Published online Oct 15, 2022. doi: 10.4251/wjgo.v14.i10.1918
Peer-review started: May 7, 2022
First decision: July 13, 2022
Revised: July 23, 2022
Accepted: September 8, 2022
Article in press: September 8, 2022
Published online: October 15, 2022
Processing time: 160 Days and 9.1 Hours
Advanced colorectal cancer (CRC) generally has poor outcomes and high mor
To confirm whether GTF3A promotes CRC progression by regulating the expression of cystatin A (Csta) gene and investigate whether GTF3A can serve as a prognostic biomarker and therapeutic target for patients with CRC.
Human tissue microarrays containing 90 pairs of CRC tissues and adjacent non-tumor tissues, and human tissue microarrays containing 20 pairs of CRC tissues, adjacent non-tumor tissues, and metastatic tissues were examined for GTF3A expression using immunohistochemistry. The survival rates of patients were analyzed. Short hairpin GTF3As and CSTAs were designed and packaged into the virus to block the expression of Gtf3a and Csta genes, respectively. In vivo tumor growth assays were performed to confirm whether GTF3A promotes CRC cell proliferation in vivo. Electrophoretic mobility shift assay and fluorescence in situ hybridization assay were used to detect the interaction of GTF3A with Csta, whereas luciferase activity assay was used to evaluate the expression of the Gtf3a and Csta genes. RNA-Sequencing (RNA-Seq) and data analyses were used to screen for target genes of GTF3A.
The expression of GTF3A was higher in CRC tissues and lymph node metastatic tissues than in the adjacent normal tissues. GTF3A was associated with CRC prognosis, and knockdown of the Gtf3a gene impaired CRC cell proliferation, invasion, and motility in vitro and in vivo. Moreover, RNA-Seq analysis revealed that GTF3A might upregulate the expression of Csta, whereas the luciferase activity assay showed that GTF3A bound to the promoter of Csta gene and increased Csta transcription. Furthermore, CSTA regulated the expression of epithelial-mesenchymal transition (EMT) markers.
GTF3A increases CSTA expression by binding to the Csta promoter, and increased CSTA level promotes CRC progression by regulating the EMT. Inhibition of GTF3A prevents CRC pro
Core Tip: Transcriptional factor III A (GTF3A) is highly expressed in colorectal cancer (CRC) tissues, and GTF3A expression is associated with CRC prognosis. GTF3A binds to the promoter of cystatin (Csta) gene to facilitate Csta transcription, which regulates the expression of epithelial-mesenchymal transition markers and promotes CRC progression. Blocking GTF3A significantly inhibits CRC cell growth. Therefore, GTF3A is a potential novel therapeutic target and prognostic biomarker for CRC.
- Citation: Wang J, Tan Y, Jia QY, Tang FQ. Transcriptional factor III A promotes colorectal cancer progression by upregulating cystatin A. World J Gastrointest Oncol 2022; 14(10): 1918-1932
- URL: https://www.wjgnet.com/1948-5204/full/v14/i10/1918.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i10.1918
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the leading cause of cancer-related death worldwide[1]. CRC has distant invasive and metastatic abilities, such as liver and lung metastases, resulting in a poor survival[2]. Clarifying the molecular mechanisms underlying CRC progression is necessary for the development of new therapeutic strategies. Transcriptional factor III A (GTF3A), an RNA polymerase III transcriptional factor, specifically binds to the internal control region of the 5S rRNA gene from nucleotides +43 to +96[3], initiating the assembly of the transcription initiation complex (5S rDNA-TFIIIA-TFIIIC2-TFIIIBbeta complex). Complex formation is not proportional to the amount of TFIIIA[4]. GTF3A is present in all human organs. GTF3A shares a common conserved transcription activating signal, nuclear localization signal, and nuclear export signal, but lacks initiated Met and accompanying conserved residues in the N-terminal regions[5]. GTF3A regulates the 5S rRNA synthesis network by binding to 5S rDNA and 5S rRNA[6]. GTF3A binds to 5S rRNA to form the 7S ribonucleoprotein particle (RNP) complex, and the complex functions as a nuclear export signal (NES) to transfer 5S rRNA to the cytoplasm depending on the NES sequence, which protects the 5S rRNA from degradation[7,8]. Several studies have suggested that 5S rRNA binds to L5 and L11 to form the 5S RNP complex, regulating the MDM2-p53 checkpoint[9-12]. Alterations in ribosome biogenesis are critical drivers of tumorgenesis, and are closely associated with increased CRC cell growth[13]. These findings suggest that GTF3A regulates CRC progression.
Cystatin A (CSTA), a cysteine proteinase inhibitor, is a type 1 cystain (stefin). CSTA is a cornified cell envelope constituent of keratinocytes that plays a critical role in epidermal development and maintenance. The high expression of CSTA is associated with the invasion and metastasis of various malignant tumors, such as pancreatic ductal adenocarcinoma[14], esophageal squamous cell carcinoma[15], lung cancer[16], hepatocellular carcinoma (HCC)[17], and nasopharyngeal carcinoma (NPC)[18]. An increasing number of studies have shown that CSTA is a potential prognostic and diagnostic biomarker for cancer progression. The activity of the Csta promoter is positively regulated by the active Ras/MEKK1/MKK7/JNK signal transduction pathway, but negatively regulated by the negative Ras/Raf-1/MEK1/ERK pathway in human keratinocytes[19]. Other cysteine protease inhibitors, cystatin SN (CST1) and cystatin S (CST4), are type 2 cystatin proteins, which enhance the metastasis of various malignant tumors and contribute to a poor patient survival[20,21]. CST1 overexpression increases cell migration and invasion by mediating the epithelial-mesenchymal transition (EMT) in breast cancer and HCC[22,23] and contributes to CRC cell proliferation[24]. CST1 is also considered an early diagnostic biomarker and potential therapeutic target in breast cancer, CRC, and gastric cancer[25,26]. In the present study, we showed that GTF3A was highly expressed in CRC, and it bound to the promoter of Csta to facilitate Csta transcription, which then regulated EMT marker expression and promoted CRC progression. Therefore, GTF3A is a potential novel therapeutic target and a prognostic biomarker for CRC.
Dulbecco’s modified Eagle’s medium (DMEM), Cell Counting Kit (CCK8), and other supplements were obtained from Life Technologies (Rockville, MD, United States). GTF3A antibody for Western blot analysis was purchased from Bethyl Laboratories, Inc (Suzhou, China). CSTA antibody was purchased from Novus (CO, United States). CST1 antibody was purchased from Invitrogen (Shanghai, China). CST4 antibody was purchased from R&D Systems (Minneapolis, MN, United States). GTF3A antibody used for the immunofluorescence assay was purchased from Bioss Antibodies (Beijing, China). The dual-luciferase reporter assay system was purchased from Promega (Madison, WI, United States). Antibodies against Snail, E-cadherin, and beta-catenin were purchased from Abcam (Cambridge, United Kingdom). Glyceraldehydes-3-phosphate dehydrogenase (GAPDH) and secondary antibodies were purchased from Proteintech Company (Wuhan, Hubei, China).
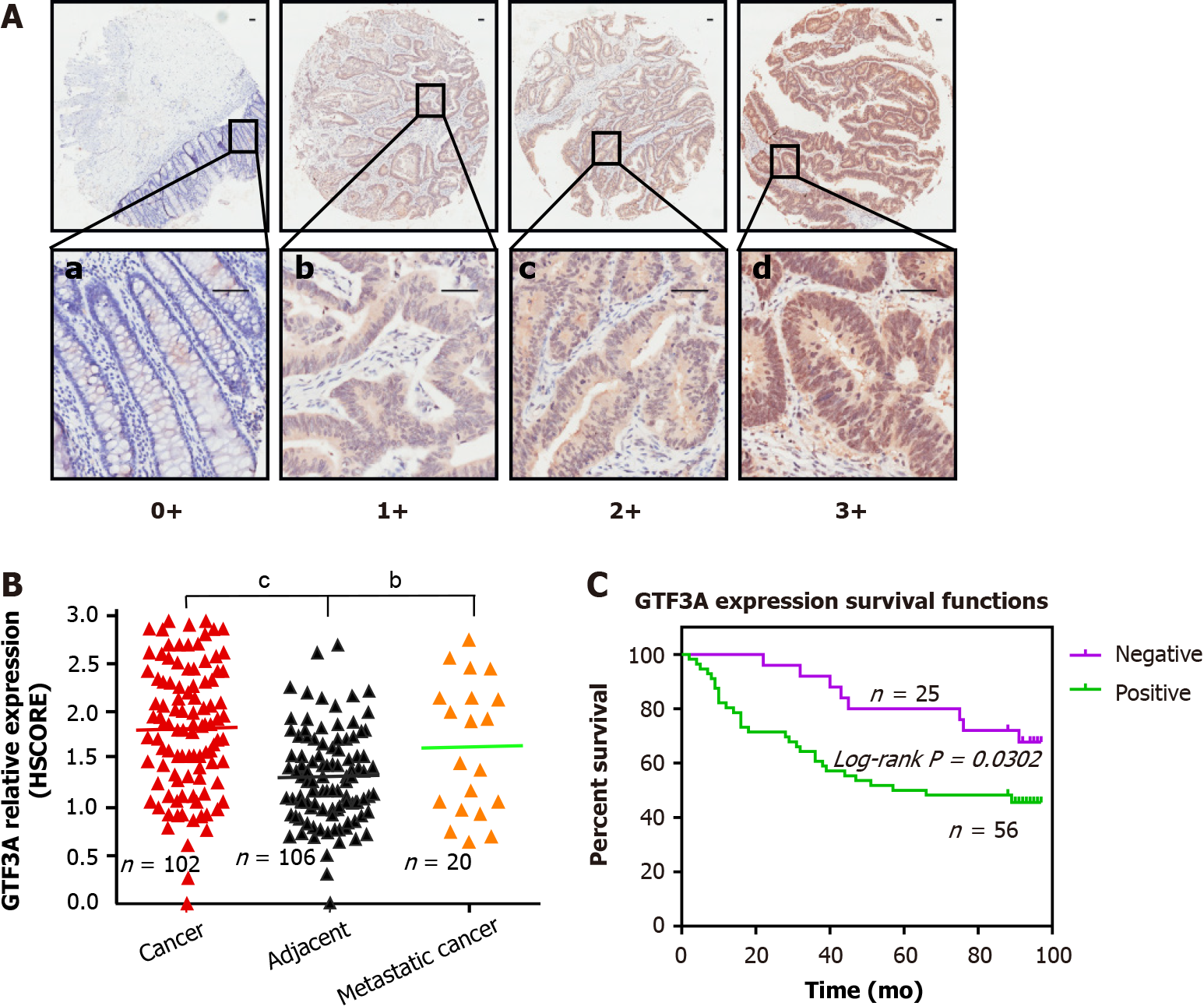
Human tissue microarrays (HCol-Ade180Sur-08) containing 90 pairs of CRC tissues and adjacent non-tumor tissues, and human tissue microarrays (HCol-Ade060Lym-01) containing 20 pairs of CRC tissues, adjacent non-tumor tissues, and lymph node metastatic tissues, were purchased from Outdo Biotech Company (Shanghai, China). Immunohistochemical (IHC) staining was performed to detect the expression of GTF3A as described previously[27]. These tissue microarrays were stained with GTF3A antibody (1:300 dilution). The use of patient materials was approved by the ethics committee of Hunan Cancer Hospital (No. KYJJ-2020-004). All IHC results were evaluated based on the semi-quantitative histological scoring (HSCORE) system using the following formula: H-SCORE = ∑ (pi × i) [“pi” represents the percentage of stained cells in an intensity area, whereas “I” represents the staining intensity (0, no labeling; 1, weak; 2, moderate; and 3, strong)].
Human CRC cell lines (HCT116, SW480, DLD1, SW620, and HT29) were obtained from the American Type Culture Collection (Manassas, VA, United States) and were purchased from the Shanghai Cell Center (Shanghai, China). All cell lines were cultured in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS) at 37 °C with 5% CO2.
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed as described previously[28]. Briefly, the total RNA of cultured cells was extracted using a total RNA kit (R6834, OMEGA), and 1 μg of DNase-treated RNA was reverse transcribed using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Shanghai, China) according to the manufacturer’s instructions. The threshold cycle (Ct) values were measured using Hieff quantitative PCR SYBR Green Mix (Yeasen, Shanghai, China) in A LightCycler 96 qPCR System (Roche). Primer sequences used are listed in Supplementary Table 1. The relative mRNA levels of each gene were normalized to those of the housekeeping gene GAPDH. Relative transcript levels were calculated as two power values of ΔCt (the differential value of Ct between GAPDH and the target cDNA).
The designed short hairpin RNAs (shRNAs) shGTF3A#1, shGTF3A#2, shGTF3A#3, shGTF3A#4, and shscramble were cloned into the lentivirus vector GV112 by Shanghai GeneKai Company (Shanghai, China). The shRNA sequences for Gtf3a are listed in Supplementary Table 2. The shRNAs for the Csta gene were designed to knock down the expression of CSTA. The shRNA sequences for Csta are shown in Supplementary Table 3. Synthetic plasmids were verified by sequence analysis and PCR and then cloned into GV493. Lipofectamine 3000 (Thermo Fisher Scientific, Carlsbad, CA, United States) was used to transfect the plasmids into HEK293T cells; 1 × 105 cells were transfected with shscramble, shGTF3A#1, shGTF3A#2, shGTF3A#3, shGTF3A#4, shCSTA#1, and shCSTA#2. The knockdown efficiency was filtered using RT-qPCR.
shGTF3As, shCSTAs, and negative control lentiviruses were packaged by the Shanghai GeneKai Company (Shanghai, China). The lentivirus titers were quantified (≥ 108 TU/mL). Following the manufacturer’s protocol, 2 × 105 of SW480 and HCT116 cells were seeded in six-well culture plates, and cultivated at 37 °C and 5% CO2. After 24 h, the cells were transfected with the appropriate amount of lentivirus at a multiplicity of infection of 20 TU/mL. After being incubated for 10 h, the culture medium was removed, and fresh DMEM containing 10% FBS was added. Next, antibiotic-free medium containing 3 μg/mL puromycin was used to screen the stable cells for 3-4 wk. Ultimately, the knockdown and negative control stable cell lines shscramble-SW480, shGTF3A#1-SW480, shGTF3A#4-SW480, shGTF3A#1-HCT116, and shGTF3A#4-HCT116 were obtained.
Western blotting was performed as previously described[29]. Briefly, 1 × 106 cells were lysed with a radioimmunoprecipitation assay lysis buffer [50 mmol/L Tris pH 7.4, 150 mmol/L NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS)] containing 100 × protease inhibitor cocktail and 100 × phosphatase inhibitor cocktail (CWBIO, Beijing, China). Following the manufacturer’s instructions, the crude lysate was centrifuged and the supernatant was collected to measure the protein concentration using the BCA Protein Assay Kit (CWBIO, Beijing, China). After being boiled at 100 °C for 5 min, 20-60 μg of protein was separated by 10% SDS- polyacrylamide gel electrophoresis and transferred to a 0.2 μm PVDF membrane (Millipore). The protein membrane was blocked with 5% non-fat milk, incubated with the primary antibody, and incubated with an appropriate peroxidase conjugated secondary antibody. The signal was detected on a gel imager using an enhanced chemiluminescence (ECL) Western blotting kit (CWBIO, Beijing, China). GAPDH was used as an internal control to verify basal expression. The ratio of specific proteins to GAPDH was calculated.
shGTF3A#1-HCT116, shGTF3A#4-HCT116, shscramble-HCT116, shGTF3A#1-SW480, shGTF3A#4-SW480, and shscramble-SW480 cells were seeded in 96-well plates, and CCK8 (ApexBio) was used to examine cell viability following the manufacturer’s protocol. Briefly, CCK8 was added to the cell plates, and incubated for 4 h. Then, the optical density at 450 nm was measured using a microplate reader. Cell viability was calculated daily for 5 d. For the colony formation assay, shGTF3A#1-HCT116, shGTF3A#4-HCT116, shscramble-HCT116, shGTF3A#1-SW480, shGTF3A#4-SW480, and shscramble-SW480 cells were seeded in 6-well plates with each well containing 1000 cells, and cultured for 16 d. The cell colonies were fixed in methanol, stained with 0.5% gentian violet, and counted automatically using a computerized microscope system.
Cell invasion and motility assays in vitro were performed using previously described methods[29]. Briefly, for the invasion assay, Matrigel (25 mg/50mL, Collaborative Biomedical Products, Bedford, MA, United States) was added to the upper chamber with 8 mm pore polycarbonate membrane filters. shGTF3A#1-HCT116, shGTF3A#4-HCT116, shscramble-HCT116, shGTF3A#1-SW480, shGTF3A#4-SW480, and shscramble-SW480 cells were seeded in the upper chamber (Neuro Probe, cabin John, MD) at a density of 1.5 × 104 cells/well in 100 μL of serum-free medium, and then incubated at 37 °C for 48 h. The bottom chamber contained standard medium with 20% FBS. Cells that invaded the lower surface of the membrane were fixed with 37% paraformaldehyde, and stained with crystal violet. Invading cells were counted under a light microscope. The motility assay was performed in a similar manner to the invasion assay without Matrigel coating.
A wound healing assay was used to measure the cell migration potential. Briefly, 2 × 105 of shGTF3A#1-HCT116, shGTF3A#4-HCT116, shscramble-HCT116, shGTF3A#1-SW480, shGTF3A#4-SW480, and shscramble-SW480 cells were seeded in 6-well plates. After the cells reached 95% confluence, the surface of the cell layer was wounded using a sterile 10 μL pipette tip. The cells were then rinsed three times with phosphate buffered saline (PBS) to move detached cells and incubated in DMEM containing 1% FBS for 48 h. Wound closure was observed under a microscope at 0, 24, and 48 h.
In vivo tumor growth assays were performed as previously described[30]. Briefly, female nude mice (aged 4-5 wk) were obtained from Hunan SJA Laboratory Animal Co. Ltd. (Changsha, China). Experiments involving animal subjects and protocols for animal studies were approved by the Laboratory Animal Research Center of Hunan Cancer Hospital (No. 2020-118). Nude mice were subcutaneously injected with 3 × 106 shGTF3A#1-SW480, shGTF3A#4-SW480, or shscramble-SW480 cells (5 mice per group). The size of the tumor that developed in the mice was measured every 3 d, and a tumor growth curve was drawn. After 4 wk, the mice were euthanized with pentobarbital sodium at 20 mg/mI, and the tumor weights were measured.
After harvesting shGTF3A#1-SW480, shGTF3A#4-SW480, and shscramble-SW480 cells, total RNA was extracted using TRIzol reagent (Invitrogen). After the rRNA was removed, the enriched longRNA (> 200 nt) was interrupted, reversely transcribed into cDNA, and repaired, and RNA sequencing (RNA-Seq) was performed to build a chain-specific database. The clean data were obtained by quality testing of Bioptic Qsep100, and then comparative mapping of genomes was acquired between clean data and the hg38 Ensemble transcriptome using HISAT2 software. The differentially expressed genes between groups were analyzed using the DESeq2 R package, and the default screening criteria were: (1) Log2 (fold change) >1; and (2) False discovery rate < 0.05.
Fluorescence in situ hybridization (FISH) assays were performed as previously described[31]. FISH probes for Csta promoter sequence were designed and synthesized by Well Bio (Guangzhou, China). The IncRNA FISH kit (C10910) was purchased from Wellbio (Guangzhou, China). The cell slides were fixed with 4% paraformaldehyde for 30 min at 25 °C, permeabilized with proteinase K for 30 min at 37 °C, and blocked for 30 min at 37 °C with 200 μL of pre-hybridization solution. After removing the pre-hybridization solution, 100 μL of the probe hybridization solution was added to the cell slides overnight at 37 °C in the dark. After being washed three times for 5 min each with 4 × SSC, 2 × SSC, 1 × SSC, and PBS, the cell slides were incubated with the appropriately diluted primary antibody (anti-GTF3A) overnight at 4 °C. After washing three times with PBS, the slides were incubated with the secondary antibody (fluorescence labelled anti-rabbit immunoglobulin G) for 90 min at 37 °C. Finally, the slides were stained with 4’,6-diamidino-2-phenylidole for 10 min at 37 °C and sealed with 90% glycerin. All images were acquired using a fluorescence microscope.
A biotin labeled Csta promoter DNA probe (5’-biotin-agctagtgacgccttttaaaacacgt ccaccattccttcctttttttc-3’) was synthesized by Sangon (Shanghai, China). Both sense and antisense strands were diluted to a concentration of 0.2 μM, mixed at 1:1, denatured at 95 °C for 5 min, maintained at 70 °C for 20 min, and formed into a double-stranded probe (diluted 10 times when used) after being cooled to room temperature. Following the instructions of the electrophoretic mobility shift assay (EMSA) kit (CWBIO, Beijing, China), the protein supernatant for each sample was separated on a 4% polyacrylamide gel, and transferred to a nylon membrane (CWBIO). The protein membrane was cross-linked using UV radiation, blocked with a confining liquid, and incubated with the indicated streptavidin-horseradish peroxidase conjugate. The signal was detected on a gel imager using the ECL Western blotting kit (CWBIO, Beijing, China).
A dual-luciferase reporter assay system (Promega) was used to detect the expression of Csta following the manufacturer’s instructions. Briefly, the Csta promoter was cloned into the firefly luciferase plasmid, and Csta promoter-luc was obtained. Then, 1 × 105 of HCT116 cells were cultured in 48-well plates. Csta promoter-luc, plasmid-Renilla, and plasmid-GTF3A were co-transfected into HCT116 cells with X-tremeGene HP. After 48 h of culture, the cells were harvested, lysed with Passive Lysis Buffer, and treated with Luciferase Assay Reagent. Firefly luciferase and renilla luciferase were measured. The ratio of firefly luciferase to renilla luciferase represents the expression of Csta.
All experiments were performed at least three times. The results are presented as the mean ± SD of three independent experiments and were analyzed using Student’s t-test. The differences between groups are reported as follows: aP < 0.05, bP < 0.01, cP < 0.001, and P > 0.05. All statistical data were calculated using GraphPad Prism software (version7.0).
To determine the expression of GTF3A in CRC tissues, two sets of CRC tissue microarray were used to detect GTF3A expression by IHC. GTF3A was found to have higher expression in the cancer tissues than in the adjacent tissues of HCol-Ade180Sur-08 microarray. To further probe whether metastatic cancers had higher expression and analyze the association of GTF3A expression with metastasis, HCol-Ade060Lym-01 microarray containing CRC cancer, metastatic tissues, and adjacent tissues was also used to detect GTF3A, and observe GTF3A expression in metastatic tissues. These two microarrays had a total of 110 cancer tissues, 110 adjacent tissues, and 20 metastatic tissues, some of which were chipped off and could not be used; 102 cases in the cancer group, 106 in the adjacent group, and 20 in the metastatic group were calculated by gray scanning and scored, and survival time and survival curve were analyzed. The expression of GTF3A in CRC and metastatic tissues was higher than that in adjacent normal tissues (Figures 1A and B, P < 0.01). To analyze whether GTF3A expression was relevant to survival time, patients with CRC were divided into GTF3A negative and positive groups, and the survival curve of CRC patients showed that the negative group had a longer overall survival than the positive group (Figure 1C, P < 0.05). These clinical data suggested that GTF3A expression is associated with CRC progression.
