypomethylation prevented the NOCs-induced cell malignant transformation and tumorigenesis.
Studies have reported that NOCs can directly act on DNA, mainly cause O6-methylguanine damage, and subsequently induce DNA mutation and double strand breaks, participating in cancer formation and progression[19,20]. The administration of MNNG can cause the destruction of pyloric mucosal structure and the occurrence of gastric adenocarcinoma in rats[24]. MNNG exposure can also induce the mutation and amplification of oncogenes participating in the occurrence of GC[26]. Moreover, the chromatin-based epigenetics regulation induced by NOCs, especially DNA methylation and histone modifications, has an essential role in cancer biology[27]. In the current study, we demonstrated that MGMT gene expression was rapidly increased after MNNG/MNU exposure, and the upregulation was continuously maintained during the early phase of cell malignant transformation. However, the extent of increased MGMT expression level was reduced progressively, leading us to speculate that the dynamic changes of MGMT expression could be involved in different steps of chemical carcinogens-induced gastric cell malignant transformation and tumorigenesis.
MGMT can remove O6-guanosine alkylation adducts caused by alkylation agents from DNA sequence in one-step reaction that restores the O6-guanosine residue to itself, consequently forming an inactive form[28]. Hence, the expression level of MGMT is fundamental for accurate DNA repair. It has been known that the transcriptional mechanism and epigenetic regulation are important to regulate the MGMT expression[1,29]. Hypoxia inducible factor 1-α can upregulate the expression level of MGMT and increase the drug resistance of glioma stem cells to temozolomide[30]. In addition, microRNAs can also bind to the 3'-untranslated region of MGMT, reduce the stability of the mRNA, and affect protein translation[31,32]. Moreover, MGMT gene promoter region lacks TATA box and CAAT box, but has rich GC sequence, which is prone to be methylated and closely related to transcriptional regulation[33]. DNA methylation in the MGMT gene has been reported in various human cancers, which can increase the sensitivity to alkylating agents in chemotherapy, influencing the tumor prognosis. However, the high level of gene methylation is usually associated with the low expression of protein level. Inhibition of MGMT protein level decreases its ability of removing O6-guanosine from the damaged DNA sites, resulting in an increase of mutation frequency and easily leading to the occurrence of tumor[34-36]. We found that MGMT upregulation was regulated by DNA hypomethylation in its gene promoter. And the subclones with high a level of MGMT showed a weak malignant proliferative activity, but with a strong anti-apoptotic effect upon exposure to DNA damage agents. This result suggested a protective effect of MGMT against NOCs-induced cell malignant transformation. Using O6-BG and by knocking MGMT down with siRNA, we showed an increased malignant proliferative ability of the transformed cells. Overexpressed MGMT decreased this effect, confirming the protected role of MGMT following chemical carcinogen exposure. In particular, the ChIP assay showed that DNMT1 was responsible for the MGMT gene promoter methylation. After 12 wk of cell transformation, the MGMT expression level was restored by recovering DNMT1 binding to the MGMT promoter region. This result suggested dynamic changes of MGMT expression, which is regulated by DNA methylation. Analysis of clinical gastric tissue samples also confirmed the dynamic changes of MGMT expression in gastric carcinogenesis. Taken together, we hypothesize that MGMT expression shows dynamic changes in gastric tumorigenesis induced by chemical carcinogens. It can be upregulated in the initiation phase for repairing the DNA damage and helping cells survive upon NOCs exposure; but in the progressive stage, it can be restored to the normal level to facilitate GC development. Hence, revealing the molecular mechanism of dynamic regulation of MGMT expression is important to help us understand the role of MGMT in GC formation and progression. However, the exact regulatory mechanisms of the dynamic changes on MGMT expression in different stages of cancer progression need to be further investigated (Supplementary Table 1).
In summary, our current study revealed the molecular mechanism of MGMT upregulation mediated by DNA hypomethylation of its gene promoter in NOCs-induced gastric cell malignant transformation, and showed the dual effects of MGMT by regulating its expression level in chemical carcinogen-induced tumorigenesis. Our findings provide a dynamic regulatory mechanism by which MGMT is implicated in cell malignant transformation and tumorigenesis induced by NOCs, and shed new light on MGMT as a potential diagnostic and therapeutic target for gastric carcinogenesis intervention by regulating aberrant epigenetic mechanisms.
