Published online Jan 15, 2022. doi: 10.4251/wjgo.v14.i1.90
Peer-review started: March 21, 2021
First decision: May 3, 2021
Revised: May 17, 2021
Accepted: December 21, 2021
Article in press: December 21, 2021
Published online: January 15, 2022
Processing time: 295 Days and 11.4 Hours
Gastric cancer (GC) poses a serious threat worldwide with unfavorable prognosis mainly due to late diagnosis and limited therapies. Therefore, precise molecular classification and search for potential targets are required for diagnosis and treatment, as GC is complicated and heterogeneous in nature. Accumulating evidence indicates that epigenetics plays a vital role in gastric carcinogenesis and progression, including histone modifications, DNA methylation and non-coding RNAs. Epigenetic biomarkers and drugs are currently under intensive evaluations to ensure efficient clinical utility in GC. In this review, key epigenetic alterations and related functions and mechanisms are summarized in GC. We focus on integration of existing epigenetic findings in GC for the bench-to-bedside translation of some pivotal epigenetic alterations into clinical practice and also describe the vacant field waiting for investigation.
Core Tip: Epigenetics plays a vital role in gastric carcinogenesis and progression. In this review, key epigenetic alterations and related functions and mechanisms are summarized in gastric cancer.
- Citation: Tang SY, Zhou PJ, Meng Y, Zeng FR, Deng GT. Gastric cancer: An epigenetic view. World J Gastrointest Oncol 2022; 14(1): 90-109
- URL: https://www.wjgnet.com/1948-5204/full/v14/i1/90.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i1.90
Gastric cancer (GC) is one of the most common malignant tumors of the digestive tract and ranks as the fifth leading cause of morbidity and second leading cause of mortality worldwide, posing a serious threat to all human beings[1]. Residents in South and East of Asia including China, Japan and Korea are reported to have a higher risk of GC[2]. Due to the unconspicuous symptoms in the early stage of GC, many patients are first diagnosed as advanced GC accompanied by tumor infiltration and metastasis. Despite of combined treatment of surgery, chemotherapy, radiotherapy, and sometimes targeted therapy and immunotherapy, GC still shows a poor prognosis with the 5-year overall survival less than 30%[3,4]. Currently routine screening for GC is endoscopy and histological examination, which is costly, invasive and often painful to patients. Therefore, development of new or alternative methods for screening, diagnosis and treatment to GC is of great clinical significance.
Epigenetics has been illustrated to be associated with the diagnosis and treatment of GC patients. GC is highly complicated and heterogeneous in nature and often genetically divided into familial and sporadic disease. Familial GC, constituting about 10% of GC patients, has a close connection to genetic alterations[5]. Sporadic GC (90% of GC) is largely related to Helicobacter pylori (H. pylori) infection and evolves in a canonical model of chronic inflammation, atrophy, intestinal metaplasia, dysplasia and finally adenocarcinoma, which is characterized by typically epigenetic alterations but scarce genetic changes across over the stages[6]. With rapid progress in epigenomics, precise molecular classification towards GC seems admirable in research and clinical medicine. In 2014, The Cancer Genome Atlas identified GC into four molecular subtypes including Epstein–Barr virus (EBV) associated, microsatellite instable (MSI), chromosomal instability (CIN), and genomically stable (GS)[7]. Apparently, GS means the genome is stable in this type of GC[8]. Among the four classes, MSI patients have the best overall prognosis and the lowest frequency of recurrence with high incidence of gene mutations and DNA methylation. Patients in EBV-subtype are associated with Epstein-Barr virus infection and have extremely high DNA methylation status. In the patients with CIN subtype, the largest proportion of GC, is more prone to chro
In this review, we mainly explore GC from an epigenetic view and summarize key epigenetic alterations and related functions and mechanisms, with special attention to histone modifications and the translational findings which guide us towards better clinical utility.
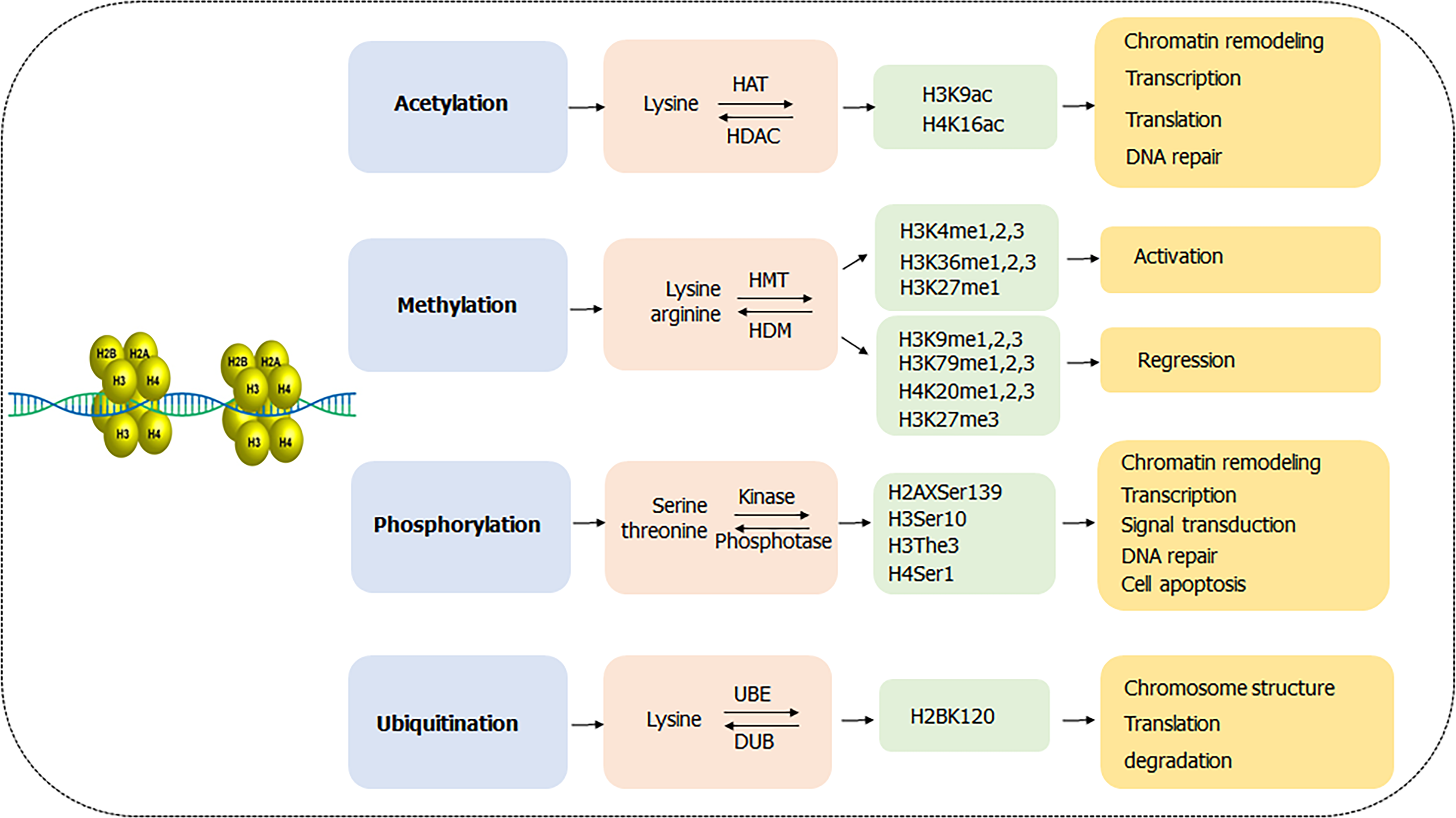
Nucleosome, as a major unit of chromatin, consists of wrapped DNA and a histone octamer formed by two copies of H2A, H2B, H3 and H4 proteins[9]. Each histone contains an accessible amino terminal tail rich in lysine, arginine, serine and threonine residues, which is often modified post-translationally and the process is called posttranslational modifications (PTMs). Studies have shown that histone PTMs in GC mainly including acetylation, methylation, phosphorylation and ubiquitination are involved in various pathophysiological cellular functions such as carcinogenesis, inflammation and epithelial-mesenchymal transition (Figure 1)[10]. In recent years, some new modifications, such as succinylation, sumoylation, butyrylation and crotonylation, have been discovered in the occurrence and progression of other gastrointestinal tumors, such as esophageal, colorectal, and hepatocarcinoma liver cancer[11-14], which provide new insights in functions and mechanisms and even therapeutic potential for cancer diagnosis and treatments. Notably, those new types of histone modifications remain a vacant field in GC and thereby it may be an innovative and interesting field to explore in the near future.
As the most common form of PTMs in GC, acetylation always occurs in N-terminal lysine residues of histone H3 and H4 and is associated with chromatin remodeling, regulation of transcription, translation and DNA repair. The acetylation of histones catalyzed by histone acetylase (HATs) transfers acetyl moieties from coenzyme A to lysine residues, opens the chromatin structure and makes it accessible to transcriptional factors, thus activating gene transcription. Instead, the histone deacetylase (HDACs) removes the acetyl groups from histone and results in repression of transcription. HATs consist of three families including GCN5, MYST and p300/CBP, while HDACs contain four classes including type I (HDAC 1,2,3,8), type II (HDAC 4,7,9,10), type III (SIRT 1-7) and type IV (HDAC 11)[15,16]. The reversible acetylation and deacetylation processes mainly facilitate GC progression by activating oncogene expression and silencing tumor suppressor gene expression.
Studies revealed that high H3K9Ac positive cells were associated with undifferentiated GC, suggesting poor prognosis of GC[17]. Further, BMP8B was highly expressed in GC tissues other than adjacent normal tissues, and reduced acetylation level of BMP8B loci on H3K9 and H4K16 influenced the development of poorly differentiated gastric tumors[18]. Many genes encoding HATs, such as KAT2B and EP300, are often genetically depleted or mutated in GC, and are significantly correlated with TNM staging[19,20]. IFN-γ-induced upregulation of histone H3 Lysine 9 acetylation (H3K9) level in gene promoter accelerates the expression of B7-H1, which contributes to tumor immune evasion in HGC-27 cells[21]. Wisnieski et al[22] demonstrated hypoacetylation of histone H3 in the initiator domain of CDKN1A decreased its mRNA level and reduced antitumor effect in GC. Besides, H. pylori-infection inhibited recruitment of HAT p300 to the p27 promoter which caused the hypoacetylation status in histone H4, then induced the downregulated p27 mRNA expression, and finally led to gastric carcinogenesis[23].
Histone methylation usually takes place on H3 and H4 Lysine or arginine residues, catalyzed by histone methyltransferases (HMTs) and reversely controlled by histone demethylases (HDMs). The methylation could be single or multiple methylations to form mono-methylation (me1), di-methylation (me2) and tri-methylation (me3), participating in the formation and maintenance of chromatin structure, DNA repair, gene inactivation and transcription[24]. Methylations on different sites have different functions in regulation of gene expression. In general, methylation of arginine residues, methylation of lysine H3K4 and H3K36, and monomethylation of H3K27 are associated with gene activation, while methylation of H3K9, H3K79 and H4K20, and dimethylation and trimethylation of H3K27 might cause gene silencing[25,26].
