Published online Nov 15, 2022. doi: 10.4251/wjgo.v14.i11.2157
Peer-review started: July 15, 2022
First decision: July 28, 2022
Revised: September 10, 2022
Accepted: October 18, 2022
Article in press: October 18, 2022
Published online: November 15, 2022
Processing time: 122 Days and 20.3 Hours
Pancreatic adenocarcinoma (PAAD) is a cancerous tumor with an extremely poor 5-year survival rate. The exploration of biomarkers for the diagnosis and treatment of PAAD is crucial in clinical practice. Krüppel-like factors (KLFs) are involved in a variety of biological functions in cells. According to multiple studies, KLF16 behave as an oncogene in prostate, breast and gastric cancers. However, no research has been done on the significance of KLF16 in PAAD.
To explore the molecular mechanisms of KLF16 in PAAD.
KLF16 was identified in the tumor specimens and normal tissues by GEPIA database and verified by quantitative real-time PCR (qRT-PCR). Knockdown or exogenous expression of KLF16, combined with in vitro and in vivo assays, was performed to show the functional significance of KLF16. The molecular mechanism of KLF16 was demonstrated by qRT-PCR, western blotting, immunoprecipitation assay and flow cytometry.
We showed that KLF16 was highly expressed in PAAD patients based on the GEPIA database. KLF16 silencing suppressed while KLF16 overexpression promoted the malignant function of PAAD cells. Based on RNA sequencing, we discovered that KLF16 potentiated the expression of SMAD6 in PAAD cells. SMAD6 transcript abundance was increased and positively correlated with KLF16 expression in PAAD samples. In addition, inhibiting SMAD6 was able to mitigate the effects of KLF16 overexpression on PAAD cell processes, suggesting the importance of SMAD6 in the development of KLF16-triggered PAAD.
KLF16/SMAD6 axis might be explored as a therapeutic target for PAAD therapy.
Core Tip: Our study provided the evidence that KLF16 acted as an oncogene in pancreatic adenocarcinoma (PAAD). We also identified that SMAD6 served as the downstream substrate of KLF16 and this signaling cascade has never been reported. This mechanism indicated a novel insight into the pathological events during the development of PAAD.
- Citation: Mi W, Zheng Z, Lu JD, Duan SQ, Zhang J, Zhang HQ, Ding YX, Yin J, Cao F, Zhang J, Li F. KLF16 promotes pancreatic adenocarcinoma cell proliferation and migration by positively regulating SMAD6. World J Gastrointest Oncol 2022; 14(11): 2157-2169
- URL: https://www.wjgnet.com/1948-5204/full/v14/i11/2157.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i11.2157
Pancreatic adenocarcinoma (PAAD) ranks fourteenth in incidence with about 496000 new cases (2.6%) per year, but seventh in mortality with roughly 466000 (4.7%) deaths per year, according to GLOBOCAN 2020[1]. Due to its dismal prognosis, experts expect that pancreatic cancer will rank third in terms of fatality by 2025[2]. Currently, the only way to improve the outcomes of this condition is to diagnose it early[3]. However, since the early signs of PAAD are not well-defined, it is difficult to diagnose, even when it progresses fast to neighboring organs[4]. As a result, new unique and useful biomarkers for the diagnosis and prognosis of PAAD are required.
Three zinc finger DNA binding domains distinguish the Krüppel-like factor (KLF) which belongs to the SP/KLF transcription factor family[5]. KLF binds to GT or GC-rich regions and regulates transcription as an activator or repressor, depending on the promoter type[6]. The KLF16 gene is located on chromosome 19 that participates in various cellular activities such as proliferation, metabolism and death[7]. Various studies on the role of KLF16 in cancer have been published in recent years with mixed results. It has been reported that KLF16 promoted progression of breast cancer via activating MAGT1[8]. Ma et al[9] found that KLF16 enhanced colorectal carcinoma progression by modulating nucleolar homeostasis and translational reprogramming. On the other hand, in some malignancies, KLF16 functions as an oncogenic suppressor. It has been reported that KLF16 suppressed human glioma cell proliferation and tumorigenesis by regulating TFAM[10]. Nevertheless, understanding the molecular mechanisms of KLF16 in PAAD is still limited.
A total of 350 cases which encompassed 171 normal and 179 tumor specimens were analyzed from the database of Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) database, which combines the TCGA database and GEO database.
Shanghai Institutes for Biological Sciences (Shanghai, China) provided the AsPC-1 and MIA PaCa-2 cell lines. Cells were incubated in Eagle’s minimal essential medium (MEM) with 10% fetal bovine serum (FBS), in a 37°C incubator supplemented with 5% CO2. L-glutamine and sodium pyruvate were both supplied by Gibco and added to the medium to ensure constant cell growth.
Cultivated cell lines were lysed in TRIzol (Ambion, Austin, TX, United States). To reverse transcribe 500 ng of RNA template, the Takara PrimeScript RT Reagent Kit was employed. The following primers were used: KLF16-F: 5'-TGGGCAAACCCTGAAGACA-3' and KLF16-R, 5’-GTTGCACAGATGGGAAGAAA-3’; SMAD6-F:5’- CCTCCCTACTCTCGGCTGTC-3’, and SMAD6-R, 5’- GGTAGCCTCCGTTTCAGTGTA-3’; and β-actin-F: 5’-CATGTACGTTGCTATCCAGGC-3’, β-actin-R, 5’-CTCCTTAATGTCACGCACGAT-3’. RT-qPCR was conducted on real-time PCR system (7500 Fast; Life Technologies Holdings Pte Ltd, Singapore). The expression of targeted genes was adjusted to β-actin.
Specific small interfering RNAs (siRNAs) targeting KLF16 (siKLF16#1, siKIF16#2) and SMAD6 (siSMAD6#1, siSMAD6#2) and negative controls siRNA (siCtrl) were provided by Genechem (Shanghai, China). For transient transfection, Invitrogen’s Lipofectamine RNAiMAX transfection reagent was employed. The sequences used were as follows: siKLF16#1 (5’-GGGUUCUUCCAAAGAACAU-3’), siKLF16#2 (5’-GGUAUCACGUGACAAUCAA-3’), siSMAD6#1 (5’-CCACATTGTCTTACACTGAAA-3’), siSMAD6#2 (5’-CTCCATCAAGGTGTTCGACTT-3’) and siCtrl (5’-UUCUCCGAACGUGUCACGU-3’).
KLF16-pCDNA3.1 was overexpressed using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, United States). shRNA targeting SMAD6 (shSMAD6) purchased from Genechem (Shanghai, China) was used for lentiviral transduction.
To lyse the cells, the RIPA lysis buffer (Beyotime, Jiangsu, China) was used. This solution contains the protease inhibitor cocktail (Roche, Basel, Switzerland). To detect protein amount present in the lysates, a Bradford reagent manufactured by Sigma was used. After that, 30 μg of protein per row was run on SDS-PAGE gels and then immunoblotted onto a 0.22 m polyvinylidene fluoride membrane (Roche). The membranes were incubated for an hour with 5% nonfat milk solution that dissolved in TBST. After adding primary antibodies against KLF16 (1:1000; sc-377519; Santa Cruz Biotechnology, Dallas, TX, United States), SMAD6 (1:1000; ab273106; Abcam), and GAPDH (1:1000; ab8245; Abcam), the membrane was incubated at 4°C overnight. After that, the secondary antibody specific to primary antibody was added. An Odessey CLx system uncovered evidence of the presence of protein bands (LI-COR, Lincoln, NE, United States).
The CCK-8 (Beyotime) was applied to determine cell proliferation rate. In all, 1 × 103 transfected cells were planted in 200 μL complete medium. A total of 20 μL of CCK-8 was used to treat the cells and cultured for a further two hours at 37°C, followed by OD450 measurement. For cell colony assay, 1.2 × 103 transfected cells were planted in six-well plates with 2 mL of complete media and cultivated for 14 d. Every 3 d, the medium was replaced. The cells were fixed with methanol and stained with 0.1% crystal violet (Solarbio, Beijing, China).
In a six-well plate, 2 × 105 AsPC-1 and MIApaca-2 cells were planted per well. Flow cytometry was performed on cells 48 h after transfection with siKLF16#1, siKLF16#2, siCtrl, or KLF16 overexpressed plasmid and Ctrl plasmid.
A cell cycle staining kit (Cat No. 4040301; Yeasen, Shanghai, China) was used to detect the cell cycle. The cells were fixed for two hours at 4°C in 75% ethanol. The cells were stained with a staining solution containing 10 μL propidium iodide and 10 μL RNaseA in a volume of 0.5 mL. The cells were stained for 30 mins at 25°C and then subjected to analysis with the Guava easyCyte HT system (Millipore) after passing through a screen with a mesh size of 400. The cell cycle was analyzed by a flow cytometry system.