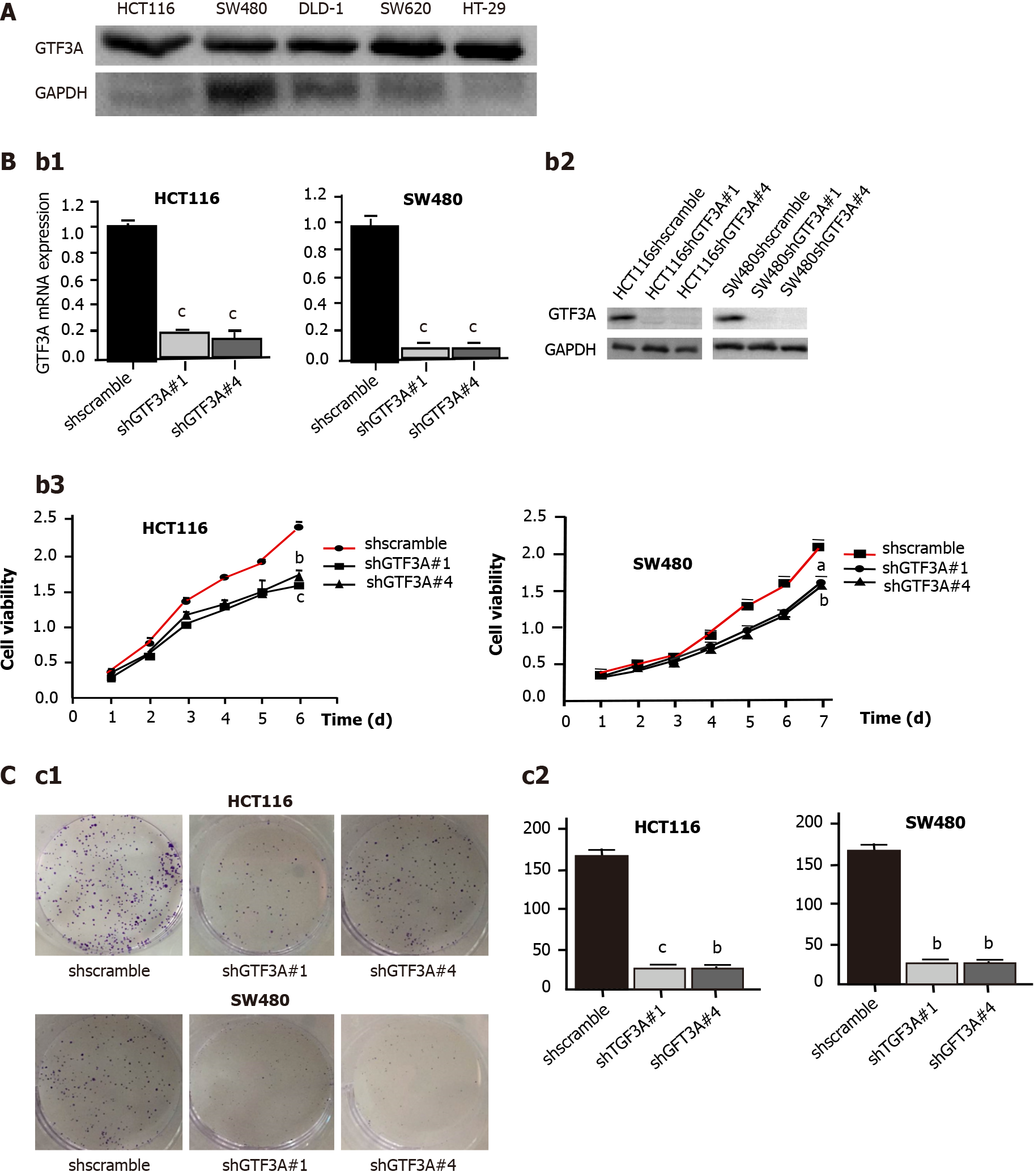
Five CRC cell lines, HCT116, SW480, DLD1, SW620, and HT-29, were used to detect the expression of GTF3A using Western blot. The results showed that SW480 cells had high expression of GTF3A, whereas HCT116, DLD1, SW620, and HT-29 cells had low expression of GTF3A (Figure 2A). To clarify the role of Gtf3a in CRC, shGTF3A#1, #2, #3, and #4 were designed and packaged into the virus. Their inhibitory effects on Gtf3a were screened, and the results showed that shGTF3A#1 and shGTF3A#4 had high knockdown efficiencies. HCT116 and SW480 cells were stably transfected with shscramble, shGTF3A#1, or shGTF3A#4. RT-qPCR was performed to evaluate knockdown efficiency, and the results showed that shGTF3A#1 and shGTF3A#4 induced effective knockdown of Gtf3a in HCT116 and SW480 cells (Figure 2B). Consistently, Western blot results showed that GTF3A protein expression was effectively decreased in shGTF3A#1 and #4-HCT116 and shGTF3A#1 and #4-SW480 cells (Figure 2B). The cell viability of HCT116 and SW480 cells was detected using the CCK8 assay after knockdown of the Gtf3a gene, and the cell proliferation of HCT116 and SW480 cells was significantly decreased in the knockdown group (Figure 2B). Furthermore, the cell colony formation assay showed that colony size and number were dramatically diminished in Gtf3a-knockdown cells (Figure 2C, P < 0.01). These data indicated that knockdown of Gtf3a inhibits the growth of CRC cells.
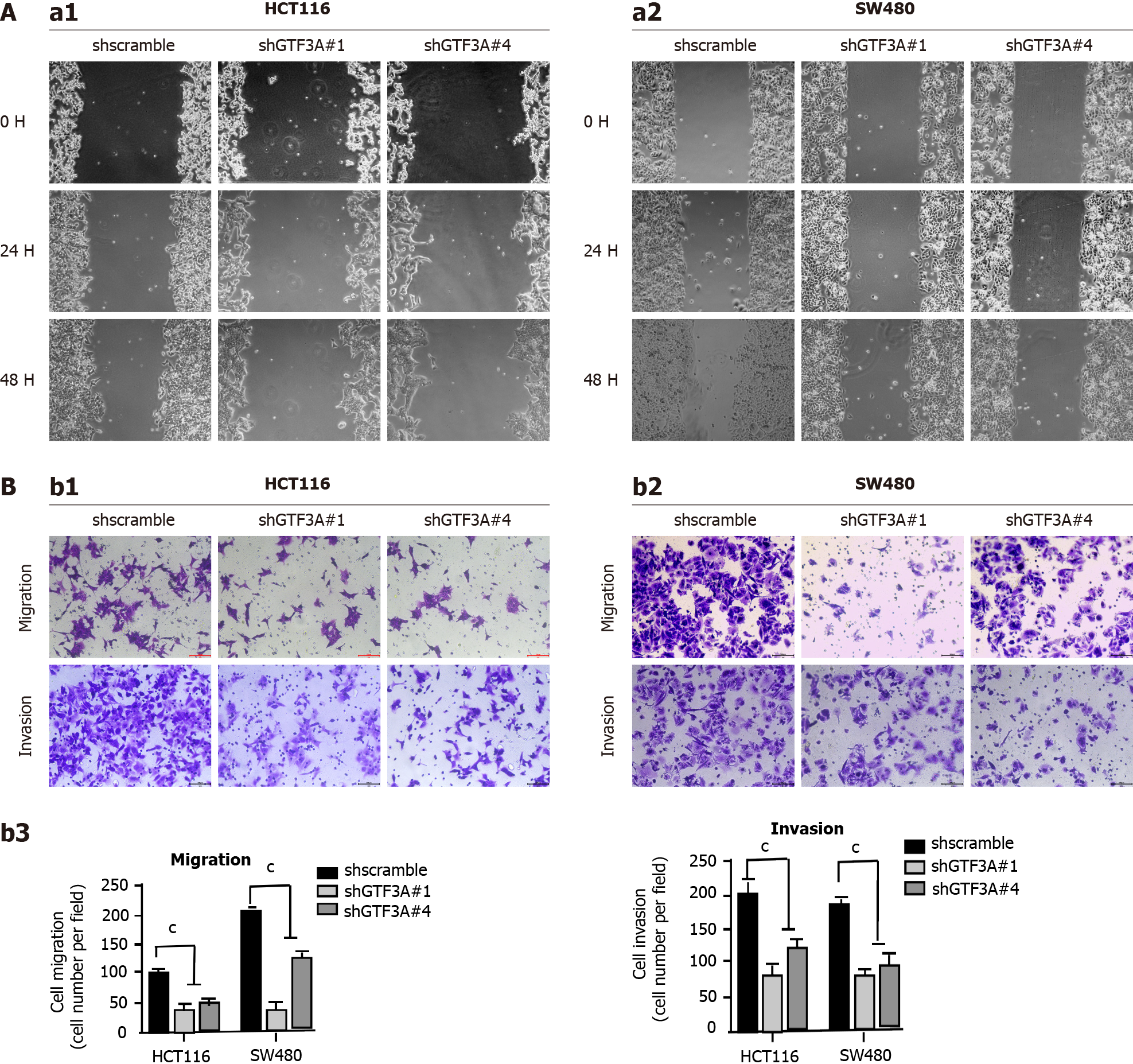
Gtf3a-knockdown SW480 and HCT116 cells were used to determine whether Gtf3a is involved in CRC cell motility and invasion. Results of the wound healing assay showed that shGTF3A#1 and #4-HCT116 and shGTF3A#1 and #4-SW480 cells had impaired migratory capability compared with shscramble cells (Figure 3A). In addition, the transwell assay showed that invasion and metastasis in the Gtf3a-knockdown groups were significantly repressed compared with those in the controls (Figure 3B), and the number of invaded or migrated cells in the knockdown groups dramatically decreased (Figure 3B). Collectively, these results highlighted that knockdown of Gtf3a suppressed CRC cell invasion and metastasis in vitro, whereas the Gtf3a gene promoted the progression of CRC.
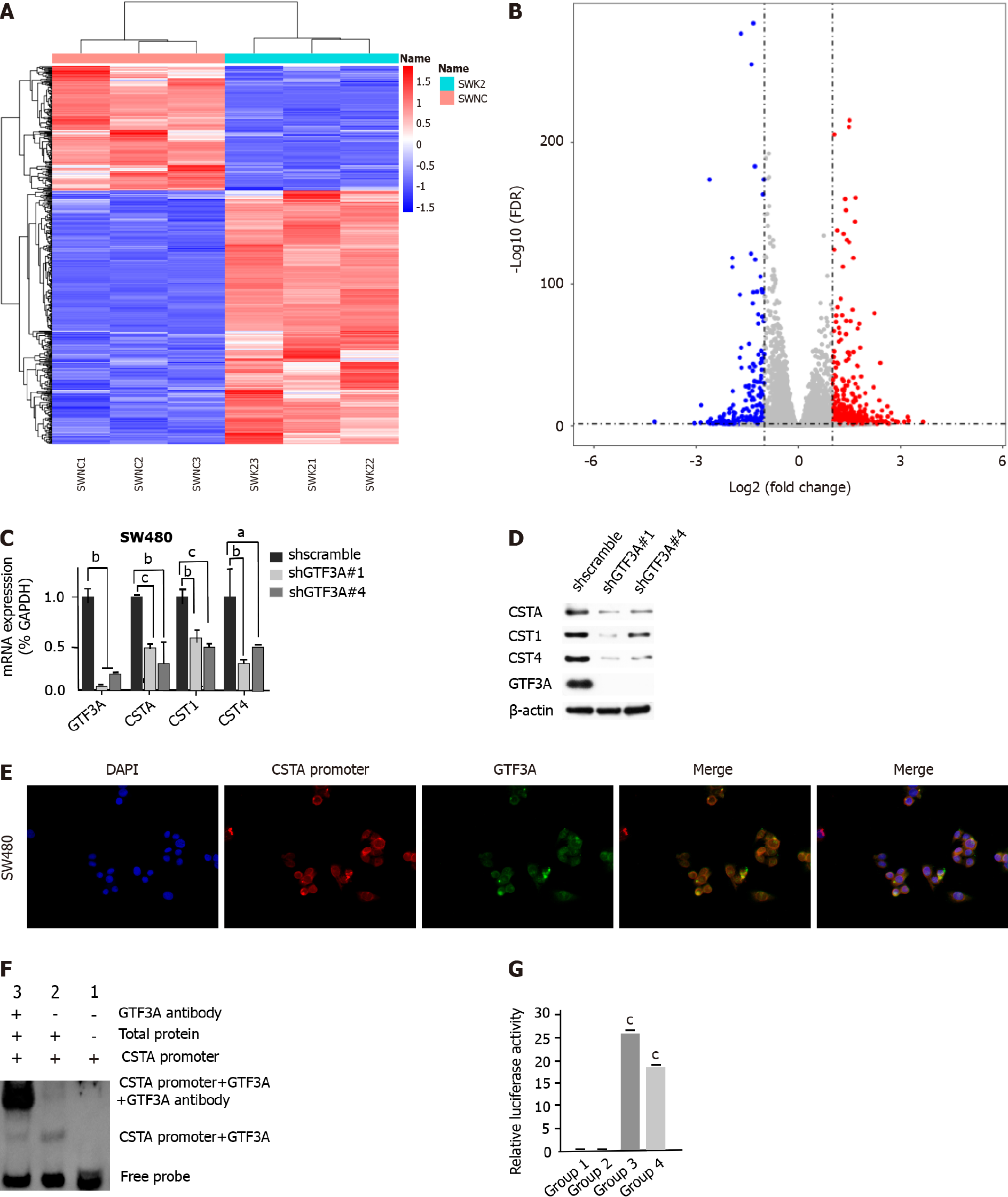
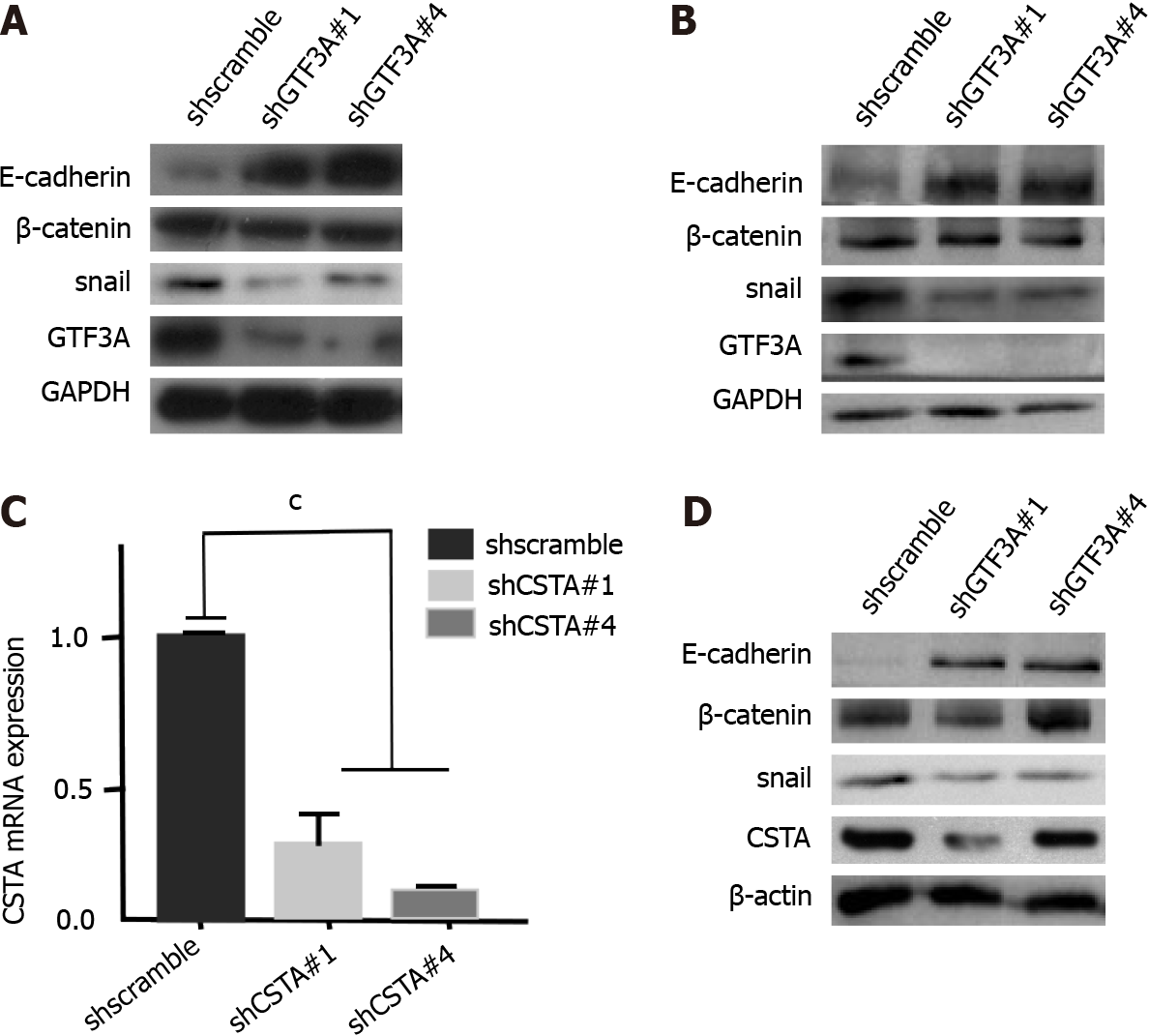
To explore the molecular mechanisms of GTF3A in CRC progression, RNA-Seq was used to screen the target genes of GTF3A, and the differential genes between SW480 Gtf3a-knockdown and scramble control cells were analyzed. The RNA-Seq results are shown in Figures 4A and 4B; several differentially expressed genes were found between the groups (Figures 4A and 4B, Supplementary Table 4). In particular, the expression of Csta/Cst1/Cst4 genes in Gtf3a-knockdown cells was dramatically lower than that in the control group (Supplementary Table 4). Csta/Cst1/Cst4 genes, members of the cystatin superfamily that encode cysteine protease inhibitors, are overexpressed in various types of cancers, subsequently promoting cancer cell metastasis and resulting in a poor prognosis[14-16]. RT-qPCR and Western blot results showed that the expression of Csta/Cst1/Cst4 genes was significantly decreased in Gtf3a-knockdown cells (Figures 4C and 4D).
The results of the RNA-Seq showed that CSTA had the largest difference after knockdown of Gtf3a in FISH experiments; the sequence probe of the Csta promoter was labeled with red fluorescence, whereas GTF3A was labeled with green fluorescence. The fluorescence staining of the GTF3A and Csta promoters was colocalized to a large extent as indicated by an orange-yellow fused fluorescence (Figure 4E), suggesting that GTF3A binds with the Csta promoter. To confirm the interaction between GTF3A and the Csta promoter, EMSA was performed to directly observe the interaction of GTF3A and the Csta promoter. The results showed that GTF3A interacted with the Csta promoter (Figure 4F). Next, a dual-luciferase assay was carried out to determine whether the interaction of GTF3A with Csta promoter increased Csta expression, and the results showed that the transcript activity of the Csta gene was significantly increased after transfection with CSTA (Figure 4G, P < 0.05). These data suggested that GTF3A binds to the Csta promoter to regulate its transcription and translation.
The above results indicated that GTF3A promoted CRC cell invasion and metastasis. To investigate the underlying mechanisms, EMT markers, such as Snail, E-cadherin, and beta-catenin, were detected in scrambled control and Gtf3a-knockdown cells. Western blot results showed that Gtf3a-knockdown HCT116 and SW480 cells had decreased Snail expression and increased E-cadherin expression compared with the scrambled control (Figures 5A and 5B). Moreover, GTF3A regulates the expression of CSTA/CST1/CST4, and CST1 promotes the migration and invasion of breast cancer cells by up-regulating E-cadherin[23]. Consistent with these results, Csta-knockdown cells were constructed (Figure 5C), and Snail, E-cadherin, and beta-catenin were detected in these cells. The cells showed down-regulated Snail expression and upregulated E-cadherin expression (Figure 5D). Collectively, GTF3A may mediate the EMT to promote CRC cell metastasis by regulating CSTA and CST1 expression.
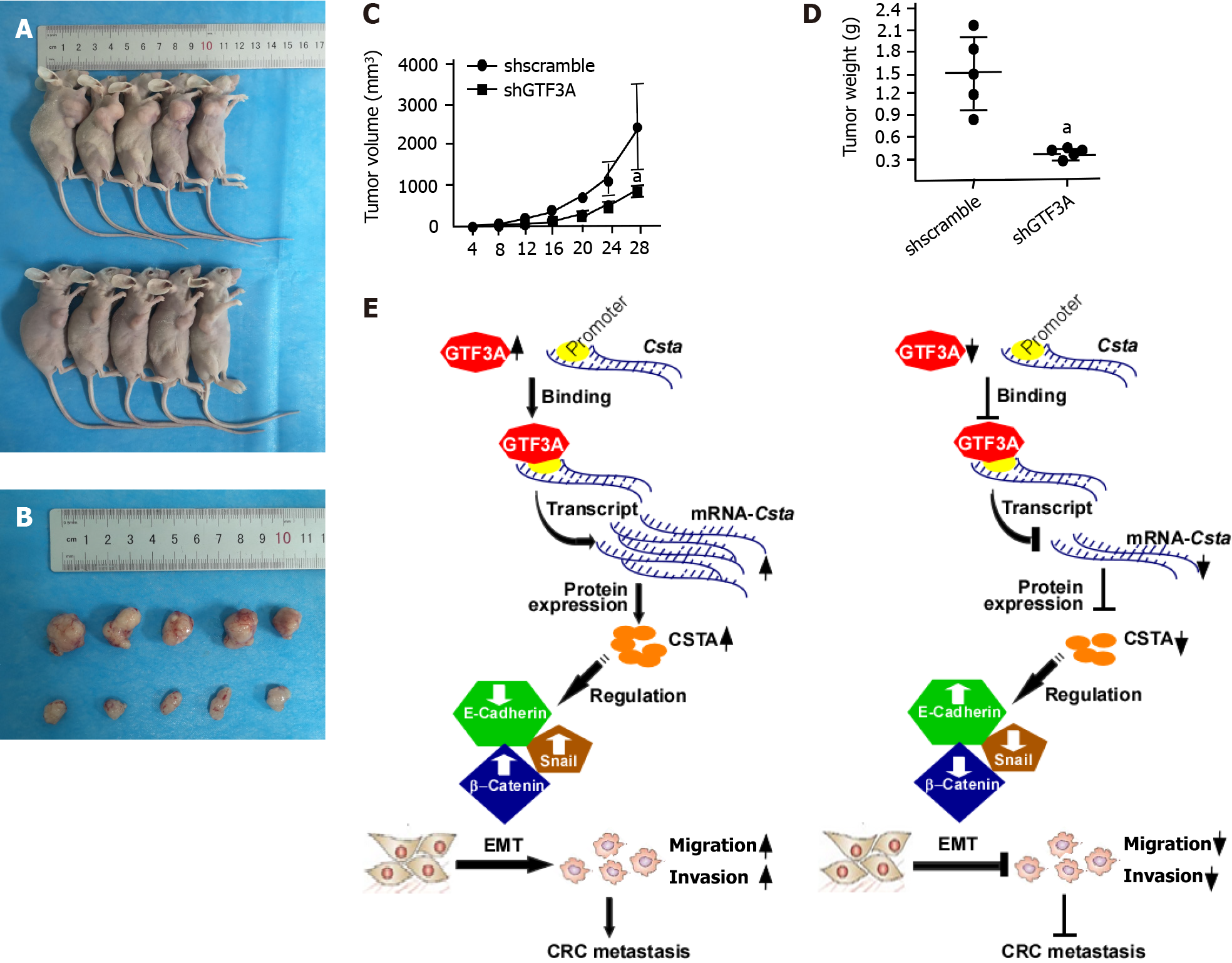
The above results showed that GTF3A promoted the proliferation, invasion, and metastasis of CRC cells in vitro. To verify whether GTF3A promotes CRC progression in vivo, nude mice were subcutaneously injected with the Gtf3a-knockdown and control cells to examine the function of Gtf3a. After 28 d, the Gtf3a-knockdown group had a significantly smaller tumor size (Figures 6A and 6B) and slower tumor growth (Figure 6C, P < 0.05) than the control group. In addition, the Gtf3a-knockdown group showed a reduction in tumor weights (Figure 6D, P < 0.05). In vivo experiments demonstrated that GTF3A promoted the growth of CRC cells.
As a transcription factor of RNA polymerase III, GTF3A is homologous to TFIIIB and TFIIIC, which guides the accurate transcription of 5S RNA genes[32]. RNA polymerase III is responsible for the transcription of non-coding genes including U6 snRNA, tRNA, and 5S RNA[33]. Deregulation of RNA polymerase III leads to the development of a large variety of human disorders[34], and upregulation of RNA polymerase III transcription has been observed in various types of cancer[35,36]. TFIIIB and TFIIIC can enhance the transcriptional activity of RNA polymerase III and mediate cellular transformation and tumor formation[37-40]. In addition, TFIIIA-mediated 5S rRNA is an essential component of 5S RNP that regulates the Hdm2-p53 checkpoint to affect ribosome biosynthesis and cancer progression[9-13], indicating that GTF3A participates in the occurrence and development of various cancer types. Overexpression of GTF3A has been observed in CRC tumors and metastatic tissues using CRC tissue arrays, and clinical data analysis suggests that GTF3A is associated with CRC progression and metastasis.
A series of in vitro and in vivo experiments was performed to examine the role of GTF3A in CRC development. The results showed that the knockdown of Gtf3a inhibited the proliferation, invasion, and metastasis of CRC cells. Generally, cancer metastasis is mostly related to the EMT[41]. We hypothesized that GTF3A promotes CRC cell metastasis by mediating the EMT. Thus, the EMT biomarkers Snail, vimentin, beta-catenin, and E-cadherin were detected. These changes in Snail and E-cadherin levels were in accordance with our hypothesis. The vimentin had no changed after knockdown of GTF3A (not shown). Both RNA-Seq and RT-qPCR showed that Csta expression was dramatically decreased in Gtf3a-knockdown cells. Furthermore, the luciferase activity assay suggested that GTF3A regulates the Csta transcription and translation by binding to the Csta promoter, therefore, Csta is a target gene of GTF3A.
CSTA is associated with invasion and metastasis in various cancer types[17,19,22], and in vitro experiments have shown that CSTA modulates the invasion and metastasis of NPC cells[18]. Based on our data, GTF3A may regulate Csta expression to mediate the EMT and promote CRC metastasis. FISH and EMSA results suggested that GTF3A binds with the promoter of the Csta gene, and the luciferase activity assay showed that GTF3A upregulated Csta transcription by binding to the Csta promoter. In addition, CSTA regulates E-cadherin and Snail expression, there mediating the EMT shift. Collectively, GTF3A upregulates CSTA expression to promote CRC metastasis by accelerating EMT shift. CST1 is associated with the progression and prognosis of various cancer types[20,25], and its overexpression modulates EMT progression by modulating the PI3K/AKT pathway in vivo and in vitro[22].
GTF3A increases Csta gene transcription and protein expression by binding to the Csta promoter, increases the expression of CSTA, enhances the EMT process, and facilitates CRC cell invasion and metastasis. However, knockdown of Gtf3a decreases CSTA expression, inhibits the EMT, and suppresses CRC cell invasion and metastasis (Figure 6E). Therefore, GTF3A is a potential novel therapeutic target and prognostic biomarker in human CRC.
Advanced colorectal cancer (CRC) generally has poor outcomes and high mortality rates. Clarifying the molecular mechanisms underlying CRC progression is necessary to develop new diagnostic and therapeutic strategies to improve CRC outcome and decrease mortality.
Transcriptional factor III A (GTF3A), an RNA polymerase III transcriptional factor, is a critical driver of tumorgenesis and aggravates CRC cell growth. The mechanism of GTF3A participating in CRC is not clear.
To confirm whether GTF3A aggravates CRC progression and investigate molecular mechanisms underlying CRC progression.
Immunohistochemistry was used to detect GTF3A expression in CRC tissues. Short hairpin GTF3As and CSTAs were designed and packaged into the virus to block the expression of Gtf3a and Csta genes. RNA sequencing and data analysis was used to screen the target genes of GTF3A. Fluorescence in situ hybridization assay was used to detect the interaction of GTF3A with Csta, and luciferase activity assay was used to evaluate the expression of Gtf3a and Csta genes.
GTF3A was highly expressed in CRC tissues and metastatic tissues, and its expression was associated with CRC prognosis. Knockdown of the Gtf3a gene impaired CRC cell proliferation, invasion, and motility in vitro and in vivo. GTF3A increased Csta transcription, and increased CSTA upregulated epithelial-mesenchymal transition (EMT) markers.
GTF3A increases CSTA expression by binding to the Csta promoter, and increased CSTA levels promote CRC progression by regulating EMT. Inhibition of GTF3A prevents CRC progression.
GTF3A may be a potential novel therapeutic target and biomarker for CRC.
The authors would like to acknowledge the members in Clinical Laboratory Center of Hunan Cancer Hospital and Xiangya Medical School of Central South University for contributions.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sahin TT, Turkey; Thummer RP, India S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Yuan YY
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25541] [Article Influence: 1824.4] [Reference Citation Analysis (7)] |
| 2. | Riihimäki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci Rep. 2016;6:29765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 420] [Cited by in RCA: 681] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 3. | Seifart KH, Wang L, Waldschmidt R, Jahn D, Wingender E. Purification of human transcription factor IIIA and its interaction with a chemically synthesized gene encoding human 5 S rRNA. J Biol Chem. 1989;264:1702-1709. [PubMed] |
| 4. | Weser S, Riemann J, Seifart KH, Meissner W. Assembly and isolation of intermediate steps of transcription complexes formed on the human 5S rRNA gene. Nucleic Acids Res. 2003;31:2408-2416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 5. | Hanas JS, Hocker JR, Cheng YG, Lerner MR, Brackett DJ, Lightfoot SA, Hanas RJ, Madhusudhan KT, Moreland RJ. cDNA cloning, DNA binding, and evolution of mammalian transcription factor IIIA. Gene. 2002;282:43-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 6. | Schulman DB, Setzer DR. Identification and characterization of transcription factor IIIA from Schizosaccharomyces pombe. Nucleic Acids Res. 2002;30:2772-2781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Fridell RA, Fischer U, Lührmann R, Meyer BE, Meinkoth JL, Malim MH, Cullen BR. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc Natl Acad Sci U S A. 1996;93:2936-2940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 88] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Layat E, Probst AV, Tourmente S. Structure, function and regulation of Transcription Factor IIIA: From Xenopus to Arabidopsis. Biochim Biophys Acta. 2013;1829:274-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237-247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 238] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 10. | Donati G, Peddigari S, Mercer CA, Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 11. | Onofrillo C, Galbiati A, Montanaro L, Derenzini M. The pre-existing population of 5S rRNA effects p53 stabilization during ribosome biogenesis inhibition. Oncotarget. 2017;8:4257-4267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Nishimura K, Kumazawa T, Kuroda T, Katagiri N, Tsuchiya M, Goto N, Furumai R, Murayama A, Yanagisawa J, Kimura K. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 2015;10:1310-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 13. | Nait Slimane S, Marcel V, Fenouil T, Catez F, Saurin JC, Bouvet P, Diaz JJ, Mertani HC. Ribosome Biogenesis Alterations in Colorectal Cancer. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Komura T, Takabatake H, Harada K, Yamato M, Miyazawa M, Yoshida K, Honda M, Wada T, Kitagawa H, Ohta T, Kaneko S, Sakai Y. Clinical features of cystatin A expression in patients with pancreatic ductal adenocarcinoma. Cancer Sci. 2017;108:2122-2129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Shiba D, Terayama M, Yamada K, Hagiwara T, Oyama C, Tamura-Nakano M, Igari T, Yokoi C, Soma D, Nohara K, Yamashita S, Dohi T, Kawamura YI. Clinicopathological significance of cystatin A expression in progression of esophageal squamous cell carcinoma. Medicine (Baltimore). 2018;97:e0357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Butler MW, Fukui T, Salit J, Shaykhiev R, Mezey JG, Hackett NR, Crystal RG. Modulation of cystatin A expression in human airway epithelium related to genotype, smoking, COPD, and lung cancer. Cancer Res. 2011;71:2572-2581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Lin YY, Chen ZW, Lin ZP, Lin LB, Yang XM, Xu LY, Xie Q. Tissue Levels of Stefin A and Stefin B in Hepatocellular Carcinoma. Anat Rec (Hoboken). 2016;299:428-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Chang KP, Wu CC, Chen HC, Chen SJ, Peng PH, Tsang NM, Lee LY, Liu SC, Liang Y, Lee YS, Hao SP, Chang YS, Yu JS. Identification of candidate nasopharyngeal carcinoma serum biomarkers by cancer cell secretome and tissue transcriptome analysis: potential usage of cystatin A for predicting nodal stage and poor prognosis. Proteomics. 2010;10:2644-2660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 19. | Takahashi H, Honma M, Ishida-Yamamoto A, Namikawa K, Kiyama H, Iizuka H. Expression of human cystatin A by keratinocytes is positively regulated via the Ras/MEKK1/MKK7/JNK signal transduction pathway but negatively regulated via the Ras/Raf-1/MEK1/ERK pathway. J Biol Chem. 2001;276:36632-36638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Cao X, Li Y, Luo RZ, Zhang L, Zhang SL, Zeng J, Han YJ, Wen ZS. Expression of Cystatin SN significantly correlates with recurrence, metastasis, and survival duration in surgically resected non-small cell lung cancer patients. Sci Rep. 2015;5:8230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Jiang J, Liu HL, Liu ZH, Tan SW, Wu B. Identification of cystatin SN as a novel biomarker for pancreatic cancer. Tumour Biol. 2015;36:3903-3910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Cui Y, Sun D, Song R, Zhang S, Liu X, Wang Y, Meng F, Lan Y, Han J, Pan S, Liang S, Zhang B, Guo H, Liu Y, Lu Z, Liu L. Upregulation of cystatin SN promotes hepatocellular carcinoma progression and predicts a poor prognosis. J Cell Physiol. 2019;234:22623-22634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 23. | Dai DN, Li Y, Chen B, Du Y, Li SB, Lu SX, Zhao ZP, Zhou AJ, Xue N, Xia TL, Zeng MS, Zhong Q, Wei WD. Elevated expression of CST1 promotes breast cancer progression and predicts a poor prognosis. J Mol Med (Berl). 2017;95:873-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 24. | Jiang J, Liu HL, Tao L, Lin XY, Yang YD, Tan SW, Wu B. Let7d inhibits colorectal cancer cell proliferation through the CST1/p65 pathway. Int J Oncol. 2018;53:781-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Yoneda K, Iida H, Endo H, Hosono K, Akiyama T, Takahashi H, Inamori M, Abe Y, Yoneda M, Fujita K, Kato S, Nozaki Y, Ichikawa Y, Uozaki H, Fukayama M, Shimamura T, Kodama T, Aburatani H, Miyazawa C, Ishii K, Hosomi N, Sagara M, Takahashi M, Ike H, Saito H, Kusakabe A, Nakajima A. Identification of Cystatin SN as a novel tumor marker for colorectal cancer. Int J Oncol. 2009;35:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Choi EH, Kim JT, Kim JH, Kim SY, Song EY, Kim JW, Yeom YI, Kim IH, Lee HG. Upregulation of the cysteine protease inhibitor, cystatin SN, contributes to cell proliferation and cathepsin inhibition in gastric cancer. Clin Chim Acta. 2009;406:45-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 27. | Hong J, Hu K, Yuan Y, Sang Y, Bu Q, Chen G, Yang L, Li B, Huang P, Chen D, Liang Y, Zhang R, Pan J, Zeng YX, Kang T. CHK1 targets spleen tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J Clin Invest. 2012;122:2165-2175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 28. | Xu S, Wu Y, Chen Q, Cao J, Hu K, Tang J, Sang Y, Lai F, Wang L, Zhang R, Li SP, Zeng YX, Yin Y, Kang T. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res. 2013;23:423-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Lu J, Li Y, Wu Y, Zhou S, Duan C, Dong Z, Kang T, Tang F. MICAL2 Mediates p53 Ubiquitin Degradation through Oxidating p53 Methionine 40 and 160 and Promotes Colorectal Cancer Malignance. Theranostics. 2018;8:5289-5306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Tang FQ, Duan CJ, Huang DM, Wang WW, Xie CL, Meng JJ, Wang L, Jiang HY, Feng DY, Wu SH, Gu HH, Li MY, Deng FL, Gong ZJ, Zhou H, Xu YH, Tan C, Zhang X, Cao Y. HSP70 and mucin 5B: novel protein targets of N,N'-dinitrosopiperazine-induced nasopharyngeal tumorigenesis. Cancer Sci. 2009;100:216-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Tang F, Zou F, Peng Z, Huang D, Wu Y, Chen Y, Duan C, Cao Y, Mei W, Tang X, Dong Z. N,N'-dinitrosopiperazine-mediated ezrin protein phosphorylation via activation of Rho kinase and protein kinase C is involved in metastasis of nasopharyngeal carcinoma 6-10B cells. J Biol Chem. 2011;286:36956-36967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Birkenmeier EH, Brown DD, Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978;15:1077-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 259] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 33. | Abascal-Palacios G, Ramsay EP, Beuron F, Morris E, Vannini A. Structural basis of RNA polymerase III transcription initiation. Nature. 2018;553:301-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 34. | Yeganeh M, Hernandez N. RNA polymerase III transcription as a disease factor. Genes Dev. 2020;34:865-882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 35. | Ramsay EP, Abascal-Palacios G, Daiß JL, King H, Gouge J, Pilsl M, Beuron F, Morris E, Gunkel P, Engel C, Vannini A. Structure of human RNA polymerase III. Nat Commun. 2020;11:6409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 36. | Cabarcas S, Schramm L. RNA polymerase III transcription in cancer: the BRF2 connection. Mol Cancer. 2011;10:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 37. | Lei J, Chen S, Zhong S. Abnormal expression of TFIIIB subunits and RNA Pol III genes is associated with hepatocellular carcinoma. Liver Res. 2017;1:112-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 38. | Zhong Q, Xi S, Liang J, Shi G, Huang Y, Zhang Y, Levy D, Zhong S. The significance of Brf1 overexpression in human hepatocellular carcinoma. Oncotarget. 2016;7:6243-6254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 39. | Bellido F, Sowada N, Mur P, Lázaro C, Pons T, Valdés-Mas R, Pineda M, Aiza G, Iglesias S, Soto JL, Urioste M, Caldés T, Balbín M, Blay P, Rueda D, Durán M, Valencia A, Moreno V, Brunet J, Blanco I, Navarro M, Calin GA, Borck G, Puente XS, Capellá G, Valle L. Association Between Germline Mutations in BRF1, a Subunit of the RNA Polymerase III Transcription Complex, and Hereditary Colorectal Cancer. Gastroenterology. 2018;154:181-194.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 40. | Gouge J, Guthertz N, Kramm K, Dergai O, Abascal-Palacios G, Satia K, Cousin P, Hernandez N, Grohmann D, Vannini A. Molecular mechanisms of Bdp1 in TFIIIB assembly and RNA polymerase III transcription initiation. Nat Commun. 2017;8:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 41. | Georgakopoulos-Soares I, Chartoumpekis DV, Kyriazopoulou V, Zaravinos A. EMT Factors and Metabolic Pathways in Cancer. Front Oncol. 2020;10:499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 228] [Article Influence: 45.6] [Reference Citation Analysis (0)] |