Specifically, repression of HDMs KDM5A and DPY300 subunits upregulated H3K4me level, inhibiting GC cell proliferation[27]. However, overexpression of HDMs LSD1 declined methylation of H3K4 in p21 promoter and repressed the transcription of p21, resulting in progression of GC[28]. An assay of familial GC patients identified INSR, FBXO24 and DOT1L as new susceptibility genes in diffuse gastric carcinoma, in which DOT1L was a histone methyltransferase involved in the mono, di and tri-methylation of H3K79, suggesting the contributing role of H3K79 in gastric carcinogenesis[29]. Methylation of H3K27 is well-investigated in GC. A paired-study of 117 GC patients showed that the level of H3K27me3 in GC and normal tissue was 56.4% and 7.25%, respectively, which negatively correlated with GC overall survival[30]. Besides, knockdown of demethylases SETDB2 was found to accelerate the expression of tumor suppressor genes WWOX and CADM1, and significantly reduced cell growth, migration and invasion in GC cells[31].
Histone phosphorylation is a dynamical process mediated by histone kinases and phosphatases, in which the phosphate group is transferred from ATP to the histone serine and threonine residues. There are several accessible sites in histone phosphorylation including H1.4 Ser27, H2AX Ser139 ( also called γ-H2AX), H3 Ser10, H3 The3 and H4 Ser1[32,33]. Particularly, histone H3 is phosphorylated at Ser10 during mitosis in all eukaryotes and induction of phosphorylation in interphase has been shown to correlate with chromosome condensation prior to mitosis[34]. Histone phosphorylation functions as a switch on chromosomal folding, compression, segregation, transcriptional regulation, cell signal transduction, cell apoptosis, and DNA damage repair[35,36].
Histone phosphorylation frequently happens in H3 and H4 with a dual role in cancer progression[32,33]. For instance, phosphorylated histone H3 at position of serine10 (H3S10) by MSK1 promoted cell proliferation during gastric tumorigenesis via the activation of downstream transcriptional factor NFATc2-related inflammatory pathway[37]. H3S10 phosphorylation also played a vital prognostic role in defining negative resection margins in GC due to its lower expression in the surgical resection margins[38]. A cohort of 122 GC patients further indicated phosphorylated histone H3 overexpression could be an independent prognostic factor[39]. Moreover, repression of Aurora B-mediated H1.4 phosphorylation at Ser27, caused by Ras-ERK1/2 signaling, evidently participated in the progression of GC[40].
Unlike the three types of histone modifications described above, histone ubiquitination always works in the crosstalk with other modifications. Histone ubiquitination often acts subsequently after histone acetylation and methylation or modifies the stability and the activity of enzymes in these acetylation and methylation processes, which endures a synergic effect on cell division, cell cycle, DNA damage and cell apoptosis in GC[41]. When the histone, usually H2A and H2B, binds to one or several ubiquitins on lysine residues, it is called mono- or poly- ubiquitination and tends to work in the following three ways: Alterations of chromosome structure, recruitment and activation of downstream proteins, and degradation in proteasome pathway[42]. Ubiquitination is a reversible process in which ubiquitin is removed from polypeptides by deubiquitinases (DUBs), a superfamily of cysteine proteases and metalloproteases that cleave ubiquitin-protein bonds[43,44].
Hahn et al[45] identified that ring finger proteins RNF20 and RNF40 constituted a heterodimeric complex that functions as the E3 ubiquitin ligase for monoubiquitination of histone H2B at lysine 120 (H2B-K120) and the tumor suppressor CDC73 exerted antitumor effect in GC through the maintenance of H2B-K120 monoubiquitination. Besides, histone ubiquitination presents a therapeutic potential in GC as the expression of ubiquitinated-H2B was significantly lower in the malignant tissues and different differentiated tumors had variant levels of H2B ubiquitination[46].
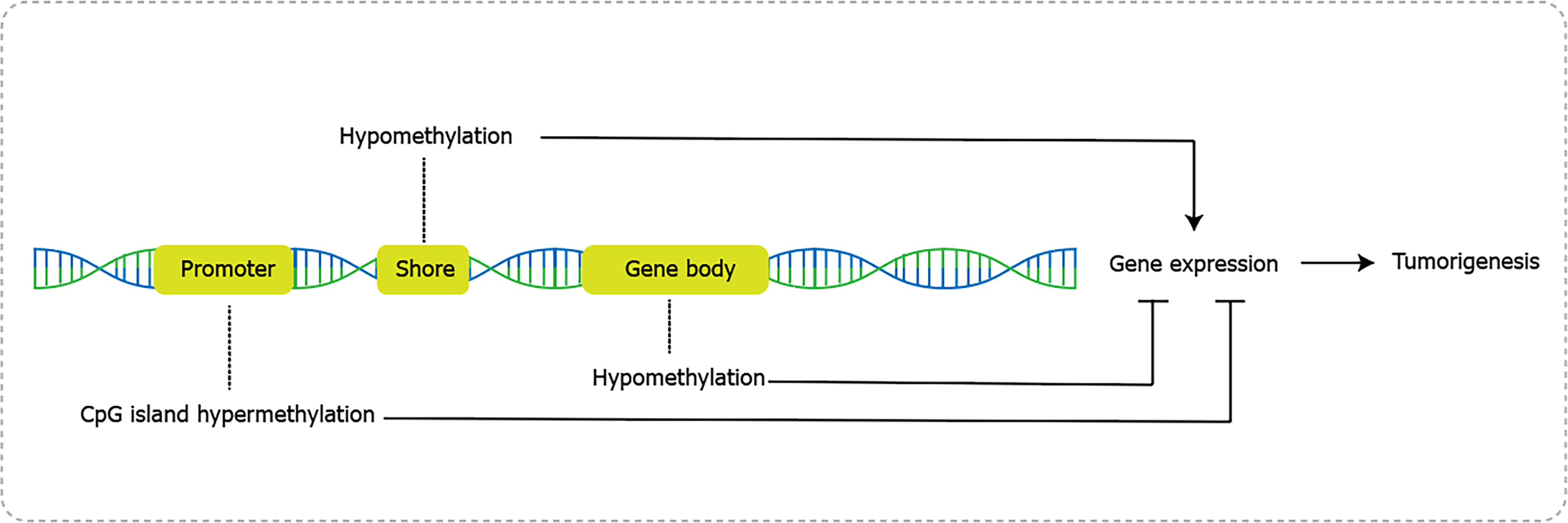
In contrast to histone methylation, DNA methylation is a more frequent and comprehensive epigenetic modification (Figure 2), mediated by DNA methyltransferase (DNMTs) and demethylases. It refers to the transfer of the methyl group (CH3) from S.adenosylmethionine to C5 and forms 5-methylcytosine[47,48]. DNA methylation occurs in the dinucleotide CpG sequence, which may form CpG islands and dispersed sequences. CpG islands exist in around 60%-70% of gene promoters in human and consist of CpG core and shore area[49]. CpG core has a specific inhibitory effect on methylation, while the shore area, also known as transitional CpG region, is variable sites for dynamical alterations between hypomethylated and hypermethylated groups. In normal cells, CpG islands are non-methylated and other CpG sequence are methylated. Once stimulated by intrinsic or extrinsic factors, the methylation status changed and caused alterations in gene transcription, and consequently lead to tumorigenesis[48].
Aberrant DNA hypermethylation usually happens in the promoter of tumor suppressor genes in GC like p16, RASSF1A and hMLH1. Hypermethylation inhibits gene transcription by reducing binding to transcription factors, thereby impeding DNA readability and resulting in gene silencing[50]. Specifically, alteration of methylation in p16 promoter inhibited the cell cycle in G1 phase and induced 5-fluorurazil chemo-resistance in GC[51]. Abnormal methylation of RASSF1A gene promoter reduced RASSF1A expression, decreased cyclin D1 accumulation, and arrested cell cycle. Consistently, GC patients presented evidently higher frequency of aberrant methylation in RASSF1A promoter than control group, indicating the potential of methylated RASSF1A promoter as a molecular marker for the diagnosis of GC[52]. In addition to methylation alterations in promoter, hypomethylation at gene body regions has a distinct association with transcription and gene hypomethylation also exerts profound effects on cancer progression[53]. For instance, hypomethylation of SAT-α and L1 was associated with shortened survival in advanced GC patients[54]. And Lineage-specific RUNX3 hypomethylation constituted the immune component in GC and was associated with the early inflammatory, preneoplastic and tumor stages[55]. Genome-wide methylation sequencing studies in GC identified both hypo- and hyper-methylation events across the genome, suggesting a dual role of global genomic methylation in the stages of gastric carcinogenesis[56].
H. pylori-induced DNA Methylation is a hot research area in the development of GC. Numerous researches revealed that H. pylori, classified as Class I carcinogen by WHO, induced and accumulated aberrant DNA methylation through continuous chronic inflammation in gastric mucosae, and such high level of epigenetic field defects increased the risk of gastric carcinogenesis[57]. For example, H. pylori infection upregulated inflammatory response genes like IL-1β, Nos2, and Tnf, and promoted the infiltration of monocytes/macrophages with residual neutrophils in noncancerous mucosae, which induced a large number of aberrant DNA methylation in tumor suppressor genes and led to malignant transformation[58]. Eradication of H. pylori had subtle influence on the decrease of DNA methylation in gerbils, while application of immunosuppressive agent (e.g., cyclosporin A) and demethylation agent (e.g., 5-Aza-2-deoxycytidine) could evidently reduce level of DNA methylation and prevent development of GC[59,60]. Moreover, high levels of DNA methylation were found in gastric biopsies of inflammatory and precancerous lesions, comparing to adjacent normal tissue, and were also correlated with a greater risk of GC incidence[61]. H. pylori-induced DNA methylation takes place in various genes involved in cell adhesion, cell cycle, DNA damage repair, inflammation, and autophagy, which allows intensive interfered targets of such epigenetic defects in diagnostic biomarker and cancer prevention[58,62].
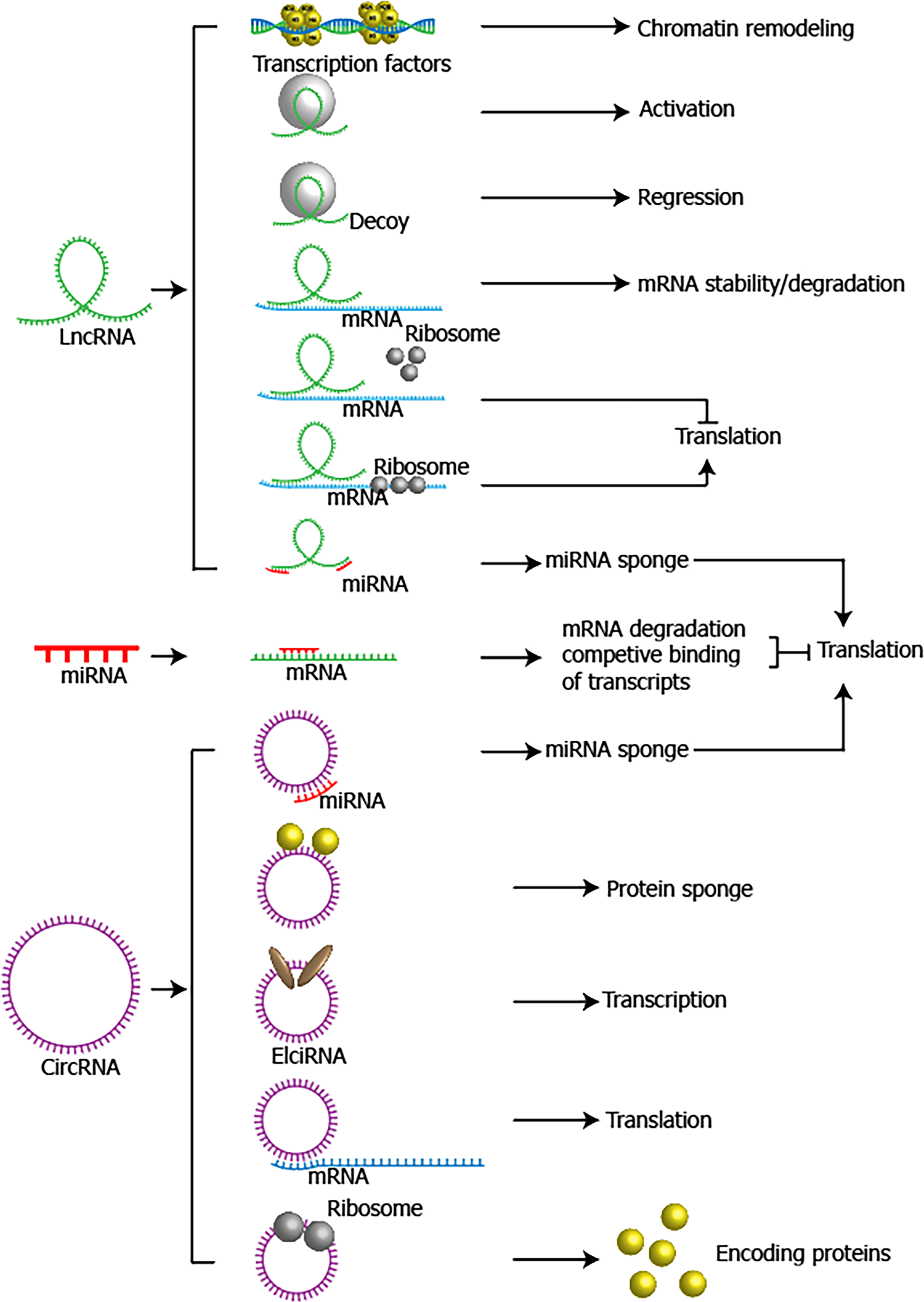
Non-coding RNAs consist of microRNAs (miRNAs), long non coding RNAs (lncRNAs), circular RNAs (circRNAs), small nucleolar RNAs (snoRNAs), small interfering RNAs (siRNAs), etc.[63]. Since the first two non-coding RNA lineage defective 4 (lin-4)[64] and lethal 7 (let-7)[65] were identified in 1993 and 2000, researchers realized that in addition to protein, some RNAs lacking of protein-coding regions, which are called non-coding RNAs, were still conserved functional molecules and required for many biological processes. Among non-coding RNAs, miRNAs, lncRNAs and circRNAs were found to have plenty of functions in GC (Figure 3), including cell proliferation, cell cycle arrest, apoptosis, migration, invasion and chemo- or radio-sensitivity[66,67].
MicroRNAs are a class of small RNAs with 18-24 nucleotides and they repress translation process and silence target gene through complementary binding with 3’untranslated terminal region (UTR) of mRNA[68]. A shaped understanding towards miRNAs has been established in the past two decades due to numerous miRNAs arrays conducted in GC. Taking the largest scale of GC miRNAs array cohort for example, a general miRNAs signature profiling was developed, in which 22 oncogenic miRNAs and 13 tumor suppressor miRNAs were identified in 353 primary Japanese gastric tumor samples. In this study, authors also revealed that different histological subtypes had different miRNA signatures[69] as diffuse-type showed 2 folds of proportion in upregulated miRNAs to intestinal-type GC. Specifically, low expression of let-7g and miR-433 and high expression of miR-214 were associated with unfavorable outcomes in GC patients[69]. MiRNAs have an edge on GC diagnosis potential over other epigenetic factors because they alter quickly and are easy to be detected in the early stage of GC. Yu et al[70] performed a miRNAs microarray in early GC mouse model and the result showed that miR200-family promoted the initiation of GC and the integration of miR200-family’s 15 target gene would provide superior predictive sensitivity and specificity for overall survival compared with each early GC indicator alone. Here we summarized the up- or down-regulated miRNAs in GC (Table 1).
| miRNAs | Expression | Targets | Functions | Ref. |
| miR-21 | Up | EMT | Tumor growth, metastasis | [89] |
| miR-183 | Up | UVRAG | Cell proliferation, autophagy, apoptosis | [90] |
| miR-765 | Up | BATF2 | Chemosensitivity | [91] |
| miR-155 | Up | TP53INP1 | Cell cycle, proliferation, migration | [92] |
| miR-130b | Up | NFκB, p65 | Cell proliferation, tumorigenesis | [93] |
| miR-92a-1-5p | Up | FOXD1 | Metaplasia | [94] |
| miR-135b | Up | FOXN3/RECK | Cell invasion, CSC-like properties | [95] |
| miR-181a-5p | Up | AKT3 | Cell proliferation, apoptosis, tumor growth | [96] |
| miR-224 | Up | PAK4 | Cell proliferation, migration | [97] |
| let-7i | Down | COL1A1 | Cell invasion, metastasis | [98] |
| miR-146a | Down | - | Cell migration | [99] |
| MiR-12129 | Down | SIRT1 | Cell cycle, proliferation | [100] |
| miR-27b | Down | NR2F2 | cell proliferation, tumor growth | [101] |
| miR-140-5p | Down | NOTCH1 | Cell proliferation, migration, apoptosis | [102] |
| miR-34a | Down | Snail | Cell proliferation, invasion | [103] |
| miR-9 | Down | TNFAIP8L3 | Cell proliferation, migration | [104] |
| miR-195 | Down | HMGB1 | Chemosensitivity | [105] |
LncRNAs are longer than 200 nucleotides and exert profound influences on multiple biological functions through regulating transcription, chromatin remodeling and post-transcriptional process[71]. They work mainly in three ways: (1) Interact with mRNA, control transcription and regulate cellular signaling pathways; (2) Act as regulators of splicing and mRNA decay; (3) work as molecular decoys for miRNAs; and (4) interact with chromatin-modifying complexes or being a scaffold to maintain the structure of nuclear speckles[72-74]. Numerous lncRNAs have been uncovered the role and related mechanisms in GC. HOTAIR is a well-studied lncRNA and it is frequently overexpressed in GC, which may play a part in metastasis through following pathways: (1) Being a sponge of miR-330[75] and miR-331-3p[76] to upregulate the downstream targets; (2) Directly silencing HOXD[76] or miR34a expression[77]; (3) Regulating Wnt/β-catenin and PI3K/Akt pathways[77]; and (4) Inducing ubiquitination of Runx3[78]. Therefore, HOTAIR was considered to be a potent diagnostic and prognostic biomarker in GC. Most of lncRNAs in GC were found to be oncogenic, like H19, MNX1-AS1, MALAT1, HULC, UCA1, etc. However, some lncRNAs like CRNDE were identified to inhibit GC progression. Here we summarized the up- or down-regulated lncRNAs and the related targets and functions in GC (Table 2).
| LncRNAs | Expression | Targets | Functions | Ref. | |
| MIAT | Up | miR-29a-3p/HDAC4 | Cell proliferation, migration and invasion | [106] | |
| PANDAR | Up | CDKN1A | Tumor growth | [107] | |
| FOXD2-AS1 | Up | EphB3 | Tumorigenesis | [108] | |
| SMARCC2 | Up | miR-551b-3p/TMPRSS4 | Cell proliferation, migration | [109] | |
| H19 | Up | miR-519d-p/LDHA | Aerobic glycolysis, proliferation, and immune escape | [110] | |
| TINCR | Up | STAU1/CDKN2B | Cell proliferation, cell cycle | [111] | |
| CCAT2 | Up | E-cadherin, LATS2 | Cell proliferation, invasion | [112] | |
| AOC4P | Up | Vimentin, MMP9 | Cell proliferation, migration, invasion | [113] | |
| CTC-497E21.4 | Up | miR-22-3p/NET1 | Cell cycle, proliferation, invasion | [114] | |
| BANCR | Up | ERK1/2, NF-κB1 | Cell proliferation, apoptosis, chemosensitivity | [115,116] | |
| HOTTIP | Up | miR-216a-5p, miR-615-3p | Chemosensitivity, cell proliferation, apoptosis | [117,118] | |
| AC100830.4, CTC-501O10.1, RP11-210K20.5 | Up | - | Differentially expressed in GC and normal tissue | [119] | |
| INHBA-AS1, CEBPA-AS1, AK001058 | Up | - | Differentially expressed in GC and normal tissue | [120] | |
| CYTOP | Up | miR-103/RAB10 | Cell proliferation, migration, apoptosis | [121] | |
| NKX2-1-AS1 | Up | SERPINE1/VEGFR-2 | Cell proliferation, angiogenesis | [122] | |
| NEAT1 | Up | miR-17-5p/TGFβR2 | Angiogenesis | [123] | |
| ZFAS1 | Up | EPAS1 | Recurrence, metastasis | [124] | |
| TSPEAR-AS2 | Up | EZH2/GJA1, miR-1207-5p/CLDN4 | Tumor progression | [125] | |
| TMEM92-AS1 | Up | YBX1/CCL5 | Tumor progression | [126] | |
| CRNDE | Down | NEDD4-1/PTEN | Chemosensitivity | [127] | |
| MEG3 | Down | miR-181a-5p/ ATP4B | Cell proliferation, migration, apoptosis | [128] | |
| PCSK2-2:1 | Down | - | Differentially expressed in GC and normal tissue | [129] | |
| GNAQ-6:1 | Down | - | Differentially expressed in GC and normal tissue | [130] | |
| CTSLP4 | Down | Hsp90α/HNRNPAB | Cell migration, invasion, EMT | [131] | |
CircRNAs are a novel class of conserved single-stranded RNA molecules derived from exonic or intronic sequences by precursor mRNA back-splicing[79]. Compared to linear RNAs, the circular structure of circRNAs confers enhanced stability to exonuclease digestion[80]. Partially similar to lncRNAs, circRNAs could also act as miRNAs sponge, regulators of alternative splicing and tools of sequestering functional proteins in gene expression and posttranscriptional modification[81]. However, some circRNAs were identified to encode functional proteins[82]. CircRNAs were reported to exert influences on tumor growth, therapeutic resistance, recurrence and metastasis[83]. GC-related sequencing data revealed a variety of circRNAs with pro- or anti-tumor roles, including CircPVT1, CircRNA_001569, CircHIPK3, etc. CiRS-7, one of the mostly investigated circRNAs, is a sponge of miR-7. MiR-7 was known as a tumor suppressor miRNA, while ciRS-7 was found to act in an oncogenic role by antagonizing miR-7-mediated PTEN/PI3K/AKT pathway in GC. Overexpression of ciRS-7 accelerated the progression of GC[84]. Undoubtedly, circRNAs are of great value in research and are emerging as a rising star in the field of cancer biology and therapy. We listed some important circRNAs, as well as their targets and functions in Table 3.
| circRNAs | Expression | Targets | Functions | Ref. |
| circFAM73A | Up | miR-490-3p/ HMGA2 | Cell proliferation, migration, CSC-like properties, chemosensitivity | [132] |
| circAFF2 | Up | miR-6894-5p/ANTXR1 | Cell proliferation, migration, invasion | [133] |
| circHIPK3 | Up | miR-637 /AKT1 | Tumorigenesis | [134] |
| circVAPA | Up | miR-125b-5p/STAT3 | Chemosensitivity | [135] |
| circMAP7D1 | Up | HER2 | Cell proliferation, apoptosis | [136] |
| circ_0006282 | Up | miR-144-5p/YWHAB | Cell proliferation, metastasis | [137] |
| circ_0081146 | Up | miR-144/ HMGB1 | Cell growth, migration, invasion | [138] |
| circ_SMAD4 | Up | miR-1276/ CTNNB1 | Tumorigenesis | [139] |
| circNEK9 | Up | miR-409-3p/MAP7 | Cell proliferation, migration, invasion | [140] |
| circ_0004104 | Up | miR-539-3p/RNF2 | Cell proliferation, metastasis, glutaminolysis | [141] |
| circPVT1 | Up | miR-152-3p | Chemosensitivity | [142] |
| hsa_circ_0023409 | Up | miR-542-3p/ IRS4 | Cell proliferation, metastasis | [143] |
| circ_0044516 | Up | miR-149-5p/HuR | Cell proliferation, migration, invasion, tumor growth | [144] |
| circLMO7 | Up | miR-30a-3p/ WNT2 | Cell growth, metastasis | [145] |
| hsa_circ_0001829 | Up | miR-155-5p/SMAD2 | Cell growth, metastasis | [146] |
| circCUL3 | Up | miR-515-5p/STAT3/HK2 | Cell proliferation, glucose consumption, lactate production, ATP quantity | [147] |
| circTMEM87A | Up | miR-142-5p/ULK1 | Cell proliferation, metastasis | [148] |
| circPTPN22 | Down | EMT | Cell proliferation, migration, EMT, invasion | [149] |
| hsa_circ_0004872 | Down | miR-224/Smad4/ADAR1 | Cell proliferation, migration, invasion, tumor growth, metastasis | [150] |
| hsa_circRNA_0009172 | Down | miR-485-3p/NTRK3 | Cell proliferation, migration, invasion, tumor growth | [151] |
| circ_002059 | Down | miR-182/ MTSS1 | Cell proliferation, migration | [152] |
| circ-ITCH | Down | miR-199a-5p/ Klotho | Metastasis | [153] |
| circCUL2 | Down | miR-142-3p/ ROCK2 | Cell transformation, chemosensitivity, tumorigenesis | [154] |
Researches on epigenetics not only revealed the underlying mechanism of cancer initiation and progression, but also provided novel diagnostic and prognostic candidate biomarkers and therapeutic targets. To the best of our knowledge, biomarkers in GC ranges from pivotal proteins, non-coding RNAs to plenty of modifications with various specificity and sensitivity, as well as epigenetic liquid biopsy, some of which have already shown favorable clinical utility (Table 4). Liquid biopsy is a simple, fast and non-invasive alternative to surgical biopsies, as blood or body fluid sample is always easy to collect. A sum of circulating tumor cells (CTCs) and cell-free nucleic acids (cfNAs) including DNA, mRNA and microRNAs could be detected in patient blood or body fluid[85]. Available information obtained from liquid biopsy could help doctors with cancer diagnosis and evaluation of clinical outcomes. Up to now, most of epigenetic liquid biopsies in GC were aberrant DNA methylations such as 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), CD40 and GHSR hypermethylation and they even could be used to identify specific cancer types[86-88]. Moreover, CTCs were often detected based on miRNA or mRNA PCR assay due to its low concentration in blood.
| Genes | Purpose | Findings | Ref. |
| RUNX3 | Diagnosis/prognosis | Methylation status correlates with liver metastasis | [155] |
| MLH1 | Diagnosis/prognosis | Methylation status correlates with tumor stage | [156] |
| RASSF1A | Diagnosis/prognosis | Methylation status correlates with advanced stage, and lymph node positivity | [157] |
| MGMT | Diagnosis/prognosis | Methylation status correlates with distant metastasis | [156] |
| ANOS1 | Diagnosis | Expression correlates with tumor progression | [158] |
| RPRML | Prognosis | Expression correlates with survival | [159] |
| CTD-2510F5.4 | Diagnosis/prognosis | Expression correlates with clinicopathological classification and survival | [160] |
| lncRNA-GC1 | Diagnosis | Circulating exosomal level correlates with early detection and disease progression | [161] |
| mesothelin | Diagnosis | Expression correlates with Peritoneal Recurrence | [162] |
| MiR-379-5pMiR-410-3p | Prognosis | Expression correlates with metastasis | [163] |
| S100A9 | Diagnosis /Prognosis | Expression correlates with tumor aggressiveness | [164] |
| Notch1/2/3/4 | Prognosis | Expression correlates with immune infiltration | [165] |
| KAT2A | Diagnosis | Expression correlates with depth of tumor invasion and tumor stage | [166] |
From the therapeutic perspective, targets involved in epigenetic modifications are potential drug targets and they are mainly divided into two groups including enzymes in histone acetylation (HAT or HDAC) and methylation (DNMT or DMT), and non-coding RNAs (miRNA or lncRNA). Some epigenetic drugs have been approved by FDA such as HDAC inhibitors (SAHA) in treatment of cutaneous T-cell lymphoma and DNMT inhibitors (vidaza, decitabine) in treatment of myelodysplatic syndromes[2]. However, most of epigenetic drugs are undergoing clinical or preclinical tests and none of them were currently ready for clinical utility in GC. As the rapid development of GC epigenetics research in recent decades, it is of great significance to integrate existing findings to ensure efficient translation applications (Table 5).
| Drugs | Targets | Status | Ref. |
| Clinical | |||
| Vorinostat + capecitabine + cisplatin | HDAC | Completed phase II test | [167] |
| Vorinostat + folinic acid+ 5‑fluorouracil+ irinotecan | HDAC | Completed phase I test | [168] |
| Azacytidine + epirubicin/oxaliplatin/capecitabine | DNMT | Completed phase I test | [169] |
| Cholecalciferol + HDACi | HDAC | Induce apoptosis in GC cells; Prevent bone loss in preliminary trials; | [170,171] |
| Preclinical | |||
| SAHA | HDAC | Suppress proliferation, induce apoptosis, chemosensitivity in GC cells | [172,173] |
| LBH589 | HDAC | Suppress proliferation, induce chemosensitivity | [174,175] |
| Resveratrol | HAT, HDAC | Suppress proliferation, invasion, tumorigenesis in GC cells | [176,177] |
| Curcumin | HAT, HDAC | Suppress viability, proliferation, migration, induce autophagy, apoptosis in GC cells | [178,179] |
| Quercetin | HAT, HDAC | Induce apoptosis, cell cycle arrest in GC cells | [180,181] |
| Garcinol | HAT, HDAC, SIRTUIN | Suppress oxidation, inflammation, tumorigenesis in GC cells | [182,183] |
| Sodium butyrate | HAT, HDAC | Induce apoptosis in GC cells | [184] |
| Tenovin 6 | SIRTUIN | Induce apoptosis, autophagy in GC cells | [185] |
| DZNEP | HMT | Suppress proliferation, apoptosis, invasion, induce apoptosis in GC cells | [186,187] |
| GSK126 | HMT | Suppress proliferation, cell cycle angiogenesis EMT, tumorigenesis in GC cells | [188,189] |
| Compound 26 | Lysine demethylase | Suppress growth, migration, invasion in GC cells | [190] |
Accumulating evidence revealed the critical role of epigenetic alterations in cancer initiation and progression. Herein, we comprehensively discussed the functions and mechanisms of epigenetic factors in GC. Drugs targeted HAT, HDAC, DNMT are undergoing preclinical and clinical trials, which is promising for improving the efficacy and survival to GC. However, epigenetic studies in GC are still challenged by lack of innovative findings in new types of histone modifications. Succinylation and sumoylation, for instance, have already been reported to participate in tumorigenesis and progression in other gastrointestinal cancers including esophageal, colorectal and liver cancer. We believe combined technologies like single cell sequencing and multiple protein omics sequencing will further broaden epigenetic investigation in gastric malignancy and GC patients will benefit from numerous epigenetic drugs in the future.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Burada F S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55774] [Article Influence: 7967.7] [Reference Citation Analysis (132)] |
| 2. | Theuer CP, Kurosaki T, Ziogas A, Butler J, Anton-Culver H. Asian patients with gastric carcinoma in the United States exhibit unique clinical features and superior overall and cancer specific survival rates. Cancer. 2000;89:1883-1892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 3. | Jin H, Pinheiro PS, Callahan KE, Altekruse SF. Examining the gastric cancer survival gap between Asians and whites in the United States. Gastric Cancer. 2017;20:573-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 4. | Cheong JH, Yang HK, Kim H, Kim WH, Kim YW, Kook MC, Park YK, Kim HH, Lee HS, Lee KH, Gu MJ, Kim HY, Lee J, Choi SH, Hong S, Kim JW, Choi YY, Hyung WJ, Jang E, Huh YM, Noh SH. Predictive test for chemotherapy response in resectable gastric cancer: a multi-cohort, retrospective analysis. Lancet Oncol. 2018;19:629-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 5. | Vogelaar IP, van der Post RS, Bisseling TM, van Krieken JHJ, Ligtenberg MJ, Hoogerbrugge N. Familial gastric cancer: detection of a hereditary cause helps to understand its etiology. Hered Cancer Clin Pract. 2012;10:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Tan P, Yeoh KG. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology. 2015;149:1153-1162.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 363] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 7. | Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, Gong L, Fu J, Jin JG, Choi MG, Sohn TS, Lee JH, Bae JM, Kim ST, Park SH, Sohn I, Jung SH, Tan P, Chen R, Hardwick J, Kang WK, Ayers M, Hongyue D, Reinhard C, Loboda A, Kim S, Aggarwal A. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1071] [Cited by in RCA: 1576] [Article Influence: 157.6] [Reference Citation Analysis (0)] |
| 8. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4845] [Article Influence: 440.5] [Reference Citation Analysis (2)] |
| 9. | Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1738] [Cited by in RCA: 1682] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 10. | Calcagno DQ, Wisnieski F, Mota ERDS, Maia de Sousa SB, Costa da Silva JM, Leal MF, Gigek CO, Santos LC, Rasmussen LT, Assumpção PP, Burbano RR, Smith MA. Role of histone acetylation in gastric cancer: implications of dietetic compounds and clinical perspectives. Epigenomics. 2019;11:349-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Yang G, Yuan Y, Yuan H, Wang J, Yun H, Geng Y, Zhao M, Li L, Weng Y, Liu Z, Feng J, Bu Y, Liu L, Wang B, Zhang X. Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Rep. 2021;22:e50967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 96] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 12. | Du L, Fakih MG, Rosen ST, Chen Y. SUMOylation of E2F1 Regulates Expression of EZH2. Cancer Res. 2020;80:4212-4223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Kishore C. Epigenetic regulation and promising therapies in colorectal cancer. Curr Mol Pharmacol. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Wan J, Liu H, Ming L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed Pharmacother. 2019;111:976-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 15. | Jenke R, Reßing N, Hansen FK, Aigner A, Büch T. Anticancer Therapy with HDAC Inhibitors: Mechanism-Based Combination Strategies and Future Perspectives. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 115] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 16. | Saha S. Histone Modifications and Other Facets of Epigenetic Regulation in Trypanosomatids: Leaving Their Mark. mBio. 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Park YS, Jin MY, Kim YJ, Yook JH, Kim BS, Jang SJ. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann Surg Oncol. 2008;15:1968-1976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 132] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 18. | Wisnieski F, Leal MF, Calcagno DQ, Santos LC, Gigek CO, Chen ES, Artigiani R, Demachki S, Assumpção PP, Lourenço LG, Burbano RR, Smith MC. BMP8B Is a Tumor Suppressor Gene Regulated by Histone Acetylation in Gastric Cancer. J Cell Biochem. 2017;118:869-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Wisnieski F, Calcagno DQ, Leal MF, Chen ES, Gigek CO, Santos LC, Pontes TB, Rasmussen LT, Payão SL, Assumpção PP, Lourenço LG, Demachki S, Artigiani R, Burbano RR, Smith MC. Differential expression of histone deacetylase and acetyltransferase genes in gastric cancer and their modulation by trichostatin A. Tumour Biol. 2014;35:6373-6381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Kim MS, Lee SH, Yoo NJ. Frameshift mutations of tumor suppressor gene EP300 in gastric and colorectal cancers with high microsatellite instability. Hum Pathol. 2013;44:2064-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Deng R, Zhang P, Liu W, Zeng X, Ma X, Shi L, Wang T, Yin Y, Chang W, Wang G, Tao K. HDAC is indispensable for IFN-γ-induced B7-H1 expression in gastric cancer. Clin Epigenetics. 2018;10:153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 22. | Wisnieski F, Calcagno DQ, Leal MF, Santos LC, Gigek CO, Chen ES, Demachki S, Artigiani R, Assumpção PP, Lourenço LG, Burbano RR, Smith MC. CDKN1A histone acetylation and gene expression relationship in gastric adenocarcinomas. Clin Exp Med. 2017;17:121-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Byun SW, Chang YJ, Chung IS, Moss SF, Kim SS. Helicobacter pylori decreases p27 expression through the delta opioid receptor-mediated inhibition of histone acetylation within the p27 promoter. Cancer Lett. 2012;326:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Michalak EM, Burr ML, Bannister AJ, Dawson MA. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat Rev Mol Cell Biol. 2019;20:573-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 369] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 25. | Song Y, Wu F, Wu J. Targeting histone methylation for cancer therapy: enzymes, inhibitors, biological activity and perspectives. J Hematol Oncol. 2016;9:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 112] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 26. | Jarrold J, Davies CC. PRMTs and Arginine Methylation: Cancer's Best-Kept Secret? Trends Mol Med. 2019;25:993-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 243] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 27. | Torres IO, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJ, Fujimori DG. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat Commun. 2015;6:6204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 28. | Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, De W, Wang C, Ji G. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 29. | Donner I, Kiviluoto T, Ristimäki A, Aaltonen LA, Vahteristo P. Exome sequencing reveals three novel candidate predisposition genes for diffuse gastric cancer. Fam Cancer. 2015;14:241-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 30. | He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, Luo GY, Zhu SL, Xie D. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prev. 2012;13:3173-3178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Nishikawaji T, Akiyama Y, Shimada S, Kojima K, Kawano T, Eishi Y, Yuasa Y, Tanaka S. Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer. Oncotarget. 2016;7:67251-67265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 32. | Elmaci İ, Altinoz MA, Sari R, Bolukbasi FH. Phosphorylated Histone H3 (PHH3) as a Novel Cell Proliferation Marker and Prognosticator for Meningeal Tumors: A Short Review. Appl Immunohistochem Mol Morphol. 2018;26:627-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 33. | Besant PG, Attwood PV. Histone H4 histidine phosphorylation: kinases, phosphatases, liver regeneration and cancer. Biochem Soc Trans. 2012;40:290-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Ajiro K, Yoda K, Utsumi K, Nishikawa Y. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J Biol Chem. 1996;271:13197-13201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 108] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Humphrey SJ, James DE, Mann M. Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol Metab. 2015;26:676-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 373] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 36. | Murakami Y. Phosphorylation of repressive histone code readers by casein kinase 2 plays diverse roles in heterochromatin regulation. J Biochem. 2019;166:3-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 37. | Qi H, Yang Z, Dai C, Wang R, Ke X, Zhang S, Xiang X, Chen K, Li C, Luo J, Shao J, Shen J. STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis. Oncogenesis. 2020;9:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 38. | Khan SA, Amnekar R, Khade B, Barreto SG, Ramadwar M, Shrikhande SV, Gupta S. p38-MAPK/MSK1-mediated overexpression of histone H3 serine 10 phosphorylation defines distance-dependent prognostic value of negative resection margin in gastric cancer. Clin Epigenetics. 2016;8:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 39. | Takahashi H, Murai Y, Tsuneyama K, Nomoto K, Okada E, Fujita H, Takano Y. Overexpression of phosphorylated histone H3 is an indicator of poor prognosis in gastric adenocarcinoma patients. Appl Immunohistochem Mol Morphol. 2006;14:296-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 40. | Xu J, Tian F, Chen X, Liu Z, Wu C, Zhao Z. Ras-ERK1/2 signaling participates in the progression of gastric cancer through repressing Aurora B-mediated H1.4 phosphorylation at Ser27. J Cell Physiol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 41. | Wang J, Qiu Z, Wu Y. Ubiquitin Regulation: The Histone Modifying Enzyme's Story. Cells. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 42. | Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1281] [Cited by in RCA: 1481] [Article Influence: 164.6] [Reference Citation Analysis (0)] |
| 43. | Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245-1256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 447] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 44. | Eletr ZM, Wilkinson KD. Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta. 2014;1843:114-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 45. | Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum Mol Genet. 2012;21:559-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 46. | Wang ZJ, Yang JL, Wang YP, Lou JY, Chen J, Liu C, Guo LD. Decreased histone H2B monoubiquitination in malignant gastric carcinoma. World J Gastroenterol. 2013;19:8099-8107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Zhang Q, Wu Y, Xu Q, Ma F, Zhang CY. Recent advances in biosensors for in vitro detection and in vivo imaging of DNA methylation. Biosens Bioelectron. 2021;171:112712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 48. | Ortiz-Barahona V, Joshi RS, Esteller M. Use of DNA methylation profiling in translational oncology. Semin Cancer Biol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 49. | Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2342] [Cited by in RCA: 2348] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 50. | Héberlé É, Bardet AF. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019;63:727-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 194] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 51. | Wang M, Li Y, Gao J, Zhou J, Gu L, Shen L, Deng D. p16 Methylation is associated with chemosensitivity to fluorouracil in patients with advanced gastric cancer. Med Oncol. 2014;31:988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 52. | Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, Amarantidis K, Chelis L, Trypsianis G, Chatzaki E, Lianidou ES, Kakolyris S. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res. 2015;778:46-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 53. | Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 859] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 54. | Kim Y, Wen X, Jeong S, Cho NY, Kim WH, Kang GH. Combinatory low methylation statuses of SAT-α and L1 are associated with shortened survival time in patients with advanced gastric cancer. Gastric Cancer. 2019;22:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 55. | Kurklu B, Whitehead RH, Ong EK, Minamoto T, Fox JG, Mann JR, Judd LM, Giraud AS, Menheniott TR. Lineage-specific RUNX3 hypomethylation marks the preneoplastic immune component of gastric cancer. Oncogene. 2015;34:2856-2866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 56. | Leodolter A, Alonso S, González B, Ebert MP, Vieth M, Röcken C, Wex T, Peitz U, Malfertheiner P, Perucho M. Somatic DNA Hypomethylation in H. pylori-Associated High-Risk Gastritis and Gastric Cancer: Enhanced Somatic Hypomethylation Associates with Advanced Stage Cancer. Clin Transl Gastroenterol. 2015;6:e85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 57. | Leung WK, Man EP, Yu J, Go MY, To KF, Yamaoka Y, Cheng VY, Ng EK, Sung JJ. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res. 2006;12:3216-3221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 58. | Perri F, Cotugno R, Piepoli A, Merla A, Quitadamo M, Gentile A, Pilotto A, Annese V, Andriulli A. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. Pylori infected patients and effect of eradication. Am J Gastroenterol. 2007;102:1361-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 143] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 59. | Niwa T, Toyoda T, Tsukamoto T, Mori A, Tatematsu M, Ushijima T. Prevention of Helicobacter pylori-induced gastric cancers in gerbils by a DNA demethylating agent. Cancer Prev Res (Phila). 2013;6:263-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 60. | Nakajima T, Enomoto S, Yamashita S, Ando T, Nakanishi Y, Nakazawa K, Oda I, Gotoda T, Ushijima T. Persistence of a component of DNA methylation in gastric mucosae after Helicobacter pylori eradication. J Gastroenterol. 2010;45:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 61. | Park JH, Park J, Choi JK, Lyu J, Bae MG, Lee YG, Bae JB, Park DY, Yang HK, Kim TY, Kim YJ. Identification of DNA methylation changes associated with human gastric cancer. BMC Med Genomics. 2011;4:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 62. | Muhammad JS, Eladl MA, Khoder G. Helicobacter pylori-induced DNA Methylation as an Epigenetic Modulator of Gastric Cancer: Recent Outcomes and Future Direction. Pathogens. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 63. | Tekcham DS, Tiwari PK. Non-coding RNAs as emerging molecular targets of gallbladder cancer. Gene. 2016;588:79-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 64. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8672] [Cited by in RCA: 8864] [Article Influence: 277.0] [Reference Citation Analysis (0)] |
| 65. | Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3462] [Cited by in RCA: 3400] [Article Influence: 136.0] [Reference Citation Analysis (0)] |
| 66. | Tsai MM, Wang CS, Tsai CY, Huang HW, Chi HC, Lin YH, Lu PH, Lin KH. Potential Diagnostic, Prognostic and Therapeutic Targets of MicroRNAs in Human Gastric Cancer. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 104] [Article Influence: 11.6] [Reference Citation Analysis (1)] |
| 67. | Wang J, Sun J, Wang J, Song Y, Gao P, Shi J, Chen P, Wang Z. Long noncoding RNAs in gastric cancer: functions and clinical applications. Onco Targets Ther. 2016;9:681-697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 68. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16069] [Article Influence: 1004.3] [Reference Citation Analysis (2)] |
| 69. | Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 676] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 70. | Yu L, Wu D, Gao H, Balic JJ, Tsykin A, Han TS, Liu YD, Kennedy CL, Li JK, Mao JQ, Tan P, Oshima M, Goodall GJ, Jenkins BJ. Clinical Utility of a STAT3-Regulated miRNA-200 Family Signature with Prognostic Potential in Early Gastric Cancer. Clin Cancer Res. 2018;24:1459-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 71. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4439] [Article Influence: 277.4] [Reference Citation Analysis (0)] |
| 72. | Marchese FP, Huarte M. Long non-coding RNAs and chromatin modifiers: their place in the epigenetic code. Epigenetics. 2014;9:21-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 73. | Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 574] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 74. | Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717-726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1198] [Cited by in RCA: 1141] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 75. | Bie L, Luo S, Li D, Wei Y, Mu Y, Chen X, Wang S, Guo P, Lu X. HOTAIR Competitively Binds MiRNA330 as a Molecular Sponge to Increase the Resistance of Gastric Cancer to Trastuzumab. Curr Cancer Drug Targets. 2020;20:700-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 76. | Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, Iijima K, Shimosegawa T, Sugamura K, Satoh K. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 212] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 77. | Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/β-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107:2620-2629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 78. | Xue M, Chen LY, Wang WJ, Su TT, Shi LH, Wang L, Zhang W, Si JM, Wang LJ, Chen SJ. HOTAIR induces the ubiquitination of Runx3 by interacting with Mex3b and enhances the invasion of gastric cancer cells. Gastric Cancer. 2018;21:756-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 79. | Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910-928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 241] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 80. | Ruan Y, Li Z, Shen Y, Li T, Zhang H, Guo J. Functions of circular RNAs and their potential applications in gastric cancer. Expert Rev Gastroenterol Hepatol. 2020;14:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 81. | Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 548] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 82. | Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of CircRNAs. Mol Cell. 2017;66:9-21.e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 988] [Cited by in RCA: 1352] [Article Influence: 169.0] [Reference Citation Analysis (0)] |
| 83. | Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;166:1055-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 154] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 84. | Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem. 2018;119:440-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 228] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 85. | Tsujiura M, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Otsuji E. Liquid biopsy of gastric cancer patients: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2014;20:3265-3286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 45] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 86. | Li W, Zhang X, Lu X, You L, Song Y, Luo Z, Zhang J, Nie J, Zheng W, Xu D, Wang Y, Dong Y, Yu S, Hong J, Shi J, Hao H, Luo F, Hua L, Wang P, Qian X, Yuan F, Wei L, Cui M, Zhang T, Liao Q, Dai M, Liu Z, Chen G, Meckel K, Adhikari S, Jia G, Bissonnette MB, Zhao Y, Zhang W, He C, Liu J. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243-1257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 264] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 87. | Amini M, Ghorban K, Mokhtarzadeh A, Dadmanesh M, Baradaran B. CD40 DNA hypermethylation in primary gastric tumors; as a novel diagnostic biomarker. Life Sci. 2020;254:117774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 88. | Amini M, Foroughi K, Talebi F, Aghagolzade Haji H, Kamali F, Jandaghi P, Hoheisel JD, Manoochehri M. GHSR DNA hypermethylation is a new epigenetic biomarker for gastric adenocarcinoma and beyond. J Cell Physiol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 89. | Xu G, Meng L, Yuan D, Li K, Zhang Y, Dang C, Zhu K. MEG3/miR21 axis affects cell mobility by suppressing epithelialmesenchymal transition in gastric cancer. Oncol Rep. 2018;40:39-48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 90. | Yuan Y, Zhang Y, Han L, Sun S, Shu Y. miR-183 inhibits autophagy and apoptosis in gastric cancer cells by targeting ultraviolet radiation resistance-associated gene. Int J Mol Med. 2018;42:3562-3570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 91. | Lin W, Miao Y, Meng X, Huang Y, Zhao W, Ruan J. miRNA-765 mediates multidrug resistance via targeting BATF2 in gastric cancer cells. FEBS Open Bio. 2020;10:1021-1030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 92. | Shi SS, Zhang HP, Yang CQ, Li LN, Shen Y, Zhang YQ. Exosomal miR-155-5p promotes proliferation and migration of gastric cancer cells by inhibiting TP53INP1 expression. Pathol Res Pract. 2020;216:152986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 93. | Ding L, Li Q, Chakrabarti J, Munoz A, Faure-Kumar E, Ocadiz-Ruiz R, Razumilava N, Zhang G, Hayes MH, Sontz RA, Mendoza ZE, Mahurkar S, Greenson JK, Perez-Perez G, Hanh NTH, Zavros Y, Samuelson LC, Iliopoulos D, Merchant JL. MiR130b from Schlafen4+ MDSCs stimulates epithelial proliferation and correlates with preneoplastic changes prior to gastric cancer. Gut. 2020;69:1750-1761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 94. | Li T, Guo H, Li H, Jiang Y, Zhuang K, Lei C, Wu J, Zhou H, Zhu R, Zhao X, Lu Y, Shi C, Nie Y, Wu K, Yuan Z, Fan DM, Shi Y. MicroRNA-92a-1-5p increases CDX2 by targeting FOXD1 in bile acids-induced gastric intestinal metaplasia. Gut. 2019;68:1751-1763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 95. | Han TS, Voon DC, Oshima H, Nakayama M, Echizen K, Sakai E, Yong ZWE, Murakami K, Yu L, Minamoto T, Ock CY, Jenkins BJ, Kim SJ, Yang HK, Oshima M. Interleukin 1 Up-regulates MicroRNA 135b to Promote Inflammation-Associated Gastric Carcinogenesis in Mice. Gastroenterology. 2019;156:1140-1155.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 96. | Lu Z, Luo T, Pang T, Du Z, Yin X, Cui H, Fang G, Xue X. MALAT1 promotes gastric adenocarcinoma through the MALAT1/miR-181a-5p/AKT3 axis. Open Biol. 2019;9:190095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 97. | Xia M, Wei J, Tong K. MiR-224 promotes proliferation and migration of gastric cancer cells through targeting PAK4. Pharmazie. 2016;71:460-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 98. | Shi Y, Duan Z, Zhang X, Wang G, Li F. Down-regulation of the let-7i facilitates gastric cancer invasion and metastasis by targeting COL1A1. Protein Cell. 2019;10:143-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 99. | Shomali N, Shirafkan N, Duijf PHG, Ghasabi M, Babaloo Z, Yousefi M, Mansoori B, Asadi M, Shanehbandi D, Baghbani E, Mohammadi A, Baradaran B. Downregulation of miR-146a promotes cell migration in Helicobacter pylori-negative gastric cancer. J Cell Biochem. 2019;120:9495-9505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 100. | Zhang W, Liao K, Liu D. MiRNA-12129 Suppresses Cell Proliferation and Block Cell Cycle Progression by Targeting SIRT1 in GASTRIC Cancer. Technol Cancer Res Treat. 2020;19:1533033820928144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 101. | Feng Q, Wu X, Li F, Ning B, Lu X, Zhang Y, Pan Y, Guan W. miR-27b inhibits gastric cancer metastasis by targeting NR2F2. Protein Cell. 2017;8:114-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 102. | Wu K, Zou J, Lin C, Jie ZG. MicroRNA-140-5p inhibits cell proliferation, migration and promotes cell apoptosis in gastric cancer through the negative regulation of THY1-mediated Notch signaling. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 103. | Zhang Y, Yuan Y, Zhang Y, Cheng L, Zhou X, Chen K. SNHG7 accelerates cell migration and invasion through regulating miR-34a-Snail-EMT axis in gastric cancer. Cell Cycle. 2020;19:142-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 104. | Fan Y, Shi Y, Lin Z, Huang X, Li J, Huang W, Shen D, Zhuang G, Liu W. miR-9-5p Suppresses Malignant Biological Behaviors of Human Gastric Cancer Cells by Negative Regulation of TNFAIP8L3. Dig Dis Sci. 2019;64:2823-2829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 105. | Wang CQ. MiR-195 reverses 5-FU resistance through targeting HMGA1 in gastric cancer cells. Eur Rev Med Pharmacol Sci. 2019;23:3771-3778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 106. | Li Y, Wang K, Wei Y, Yao Q, Zhang Q, Qu H, Zhu G. lncRNA-MIAT regulates cell biological behaviors in gastric cancer through a mechanism involving the miR-29a-3p/HDAC4 axis. Oncol Rep. 2017;38:3465-3472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 107. | Liu J, Ben Q, Lu E, He X, Yang X, Ma J, Zhang W, Wang Z, Liu T, Zhang J, Wang H. Long noncoding RNA PANDAR blocks CDKN1A gene transcription by competitive interaction with p53 protein in gastric cancer. Cell Death Dis. 2018;9:168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 108. | Xu TP, Wang WY, Ma P, Shuai Y, Zhao K, Wang YF, Li W, Xia R, Chen WM, Zhang EB, Shu YQ. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene. 2018;37:5020-5036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 109. | Yuan H, Chen Z, Bai S, Wei H, Wang Y, Ji R, Guo Q, Li Q, Ye Y, Wu J, Zhou Y, Qiao L. Molecular mechanisms of lncRNA SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett. 2018;418:84-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 110. | Sun L, Li J, Yan W, Yao Z, Wang R, Zhou X, Wu H, Zhang G, Shi T, Chen W. H19 promotes aerobic glycolysis, proliferation, and immune escape of gastric cancer cells through the microRNA-519d-3p/lactate dehydrogenase A axis. Cancer Sci. 2021;112:2245-2259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 111. | Xu TP, Wang YF, Xiong WL, Ma P, Wang WY, Chen WM, Huang MD, Xia R, Wang R, Zhang EB, Liu YW, De W, Shu YQ. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 112. | Wang YJ, Liu JZ, Lv P, Dang Y, Gao JY, Wang Y. Long non-coding RNA CCAT2 promotes gastric cancer proliferation and invasion by regulating the E-cadherin and LATS2. Am J Cancer Res. 2016;6:2651-2660. [PubMed] |
| 113. | Zhang K, Lu C, Huang X, Cui J, Li J, Gao Y, Liang W, Liu Y, Sun Y, Liu H, Wei B, Chen L. Long noncoding RNA AOC4P regulates tumor cell proliferation and invasion by epithelial-mesenchymal transition in gastric cancer. Therap Adv Gastroenterol. 2019;12:1756284819827697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 114. | Zong W, Feng W, Jiang Y, Ju S, Cui M, Jing R. Evaluating the diagnostic and prognostic value of serum long non-coding RNA CTC-497E21.4 in gastric cancer. Clin Chem Lab Med. 2019;57:1063-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 115. | Miao X, Liu Y, Fan Y, Wang G, Zhu H. LncRNA BANCR Attenuates the Killing Capacity of Cisplatin on Gastric Cancer Cell Through the ERK1/2 Pathway. Cancer Manag Res. 2021;13:287-296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 116. | Zhang ZX, Liu ZQ, Jiang B, Lu XY, Ning XF, Yuan CT, Wang AL. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-κB1. Biochem Biophys Res Commun. 2015;465:225-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 117. | Xiao ZS, Long H, Zhao L, Li HX, Zhang XN. LncRNA HOTTIP promotes proliferation and inhibits apoptosis of gastric carcinoma cells via adsorbing miR-615-3p. Eur Rev Med Pharmacol Sci. 2020;24:6692-6698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 118. | Zhao R, Zhang X, Zhang Y, Yang Y, Sun Y, Zheng X, Qu A, Umwali Y. HOTTIP Predicts Poor Survival in Gastric Cancer Patients and Contributes to Cisplatin Resistance by Sponging miR-216a-5p. Front Cell Dev Biol. 2020;8:348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 119. | Liu J, Wang J, Song Y, Ma B, Luo J, Ni Z, Gao P, Sun J, Zhao J, Chen X, Wang Z. A panel consisting of three novel circulating lncRNAs, is it a predictive tool for gastric cancer? J Cell Mol Med. 2018;22:3605-3613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 120. | Ke D, Li H, Zhang Y, An Y, Fu H, Fang X, Zheng X. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8:21516-21525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 121. | Wei F, Wang Y, Zhou Y, Li Y. Long noncoding RNA CYTOR triggers gastric cancer progression by targeting miR-103/RAB10. Acta Biochim Biophys Sin (Shanghai). 2021;53:1044-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 122. | Teng F, Zhang JX, Chen Y, Shen XD, Su C, Guo YJ, Wang PH, Shi CC, Lei M, Cao YO, Liu SQ. LncRNA NKX2-1-AS1 promotes tumor progression and angiogenesis via upregulation of SERPINE1 expression and activation of the VEGFR-2 signaling pathway in gastric cancer. Mol Oncol. 2021;15:1234-1255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 123. | Xu Y, Li Y, Qiu Y, Sun F, Zhu G, Sun J, Cai G, Lin W, Fu Y, Wu H, Jiang S, Wen Z, Feng F, Luo J, Yang Y, Zhang Q. LncRNA NEAT1 Promotes Gastric Cancer Progression Through miR-17-5p/TGFβR2 Axis Up-Regulated Angiogenesis. Front Cell Dev Biol. 2021;9:705697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 124. | Zhu T, Wang Z, Wang G, Hu Z, Ding H, Li R, Sun J. Long non-coding RNA ZFAS1 promotes the expression of EPAS1 in gastric cardia adenocarcinoma. J Adv Res. 2021;28:7-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 125. | Ma ZH, Shuai Y, Gao XY, Yan Y, Wang KM, Wen XZ, Ji JF. BTEB2-Activated lncRNA TSPEAR-AS2 Drives GC Progression through Suppressing GJA1 Expression and Upregulating CLDN4 Expression. Mol Ther Nucleic Acids. 2020;22:1129-1141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 126. | Song S, He X, Wang J, Song H, Wang Y, Liu Y, Zhou Z, Yu Z, Miao D, Xue Y. A novel long noncoding RNA, TMEM92-AS1, promotes gastric cancer progression by binding to YBX1 to mediate CCL5. Mol Oncol. 2021;15:1256-1273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 127. | Xin L, Zhou LQ, Liu C, Zeng F, Yuan YW, Zhou Q, Li SH, Wu Y, Wang JL, Wu DZ, Lu H. Transfer of LncRNA CRNDE in TAM-derived exosomes is linked with cisplatin resistance in gastric cancer. EMBO Rep. 2021;22:e52124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 80] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 128. | Ding L, Tian Y, Wang L, Bi M, Teng D, Hong S. Hypermethylated long noncoding RNA MEG3 promotes the progression of gastric cancer. Aging (Albany NY). 2019;11:8139-8155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 129. | Cai C, Zhang H, Zhu Y, Zheng P, Xu Y, Sun J, Zhang M, Lan T, Gu B, Li S, Ma P. Serum Exosomal Long Noncoding RNA pcsk2-2:1 As A Potential Novel Diagnostic Biomarker For Gastric Cancer. Onco Targets Ther. 2019;12:10035-10041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 130. | Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J, Shi L, Ma P, Xu Y, Zheng P. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta. 2020;501:252-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 131. | Pan T, Yu Z, Jin Z, Wu X, Wu A, Hou J, Chang X, Fan Z, Li J, Yu B, Li F, Yan C, Yang Z, Zhu Z, Liu B, Su L. Tumor suppressor lnc-CTSLP4 inhibits EMT and metastasis of gastric cancer by attenuating HNRNPAB-dependent Snail transcription. Mol Ther Nucleic Acids. 2021;23:1288-1303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 132. | Xia Y, Lv J, Jiang T, Li B, Li Y, He Z, Xuan Z, Sun G, Wang S, Li Z, Wang W, Wang L, Xu Z. CircFAM73A promotes the cancer stem cell-like properties of gastric cancer through the miR-490-3p/HMGA2 positive feedback loop and HNRNPK-mediated β-catenin stabilization. J Exp Clin Cancer Res. 2021;40:103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 133. | Bu X, Chen Z, Zhang A, Zhou X, Zhang X, Yuan H, Zhang Y, Yin C, Yan Y. Circular RNA circAFF2 accelerates gastric cancer development by activating miR-6894-5p and regulating ANTXR 1 expression. Clin Res Hepatol Gastroenterol. 2021;45:101671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 134. | Yang D, Hu Z, Zhang Y, Zhang X, Xu J, Fu H, Zhu Z, Feng D, Cai Q. CircHIPK3 Promotes the Tumorigenesis and Development of Gastric Cancer Through miR-637/AKT1 Pathway. Front Oncol. 2021;11:637761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 135. | Deng P, Sun M, Zhao WY, Hou B, Li K, Zhang T, Gu F. Circular RNA circVAPA promotes chemotherapy drug resistance in gastric cancer progression by regulating miR-125b-5p/STAT3 axis. World J Gastroenterol. 2021;27:487-500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 136. | Yang H, Wu Z, Liu X, Chen M, Zhang X, Jiang Y. NFIB promotes the progression of gastric cancer by upregulating circMAP7D1 to stabilize HER2 mRNA. Mol Med Rep. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 137. | Hua Y, Wang H, Wu X, Yang L, Wang C, Li X, Jin Y, Li M, Wang L, Dong C, Yin F. Circular RNA Circ_0006282 Promotes Cell Proliferation and Metastasis in Gastric Cancer by Regulating MicroRNA-144-5p/Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein β Axis. Cancer Manag Res. 2021;13:815-827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 138. | Xu Q, Liao B, Hu S, Zhou Y, Xia W. Circular RNA 0081146 facilitates the progression of gastric cancer by sponging miR-144 and up-regulating HMGB1. Biotechnol Lett. 2021;43:767-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 139. | Wang L, Li B, Yi X, Xiao X, Zheng Q, Ma L. Circ_SMAD4 promotes gastric carcinogenesis by activating wnt/β-catenin pathway. Cell Prolif. 2021;54:e12981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 140. | Yu L, Xie J, Liu X, Yu Y, Wang S. Plasma Exosomal CircNEK9 Accelerates the Progression of Gastric Cancer via miR-409-3p/MAP7 Axis. Dig Dis Sci. 2021;66:4274-4289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 141. | Yue F, Peng K, Zhang L, Zhang J. Circ_0004104 Accelerates the Progression of Gastric Cancer by Regulating the miR-539-3p/RNF2 Axis. Dig Dis Sci. 2021;66:4290-4301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 142. | Wang X, Zhang Y, Li W, Liu X. Knockdown of cir_RNA PVT1 Elevates Gastric Cancer Cisplatin Sensitivity via Sponging miR-152-3p. J Surg Res. 2021;261:185-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 143. | Li J, Yang Y, Xu D, Cao L. hsa_circ_0023409 Accelerates Gastric Cancer Cell Growth and Metastasis Through Regulating the IRS4/PI3K/AKT Pathway. Cell Transplant. 2021;30:963689720975390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 144. | Yang Y, Cai B, Shi X, Duan C, Tong T, Yu C. circ_0044516 functions in the progression of gastric cancer by modulating MicroRNA-149-5p/HuR axis. Mol Cell Biochem. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 145. | Cao J, Zhang X, Xu P, Wang H, Wang S, Zhang L, Li Z, Xie L, Sun G, Xia Y, Lv J, Yang J, Xu Z. Circular RNA circLMO7 acts as a microRNA-30a-3p sponge to promote gastric cancer progression via the WNT2/β-catenin pathway. J Exp Clin Cancer Res. 2021;40:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 146. | Niu Q, Dong Z, Liang M, Luo Y, Lin H, Lin M, Zhong X, Yao W, Weng J, Zhou X. Circular RNA hsa_circ_0001829 promotes gastric cancer progression through miR-155-5p/SMAD2 axis. J Exp Clin Cancer Res. 2020;39:280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 147. | Pu Z, Xu M, Yuan X, Xie H, Zhao J. Circular RNA circCUL3 Accelerates the Warburg Effect Progression of Gastric Cancer through Regulating the STAT3/HK2 Axis. Mol Ther Nucleic Acids. 2020;22:310-318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 148. | Wang H, Sun G, Xu P, Lv J, Zhang X, Zhang L, Wang S, Cao J, Xia Y, Xuan Z, Li B, Huang X, Jiang T, Fang L, Xu Z. Circular RNA TMEM87A promotes cell proliferation and metastasis of gastric cancer by elevating ULK1 via sponging miR-142-5p. J Gastroenterol. 2021;56:125-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 149. | Ma S, Kong S, Gu X, Xu Y, Tao M, Shen L, Shen X, Ju S. As a biomarker for gastric cancer, circPTPN22 regulates the progression of gastric cancer through the EMT pathway. Cancer Cell Int. 2021;21:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 150. | Ma C, Wang X, Yang F, Zang Y, Liu J, Xu X, Li W, Jia J, Liu Z. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020;19:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 151. | Wang H, Wang N, Zheng X, Wu L, Fan C, Li X, Ye K, Han S. Circular RNA hsa_circ_0009172 suppresses gastric cancer by regulation of microRNA-485-3p-mediated NTRK3. Cancer Gene Ther. 2021;28:1312-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 152. | Li T, Zuo X, Meng X. Circ_002059 suppresses cell proliferation and migration of gastric cancer via miR-182/MTSS1 axis. Acta Biochim Biophys Sin (Shanghai). 2021;53:454-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 153. | Wang Y, Wang H, Zheng R, Wu P, Sun Z, Chen J, Zhang L, Zhang C, Qian H, Jiang J, Xu W. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle. 2021;20:522-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 154. | Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, Li X, Dang Y, Zhang G. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 175] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 155. | Sakakura C, Hamada T, Miyagawa K, Nishio M, Miyashita A, Nagata H, Ida H, Yazumi S, Otsuji E, Chiba T, Ito K, Ito Y. Quantitative analysis of tumor-derived methylated RUNX3 sequences in the serum of gastric cancer patients. Anticancer Res. 2009;29:2619-2625. [PubMed] |
| 156. | Kolesnikova EV, Tamkovich SN, Bryzgunova OE, Shelestyuk PI, Permyakova VI, Vlassov VV, Tuzikov AS, Laktionov PP, Rykova EY. Circulating DNA in the blood of gastric cancer patients. Ann N Y Acad Sci. 2008;1137:226-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 157. | Pimson C, Ekalaksananan T, Pientong C, Promthet S, Putthanachote N, Suwanrungruang K, Wiangnon S. Aberrant methylation of PCDH10 and RASSF1A genes in blood samples for non-invasive diagnosis and prognostic assessment of gastric cancer. PeerJ. 2016;4:e2112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 158. | Kanda M, Shimizu D, Fujii T, Sueoka S, Tanaka Y, Ezaka K, Takami H, Tanaka H, Hashimoto R, Iwata N, Kobayashi D, Tanaka C, Yamada S, Nakayama G, Sugimoto H, Koike M, Fujiwara M, Kodera Y. Function and diagnostic value of Anosmin-1 in gastric cancer progression. Int J Cancer. 2016;138:721-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 159. | Alarcón MA, Olivares W, Córdova-Delgado M, Muñoz-Medel M, de Mayo T, Carrasco-Aviño G, Wichmann I, Landeros N, Amigo J, Norero E, Villarroel-Espíndola F, Riquelme A, Garrido M, Owen GI, Corvalán AH. The Reprimo-Like Gene Is an Epigenetic-Mediated Tumor Suppressor and a Candidate Biomarker for the Non-Invasive Detection of Gastric Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 160. | Nanishi K, Konishi H, Shoda K, Arita T, Kosuga T, Komatsu S, Shiozaki A, Kubota T, Fujiwara H, Okamoto K, Ichikawa D, Otsuji E. Circulating circERBB2 as a potential prognostic biomarker for gastric cancer: An investigative study. Cancer Sci. 2020;111:4177-4186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 161. | Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H, Zhang K, Li J, Chang R, Xie T, Wang X, Li B, Chen Y, Yang Y, Chen L. Circulating Exosomal Gastric Cancer-Associated Long Noncoding RNA1 as a Biomarker for Early Detection and Monitoring Progression of Gastric Cancer: A Multiphase Study. JAMA Surg. 2020;155:572-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 127] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 162. | Shin SJ, Park S, Kim MH, Nam CM, Kim H, Choi YY, Jung MK, Choi HJ, Rha SY, Chung HC. Mesothelin Expression Is a Predictive Factor for Peritoneal Recurrence in Curatively Resected Stage III Gastric Cancer. Oncologist. 2019;24:e1108-e1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 163. | Liu X, Chu KM. Exosomal miRNAs as circulating biomarkers for prediction of development of haematogenous metastasis after surgery for stage II/III gastric cancer. J Cell Mol Med. 2020;24:6220-6232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (2)] |
| 164. | Zhao Z, Zhang C, Zhao Q. S100A9 as a novel diagnostic and prognostic biomarker in human gastric cancer. Scand J Gastroenterol. 2020;55:338-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 165. | Hu J, Yu J, Gan J, Song N, Shi L, Liu J, Zhang Z, Du J. Notch1/2/3/4 are prognostic biomarker and correlated with immune infiltrates in gastric cancer. Aging (Albany NY). 2020;12:2595-2609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 166. | Meng X, Zhao Y, Liu J, Wang L, Dong Z, Zhang T, Gu X, Zheng Z. Comprehensive analysis of histone modification-associated genes on differential gene expression and prognosis in gastric cancer. Exp Ther Med. 2019;18:2219-2230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 167. | Yoo C, Ryu MH, Na YS, Ryoo BY, Lee CW, Kang YK. Vorinostat in combination with capecitabine plus cisplatin as a first-line chemotherapy for patients with metastatic or unresectable gastric cancer: phase II study and biomarker analysis. Br J Cancer. 2016;114:1185-1190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 168. | Yoo C, Ryu MH, Na YS, Ryoo BY, Lee CW, Maeng J, Kim SY, Koo DH, Park I, Kang YK. Phase I and pharmacodynamic study of vorinostat combined with capecitabine and cisplatin as first-line chemotherapy in advanced gastric cancer. Invest New Drugs. 2014;32:271-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 169. | Schneider BJ, Shah MA, Klute K, Ocean A, Popa E, Altorki N, Lieberman M, Schreiner A, Yantiss R, Christos PJ, Palmer R, You D, Viale A, Kermani P, Scandura JM. Phase I Study of Epigenetic Priming with Azacitidine Prior to Standard Neoadjuvant Chemotherapy for Patients with Resectable Gastric and Esophageal Adenocarcinoma: Evidence of Tumor Hypomethylation as an Indicator of Major Histopathologic Response. Clin Cancer Res. 2017;23:2673-2680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 170. | Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, Wu DL, Ho IHT, Wu JCY, Cheung CKY, Zhang YC, Lau AHY, Ashktorab H, Smoot DT, Fang EF, Chan MTV, Gin T, Gong W, Wu WKK, Cho CH. Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy. 2019;15:707-725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 171. | Ha J, Lee JM, Lim Y, Kim MK, Kwon HS, Song KH, Jeon HM, Kang MI, Baek KH. Effect of bisphosphonate on the prevention of bone loss in patients with gastric cancer after gastrectomy: A randomized controlled trial. Bone. 2020;130:115138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 172. | Seah KS, Loh JY, Nguyen TTT, Tan HL, Hutchinson PE, Lim KK, Dymock BW, Long YC, Lee EJD, Shen HM, Chen ES. SAHA and cisplatin sensitize gastric cancer cells to doxorubicin by induction of DNA damage, apoptosis and perturbation of AMPK-mTOR signalling. Exp Cell Res. 2018;370:283-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 173. | Xiong K, Zhang H, Du Y, Tian J, Ding S. Identification of HDAC9 as a viable therapeutic target for the treatment of gastric cancer. Exp Mol Med. 2019;51:1-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 174. | Regel I, Merkl L, Friedrich T, Burgermeister E, Zimmermann W, Einwächter H, Herrmann K, Langer R, Röcken C, Hofheinz R, Schmid R, Ebert MP. Pan-histone deacetylase inhibitor panobinostat sensitizes gastric cancer cells to anthracyclines via induction of CITED2. Gastroenterology. 2012;143:99-109.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 175. | Singh A, Patel P, Jageshwar, Patel VK, Jain DK, Kamal M, Rajak H. The Safety, Efficacy and Therapeutic Potential of Histone Deacetylase Inhibitors with Special Reference to Panobinostat in Gastrointestinal Tumors: A Review of Preclinical and Clinical Studies. Curr Cancer Drug Targets. 2018;18:720-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 176. | Kim S, Kim W, Kim DH, Jang JH, Kim SJ, Park SA, Hahn H, Han BW, Na HK, Chun KS, Choi BY, Surh YJ. Resveratrol suppresses gastric cancer cell proliferation and survival through inhibition of PIM-1 kinase activity. Arch Biochem Biophys. 2020;689:108413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 177. | Yang T, Zhang J, Zhou J, Zhu M, Wang L, Yan L. Resveratrol inhibits Interleukin-6 induced invasion of human gastric cancer cells. Biomed Pharmacother. 2018;99:766-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 178. | Barati N, Momtazi-Borojeni AA, Majeed M, Sahebkar A. Potential therapeutic effects of curcumin in gastric cancer. J Cell Physiol. 2019;234:2317-2328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 179. | Li W, Zhou Y, Yang J, Li H, Zhang H, Zheng P. Curcumin induces apoptotic cell death and protective autophagy in human gastric cancer cells. Oncol Rep. 2017;37:3459-3466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 180. | Shang HS, Lu HF, Lee CH, Chiang HS, Chu YL, Chen A, Lin YF, Chung JG. Quercetin induced cell apoptosis and altered gene expression in AGS human gastric cancer cells. Environ Toxicol. 2018;33:1168-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 181. | Hemati M, Haghiralsadat F, Jafary F, Moosavizadeh S, Moradi A. Targeting cell cycle protein in gastric cancer with CDC20siRNA and anticancer drugs (doxorubicin and quercetin) co-loaded cationic PEGylated nanoniosomes. Int J Nanomedicine. 2019;14:6575-6585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 182. | Liu C, Ho PC, Wong FC, Sethi G, Wang LZ, Goh BC. Garcinol: Current status of its anti-oxidative, anti-inflammatory and anti-cancer effects. Cancer Lett. 2015;362:8-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 141] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 183. | Zheng Y, Guo C, Zhang X, Wang X, Ma A. Garcinol acts as an antineoplastic agent in human gastric cancer by inhibiting the PI3K/AKT signaling pathway. Oncol Lett. 2020;20:667-676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 184. | Shin H, Lee YS, Lee YC. Sodium butyrate-induced DAPK-mediated apoptosis in human gastric cancer cells. Oncol Rep. 2012;27:1111-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 185. | Ke X, Qin Q, Deng T, Liao Y, Gao SJ. Heterogeneous Responses of Gastric Cancer Cell Lines to Tenovin-6 and Synergistic Effect with Chloroquine. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 186. | Huang R, Jin X, Gao Y, Yuan H, Wang F, Cao X. DZNep inhibits Hif-1α and Wnt signalling molecules to attenuate the proliferation and invasion of BGC-823 gastric cancer cells. Oncol Lett. 2019;18:4308-4316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 187. | Clermont PL, Fornaro L, Crea F. Elevated expression of a pharmacologic Polycomb signature predicts poor prognosis in gastric and breast cancer. Epigenomics. 2017;9:1329-1335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 188. | Liu S, Rong G, Li X, Geng L, Zeng Z, Jiang D, Yang J, Wei Y. Diosgenin and GSK126 Produce Synergistic Effects on Epithelial-Mesenchymal Transition in Gastric Cancer Cells by Mediating EZH2 via the Rho/ROCK Signaling Pathway. Onco Targets Ther. 2020;13:5057-5067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 189. | Chen YT, Zhu F, Lin WR, Ying RB, Yang YP, Zeng LH. The novel EZH2 inhibitor, GSK126, suppresses cell migration and angiogenesis via down-regulating VEGF-A. Cancer Chemother Pharmacol. 2016;77:757-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 190. | Zheng YC, Duan YC, Ma JL, Xu RM, Zi X, Lv WL, Wang MM, Ye XW, Zhu S, Mobley D, Zhu YY, Wang JW, Li JF, Wang ZR, Zhao W, Liu HM. Triazole-dithiocarbamate based selective lysine specific demethylase 1 (LSD1) inactivators inhibit gastric cancer cell growth, invasion, and migration. J Med Chem. 2013;56:8543-8560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 191] [Article Influence: 15.9] [Reference Citation Analysis (0)] |