O6-methylguanine-DNA methyltransferase (MGMT) is a specific enzyme that repairs the mispairing base O6-methyl-guanine induced by methylating environmental and experimental carcinogens. The N-nitroso compounds (NOCs) N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) and N-methyl-N-nitroso-urea (MNU) are monofunctional alkylating agents which can directly bind to the DNA and induce the formation of O6-methylguanine adducts to promote gene mutation and tumorigenesis. They are widely accepted chemical carcinogens for studying the mechanisms of mutagenesis and carcinogenesis induced by NOCs.
The underlying regulatory mechanism of MGMT involved in NOCs-induced tumorigenesis, especially in the initiation phase, remains largely unclear.
To investigate the molecular regulatory mechanism of MGMT in NOCs-induced gastric cell malignant transformation and tumorigenesis.
We established a gastric epithelial cell malignant transformation model induced by MNNG or MNU treatment. Cell proliferation, colony formation, soft agar, cell migration, and xenograft assays were used to verify the malignant phenotype. By using quantitative real-time polymerase chain reaction (qPCR) and Western blot analysis, we detected the MGMT expression in malignant transformed cells. We also confirmed the MGMT expression in clinical early stage gastric tumor tissues by qPCR and immunohistochemistry. MGMT gene promoter DNA methylation level was analyzed by methylation-specific PCR and bisulfite sequencing PCR. The effect of MGMT in cell malignant transformation was analyzed by colony formation and soft agar assays.
MGMT expression was upregulated in NOCs-induced gastric cell malignant transformation and in clinical early stage gastric cancer tissues. The upregulation of MGMT was regulated by the hypo
The upregulation of MGMT expression is mediated by the hypomethylation of its DNA promoter in NOCs-induced gastric cell malignant transformation.
The findings provide a dynamic regulatory mechanism of MGMT expression in cell malignant transformation and tumorigenesis induced by NOCs, supporting that MGMT might be a potential diagnostic and therapeutic target for gastric carcinogenesis.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Bai G, Batool SN S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Yuan YY
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55774] [Article Influence: 7967.7] [Reference Citation Analysis (132)] |
| 2. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13205] [Article Influence: 1467.2] [Reference Citation Analysis (3)] |
| 3. | Padmanabhan N, Ushijima T, Tan P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat Rev Gastroenterol Hepatol. 2017;14:467-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 4. | Yamashita S, Kishino T, Takahashi T, Shimazu T, Charvat H, Kakugawa Y, Nakajima T, Lee YC, Iida N, Maeda M, Hattori N, Takeshima H, Nagano R, Oda I, Tsugane S, Wu MS, Ushijima T. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc Natl Acad Sci U S A. 2018;115:1328-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 5. | Tie J, Zhang X, Fan D. Epigenetic roles in the malignant transformation of gastric mucosal cells. Cell Mol Life Sci. 2016;73:4599-4610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Klutstein M, Nejman D, Greenfield R, Cedar H. DNA Methylation in Cancer and Aging. Cancer Res. 2016;76:3446-3450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 603] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 7. | Takeshima H, Ushijima T. Accumulation of genetic and epigenetic alterations in normal cells and cancer risk. NPJ Precis Oncol. 2019;3:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 162] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 8. | Zheng SC, Widschwendter M, Teschendorff AE. Epigenetic drift, epigenetic clocks and cancer risk. Epigenomics. 2016;8:705-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 9. | Christmann M, Verbeek B, Roos WP, Kaina B. O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: enzyme activity, promoter methylation and immunohistochemistry. Biochim Biophys Acta. 2011;1816:179-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Mur P, Rodríguez de Lope Á, Díaz-Crespo FJ, Hernández-Iglesias T, Ribalta T, Fiaño C, García JF, Rey JA, Mollejo M, Meléndez B. Impact on prognosis of the regional distribution of MGMT methylation with respect to the CpG island methylator phenotype and age in glioma patients. J Neurooncol. 2015;122:441-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Wick W, Platten M. Understanding and targeting alkylator resistance in glioblastoma. Cancer Discov. 2014;4:1120-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 584] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 13. | Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem Cell Biol. 2005;83:429-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Fornaro L, Vivaldi C, Caparello C, Musettini G, Baldini E, Masi G, Falcone A. Pharmacoepigenetics in gastrointestinal tumors: MGMT methylation and beyond. Front Biosci (Elite Ed). 2016;8:170-180. [PubMed] |
| 15. | Alizadeh Naini M, Kavousipour S, Hasanzarini M, Nasrollah A, Monabati A, Mokarram P. O6-Methyguanine-DNA Methyl Transferase (MGMT) Promoter Methylation in Serum DNA of Iranian Patients with Colorectal Cancer. Asian Pac J Cancer Prev. 2018;19:1223-1227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 16. | Gu C, Lu J, Cui T, Lu C, Shi H, Xu W, Yuan X, Yang X, Huang Y, Lu M. Association between MGMT promoter methylation and non-small cell lung cancer: a meta-analysis. PLoS One. 2013;8:e72633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Yousuf A, Bhat MY, Pandith AA, Afroze D, Khan NP, Alam K, Shah P, Shah MA, Mudassar S. MGMT gene silencing by promoter hypermethylation in gastric cancer in a high incidence area. Cell Oncol (Dordr). 2014;37:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Oldrini B, Vaquero-Siguero N, Mu Q, Kroon P, Zhang Y, Galán-Ganga M, Bao Z, Wang Z, Liu H, Sa JK, Zhao J, Kim H, Rodriguez-Perales S, Nam DH, Verhaak RGW, Rabadan R, Jiang T, Wang J, Squatrito M. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat Commun. 2020;11:3883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 136] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 19. | Mirvish SS. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 1995;93:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 593] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 20. | Loh YH, Jakszyn P, Luben RN, Mulligan AA, Mitrou PN, Khaw KT. N-Nitroso compounds and cancer incidence: the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am J Clin Nutr. 2011;93:1053-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 143] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Chen K, Zhang S, Ke X, Qi H, Shao J, Shen J. Biphasic reduction of histone H3 phosphorylation in response to N-nitroso compounds induced DNA damage. Biochim Biophys Acta. 2016;1860:1836-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Qi H, Yang Z, Dai C, Wang R, Ke X, Zhang S, Xiang X, Chen K, Li C, Luo J, Shao J, Shen J. STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis. Oncogenesis. 2020;9:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 23. | Higginson J, DeVita VT Jr. IARC monographs on the evaluation of carcinogenic risk of chemicals to humans. Am Ind Hyg Assoc J. 1980;41:A26, A28, A30 passim. [PubMed] |
| 24. | Sugimura T, Fujimura S. Tumour production in glandular stomach of rat by N-methyl-N'-nitro-N-nitrosoguanidine. Nature. 1967;216:943-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 231] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Kwak H, Lee M, Cho M. Interrelationship of apoptosis, mutation, and cell proliferation in N-methyl-N'-nitro-N-nitrosoguanidine (MNNG)-induced medaka carcinogenesis model. Aquat Toxicol. 2000;50:317-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 26. | Arber N, Hibshoosh H, Moss SF, Sutter T, Zhang Y, Begg M, Wang S, Weinstein IB, Holt PR. Increased expression of cyclin D1 is an early event in multistage colorectal carcinogenesis. Gastroenterology. 1996;110:669-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 219] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 27. | Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243-251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 722] [Cited by in RCA: 610] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 28. | Iyama T, Wilson DM 3rd. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst). 2013;12:620-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 499] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 29. | Bocangel D, Sengupta S, Mitra S, Bhakat KK. p53-Mediated down-regulation of the human DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) via interaction with Sp1 transcription factor. Anticancer Res. 2009;29:3741-3750. [PubMed] |
| 30. | Pistollato F, Abbadi S, Rampazzo E, Persano L, Della Puppa A, Frasson C, Sarto E, Scienza R, D'avella D, Basso G. Intratumoral hypoxic gradient drives stem cells distribution and MGMT expression in glioblastoma. Stem Cells. 2010;28:851-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 217] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 31. | Wang J, Sai K, Chen FR, Chen ZP. miR-181b modulates glioma cell sensitivity to temozolomide by targeting MEK1. Cancer Chemother Pharmacol. 2013;72:147-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Jesionek-Kupnicka D, Braun M, Trąbska-Kluch B, Czech J, Szybka M, Szymańska B, Kulczycka-Wojdala D, Bieńkowski M, Kordek R, Zawlik I. MiR-21, miR-34a, miR-125b, miR-181d and miR-648 Levels inversely correlate with MGMT and TP53 expression in primary glioblastoma patients. Arch Med Sci. 2019;15:504-512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 33. | Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121:651-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 34. | Butler M, Pongor L, Su YT, Xi L, Raffeld M, Quezado M, Trepel J, Aldape K, Pommier Y, Wu J. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer. 2020;6:380-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 191] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 35. | Yu W, Zhang L, Wei Q, Shao A. O6-Methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front Oncol. 2019;9:1547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 165] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 36. | Bouras E, Karakioulaki M, Bougioukas KI, Aivaliotis M, Tzimagiorgis G, Chourdakis M. Gene promoter methylation and cancer: An umbrella review. Gene. 2019;710:333-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |