Published online Nov 15, 2022. doi: 10.4251/wjgo.v14.i11.2108
Peer-review started: June 25, 2022
First decision: August 30, 2022
Revised: September 13, 2022
Accepted: October 19, 2022
Article in press: October 19, 2022
Published online: November 15, 2022
Processing time: 142 Days and 21.9 Hours
The incidence of colorectal cancer in humans is high, and it is in the top five for cancer-related morbidity and mortality. It is one of the main threats to human health. The function of long noncoding RNAs in tumor occurrence and deve
To find out if long noncoding RNA RP5-881L22.5 played a role in the patho
We analyzed the transcriptome data and clinical data in The Cancer Genome Atlas-colon adenocarcinoma. The CIRBERSORT algorithm was applied to evaluate these tumor-infiltrating immune cells in The Cancer Genome Atlas-colon adenocarcinoma cancer tissue samples. Using the “estimate” package in R, we assessed the tumor immune microenvironment. The expression level of RP5-881L22.5 in tumor tissue and adjacent normal tissue samples from 4 pairs of colorectal cancer patients was determined by quantitative reverse transcription PCR. Colorectal cancer cells were tested for invasiveness using a transwell invasion assay after RP5-881L22.5 expression was knocked down.
The expression of lncRNA RP5-881L22.5 was related to the clinical characteristics of the tumors, and it was negatively related to the infiltration level of immune cells in the tumor microenvironment and the expression of T cell inhibitory receptors. A major function of its coexpressed mRNA was to regulate tumor immunity, such as the immune response. When quantitative reverse transcription PCR was performed on tumor tissues from 4 pairs of colorectal cancer patients, the results showed that RP5-881L22.5 was highly expressed. Subsequently, knocking down the expression of RP5-881L22.5, the invasiveness of colorectal cancer cell lines was reduced, and the apoptosis rate was increased.
RP5-881L22.5 plays a crucial role in the microenvironment of tumors as well as in the pathogenesis of colorectal cancer. The relationship between RP5-881L22.5 and the tumor immune microenvironment deserves further study.
Core Tip: Long noncoding RNA RP5-881L22.5 is related to the clinical characteristics of tumors, and it is negatively related to the infiltration level of immune cells in the tumor microenvironment and the expression of T cell inhibitory receptors. RP5-881L22.5 may play an important role in the tumor immune microenvironment as well as in the pathogenesis of colorectal cancer. The relationship between RP5-881L22.5 and the tumor immune microenvironment deserves further study.
- Citation: Zong H, Zou JQ, Huang JP, Huang ST. Potential role of long noncoding RNA RP5-881L22.5 as a novel biomarker and therapeutic target of colorectal cancer. World J Gastrointest Oncol 2022; 14(11): 2108-2121
- URL: https://www.wjgnet.com/1948-5204/full/v14/i11/2108.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i11.2108
Among all malignant tumors, colorectal cancer is the most common worldwide. In total, 1148515 new cases were diagnosed worldwide in 2020[1]. Although the diagnosis and treatment of colorectal cancer have been continuously improved in recent years, the 5-year survival rate of colorectal cancer patients has not changed significantly. Greater than 20% of patients have already experienced metastasis at diagnosis, and approximately 50% of patients experience metastases during treatment[2]. Therefore, more research on the pathogenesis, early diagnosis, early treatment and prognosis of colorectal cancer is needed.
With the progression of research, it is increasingly recognized that tumors are a systemic disease, and the tumor immune microenvironment is a key factor involved in the pathogenesis and mechanism of tumors[3,4]. Innate immunity and adaptive immunity run through the whole disease process[5,6]. In the early stage of tumorigenesis, natural killer (NK) cells can recognize specific ligands on the surface of tumor cells, even if there are only a few tumor cells[3]. On the other hand, the activation of macrophages and dendritic cells, especially T cells and B cells, in the tumor immune microenvironment produces a large amount of additional cytokines, which further promotes the activation of CD8+ cytotoxic T cells and promotes immune memory[7]. However, given the long-term presence of tumors in the body, tumor cells can resist or suppress anti-tumor immune responses, leading to immune evasion and promoting tumor progression, which also represents a challenge for immunotherapy[8].
Long noncoding RNAs (lncRNAs) have a length over 200 nucleotides and are involved in many normal human physiological functions and pathogenic processes of disease[9,10]. In addition, extensive studies have found that lncRNAs have a pivotal function in the formation of the tumor immune microenvironment[11]. Therefore, we designed this study to explore whether lncRNAs can be used as molecular markers for a more accurate prediction of colorectal cancer prognosis and whether the molecular mechanisms of lncRNAs are the reasons for the differences in colorectal cancer prognosis as well as their impact on the tumor immune microenvironment.
We downloaded the transcriptome sequencing data and clinical data from the Colon Adenocarcinoma (COAD) project in The Cancer Genome Atlas (TCGA) from the UCSC Xena (https://xenabrowser.net/) website[12,13]. The data included HTseq-FPKM data [log2 (FPKM+1)] (https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-COAD.htseq_fpkm.tsv.gz), survival data (https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-COAD.survival.tsv) and clinical data (Phenotype including pathological staging and other data) (https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-COAD.GDC_phenotype.tsv.gz). The dataset contains 512 colon cancer tissues and adjacent normal tissues, of which 448 colon cancer tissues have complete survival data and pathological stage data (tumor stage was determined according to the UICC and AJCC TNM classification system).
According to the human reference genome GRch38 file (release 22) provided on the GENCODE website (https://www.gencodegenes.org), we converted gene IDs from Ensembl to symbols. Then, according to the annotation files provided by GENCODE, we extracted the mRNA and lncRNA expression datasets from the TCGA-COAD sequencing data[14].
Genes with extremely low expression levels [log2 (FKPM+1) < 0.5] were removed, and the differential expression of HTseq-FPKM data was analyzed with “limma” (R language, V4.1.1)[15]. We compared mRNA and lncRNA expression differences between 471 COAD tumor specimens and 41 adjacent normal tissue specimens. The lncRNA RP5-881L22.5 was selected as the research object, and a comparison of RP5-881L22.5 expression across different stages of COAD was conducted. Kaplan-Meier survival curves were drawn for patients grouped by different expression levels, and the survival differences of all patients grouped by expression levels as well as the effect of the expression differences of RP5-881L22.5 in different stages on survival were compared.
The expression of RP5-881L22.5 in 33 cancers in TCGA was acquired from the GEPIA2 (http://gepia2.cancer-pku.cn/). The differential expression of RP5-881L22.5 between colon cancer tissue (TCGA-COAD) and normal colon tissue (data involved the TCGA-COAD project and Genotype-Tissue Expression (GTEx) database) were analyzed[16,17].
We downloaded the benchmark database file (LM22.txt) of 22 tumor-infiltrating immune cells (CD8+ T cells, CD4 naive T cells, regulatory reg cells, naïve B cells, memory B cells, plasma cells, CD4 memory resting T cells, CD4 memory activated T cells, follicular helper T cells, gamma delta T cells, resting NK cells, activated NK cells, monocytes, macrophages M0/M1/M2, resting dendritic cells, activated dendritic cells, resting mast cells, activated mast cells, eosinophils and neutrophils). The CIRBERSORT algorithm was applied to evaluate these tumor-infiltrating immune cells in TCGA-COAD cancer tissue samples[18]. Using the “estimate” package in the R language, we evaluated the tumor immune microenvironment[19]. All TCGA-COAD cancer tissue samples were divided into high expression and low expression groups according to the median expression of RP5-881L22.5, and the differences in immune components and immune cell infiltration between the two groups were compared. In addition, the expression levels of the coinhibitory receptors LAG3, CTLA4, HAVCR2, TIGIT and CD244 were compared between the two groups[20].
Pearson correlation analysis was performed using the “cor.test” function of the basic R package. Among the differentially expressed mRNAs (logFC > 1 or logFC < -1, and adj.P.val < 0.05), the mRNAs coexpressed with RP5-881L22.5 were screened out. Gene set enrichment analysis of coexpressed mRNAs was performed using the “clusterProfiler” package in R language[21].
Colorectal cancer tissue specimens were obtained from the radical surgery specimens of colorectal cancer patients admitted to the Gastrointestinal Surgery Department of the Second Affiliated Hospital of Southern University of Science and Technology (Shenzhen Third People's Hospital) in August 2021. All patients were diagnosed by pathological diagnosis of colonoscopy biopsy: (1) Patients with acute infection, such as intestinal obstruction or intestinal perforation, were excluded; (2) Patients with HIV infection, autoimmune diseases, inflammatory bowel disease and other immune system-related diseases were excluded; (3) Preoperative neoadjuvant treatment patients were excluded; and (4) Patients with severe heart and lung insufficiency who could not tolerate surgery were excluded. Finally, eight tissue samples were obtained, which were colorectal cancer (3 cases of colon cancer, 1 case of rectal cancer), and four tumor tissues and four adjacent normal intestinal tissues were saved and transferred to a -80 °C freezer for storage. The experiment was ethically approved by the Ethics Committee of Shenzhen Third People’s Hospital (Shenzhen, China; license no. HE2022177).
The colorectal cancer cell line DLD-1 was obtained from ATCC (Manassas, VA, United States). 1640 medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin and 3% glucose (HyClone, Logan, UT, United States) was used to incubate DLD-1 cells. Control siRNA and RP5-881L22.5 siRNA were purchased from RiboBio Co., Ltd. (GuangZhou, China). Lipofectamine 2000 Transfection Reagent (Invitrogen, CA, United States) was used to transfect siRNA into DLD-1 cells.
TRIzol reagent (ELK Biotechnology, Wuhan, China) was used to isolate total RNA from colorectal cancer tissues. On 1% agarose gels, degradation and contamination of RNA were monitored after isolation. Synthesis of first-strand cDNA was performed using an M-MLV Reverse Transcriptase kit (ELK Biotechnology, Wuhan, China). Quantitative real-time PCR was completed on a StepOne™ Real-Time PCR machine (Life Technologies, CA, United States) with 3 replicate per sample using QuFast SYBR Green PCR Master Mix kit (ELK Biotechnology, Wuhan, China). GAPDH was used as an endogenous control gene. The reactions were incubated at 95 °C for 1 min, followed by 40 cycles at 95 °C for 15 s, 58 °C for 20 s and 72 °C for 45 s. Calculation of relative expression levels was performed using the 2−ΔΔCT value relative quantification. The primer sequences were as follows: RP5-881L22.5 (ENSG00000226812) Sense: 5’- TATTGAGCACCTACTATGTACCAGG -3’ and Antisense: 5’- GTTAGAGCTCAGTCTCTCACAGCTC -3’; and GAPDH Sense: 5’- CATCATCCCTGCCTCTACTGG -3’ and Antisense: 5’- GTGGGTGTCGCTGTTGAAGTC -3’.
To prepare the transwell chamber for use, 50 μL of Matrigel (Corning, United States) and medium were diluted 1:3 and added to the chamber. Cell suspension (1 mL) was centrifuged at 1500 rpm for 5 min after being diluted to 105 cells/mL. The cell suspension was pipetted into a transwell chamber with 1 mL of serum-free medium. In a 24-well plate, 500 μL of complete medium containing 10% fetal bovine serum was added, and the chamber was inserted. The plate was incubated at 37 °C in a CO2 (5% content) incubator for 24 h (adjusted according to the experiment). A 0.5% crystal violet solution (1:4) was diluted into a 0.1% crystal violet staining solution with PBS solution. The medium was washed with PBS, and the glue and cells in the upper chamber were wiped off with a cotton swab, fixed with paraformaldehyde for 20 min, washed twice with PBS, stained with crystal violet for 10 min, and washed to remove the crystal violet on the surface. The side devoid of cell seeding was photographed under an inverted microscope (OLYMPUS).
Precooled PBS was added at 4 °C, and an appropriate amount of binding buffer was diluted for use. After washing with PBS once, the cells in the 6-well plate were digested with 400 μL of 0.25% trypsin. The digestion was terminated by adding complete medium once the cells had become round and some had fallen off. We collected samples in 1.5 mL EP tubes and centrifuged them at 300 × g for 5 min, discarding the supernatant. One milliliter of PBS was added to resuspend the cells, they were centrifuged at 300 × g for 5 min, and the supernatant was discarded. A 200 μL solution of Binding Buffer was used to resuspend the pellet. Following this, 5 μL of Annexin V-FITC (Sungene Biotech, Tianjin, China) were added, mixed well and incubated for 10 min in the dark. Next, 5 μL propidium iodide was added, mixed well and incubated in the dark for 5 min. On-board inspection was performed within 1 h.
Comparing survival rates between groups was carried out using Kaplan-Meier survival curves and log-rank tests. The immune scores, the infiltration of immune cells and the expression levels of each gene were compared between groups using Wilcoxon tests. Differences were considered statistically significant when P < 0.05. All statistical procedures and graphs were completed using R programmer language (V4.1.1).
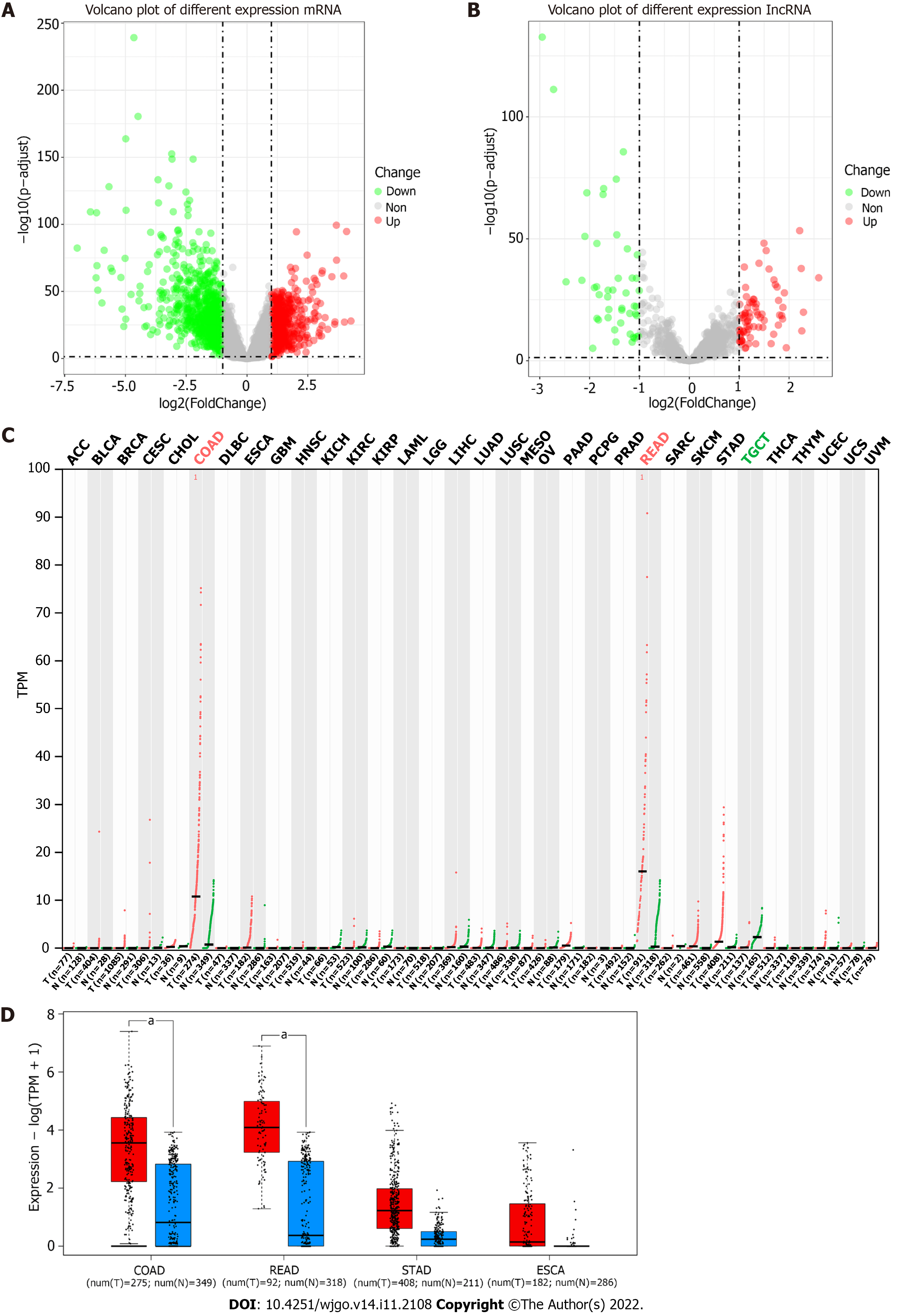
According to the GENCODE annotation file, we identified a total of 19712 mRNAs and 15878 lncRNAs in TCGA-COAD database. Genes with very low expression levels [log2 (FKPM+1) < 0.5] were deleted, and differential expression analysis was performed on the remaining 13285 mRNAs and 1850 lncRNAs. There were 1612 differentially expressed mRNAs and 122 differentially expressed lncRNAs (logFC > 1 or < -1 and adjusted P value < 0.05) (Figure 1A and B).
By querying the expression of RP5-881L22.5 in all tumor tissues from the TCGA database on the GEPIA2 website, we found that among all 33 malignant tumors, only COAD and READ had significant differential expression (log2FC > 1.0, q value < 0.01) (Figure 1C) between cancer and normal tissues. Additionally, we found similar differences in gastric and esophageal cancers, both of which are malignant tumors of the digestive tract (Figure 1D).
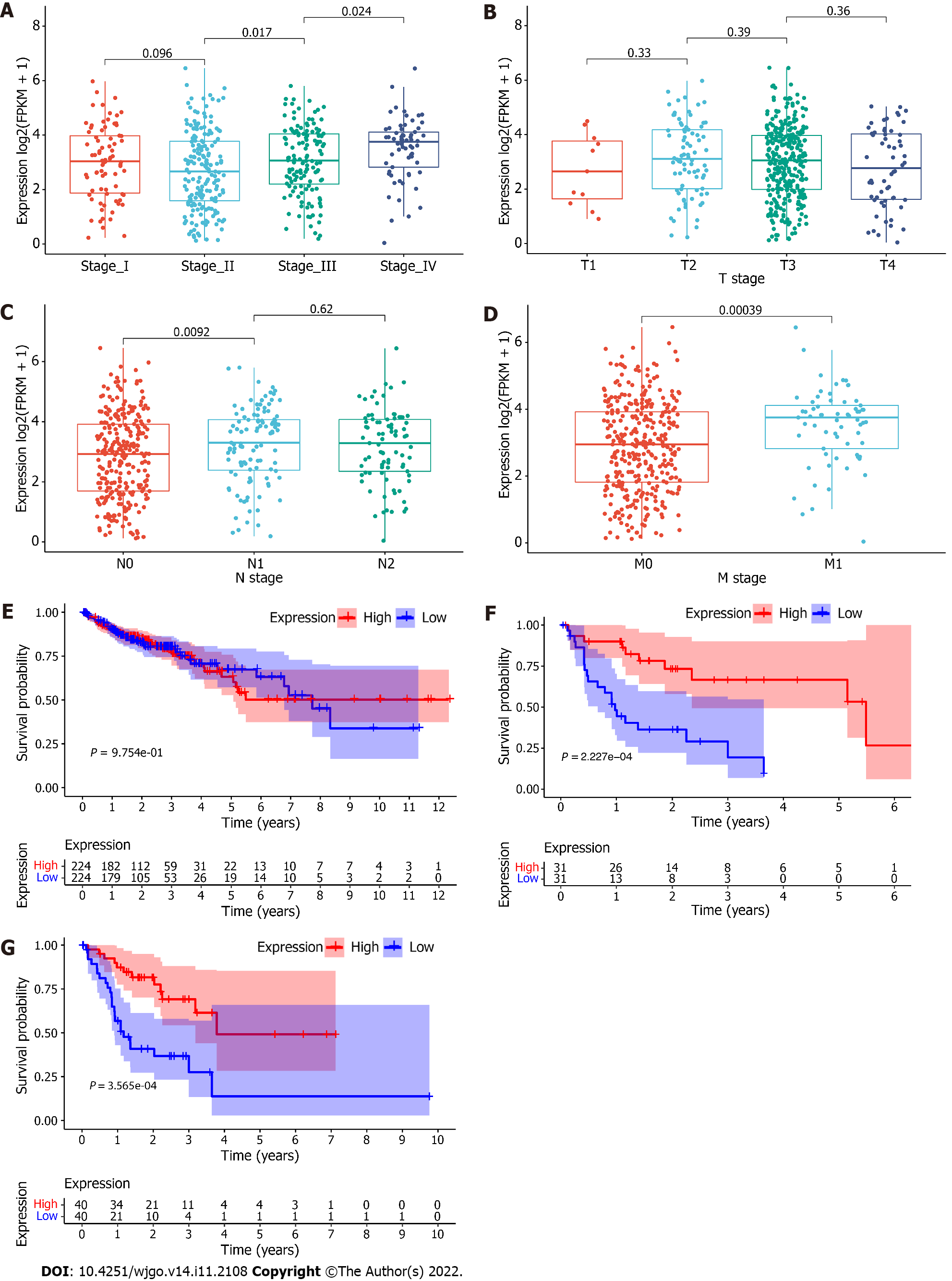
Based on the median expression of RP5-881L22.5 in 448 colon cancer tissue, we divided all samples into two groups with 224 samples in each. The 62 specimens of patients in stage IV were divided into two groups according to the median expression of RP5-881L22.5 with 31 samples in each group. Eighty specimens of patients in the N2 stage were divided into two groups according to the median expression of RP5-881L22.5 with 40 samples in each group. A positive correlation was found between RP5-881L22.5 expression and TNM stage (UICC and AJCC TNM staging system) as well as N stage and M stage, although no association was found with T stage (Figure 2A-D). However, after dividing all COAD samples into high and low groups according to RP5-881L22.5 expression levels, no difference in survival was noted between the two groups. For stage IV patients and patients with lymph node metastasis stage N2, high expression was associated with better survival. No differences in survival were noted among the other stages (Figure 2E-G).
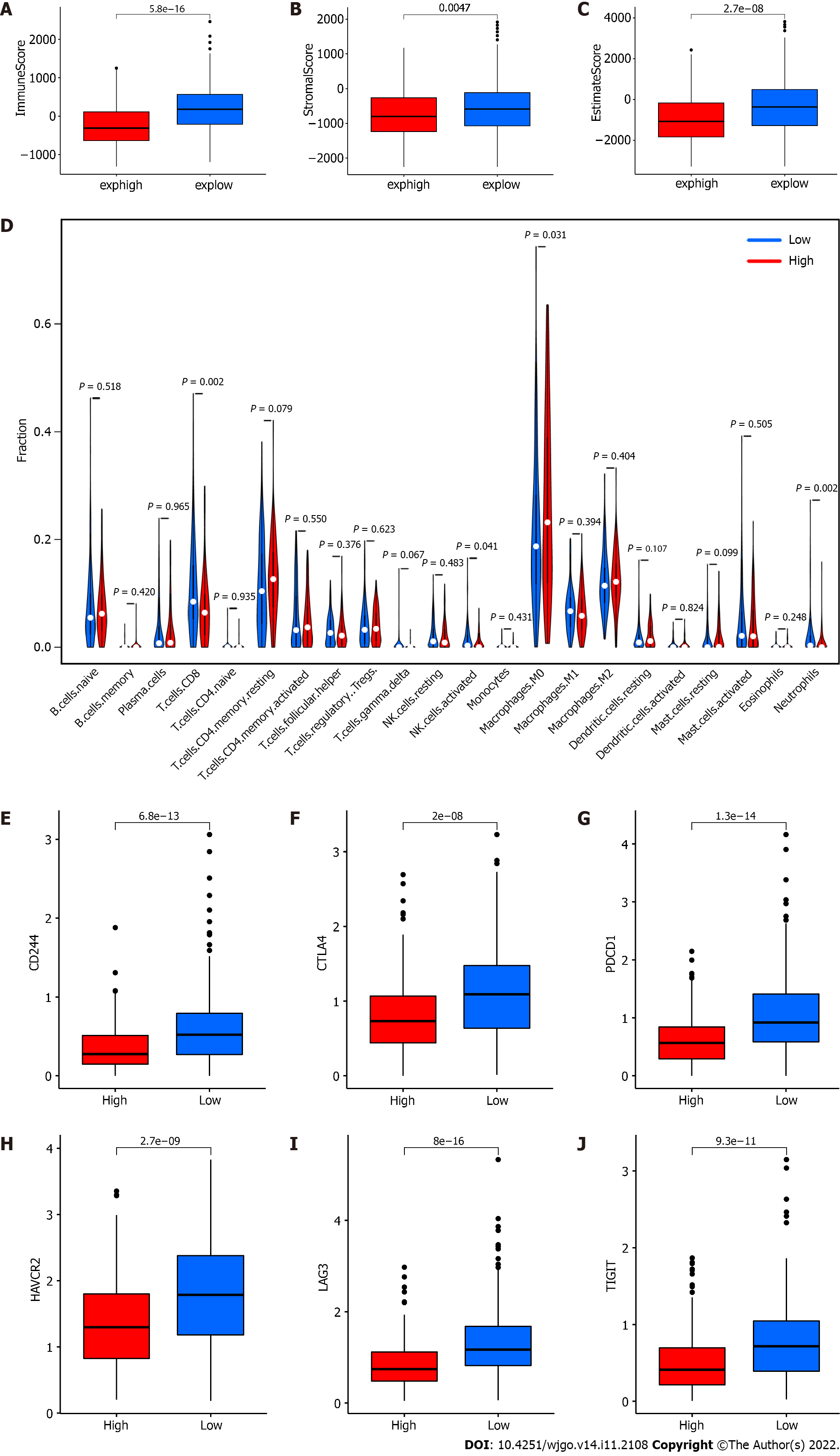
To explore whether the expression of RP5-881L22.5 is related to the tumor immune microenvironment, we divided all samples from TCGA-COAD into high and low groups according to the median expression of RP5-881L22.5 and compared the immune microenvironment differences. According to our findings, the immune score and tumor stroma score in the high expression group were lower than those in the low expression group, and the differences were significant (Figure 3A-C). Comparing the infiltration of 22 types of immune cells in the high and low groups, four types of immune cells showed significant differences, including CD8 T cells, M0 macrophages, activated NK cells and neutrophils (P < 0.05) (Figure 3D). T cell exhaustion is one of the reasons for the formation of a tumor suppressive immune microenvironment, and the coinhibitory receptors PDCD1 (PD-1), LAG3, CTLA4, HAVCR2, TIGIT, CD244, etc, related to T cell exhaustion were also significantly different between the two groups. The expression levels of these genes in the RP5-881L22.5 high expression group were lower, and statistically significant differences were found (Figure 3E-J).
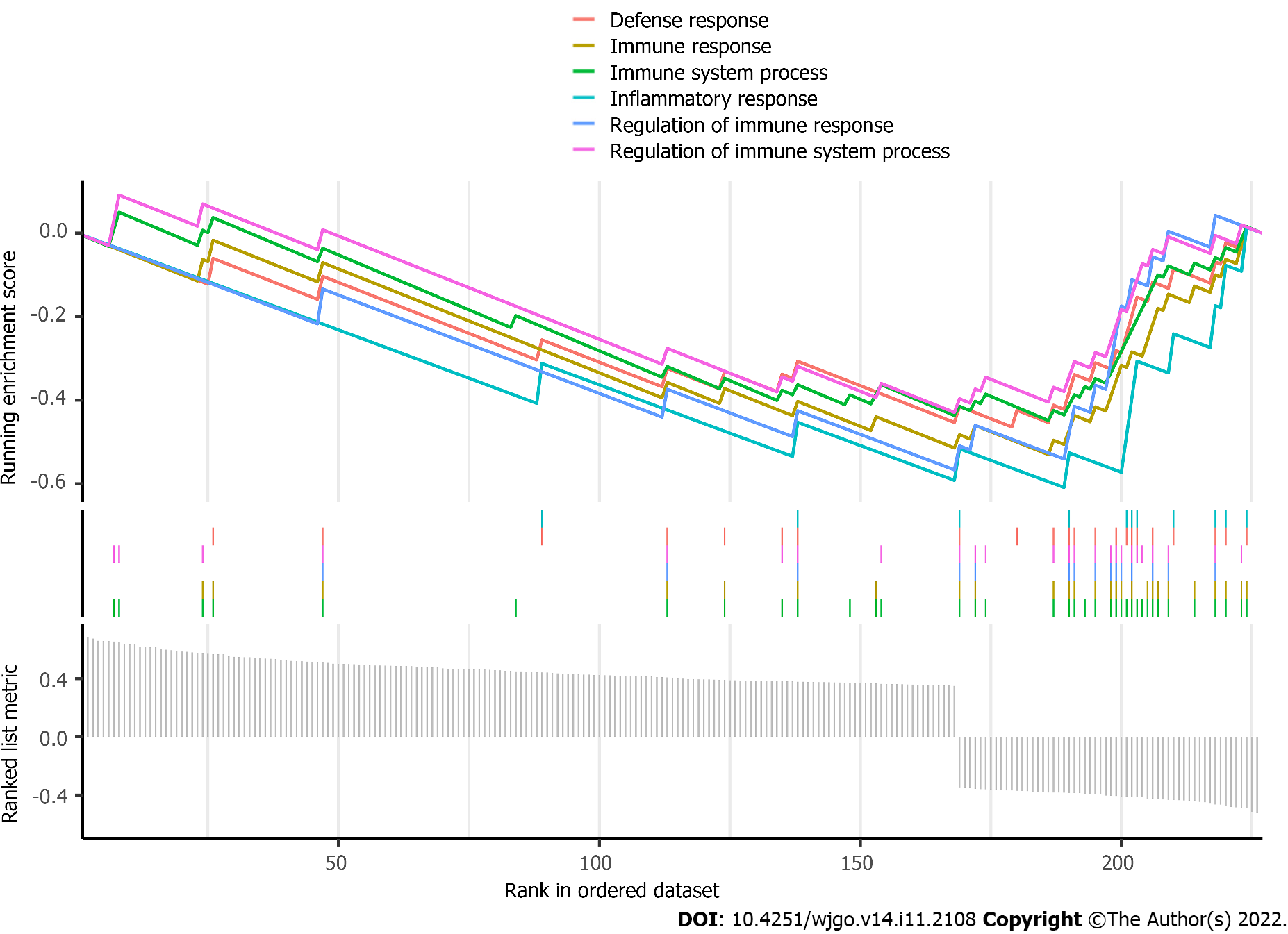
Afterward, coexpression analysis was performed on the transcriptome data from TCGA-COAD, and the genes coexpressed with RP5-881L22.5 among the differentially expressed mRNAs were selected. Through gene set enrichment analysis, it was found that the genes coexpressed with RP5-881L22.5 among the differentially expressed mRNAs were mainly involved in immune biological processes or pathways such as the immune response (Figure 4).
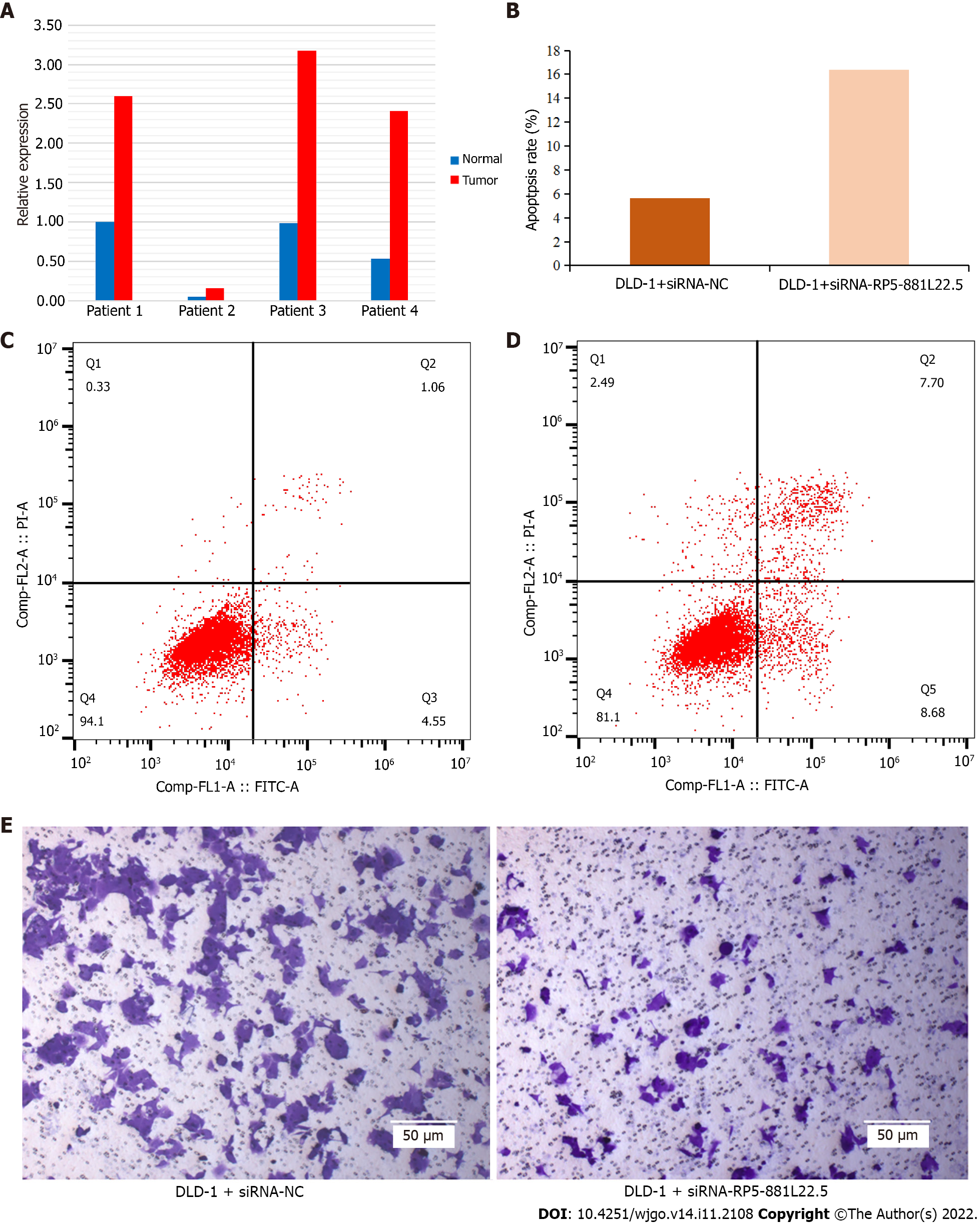
Four pairs of tissue samples from colorectal cancer patients were assessed by quantitative reverse transcription PCR. We found that RP5-881L22.5 was generally highly expressed in tumor tissues compared with adjacent normal tissues (Figure 5A), which was consistent with the aforementioned bioinformatics analysis results. The apoptosis assay showed that the apoptosis rate of colorectal cancer cells increased after RP5-881L22.5 knockdown (Figure 5B-D). The transwell invasion assay revealed reduced invasiveness (Figure 5E). These results indicated that RP5-881L22.5 is a significant contributor to the occurrence and development of colorectal cancer.
LncRNAs play important functions in many regulatory mechanisms, such as transcriptional silencing, transcriptional activation, chromosome modification and intranuclear transport and play important roles in the occurrence and development of cancer[22,23]. Liu et al[24] found that three lncRNAs, LINC00114, LINC00261 and HOTAIR, can accurately judge the prognosis of colorectal cancer. Zhang et al[25] found that lncRNA-NEAT1 interacts with DDX5 to activate Wnt/β-catenin signaling, thereby promoting the progression of colorectal cancer. Ni et al[26] found that lncRNA-GAS5 controls colorectal cancer progression by regulating the phosphorylation and interpretation of YAP. This process is also negatively regulated by the m6A methylation reader protein YTHDF3.
We observed significant differences in lncRNA expression between colon cancer tumor tissues and normal colon tissues based on the analysis of TCGA dataset. This expression difference was also observed in rectal cancer, gastric cancer and esophageal cancer. Moreover, its expression was correlated with clinical features, such as the pathological stage of patients, and exhibited a significant correlation with survival in specific populations. These findings illustrate that RP5-881L22.5 may be a unique molecular marker in digestive tract cancer and may be involved in some important processes in the pathogenic process of digestive tract cancer. Subsequent clinical specimen detection and cell function experiments also verified this conclusion. All of these findings indicate that this lncRNA is likely to be related to the pathogenic process of colorectal cancer.
In addition to tumor cell characteristics, the microenvironment of the tumor also plays a substantial role in cancer[27]. Many lncRNAs show strong cell type-specific expression phenomena. Most mRNAs are expressed in the vast majority of cell types, whereas more than half of the cells contain only a few lncRNAs[28]. Cells with malignant characteristics had 9% of lncRNAs, lymphocytes had 11%, myeloid cells had 6%, and epithelial cells had 5%[29]. In particular, immune-specific lncRNAs recruit proteins to specific genomic loci to regulate target gene expression epigenetically and transcriptionally in most cases, thereby controlling the activity and differentiation of immune cells. For example, H19, ROCKI, lnc13 and HOXA-AS2 regulate target genes in immune cells by exerting protein recruitment functions or controlling chromatin accessibility[30-33]. At the same time, the tumor immune microenvironment can also be regulated by lncRNAs by targeting macrophages, dendritic cells, regulatory T cells and CD8+ T lymphocytes through a competing endogenous RNA mechanism, such as the SNHG1/miR-448/IDO regulator network[34], the SNHG16/miR-16-5p/SMAD5 regulator network[35], the FENDR/miR-423-5p/GADD45B regulator network[36], the SBF2-AS1miR-122-5p/XIAP regulator network[37] and the NEAT1/miR-155/Tim-3 regulator network[38].
Of course, in addition to their role in immune cells, lncRNAs also modulate the presentation of antigen or PD-L1 in tumor cells[39,40]. Li et al[41] identified a lncRNA, lncRNA inducing major histocompatibility complex I and immunogenicity of tumor, in humans and mice. They proposed the lncRNA inducing major histocompatibility complex I and immunogenicity of tumor-GBP-HSF1 axis as a therapeutic target for immunotherapy to modulate major histocompatibility complex I expression based on experimental validation in vivo and in vitro. In conclusion, increasing evidence indicates that the key mechanisms by which lncRNAs regulate tumor immunity involve various aspects, such as antigen presentation and T cell exhaustion.
In our study, RP5-881L22.5 expression in colorectal cancer was also strongly related to the tumor immune microenvironment. There was a negative correlation between its expression and the presence of CD8+ T lymphocytes and activated NK cells in the tumor microenvironment, and it was negatively correlated with the expression of various coinhibitory receptors on the surface of T lymphocytes. In addition, RP5-881L22.5 coexpressed genes were more involved in the pathways of tumor immune microenvironment formation, such as immune response and immune signal transmission.
In particular, TCGA data analysis revealed that RP5-881L22.5 expression was related to some clinical features of colorectal cancer patients but not to the T stage of the tumor, and no difference in survival was noted between the high expression group and the low expression group. Furthermore, a subset of advanced stage tumors showed better survival despite high marker expression. The expression of inhibitory receptors in the RP5-881L22.5 high expression group decreased. Therefore, T cells showed relatively stronger antitumor immune activity in the high expression group. This finding may explain why advanced tumors with higher RP5-881L22.5 expression showed a better survival rate. Further research on immunotherapy of advanced tumors using RP5-881L22.5 as a marker should be explored.
RP5-881L22.5 expression levels were significantly different between colorectal cancer tissues and nontumor tissues , and RP5-881L22.5 expression in tumor samples was considerably higher. RP5-881L22.5 expression levels were significantly correlated with clinicopathological stage and could predict prognosis for colorectal cancer. Moreover, RP5-881L22.5 showed an obvious immune correlation in colorectal cancer and might be a key molecule in the formation of the immunosuppressive microenvironment, which deserves further research. The relationship between RP5-881L22.5 and the tumor immune microenvironment, as well as the different prognostic differences it represents in tumors with different clinical characteristics, needs to further investigated in clinical tissue samples.
Colorectal cancer is one of the most common malignant tumors in the world. Long noncoding RNAs (lncRNAs) are involved in many normal human physiological functions and pathogenic processes of diseases.
LncRNAs may be used as molecular markers for more accurate prediction of colorectal cancer prognosis. The molecular mechanisms of lncRNAs involved can impact colorectal cancer prognosis and impact the tumor immune microenvironment. Further research into these questions is needed.
To explore the differential expression analysis of mRNAs and lncRNAs in the colorectal cancer and to determine the association between RP5-881L22.5 expression and the tumor immune microenvironment.
We analyzed the immune cell microenvironment through the bioinformatics combined with clinical data and cell experiments to verify the results.
RP5-881 L22.5 expression led to a difference in prognosis, which warrants further research on immunotherapy of advanced tumors with RP5-881 L22.5 as a marker.
RP5-881 L22.5 plays an important role in the pathogenesis of colorectal cancer.
RP5-881 L22.5 may be an important research target for the treatment of colorectal cancer.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Batyrbekov K, Kazakhstan; Ezenkwa US; Manojlovic N, Serbia S-Editor: Chen YL L-Editor: Filipodia P-Editor: Li X
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64442] [Article Influence: 16110.5] [Reference Citation Analysis (176)] |
| 2. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 11920] [Article Influence: 2980.0] [Reference Citation Analysis (4)] |
| 3. | Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557-4566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 750] [Cited by in RCA: 2136] [Article Influence: 356.0] [Reference Citation Analysis (2)] |
| 4. | Pansy K, Uhl B, Krstic J, Szmyra M, Fechter K, Santiso A, Thüminger L, Greinix H, Kargl J, Prochazka K, Feichtinger J, Deutsch AJ. Immune Regulatory Processes of the Tumor Microenvironment under Malignant Conditions. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 5. | Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2228] [Cited by in RCA: 3058] [Article Influence: 254.8] [Reference Citation Analysis (0)] |
| 6. | Jordan SC. Innate and adaptive immune responses to SARS-CoV-2 in humans: relevance to acquired immunity and vaccine responses. Clin Exp Immunol. 2021;204:310-320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 7. | Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, Li J, Li F, Tan HB. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 936] [Article Influence: 156.0] [Reference Citation Analysis (0)] |
| 8. | O'Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019;16:151-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 1197] [Article Influence: 199.5] [Reference Citation Analysis (0)] |
| 9. | Statello L, Guo C J, Chen L L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96-118. |
| 10. | Arraiano CM. Regulatory noncoding RNAs: functions and applications in health and disease. FEBS J. 2021;288:6308-6309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Pi YN, Qi WC, Xia BR, Lou G, Jin WL. Long Non-Coding RNAs in the Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential. Front Immunol. 2021;12:697083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 12. | Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, Zhu J, Haussler D. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2228] [Cited by in RCA: 2557] [Article Influence: 511.4] [Reference Citation Analysis (0)] |
| 13. | Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19:A68-A77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 908] [Cited by in RCA: 2030] [Article Influence: 203.0] [Reference Citation Analysis (0)] |
| 14. | Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766-D773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2216] [Cited by in RCA: 2084] [Article Influence: 347.3] [Reference Citation Analysis (0)] |
| 15. | Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16184] [Cited by in RCA: 25600] [Article Influence: 2560.0] [Reference Citation Analysis (0)] |
| 16. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3057] [Article Influence: 509.5] [Reference Citation Analysis (0)] |
| 17. | eGTEx Project. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet. 2017;49:1664-1670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 140] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 18. | Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4763] [Cited by in RCA: 8878] [Article Influence: 887.8] [Reference Citation Analysis (0)] |
| 19. | Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3056] [Cited by in RCA: 6426] [Article Influence: 584.2] [Reference Citation Analysis (0)] |
| 20. | Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2189] [Cited by in RCA: 3204] [Article Influence: 320.4] [Reference Citation Analysis (0)] |
| 21. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 4723] [Article Influence: 1180.8] [Reference Citation Analysis (0)] |
| 22. | Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2088] [Cited by in RCA: 2156] [Article Influence: 269.5] [Reference Citation Analysis (0)] |
| 23. | Lorenzi L, Avila Cobos F, Decock A, Everaert C, Helsmoortel H, Lefever S, Verboom K, Volders PJ, Speleman F, Vandesompele J, Mestdagh P. Long noncoding RNA expression profiling in cancer: Challenges and opportunities. Genes Chromosomes Cancer. 2019;58:191-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 24. | Liu S, Cao Q, An G, Yan B, Lei L. Identification of the 3-lncRNA Signature as a Prognostic Biomarker for Colorectal Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 25. | Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, Xu M, Sun H, Liu C, Wei P, Du X. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11:113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 283] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 26. | Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 454] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 27. | Grassi L, Izuogu OG, Jorge NAN, Seyres D, Bustamante M, Burden F, Farrow S, Farahi N, Martin FJ, Frankish A, Mudge JM, Kostadima M, Petersen R, Lambourne JJ, Rowlston S, Martin-Rendon E, Clarke L, Downes K, Estivill X, Flicek P, Martens JHA, Yaspo ML, Stunnenberg HG, Ouwehand WH, Passetti F, Turro E, Frontini M. Cell type-specific novel long non-coding RNA and circular RNA in the BLUEPRINT hematopoietic transcriptomes atlas. Haematologica. 2021;106:2613-2623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 612] [Article Influence: 76.5] [Reference Citation Analysis (0)] |
| 29. | Park EG, Pyo SJ, Cui Y, Yoon SH, Nam JW. Tumor immune microenvironment lncRNAs. Brief Bioinform. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 30. | Zhou J, Xu J, Zhang L, Liu S, Ma Y, Wen X, Hao J, Li Z, Ni Y, Li X, Zhou F, Li Q, Wang F, Wang X, Si Y, Zhang P, Liu C, Bartolomei M, Tang F, Liu B, Yu J, Lan Y. Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals that H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development. Cell Stem Cell. 2019;24:285-298.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 31. | Zhang Q, Chao TC, Patil VS, Qin Y, Tiwari SK, Chiou J, Dobin A, Tsai CM, Li Z, Dang J, Gupta S, Urdahl K, Nizet V, Gingeras TR, Gaulton KJ, Rana TM. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 32. | Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, Schneider R, Kiledjian M, Bilbao JR, Ghosh S. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 196] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 33. | Zhao H, Zhang X, Frazão JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem. 2013;114:2375-2383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 34. | Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol. 2018;118:24-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 35. | Ni C, Fang QQ, Chen WZ, Jiang JX, Jiang Z, Ye J, Zhang T, Yang L, Meng FB, Xia WJ, Zhong M, Huang J. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct Target Ther. 2020;5:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 193] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 36. | Yu Z, Zhao H, Feng X, Li H, Qiu C, Yi X, Tang H, Zhang J. Long Non-coding RNA FENDRR Acts as a miR-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids. 2019;17:516-529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 37. | Yin Z, Zhou Y, Ma T, Chen S, Shi N, Zou Y, Hou B, Zhang C. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J Cell Mol Med. 2020;24:5028-5038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 38. | Zhang M, Zheng Y, Sun Y, Li S, Chen L, Jin X, Hou X, Liu X, Chen Q, Li J, Liu M, Zheng X, Zhang Y, Wu J, Yu B. Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome. Theranostics. 2019;9:3425-3442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 39. | Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, Egranov SD, Zhang Y, Xia W, Gong J, Pan Y, Chatterjee SS, Yao J, Evans KW, Nguyen TK, Park PK, Liu J, Coarfa C, Donepudi SR, Putluri V, Putluri N, Sreekumar A, Ambati CR, Hawke DH, Marks JR, Gunaratne PH, Caudle AS, Sahin AA, Hortobagyi GN, Meric-Bernstam F, Chen L, Yu D, Hung MC, Curran MA, Han L, Lin C, Yang L. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. 2019;20:835-851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 256] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 40. | Jiang W, Pan S, Chen X, Wang ZW, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer. 2021;20:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 41. | Li G, Kryczek I, Nam J, Li X, Li S, Li J, Wei S, Grove S, Vatan L, Zhou J, Du W, Lin H, Wang T, Subramanian C, Moon JJ, Cieslik M, Cohen M, Zou W. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat Cell Biol. 2021;23:526-537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 119] [Article Influence: 29.8] [Reference Citation Analysis (0)] |