The Annexin V Apoptosis Detection Kit (Cat No. 88-8007; Invitrogen, Waltham, MA, United States) was used to examine apoptosis. The cells (1 to 5 × 105) were collected in 1X binding buffer and were incubated with staining solution in the dark for 15 min. The samples were analyzed using the Guava easyCyte HT system (Millipore, Burlington, MA, United States) within 1 h.
5.0 × 104 AsPC-1 or MIApaca-2 cells in 300 μL medium were planted in the upper chambers of 24-well Corning® FluoroBlokTM Cell Culture Inserts (Corning, NY, United States). The bottom chamber was added to 600 μL medium which contained 10% FBS. The migrating cells were stained by 0.1% crystal violet. Images of migrating cells were collected under a microscope.
RNA sequencing (RNA-seq) was carried out on MIA PaCa-2 cells using the Illumina HiSeq TM 4000 (Illumina, San Diego, CA, United States. The sequencing reads were processed using the Fast-QC program. The investigation of the signaling pathway was carried out by KEGG after the KLF16 knockdown.
Vital River Company (Beijing, China) provided the Balb/c nude mice (4-6 wk, female). MIA PaCa-2 cells were subcutaneously implanted into three different groups of nude mice (n = 5). On days 20, 25, 29, 33, 38, and 43, the tumor’s volume was assessed. Tumor volume = 4/3 × π × [(length (l) + width (w)) / 4]3[11]. The mice were euthanized after 43 d, and the tumor tissues were surgically removed and studied. Every experiment that included animals was carried out in compliance with the Principles on the Protection of Experimental Animals that are outlined by the Beijing Friendship Hospital.
Data were presented as mean standard deviation and analyzed by using GraphPad Prism 8.0 software (La Jolla, CA, United States). When comparing the difference between one or more groups, Student’s t test or one-way ANOVA followed by Tukey’s multiple comparison test was applied. Statistical significance was considered when P < 0.05.
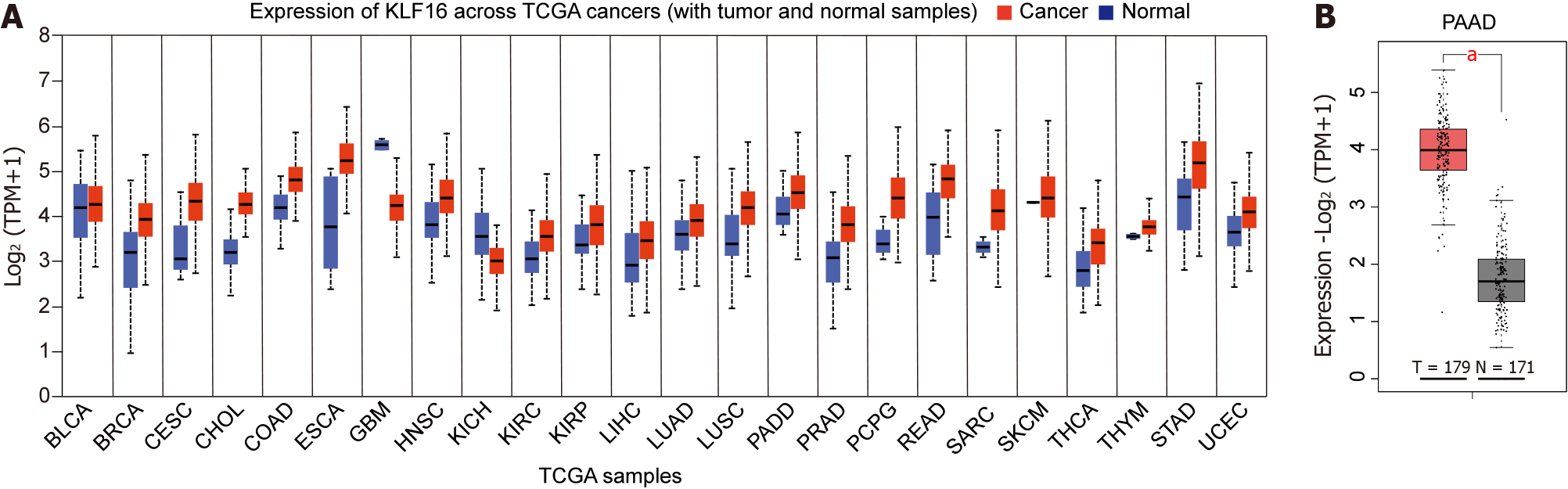
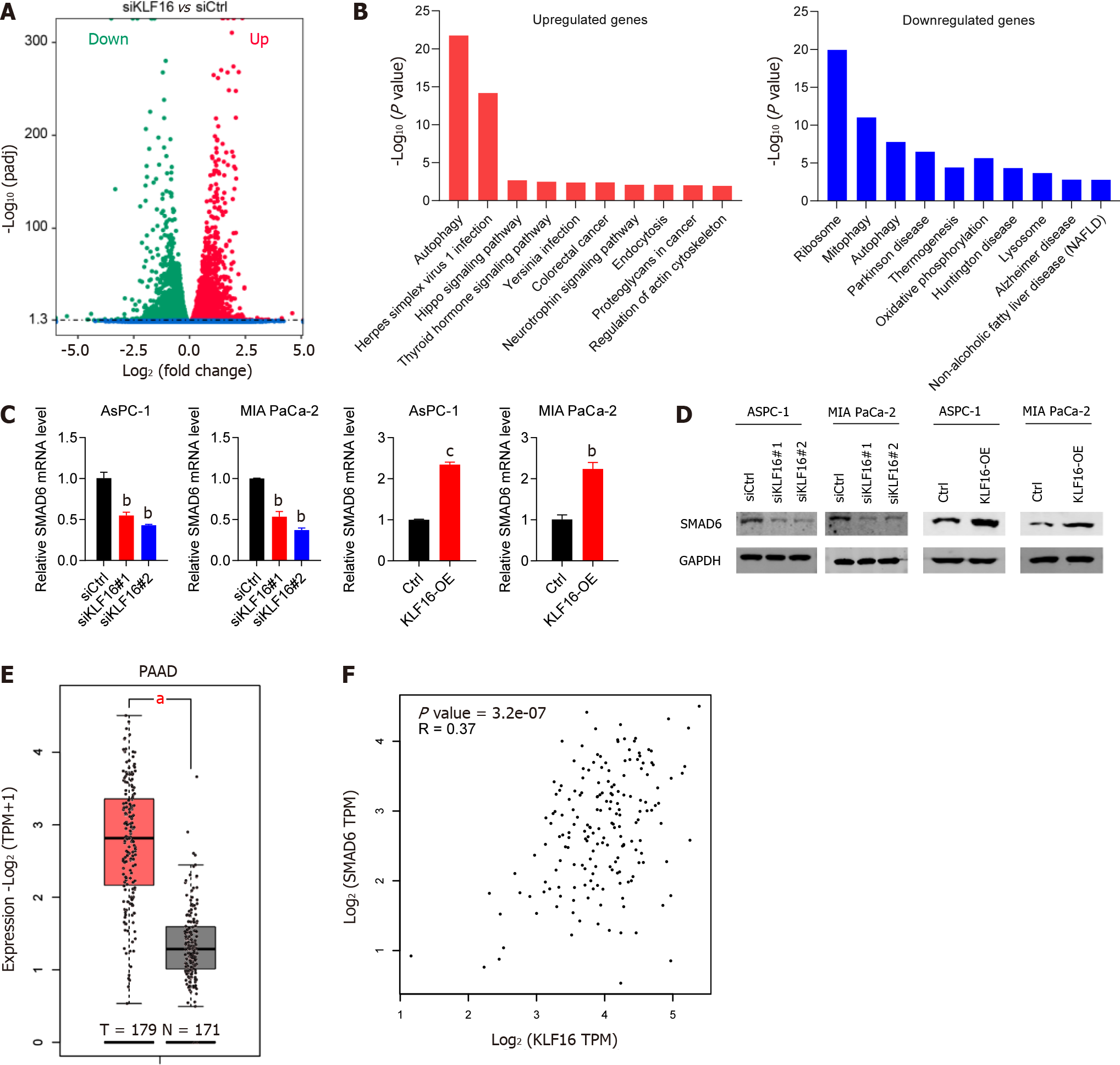
To examine the functions of KLF16 in PAAD, we interrogated the GEPIA database (http://gepia.cancer-pku.cn) and assessed the expression of KLF16 across different tumor histocytes. We found that KLF16 was overexpressed in almost all tumors (Figure 1A). More importantly, we found that KLF16 upregulation was observed in 179 PAAD patients compared with 171 healthy subjects (P < 0.05; Figure 1B).
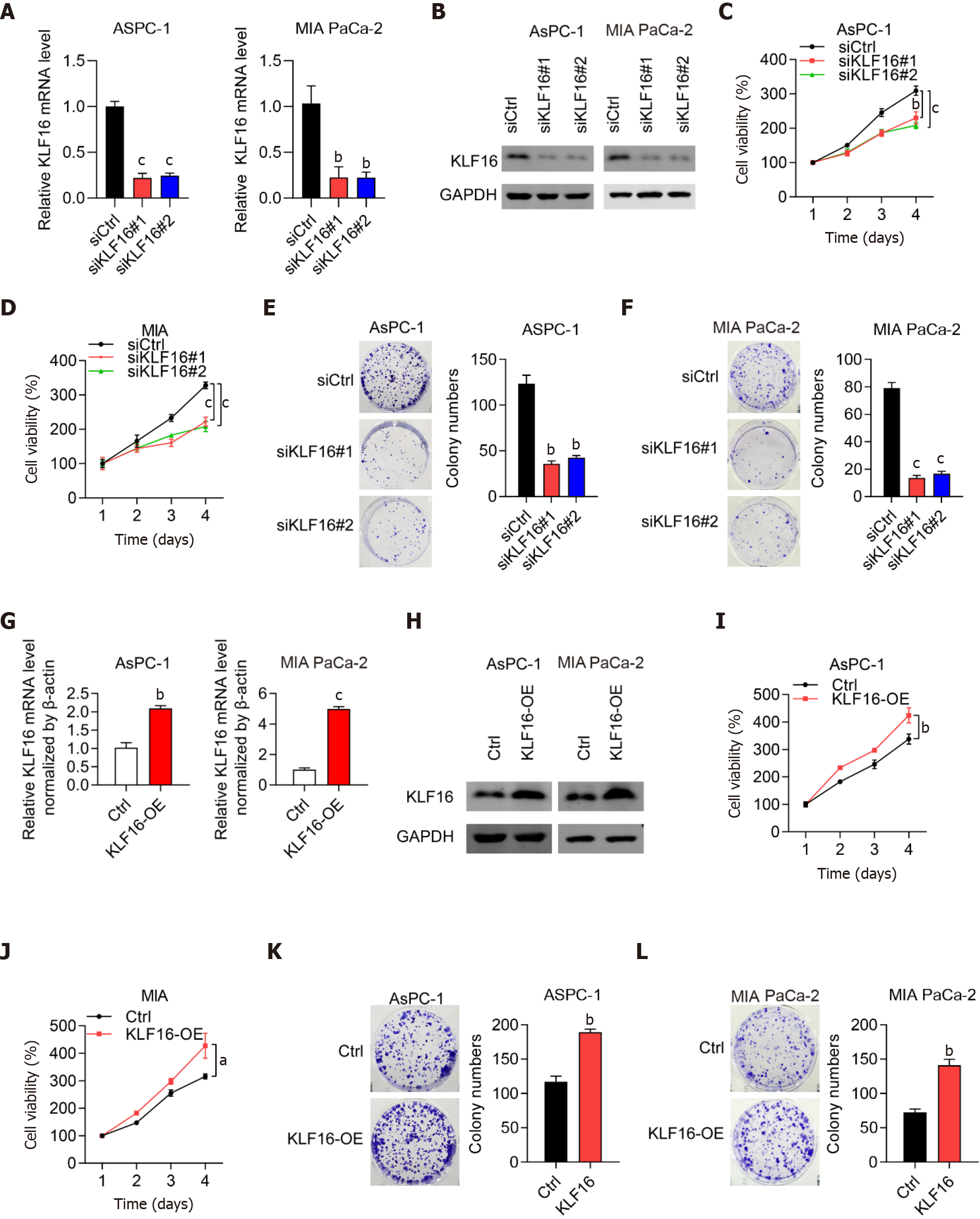
To examine if KLF16 could influence the progression of PAAD cells, we performed knockdown and overexpression studies. Firstly, we showed that KLF16 expression was significantly reduced in PAAD cells transfected with siRNAs against KLF16, indicating that the knockdown was effective (Figure 2A and 2B). The number of cell colonies and viability of PAAD cells were significantly reduced when KLF16 was knocked down (Figure 2C-F). Then overexpression experiments were conducted. The results showed that KLF16 overexpression was successful (Figure 2G and 2H). When KLF16 was overexpressed in AsPC-1 and MIA PaCa-2 cells, both the cell viability and the number of cell colonies considerably increased (Figure 2I-L). These findings indicated that KLF16 accelerated the proliferation of PAAD cells.
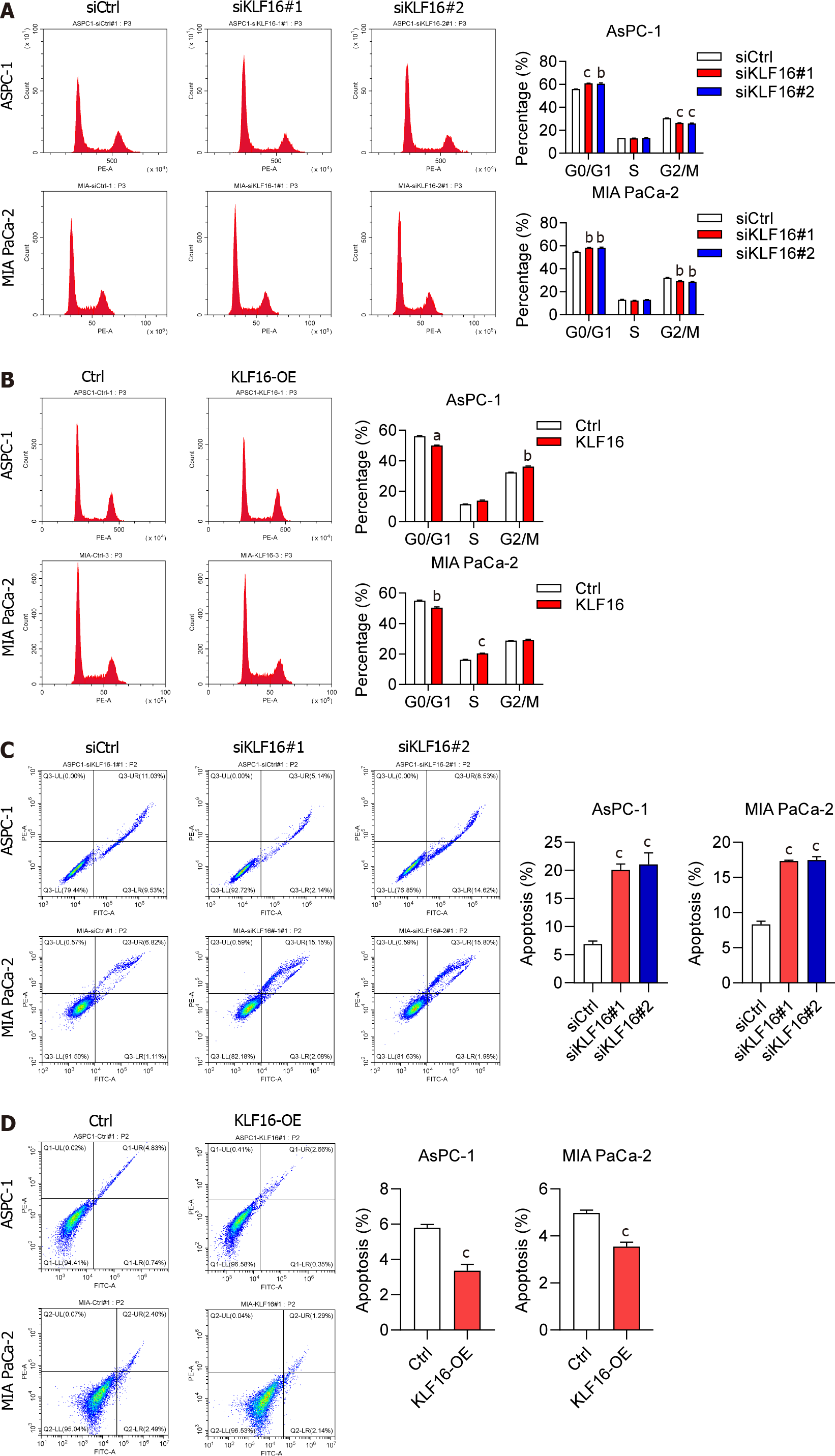
We then investigated the impact of KLF16 on the PAAD cell cycle as well as the apoptotic process. Following KLF16 knockdown, an enhanced number of the cells at G0/G1 phase and a reduced number of the cells at G2/M phase were observed (Figure 3A). When KLF16 was overexpressed in cells, the opposite effects were found (Figure 3B). Apoptosis in the cells was inhibited after KLF16 overexpression, whereas it was significantly promoted by KLF16 downregulation (Figure 3C and 3D).
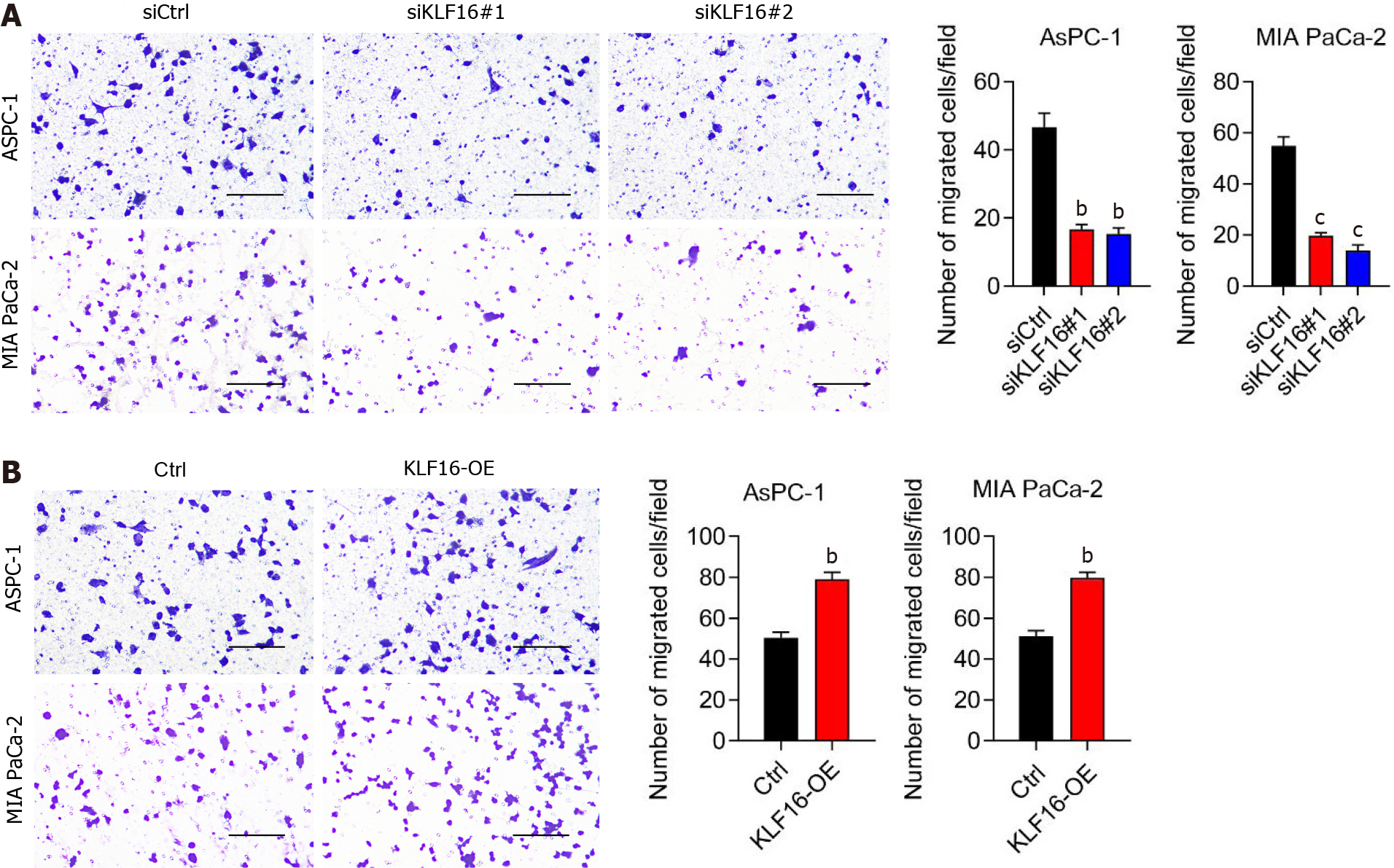
Following that, we investigated whether KLF16 influenced the migration of cells. After KLF16 was knocked down, the amount of cell migration was dramatically reduced (Figure 4A). When KLF16 was overexpressed, the number of migrating cells increased (Figure 4B). According to these findings, KLF16 could regulate the migration of the PAAD cell.
RNA-seq studies were carried out so that we could determine the pathways that KLF16 is responsible for in the development of PAAD. As shown in Figure 5A, the suppression of KLF16 in MIA PaCa-2 cells resulted in downregulation and upregulation of several genes. Figure 5B displays the top ten pathways that were discovered by cluster analysis to be associated with these genes. Autophagy-related genes were found to be among the most highly upregulated genes, while ribosome-related genes were the most highly downregulated. A highly expressed SMAD6 gene that had not been characterized before was found in PAAD. As a result, we investigated the connection between KLF16 and SMAD6. After KLF16 was either knocked down or overexpressed, respectively, we found that there was either a considerable rise or reduction in the levels of SMAD6 (Figure 5C and 5D). In addition, SMAD6 expression was increased in PAAD tissues retrieved from the TCGA database (Figure 5E). According to the findings of the study of correlation, the level of expression of KLF16 was shown to have a positive association with the level of expression of SMAD6 (r = 0.37; Figure 5F). Collectively, KLF16 promoted the expression of SMAD6 in PAAD cells and tissues.
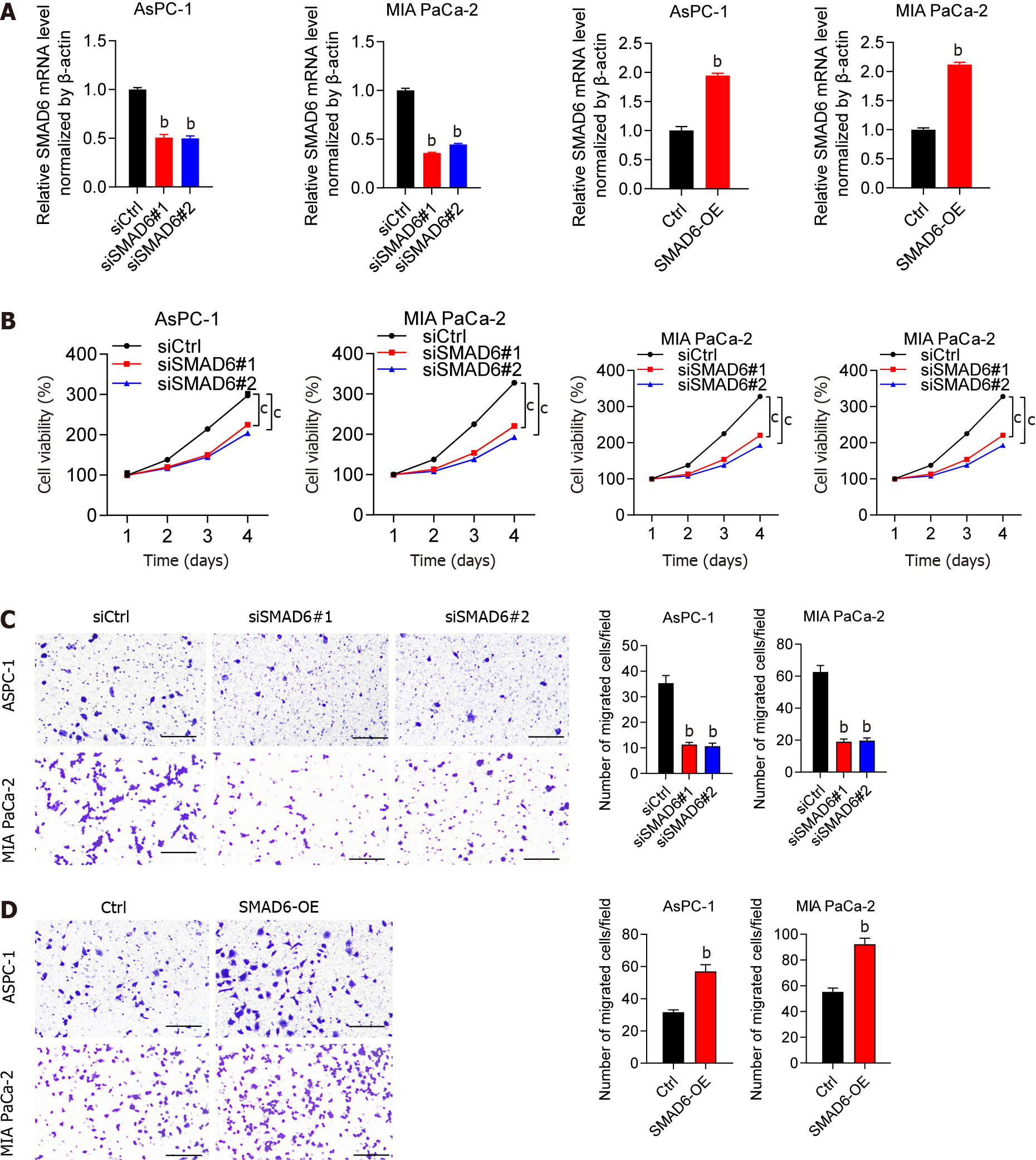
Following that, we explored the impact of SMAD6 on PAAD cells. qPCR was used to test the efficiency of SMAD6 knockdown or overexpression, as shown in Figure 6A. The ability of PAAD cells was suppressed by SMAD6 downregulation, while it was increased when SMAD6 was overexpressed (Figure 6B). This oncogenic function of SMAD6 was further validated by the findings that the number of migrating cells was greatly inhibited by downregulation of SMAD6 (Figure 6C) but was stimulated after SMAD6 overexpression (Figure 6D). According to these findings, SMAD6 was responsible for PAAD cell proliferation and migration.
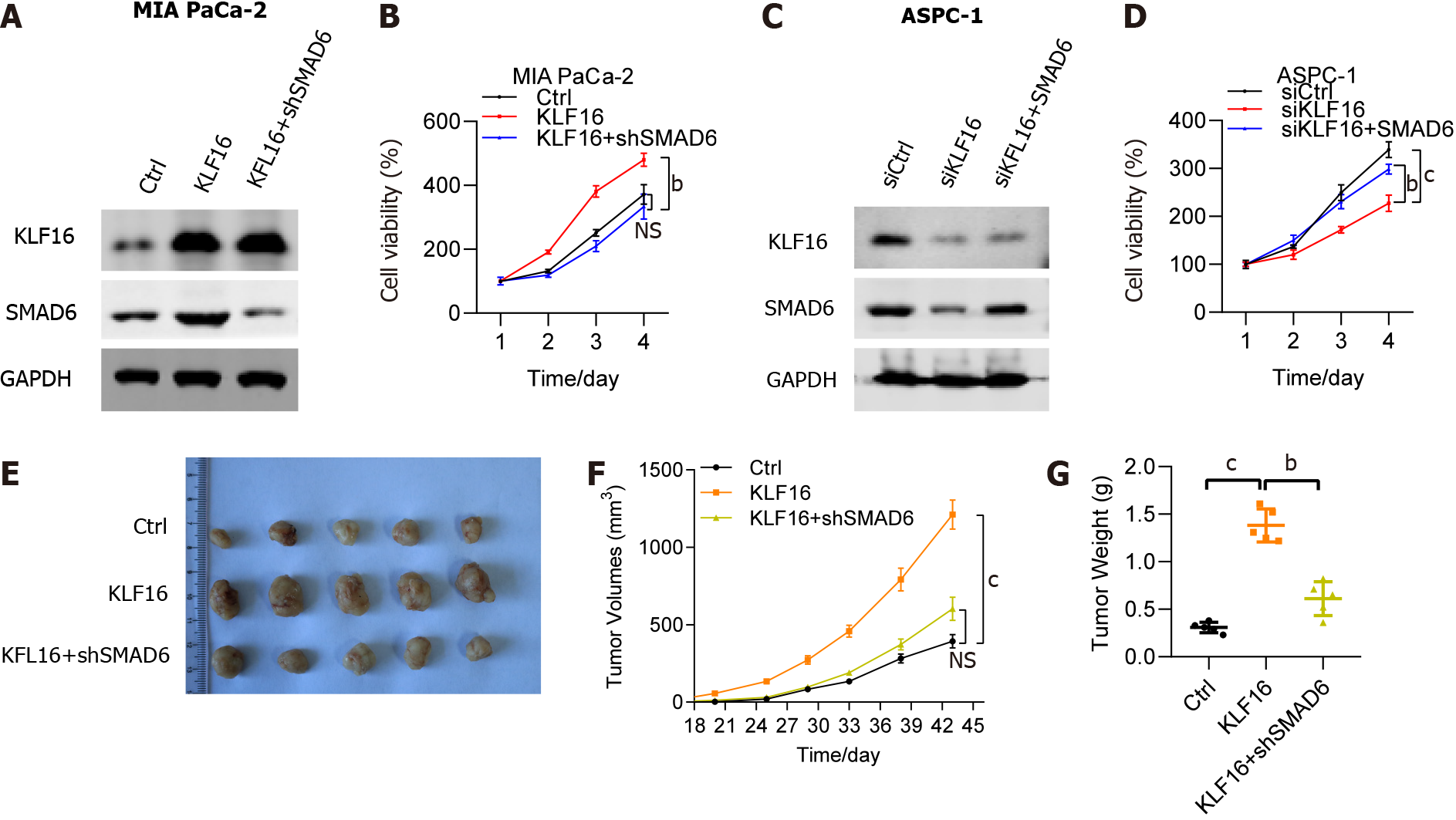
To understand how SMAD6 interacted with KLF16 overexpression, we investigated the impact of SMAD6 knockdown on KLF16-mediated PAAD. The protein level of SMAD6 was moderately elevated in MIA PaCa-2 cells transfected with KLF16 and was drastically reduced in cells transfected with KLF16+shSMAD6 (Figure 7A). The viability of MIA PaCa-2 cells improved when KLF16 was overexpressed, but SMAD6 knockdown had a significant adverse effect on that viability (Figure 7B). Moreover, we overexpressed SMAD6 in the KLF16 knockdown AsPC1 cells. SMAD6 was significantly downregulated in the KLF16 knockdown cells, while the protein level of SMAD6 was rescued after SMAD6 overexpression (Figure 7C). The viability of AsPC1 cells was impaired when KLF16 was knocked down, but SMAD6 overexpression had a significant adverse effect on that viability (Figure 7D). In vivo tumor transplantation investigations validated these results as well, showing that the tumor size and volume of xenografts increased after KLF16 overexpression in nude mice injected with MIA PaCa-2 cells, but decreased following SMAD6 knockdown (Figure 7E-G). Overall, SMAD6 knockdown rescued the effects of KLF16 overexpression in PAAD cells.
KLF16 was highly expressed in PAAD patients based on the GEPIA database. KLF16 silencing suppressed, while KLF16 overexpression promoted the malignant function of PAAD cells. Based on RNA sequencing, we discovered that KLF16 potentiated the expression of SMAD6 in PAAD cells. SMAD6 transcript abundance was increased and positively correlated with KLF16 expression in PAAD samples, and inhibiting SMAD6 was able to mitigate the effects of KLF16 overexpression on PAAD cell processes, suggesting the importance of SMAD6 in the development of KLF16-triggered PAAD.
Pancreatic adenocarcinoma, a very aggressive tumor of the digestive system, is difficult to diagnose and treat. PAAD is also known as pancreatic ductal adenocarcinoma, a kind of pancreatic cancer that accounts for the vast majority of cases (95%)[12]. Accumulation of genetic alterations, such as KRAS, CDKN2A/P16, TP53 and SMAD4 contributes to the progression of PAAD[13,14]. These abnormalities may be used to develop a novel effective therapy targeting PAAD.
KLFs refer to zinc finger transcription factors involved in various developmental processes through enhancing and/or repressing the expression of several genes[15]. KLF3, KLF4, KLF12 and KLF15 are only a few of the KLFs that participate in the formation and advancement of PAAD. KLF3 is a pancreatic cancer cell growth inhibitor, according to the study conducted by Wan et al[16]. KLF12 was found to be a miR-137 target and inhibited the cancer stem cell phenotype in pancreatic cancer cells[17]. In contrast, Zhu et al[18] found that downregulated Caveolin-1 by KLF4 maintained pancreatic cancer epithelial-mesenchymal transition and metastasis. As a member of KLFs, KLF16’s function in PAAD is less clear. During our research, we observed that pancreatic adenocarcinoma samples and cells had elevated levels of KLF16. By completing knockdown and overexpression studies on KLF16, we were able to show that it controlled PAAD cell malignancy. These studies were carried out using PAAD cells.
SMAD6, a member of the SMAD family, was discovered in the 1990s and serves as a key mediator of the TGF signaling pathway[19,20]. In mammalian cells, eight SMAD have been discovered[21]. SMAD4 Loss predicts poor prognosis in PAAD[14], while SMAD7 has been discovered to be adversely regulated in PAAD[22]. SMAD6 was shown to be downregulated in patients with colorectal cancer[23], while its upregulation was associated with poor patient survival from lung cancer[24]. Experiments using RNA sequencing were carried out so that we could determine the specific mechanism by which KLF16 is involved in the development of PAAD. It has been reported that KLF16 acted as a transcriptional repressor[25]. In the present study, KLF16 expression was discovered to have a favorable positive correlation with SMAD6, which was considerably upregulated. In this context, it was suggested that SMAD6 is not the directed target gene of KLF16. Hence, the role of KLF16 in regulating SMAD6 expression required further experimentation. Finally, we showed that SMAD6 expression was elevated in PAAD samples as well. SMAD6 knockdown inhibited PAAD cell proliferation and migration, but SMAD6 overexpression increased PAAD cell advancement. In addition, inhibiting SMAD6 in PAAD cells reversed the effects of KLF16 overexpression. Overall, KLF16 is an oncogenic protein in PAAD that positively influence SMAD6 expression.
Although this study can fully explain the mechanism of KLF16 on the proliferation and migration of PAAD cells, the lack of prognosis information of patients makes this study limited to a certain extent. It will be further verified in human pancreatic cancer tissues in the future, so as to better support the experimental results and enhance the credibility of the study.
The role that KLF16 and SMAD6 play in PAAD was investigated in this work. Both genes carried out their oncogenic functions and contributed to the development of PAAD. Furthermore, we identified a strong correlation between KLF16 and SMAD6 expression levels. KLF16 and SMAD6 appear to have the potential to act as both a novel prognostic marker and a potential therapeutic target for PAAD, based on our findings.
Pancreatic adenocarcinoma (PAAD) is a cancerous tumor with an extremely poor 5-year survival rate. The exploration of biomarkers for the diagnosis and treatment of PAAD is crucial in clinical practice.
KLF16 behaves as an oncogene in prostate, breast and gastric cancers. However, no research has been done on the significance of Krüppel-like factor 16 (KLF16) in PAAD.
This study aimed to explore the molecular mechanisms of KLF16 in PAAD.
KLF16 was identified in the tumor specimens and normal tissues by GEPIA database and verified by quantitative real-time PCR (qRT-PCR). Knockdown or exogenous expression of KLF16, combined with in vitro and in vivo assays, was performed to show the functional significance of KLF16. The molecular mechanism of KLF16 was demonstrated by qRT-PCR, western blotting, immunoprecipitation assay and flow cytometry.
KLF16 was highly expressed in PAAD patients based on the GEPIA database. KLF16 silencing suppressed, while KLF16 overexpression promoted the malignant function of PAAD cells. Based on RNA sequencing, we discovered that KLF16 potentiated the expression of SMAD6 in PAAD cells. SMAD6 transcript abundance was increased and positively correlated with KLF16 expression in PAAD samples. In addition, inhibiting SMAD6 was able to mitigate the effects of KLF16 overexpression on PAAD cell processes, suggesting the importance of SMAD6 in the development of KLF16-triggered PAAD.
KLF16/SMAD6 axis might be explored as a therapeutic target for PAAD therapy.
KLF16 and SMAD6 appear to have the potential to act as both a novel prognostic marker and a potential therapeutic target for PAAD, based on our findings.
We would like to thank the Clinical Data and Biobank Resource of Beijing Friendship Hospital for their help with data processing.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Miceli V, Italy; Tantau AI, Romania S-Editor: Wu YXJ L-Editor: Filipodia P-Editor: Yuan YY
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64718] [Article Influence: 16179.5] [Reference Citation Analysis (177)] |
| 2. | Ferlay J, Partensky C, Bray F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol. 2016;55:1158-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 312] [Article Influence: 34.7] [Reference Citation Analysis (1)] |
| 3. | Ottenhof NA, Milne AN, Morsink FH, Drillenburg P, Ten Kate FJ, Maitra A, Offerhaus GJ. Pancreatic intraepithelial neoplasia and pancreatic tumorigenesis: of mice and men. Arch Pathol Lab Med. 2009;133:375-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 4. | Goral V. Pancreatic Cancer: Pathogenesis and Diagnosis. Asian Pac J Cancer Prev. 2015;16:5619-5624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 174] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 5. | Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991-3000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 491] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 6. | Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996-2001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 318] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 7. | Sun N, Shen C, Zhang L, Wu X, Yu Y, Yang X, Yang C, Zhong C, Gao Z, Miao W, Yang Z, Gao W, Hu L, Williams K, Liu C, Chang Y, Gao Y. Hepatic Krüppel-like factor 16 (KLF16) targets PPARα to improve steatohepatitis and insulin resistance. Gut. 2021;70:2183-2195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 8. | Bang S, Li J, Zhang M, Cui R, Wu X, Xin Z, Ma D, Zhang J, Zhang H. The Clinical Relevance and Function of Krüppel-Like Factor 16 in Breast Cancer. Cancer Manag Res. 2020;12:6373-6383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Ma XD, Xu SD, Hao SH, Han K, Chen JW, Ling H, Chen RX, Jin XH, Cao JH, Lin JL, Ou QJ, Fang YJ, Pan ZZ, Xie D, Wang FW. KLF16 enhances stress tolerance of colorectal carcinomas by modulating nucleolar homeostasis and translational reprogramming. Mol Ther. 2022;30:2828-2843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 10. | Chen X, Li S, Ke Y, Wu S, Huang T, Hu W, Fu H, Guo X. KLF16 suppresses human glioma cell proliferation and tumourigenicity by targeting TFAM. Artif Cells Nanomed Biotechnol. 2018;46:608-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Lee MD, She Y, Soskis MJ, Borella CP, Gardner JR, Hayes PA, Dy BM, Heaney ML, Philips MR, Bornmann WG, Sirotnak FM, Scheinberg DA. Human mitochondrial peptide deformylase, a new anticancer target of actinonin-based antibiotics. J Clin Invest. 2004;114:1107-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 12. | Tanaka S. Molecular Pathogenesis and Targeted Therapy of Pancreatic Cancer. Ann Surg Oncol. 2016;23 Suppl 2:S197-S205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2129] [Cited by in RCA: 2115] [Article Influence: 151.1] [Reference Citation Analysis (3)] |
| 14. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3216] [Cited by in RCA: 3027] [Article Influence: 178.1] [Reference Citation Analysis (0)] |
| 15. | Swamynathan SK. Krüppel-like factors: three fingers in control. Hum Genomics. 2010;4:263-270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 16. | Wan Y, Luo H, Yang M, Tian X, Peng B, Zhan T, Chen X, Ding Y, He J, Cheng X, Huang X, Zhang Y. miR-324-5p Contributes to Cell Proliferation and Apoptosis in Pancreatic Cancer by Targeting KLF3. Mol Ther Oncolytics. 2020;18:432-442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 17. | He Z, Guo X, Tian S, Zhu C, Chen S, Yu C, Jiang J, Sun C. MicroRNA-137 reduces stemness features of pancreatic cancer cells by targeting KLF12. J Exp Clin Cancer Res. 2019;38:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 18. | Zhu Z, Yu Z, Wang J, Zhou L, Zhang J, Yao B, Dou J, Qiu Z, Huang C. Krüppel-Like Factor 4 Inhibits Pancreatic Cancer Epithelial-to-Mesenchymal Transition and Metastasis by Down-Regulating Caveolin-1 Expression. Cell Physiol Biochem. 2018;46:238-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 470] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 20. | Derynck R, Gelbart WM, Harland RM, Heldin CH, Kern SE, Massagué J, Melton DA, Mlodzik M, Padgett RW, Roberts AB, Smith J, Thomsen GH, Vogelstein B, Wang XF. Nomenclature: vertebrate mediators of TGFbeta family signals. Cell. 1996;87:173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 138] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 21. | Singh P, Wig JD, Srinivasan R. The Smad family and its role in pancreatic cancer. Indian J Cancer. 2011;48:351-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Wang Y, Yang Y, Wang Y, Li X, Xiao Y, Wang W. High Cancer Susceptibility Candidate 8 Expression Is Associated With Poor Prognosis of Pancreatic Adenocarcinoma: Validated Analysis Based on Four Cancer Databases. Front Cell Dev Biol. 2020;8:392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Zhang Z, Wang L, Wang Q, Zhang M, Wang B, Jiang K, Ye Y, Wang S, Shen Z. Molecular Characterization and Clinical Relevance of RNA Binding Proteins in Colorectal Cancer. Front Genet. 2020;11:580149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Jeon HS, Dracheva T, Yang SH, Meerzaman D, Fukuoka J, Shakoori A, Shilo K, Travis WD, Jen J. SMAD6 contributes to patient survival in non-small cell lung cancer and its knockdown reestablishes TGF-beta homeostasis in lung cancer cells. Cancer Res. 2008;68:9686-9692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Yao W, Jiao Y, Zhou Y, Luo X. KLF13 suppresses the proliferation and growth of colorectal cancer cells through transcriptionally inhibiting HMGCS1-mediated cholesterol biosynthesis. Cell Biosci. 2020;10:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |