Published online Jul 15, 2021. doi: 10.4251/wjgo.v13.i7.673
Peer-review started: February 20, 2021
First decision: March 29, 2021
Revised: April 6, 2021
Accepted: June 17, 2021
Article in press: June 17, 2021
Published online: July 15, 2021
Processing time: 140 Days and 1.3 Hours
Tumor-specific neoantigens, which are expressed on tumor cells, can induce an effective antitumor cytotoxic T-cell response and mediate tumor regression. Among tumor immunotherapies, neoantigen vaccines are in early human clinical trials and have demonstrated substantial efficiency. Compared with more neoantigens in melanoma, the paucity and inefficient identification of effective neoantigens in hepatocellular carcinoma (HCC) remain enormous challenges in effectively treating this malignancy. In this review, we highlight the current development of HCC neoantigens in its generation, screening, and identification. We also discuss the possibility that there are more effective neoantigens in hepatitis B virus (HBV)-related HCC than in non-HBV-related HCC. In addition, since HCC is an immunosuppressive tumor, strategies that reverse immunosuppression and enhance the immune response should be considered for the practical exploitation of HCC neoantigens. In summary, this review offers some strategies to solve existing problems in HCC neoantigen research and provide further insights for immunotherapy.
Core Tip: In previous studies of tumor immunotherapy strategies targeting neoantigens, the unique events generating the neoantigens in each tumor have not been well studied. In this paper, we analyze the generation events of hepatocellular carcinoma (HCC) neoantigens. We also provide some methods for screening and identifying HCC neoantigens and clinical strategies for exploiting HCC neoantigens.
- Citation: Chen P, Fang QX, Chen DB, Chen HS. Neoantigen vaccine: An emerging immunotherapy for hepatocellular carcinoma. World J Gastrointest Oncol 2021; 13(7): 673-683
- URL: https://www.wjgnet.com/1948-5204/full/v13/i7/673.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i7.673
Hepatocellular carcinoma (HCC) is the most common type of primary malignant liver cancer. It is also one of the most common malignant tumors with high mortality in the world. Due to the influence of risk factors such as viral hepatitis, alcoholism, and obesity, the incidence rate of HCC continues to rise. Despite progress in treatments for HCC, such as surgery, percutaneous intratumoral injection of absolute ethanol, radiofrequency ablation, hepatic artery embolization, liver transplantation, and targeted therapy, the prognosis remains poor because of the high recurrence rate and high metastasis rate[1,2].
In recent years, immunotherapy holds great promise to patients with HCC. Immune checkpoint inhibitor (ICI) therapy[3] and adoptive cell therapy, represented by anti-programmed cell death 1 (PD-1) antibody therapy and chimeric antigen receptor T cell (CAR-T) therapy[4], have made a major contribution to HCC immunotherapy. However, there remain limitations in these immunotherapies. Anti-PD-1 antibody therapy induces severe side effects in most patients and the beneficiary group is limited; CAR-T therapy has low effectiveness in solid tumors. These shortcomings limited wide clinical application of HCC immunotherapy, and new therapeutic strategies are needed to remedy these obstacles. The study of HCC neoantigens and personalized neoantigen vaccines is a promising direction. Here, we review the current research related to HCC neoantigens. Specifically, we discuss the methods for screening and identifying HCC neoantigens and strategies for exploiting HCC neoantigens in immunotherapy.
Neoantigens are novel peptides that are generated by special events, such as gene mutation, alternative splicing, and virus integration in tumor cells. They not only have good tumor specificity but also a high degree of immunogenicity. Therefore, neoantigens can be well recognized and attacked by the immune system in humans[5-8].
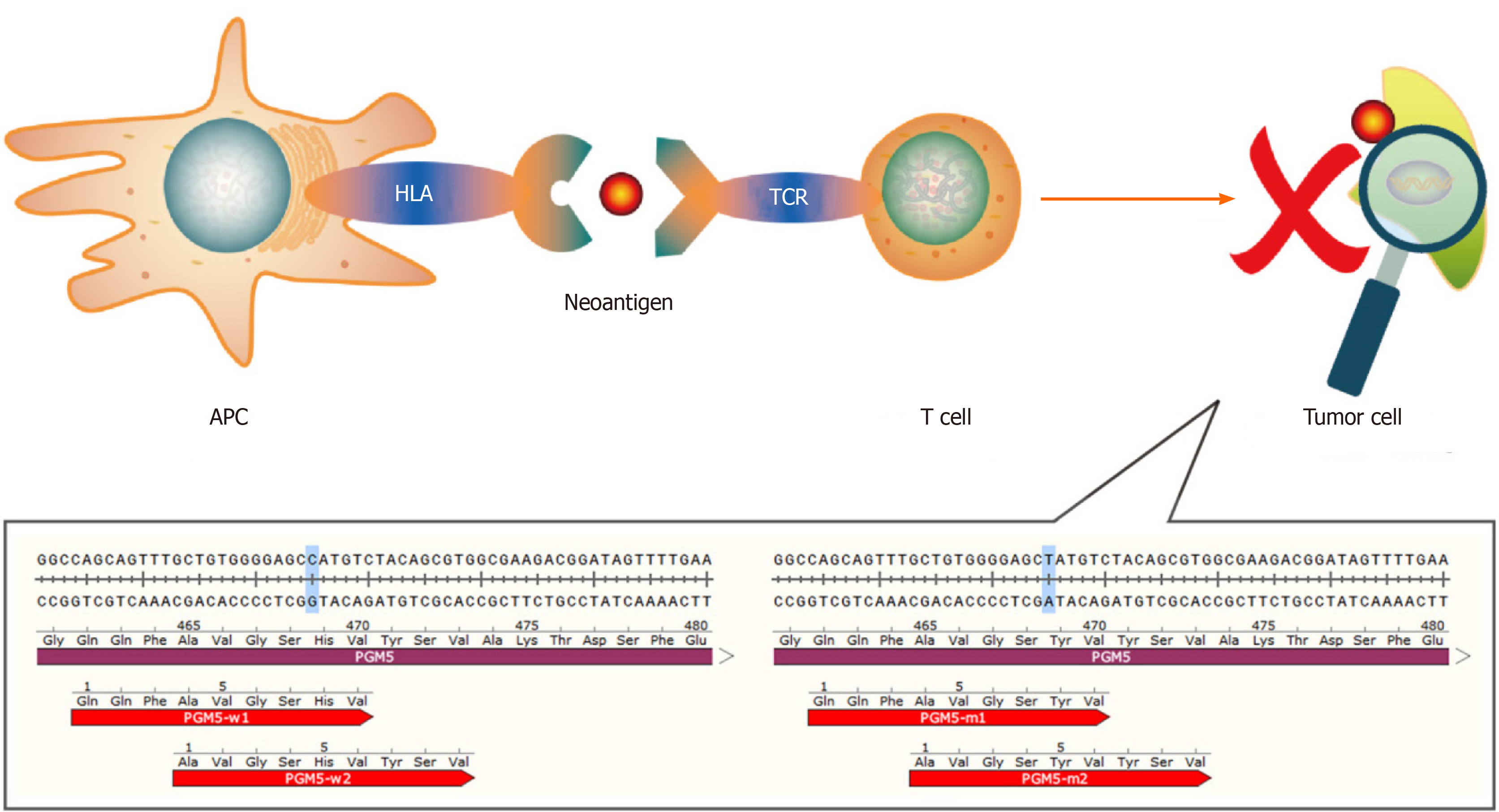
The neoantigens that have been identified are mainly produced by gene mutations. Most mutations are single-base mutations and produce single neoantigens whose lengths range from 7 to 11 amino acids[9,10]. However, there are also some special cases. For example, mutation of C to T in the PGM5 gene replaces the histidine residue with tyrosine. As such, two 9-peptide neoantigens are generated under the action of the proteasome[11] (Figure 1). Tumor that have higher tumor mutation burden (TMB) can produce more neoantigens as a result of mutations than those with lower TMB. A study showed that melanoma has the highest TMB among 30 different types of tumors, and it is generally believed that melanoma has the most mutation-induced neoantigens. The TMB of HCC ranks 12th, and the median TMB in HCC is approximately 2.0 mutations/megabase[12]. A study found some mutation-induced neoantigens in 5 cases of melanoma but failed in 16 cases of HCC with the same workflow[13]. The above results suggest that the process used for detection of mutation-induced neoantigens in melanoma may not be ideal for HCC mutation-induced neoantigens. And it is necessary to optimize this workflow according to the characteristics of HCC when we use it in HCC. We have an in-depth discussion in this review.
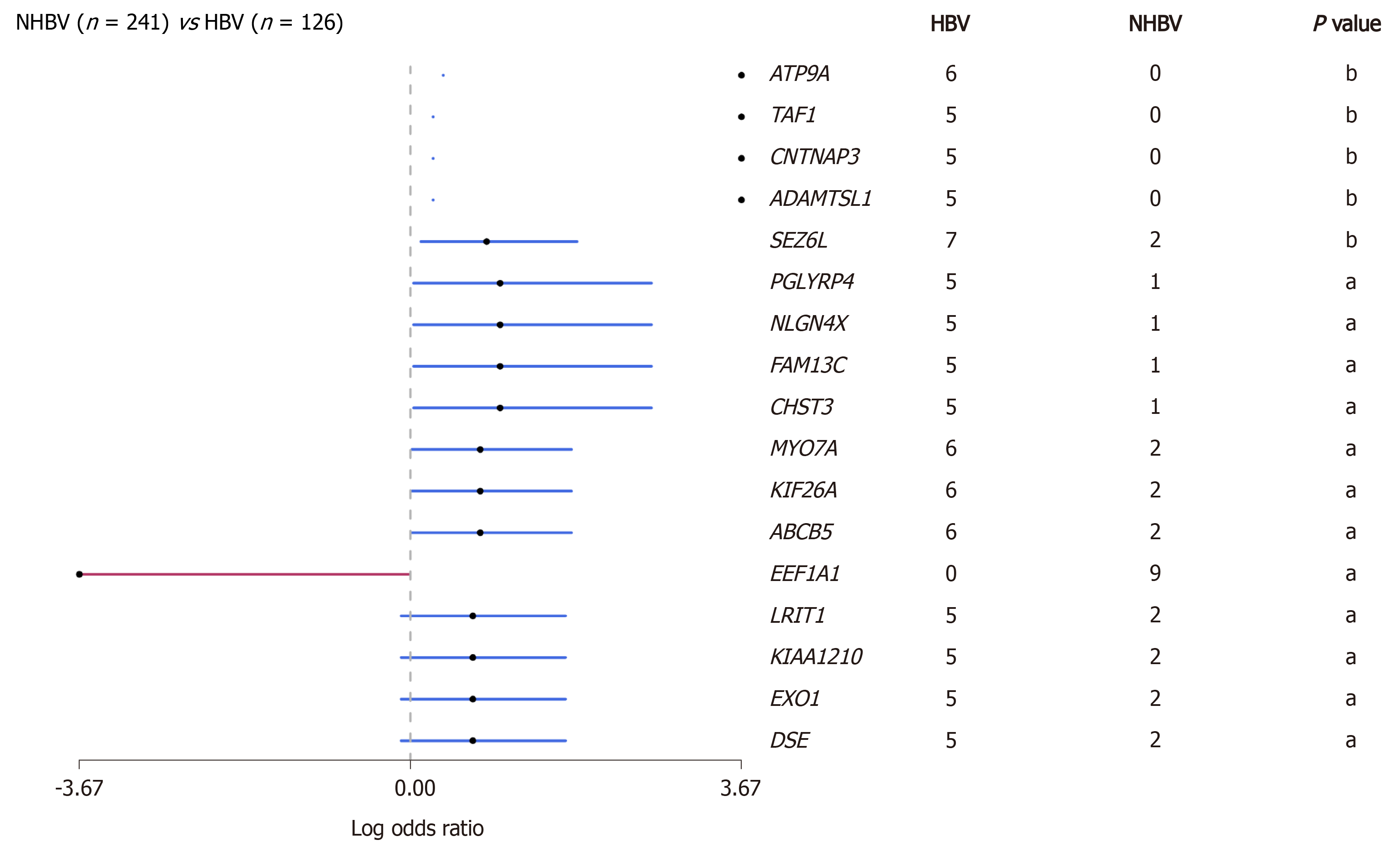
Hepatitis B virus (HBV) infection is the main cause of liver cirrhosis and HCC. HBV-related HCC accounts for the majority of HCC cases in the world, especially in Asians[14,15]. Therefore, the neoantigen spectrum of HCC in Asians may be quite different from that in other populations. Due to long-term viral infection that may have an impact on tumor mutation patterns, we conducted a preliminary analysis of the HBV-related HCC genome using data from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/tcga) and found that the mutated genes in this cohort were significantly different from those in a non-HBV-related HCC cohort (Figure 2). A total of 1973 mutation-induced peptides were identified in 159 cases of HBV-related HCC, including some mutation-induced neoantigens[16]. It gives us a clue that there could be more effective mutation-induced neoantigens in HBV-related HCC than in non-HBV-related HCC.
In addition, there are many events of alternative splicing and virus integration in HCC. These events are also likely to produce neoantigens, but they have not been fully studied. There are more alternative splicing events in tumors vs normal tissue. Among them, retained intron (RI) is the most worthy of attention. The neoantigens produced from the specific RI in the tumor may have great immunogenicity[17-19]. One of the most noteworthy virus integration is HBV integration. By inserting a promoter into the noncoding region, HBV can transcribe not only its own genes but also part of the noncoding region sequence, resulting in the production of viral peptides and the peptides derived from noncoding region. This integration of HBV is an early driver event and remains extremely stable during HCC progression[20]. Therefore, if this mechanism can produce neoantigens, it will be a very good target for HCC immunotherapy. In addition, adeno-associated virus 2 (AAV2) also can integrate into the genome in HCC. Moreover, the positions of virus integration are relatively located in the same sites. For example, both HBV integration and AAV2 integration have been found in the CCNA2 gene in HCC, and all the integration sites were located in the intron region between the second exon and the third exon of the gene[21,22]. Therefore, through systematic analysis of these integration events, we may be able to find some neoantigens that exist in the early dominant clones and remain relatively stable throughout virus-related HCC progression.
Unfortunately, the research on HCC neoantigens is mostly limited to a single sample of the HCC tumor now. In comparison, the sampling of circulating tumor cells (CTCs) in human peripheral blood is more convenient. A large amount of biological information such as gene mutations can be obtained from CTCs[23]. The detection of genes related to HCC in CTCs or circulating tumor DNA is also helpful for obtaining the neoantigen spectrum of inoperable advanced HCC or unresectable metastatic HCC. It is also beneficial for obtaining the neoantigen spectrum of potential recurrent and metastatic lesions (earlier than could be achieved by imaging)[24-27]. In addition, there may be some tumor cells in the ascites of patients with HCC. Whether the HCC neoantigen spectrum can be determined from the ascites is also worth studying.
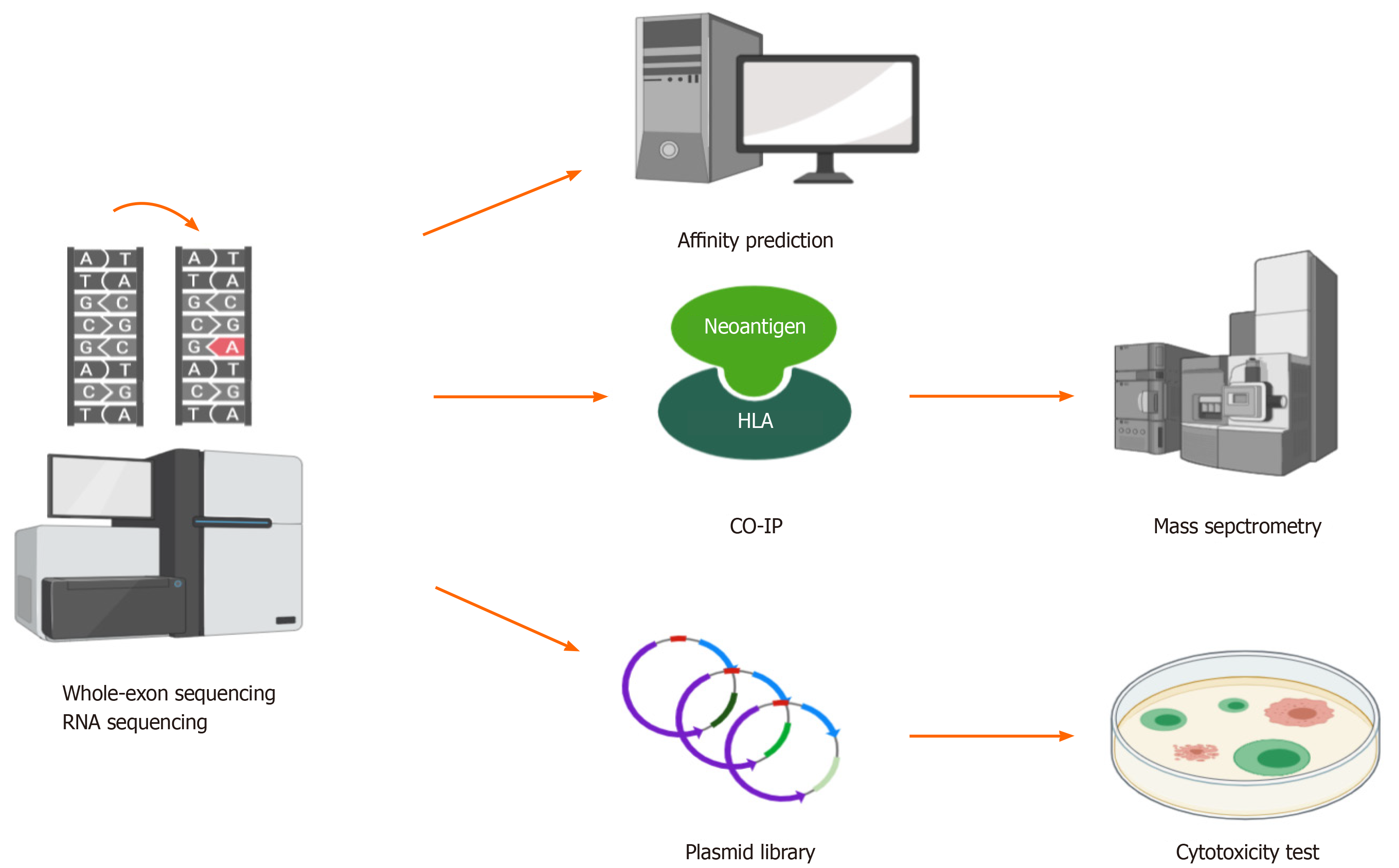
The most challenging process of neoantigen immunotherapy is the screening and identification of neoantigens. At present, there are three classical methods that can be used to screen and identify HCC neoantigens (Figure 3).
The first method is predicting neoantigens with a computer algorithm. For mutation-induced neoantigens, accurate prediction requires a large amount of data and many calculations, which mainly require the following aspects: (1) Accurate identification of all kinds of mutation sites from billions of bases; (2) Calculation of the frequency and expression level of mutation sites; (3) Accurate identification of patient alleles from thousands of human leukocyte antigen (HLA) alleles; and (4) Evaluation of the immunogenicity of the antigen peptides produced by each mutation site by bioinformatics and artificial intelligence (AI) algorithms. However, accurate prediction is difficult. A large number of tools and algorithms have been developed to predict the affinity between antigen peptides and HLA molecules, but their accuracy is still the main bottleneck of all kinds of methods. Khodadoust et al[28] predicted 108 candidate peptides in eight tumor patients by means of this approach. After verification experiments with peptide-HLA tetramer T-cell responses against predicted neoantigens, it was found that all of the predictions had failed. The accuracy of computer prediction of neoantigens needs to be improved. Since the quantity and quality of overall gene mutations as neoantigens in HCC are not high, the previously mentioned sources of neoantigens, that is, virus infection, virus integration, and alternative splicing, should be taken into account when making predictions. Deeply comparing the affinity of HLAs between candidate peptides and wild-type peptides, excluding the loss of HLA heterozygosity early, investigating the structure and properties of unique amino acid, and using AI tools to reconstruct and analyze the three-dimensional structure of HLA molecules, peptides, and T cell receptors, are better for screening and identifying HCC neoantigens than traditional prediction algorithms[29-33].
The second method is the combination of coimmunoprecipitation and mass spectrometry to screen and identify HCC neoantigens. There is experimental evidence supporting this combination at the peptide level, and its accuracy is higher than that of computer algorithms. The combination can be used to establish a high-affinity peptide database of high-frequency HLAs in human populations. The limitation of this method is that experiments are complex with a long period[34-36].
The two methods introduced above mainly depend on the affinity between candidate peptides and HLA molecules. However, not all candidate peptides with high affinity can induce a strong immune response. Therefore, functional experiments, in which neoantigens are screened and identified based on their susceptibility to killing by specific T cells, need to be performed.
Thus, the third method is constructing a library and performing cytotoxic experiments to screen and identify neoantigens. This method is superior at present. Lu et al[37] cultured and activated tumor-infiltrating lymphocytes (TILs) from patients as effector cells and constructed target cells transfected tandem minigene libraries. Through detecting the secretion of interferon-γ (IFN-γ) in cytotoxic experiments, they successfully obtained two melanoma neoantigens[37]. Since TILs from the HCC tissue samples could also be cultured and activated[38], this method is also feasible for the screening and identification of HCC neoantigens. However, there are some shortcomings of this method. On the one hand, it is inefficient to monitor the immune response by detecting the secretion of IFN-γ based on enzyme linked immunosorbent assay (ELISA), and the experimental steps are cumbersome. On the other hand, some nonnatural genes can be artificially generated by stringing genes together, which can produce systematic errors. Recently, Kula et al[39] established a high-throughput platform called T-Scan to screen and identify antigens that can be specifically recognized by T cells. T-Scan uses lentiviral delivery of antigen libraries into cells for endogenous processing and presentation on HLA molecules. Target cells functionally recognized by T cells are isolated using a reporter for granzyme B activity, and the antigens mediating recognition are identified by next-generation sequencing[39]. Such a platform can be used as a more optimized screening and identification tool for HCC neoantigens.
If HCC tumors can be induced to have more neoantigens, we could screen and identify more quality epitopes. Russo et al[40] found that targeted inhibition of epidermal growth factor receptor/B-raf could reduce gene repair and increase the mutation of genes and antigens in colorectal cancers[40]. In the treatment of non-small-cell lung cancer, Formenti et al[41] found that radiotherapy could not only promote the activation of antitumor T cells but also partly induce the exposure of immunogenic mutations[41]. In addition, another study showed that combination treatment with sialic acid and a histone deacetylase inhibitor in neuroblastoma could upregulate the expression of the GD2 antigen, which is the main target of neuroblastoma immunotherapy[42]. Whether the above strategies that could improve antigen exposure and antigen expression can be used to increase the number and quality of HCC neoan
Dong et al[43] screened and identified some HCC neoantigens and evaluated the proportion and immune function of T cells recognizing them by peptide-HLA tetramer T-cell responses against predicted neoantigens and ELISA in vitro, which confirmed that HCC neoantigen-specific T cells have good killing function in vitro[43]. TP53 is the most commonly mutated gene in HCC. Yang et al[4] found that patients with a TP53 neoantigen had longer overall survival, higher immune score, better prognosis, higher cytotoxic lymphocyte (CTL) infiltration, and higher cytolytic activity score than patients without. However, the prognosis of the patients was not correlated with TMB or neoantigen burden[44]. Moreover, a study analyzed 115 cases from the TCGA database and found that somatic mutations and neoantigen burden were not associated with progression-free survival in HCC patients who did not receive immunotherapy. In patients with high expression of granzyme A, a direct correlation between the number and quality of neoantigens and survival is observed. Such evidence shows that neoantigens are useful only when the cytotoxic activity of TILs is high[45]. The same results may be observed in cases of high expression of granzyme B and IFN-γ, which may be closely related to the fact that HCC is an immunosuppressive tumor[46]. TILs in HCC often show an exhausted phenotype with low cytotoxic activity[47]. An increasing number of genes related to TIL function exhaustion have been found, such as PD-1[48], programmed death-ligand 1[49], and CTLA4[50]. Various targeted inhibitors aimed at these genes have been developed. Recently, nivolumab, which is a fully human immunoglobulin G4 anti-PD-1 monoclonal antibody, was approved by the Food and Drug Administration for liver cancer as a second line treatment after failure of sorafenib based on the data of the multi-cohort phase 1/2 trial CheckMate-040[51]. Using combination treatment with interleukin-2 (IL-2)[52] and other cytokines, TIL function exhaustion can be reversed.
To further maximize the effect of neoantigen vaccines, researchers have developed a variety of new technologies. The latest research has shown that expressing neoantigens in chimeric HBV core antigens is a promising option that can selectively induce tumor-specific effector CD8+ T cell activation through DNA vaccination. In this study, the researchers constructed a Db/Sp244-252/R251H neoantigen epitope (changing an amino acid site) in the EndoB2-Sp protein and found that a single injection of the EndoB2-Sp expression vector into C57Bl/6j mice could effectively induce activation of IFN-γ+CD8+ T cells specifically targeting the neoantigen epitope (Db/Sp244-252/R251H). The expression of Db/Sp244-252/R251H within the core antigen of assembly deficient HBV induced a considerable number of CD8+ T cells specifically targeting Db/Sp244-252/R251H compared with the EndoB2-Sp vaccine[53]. Zhang et al[54] used the low-toxicity cholesterol-modified antimicrobial peptide DP7 (DP7-C), which has dual functions as a carrier and an immune adjuvant, to improve dendritic cell (DC)-based vaccine efficacy. It achieved promising antitumor effects in a mouse model. In addition, after stimulation with DP7-C, the antigen uptake efficiency of monocyte-derived DCs (MoDCs) in patients with advanced lung cancer increased from 14%-40% to 88%-98%, the antigen presentation efficiency increased from approximately 15% to approximately 65%, and the proportion of mature MoDCs increased from approximately 20% to approximately 60%[54]. The antigen presentation strategy targeting mannose receptor on the surface of DC cells can also greatly increase the efficiency of antigen presentation, and clinical studies have shown that it has good safety and efficacy[55]. In addition, Ni et al[56] invented a bi-adjuvant neoantigen nanovaccine (banNV). In this formulation, two adjuvants were added to the original single neoantigen vaccine: The Toll-like receptor (TLR) 7/8 agonist R848 and the TLR9 agonist CpG. The immunogenicity of the neoantigen was increased, and the acute systemic toxicity was reduced. In combination with anti-PD-1 therapy, banNV achieved a superior effect on colorectal cancer[56]. We can also use the above new techniques to exploit application of neoantigens in HCC immunotherapy.
Therefore, on the one hand, efficiently screening and identifying quality neoantigens in HCC, evaluating the existing HCC-related antigens, and enhancing their quantity and quality by means of some strategies that can improve antigen exposure and antigen expression are goals for future studies. Such studies might provide a series of antigen targets with relatively high-level immunogenicity for accurate treatment of HCC. On the other hand, ICIs, some cytokines, such as IL-2, and a variety of new technologies should be applied to activate and expand TILs to generate sufficient numbers of powerful T cells, which will enable achievement of effective control of HCC in humans. This is a comprehensive strategy for the exploitation of neoantigens in HCC immunotherapy in the future.
According to ClinicalTraials.gov (https://clinicaltrials.gov/ct2/home), there are only five clinical studies related to HCC neoantigen vaccines, and no promising results have yet been generated (Table 1). With the rapid development of high-throughput sequencing and bioinformatics technologies, more efforts exploring neoantigens should be made to achieve effective treatment of HCC. A key requirement of such research is determining how to establish effective methods to screen and identify a large number of quality HCC neoantigens. To improve the accuracy of the screening and identification of HCC neoantigens, we should adopt more advanced HCC neoantigen prediction algorithms and more sensitive and less time-consuming methods of screening and identification in vitro. A personalized method of screening and identification and a universal library of HCC neoantigens should be established, and a unique HCC neoantigen therapeutic strategy should be adopted to open a new avenue for HCC immunotherapy.
| Tumor | Interventions | Phase | Number enrolled | NCT number | Initial date | Sponsor/collaborators |
| Hepatocellular carcinoma | Neoantigen DNA vaccine (GNOS-PV02), plasmid encoded IL-12 (INO-9012), pembrolizumab (MK-3475) | Ⅰ/Ⅱ | 24 | NCT04251117 | January 31, 2020 | Geneos Therapeutics |
| Fibrolamellar hepatocellular carcinoma | DNAJB1-PRKACA peptide vaccine, nivolumab, ipilimumab | Ⅰ | 12 | NCT04248569 | January 30, 2020 | Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Bristol-Myers Squibb, Fibrolamellar Cancer Foundation |
| Gastric cancer, hepatocellular carcinoma, non-small-cell lung cancer, colorectal cancer | Neoantigen-primed DC vaccine | Ⅰ | 80 | NCT04147078 | October 31, 2019 | Sichuan University |
| Hepatocellular carcinoma | Neoantigen-based DC vaccine, microwave ablation | Ⅰ | 24 | NCT03674073 | September 17, 2018 | Chinese PLA General Hospital, Likang Life Sciences Holdings Limited |
| Hepatocellular carcinoma | NRT radiotherapy | Ⅰ/Ⅱ | 40 | NCT03199807 | June 27, 2017 | The Affiliated Nanjing Drum Tower Hospital of Nanjing University Medical School |
There are a variety of events leading to the generation of HCC neoantigens. The study of HCC neoantigens should not only focus on mutation-induced neoantigens but also consider neoantigens generated by multiple paths and the characteristics of HCC. The screening and identification methods used for HCC neoantigens should be optimized considering the above factors. In addition, since HCC is an immunosuppressive tumor, strategies that reverse immunosuppression and enhance the immune response should be considered for the practical exploitation of HCC neoantigens.
We thank Xing-Wang Xie (Peking University People’s Hospital) and Wei-Jia Liao (Affiliated Hospital of Guilin Medical University) for helpful suggestions.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kim IH, Sahin TT S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Li JH
| 1. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 4899] [Article Influence: 699.9] [Reference Citation Analysis (1)] |
| 2. | Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1933] [Cited by in RCA: 1873] [Article Influence: 208.1] [Reference Citation Analysis (4)] |
| 3. | Iñarrairaegui M, Melero I, Sangro B. Immunotherapy of Hepatocellular Carcinoma: Facts and Hopes. Clin Cancer Res. 2018;24:1518-1524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 4. | Guo J, Tang Q. Recent updates on chimeric antigen receptor T cell therapy for hepatocellular carcinoma. Cancer Gene Ther. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | Fritsch EF, Burkhardt UE, Hacohen N, Wu CJ. Personal Neoantigen Cancer Vaccines: A Road Not Fully Paved. Cancer Immunol Res. 2020;8:1465-1469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 6. | Kishton RJ, Lynn RC, Restifo NP. Strength in Numbers: Identifying Neoantigen Targets for Cancer Immunotherapy. Cell. 2020;183:591-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 7. | Jou J, Harrington KJ, Zocca MB, Ehrnrooth E, Cohen EEW. The Changing Landscape of Therapeutic Cancer Vaccines-Novel Platforms and Neoantigen Identification. Clin Cancer Res. 2021;27:689-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 8. | Leng Q, Tarbe M, Long Q, Wang F. Pre-existing heterologous T-cell immunity and neoantigen immunogenicity. Clin Transl Immunology. 2020;9:e01111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, Chen C, Olive O, Carter TA, Li S, Lieb DJ, Eisenhaure T, Gjini E, Stevens J, Lane WJ, Javeri I, Nellaiappan K, Salazar AM, Daley H, Seaman M, Buchbinder EI, Yoon CH, Harden M, Lennon N, Gabriel S, Rodig SJ, Barouch DH, Aster JC, Getz G, Wucherpfennig K, Neuberg D, Ritz J, Lander ES, Fritsch EF, Hacohen N, Wu CJ. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2010] [Cited by in RCA: 2053] [Article Influence: 256.6] [Reference Citation Analysis (0)] |
| 10. | Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, Bukur V, Tadmor AD, Luxemburger U, Schrörs B, Omokoko T, Vormehr M, Albrecht C, Paruzynski A, Kuhn AN, Buck J, Heesch S, Schreeb KH, Müller F, Ortseifer I, Vogler I, Godehardt E, Attig S, Rae R, Breitkreuz A, Tolliver C, Suchan M, Martic G, Hohberger A, Sorn P, Diekmann J, Ciesla J, Waksmann O, Brück AK, Witt M, Zillgen M, Rothermel A, Kasemann B, Langer D, Bolte S, Diken M, Kreiter S, Nemecek R, Gebhardt C, Grabbe S, Höller C, Utikal J, Huber C, Loquai C, Türeci Ö. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1278] [Cited by in RCA: 1702] [Article Influence: 212.8] [Reference Citation Analysis (0)] |
| 11. | Strønen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, Donia M, Böschen ML, Lund-Johansen F, Olweus J, Schumacher TN. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352:1337-1341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 350] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 12. | Lu L, Jiang J, Zhan M, Zhang H, Wang QT, Sun SN, Guo XK, Yin H, Wei Y, Liu JO, Li SY, Li Y, He YW. Targeting Neoantigens in Hepatocellular Carcinoma for Immunotherapy: A Futile Strategy? Hepatology. 2021;73:414-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 13. | Löffler MW, Mohr C, Bichmann L, Freudenmann LK, Walzer M, Schroeder CM, Trautwein N, Hilke FJ, Zinser RS, Mühlenbruch L, Kowalewski DJ, Schuster H, Sturm M, Matthes J, Riess O, Czemmel S, Nahnsen S, Königsrainer I, Thiel K, Nadalin S, Beckert S, Bösmüller H, Fend F, Velic A, Maček B, Haen SP, Buonaguro L, Kohlbacher O, Stevanović S, Königsrainer A; HEPAVAC Consortium; Rammensee HG. Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma. Genome Med. 2019;11:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 14. | Samji H, Yu A, Kuo M, Alavi M, Woods R, Alvarez M, Dore GJ, Tyndall M, Krajden M, Janjua NZ; BC Hepatitis Testers Cohort Team. Late hepatitis B and C diagnosis in relation to disease decompensation and hepatocellular carcinoma development. J Hepatol. 2017;67:909-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Lin L, Yan L, Liu Y, Qu C, Ni J, Li H. The Burden and Trends of Primary Liver Cancer Caused by Specific Etiologies from 1990 to 2017 at the Global, Regional, National, Age, and Sex Level Results from the Global Burden of Disease Study 2017. Liver Cancer. 2020;9:563-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 16. | Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, Huang C, Li J, Dong X, Zhou Y, Liu Q, Ma L, Wang X, Zhou J, Liu Y, Boja E, Robles AI, Ma W, Wang P, Li Y, Ding L, Wen B, Zhang B, Rodriguez H, Gao D, Zhou H, Fan J. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 2019; 179: 561-577. e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 617] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 17. | Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O, Sander C; Cancer Genome Atlas Research Network; Rätsch G. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell 2018; 34: 211-224. e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 694] [Cited by in RCA: 638] [Article Influence: 91.1] [Reference Citation Analysis (0)] |
| 18. | Robinson TJ, Freedman JA, Al Abo M, Deveaux AE, LaCroix B, Patierno BM, George DJ, Patierno SR. Alternative RNA Splicing as a Potential Major Source of Untapped Molecular Targets in Precision Oncology and Cancer Disparities. Clin Cancer Res. 2019;25:2963-2968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 19. | Lee SE, Alcedo KP, Kim HJ, Snider NT. Alternative Splicing in Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol. 2020;10:699-712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 20. | Duan M, Hao J, Cui S, Worthley DL, Zhang S, Wang Z, Shi J, Liu L, Wang X, Ke A, Cao Y, Xi R, Zhang X, Zhou J, Fan J, Li C, Gao Q. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 2018;28:359-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 21. | Chiu YT, Wong JK, Choi SW, Sze KM, Ho DW, Chan LK, Lee JM, Man K, Cherny S, Yang WL, Wong CM, Sham PC, Ng IO. Novel pre-mRNA splicing of intronically integrated HBV generates oncogenic chimera in hepatocellular carcinoma. J Hepatol. 2016;64:1256-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Nault JC, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, Letouzé E, Pilati C, Verret B, Blanc JF, Balabaud C, Calderaro J, Laurent A, Letexier M, Bioulac-Sage P, Calvo F, Zucman-Rossi J. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 376] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 23. | Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee GS, Li T, Chabot MS, Ly A, Taplin ME, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS, Love JC. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 449] [Cited by in RCA: 434] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 24. | Radovich M, Jiang G, Hancock BA, Chitambar C, Nanda R, Falkson C, Lynce FC, Gallagher C, Isaacs C, Blaya M, Paplomata E, Walling R, Daily K, Mahtani R, Thompson MA, Graham R, Cooper ME, Pavlick DC, Albacker LA, Gregg J, Solzak JP, Chen YH, Bales CL, Cantor E, Shen F, Storniolo AMV, Badve S, Ballinger TJ, Chang CL, Zhong Y, Savran C, Miller KD, Schneider BP. Association of Circulating Tumor DNA and Circulating Tumor Cells After Neoadjuvant Chemotherapy With Disease Recurrence in Patients With Triple-Negative Breast Cancer: Preplanned Secondary Analysis of the BRE12-158 Randomized Clinical Trial. JAMA Oncol. 2020;6:1410-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 212] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 25. | Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019;19:553-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 398] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 26. | Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827-1840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 326] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 27. | Cabel L, Proudhon C, Romano E, Girard N, Lantz O, Stern MH, Pierga JY, Bidard FC. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol. 2018;15:639-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 155] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 28. | Khodadoust MS, Olsson N, Wagar LE, Haabeth OA, Chen B, Swaminathan K, Rawson K, Liu CL, Steiner D, Lund P, Rao S, Zhang L, Marceau C, Stehr H, Newman AM, Czerwinski DK, Carlton VE, Moorhead M, Faham M, Kohrt HE, Carette J, Green MR, Davis MM, Levy R, Elias JE, Alizadeh AA. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature. 2017;543:723-727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 205] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 29. | Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol. 2017;199:3360-3368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 1012] [Article Influence: 126.5] [Reference Citation Analysis (0)] |
| 30. | Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449-W454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 958] [Cited by in RCA: 1203] [Article Influence: 240.6] [Reference Citation Analysis (0)] |
| 31. | Rech AJ, Balli D, Mantero A, Ishwaran H, Nathanson KL, Stanger BZ, Vonderheide RH. Tumor Immunity and Survival as a Function of Alternative Neopeptides in Human Cancer. Cancer Immunol Res. 2018;6:276-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 32. | Newey A, Griffiths B, Michaux J, Pak HS, Stevenson BJ, Woolston A, Semiannikova M, Spain G, Barber LJ, Matthews N, Rao S, Watkins D, Chau I, Coukos G, Racle J, Gfeller D, Starling N, Cunningham D, Bassani-Sternberg M, Gerlinger M. Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK-inhibitor treatment. J Immunother Cancer. 2019;7:309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 33. | Devlin JR, Alonso JA, Ayres CM, Keller GLJ, Bobisse S, Vander Kooi CW, Coukos G, Gfeller D, Harari A, Baker BM. Structural dissimilarity from self drives neoepitope escape from immune tolerance. Nat Chem Biol. 2020;16:1269-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 34. | Vizcaíno JA, Kubiniok P, Kovalchik KA, Ma Q, Duquette JD, Mongrain I, Deutsch EW, Peters B, Sette A, Sirois I, Caron E. The Human Immunopeptidome Project: A Roadmap to Predict and Treat Immune Diseases. Mol Cell Proteomics. 2020;19:31-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 35. | Kote S, Pirog A, Bedran G, Alfaro J, Dapic I. Mass Spectrometry-Based Identification of MHC-Associated Peptides. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 36. | Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, Sidney J, Stevens J, Lane W, Zhang GL, Eisenhaure TM, Clauser KR, Hacohen N, Rooney MS, Carr SA, Wu CJ. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity. 2017;46:315-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 458] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 37. | Lu YC, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y, Rosenberg SA, Robbins PF. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20:3401-3410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 315] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 38. | Wang WC, Zhang ZQ, Li PP, Ma JY, Chen L, Qian HH, Shi LH, Yin ZF, Sun B, Zhang XF. Anti-tumor activity and mechanism of oligoclonal hepatocellular carcinoma tumor-infiltrating lymphocytes in vivo and in vitro. Cancer Biol Ther. 2019;20:1187-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 39. | Kula T, Dezfulian MH, Wang CI, Abdelfattah NS, Hartman ZC, Wucherpfennig KW, Lyerly HK, Elledge SJ. T-Scan: A Genome-wide Method for the Systematic Discovery of T Cell Epitopes. Cell 2019; 178: 1016-1028. e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 168] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 40. | Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magrì A, Novara L, Sarotto I, Nagel ZD, Piett CG, Amatu A, Sartore-Bianchi A, Siena S, Bertotti A, Trusolino L, Corigliano M, Gherardi M, Lagomarsino MC, Di Nicolantonio F, Bardelli A. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:1473-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 298] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 41. | Formenti SC, Rudqvist NP, Golden E, Cooper B, Wennerberg E, Lhuillier C, Vanpouille-Box C, Friedman K, Ferrari de Andrade L, Wucherpfennig KW, Heguy A, Imai N, Gnjatic S, Emerson RO, Zhou XK, Zhang T, Chachoua A, Demaria S. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat Med. 2018;24:1845-1851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 591] [Cited by in RCA: 702] [Article Influence: 100.3] [Reference Citation Analysis (0)] |
| 42. | van den Bijgaart RJE, Kroesen M, Wassink M, Brok IC, Kers-Rebel ED, Boon L, Heise T, van Scherpenzeel M, Lefeber DJ, Boltje TJ, den Brok MH, Hoogerbrugge PM, Büll C, Adema GJ. Combined sialic acid and histone deacetylase (HDAC) inhibitor treatment up-regulates the neuroblastoma antigen GD2. J Biol Chem. 2019;294:4437-4449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 43. | Dong LQ, Peng LH, Ma LJ, Liu DB, Zhang S, Luo SZ, Rao JH, Zhu HW, Yang SX, Xi SJ, Chen M, Xie FF, Li FQ, Li WH, Ye C, Lin LY, Wang YJ, Wang XY, Gao DM, Zhou H, Yang HM, Wang J, Zhu SD, Wang XD, Cao Y, Zhou J, Fan J, Wu K, Gao Q. Heterogeneous immunogenomic features and distinct escape mechanisms in multifocal hepatocellular carcinoma. J Hepatol. 2020;72:896-908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 133] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 44. | Yang H, Sun L, Guan A, Yin H, Liu M, Mao X, Xu H, Zhao H, Lu X, Sang X, Zhong S, Chen Q, Mao Y. Unique TP53 neoantigen and the immune microenvironment in long-term survivors of Hepatocellular carcinoma. Cancer Immunol Immunother. 2021;70:667-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 45. | Mauriello A, Zeuli R, Cavalluzzo B, Petrizzo A, Tornesello ML, Buonaguro FM, Ceccarelli M, Tagliamonte M, Buonaguro L. High Somatic Mutation and Neoantigen Burden Do Not Correlate with Decreased Progression-Free Survival in HCC Patients not Undergoing Immunotherapy. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 46. | Willimsky G, Schmidt K, Loddenkemper C, Gellermann J, Blankenstein T. Virus-induced hepatocellular carcinomas cause antigen-specific local tolerance. J Clin Invest. 2013;123:1032-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 47. | Flecken T, Schmidt N, Hild S, Gostick E, Drognitz O, Zeiser R, Schemmer P, Bruns H, Eiermann T, Price DA, Blum HE, Neumann-Haefelin C, Thimme R. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology. 2014;59:1415-1426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 288] [Cited by in RCA: 298] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 48. | Hughes PE, Caenepeel S, Wu LC. Targeted Therapy and Checkpoint Immunotherapy Combinations for the Treatment of Cancer. Trends Immunol. 2016;37:462-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 217] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 49. | Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, Li D, Verret W, Xu DZ, Hernandez S, Liu J, Huang C, Mulla S, Wang Y, Lim HY, Zhu AX, Cheng AL; IMbrave150 Investigators. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med. 2020;382:1894-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2542] [Cited by in RCA: 4695] [Article Influence: 939.0] [Reference Citation Analysis (2)] |
| 50. | Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res. 2013;19:5300-5309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 534] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 51. | Finkelmeier F, Waidmann O, Trojan J. Nivolumab for the treatment of hepatocellular carcinoma. Expert Rev Anticancer Ther. 2018;18:1169-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 52. | Leko V, McDuffie LA, Zheng Z, Gartner JJ, Prickett TD, Apolo AB, Agarwal PK, Rosenberg SA, Lu YC. Identification of Neoantigen-Reactive Tumor-Infiltrating Lymphocytes in Primary Bladder Cancer. J Immunol. 2019;202:3458-3467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 53. | Stifter K, Dekhtiarenko I, Krieger J, Tissot AC, Seufferlein T, Wagner M, Schirmbeck R. A tumor-specific neoepitope expressed in homologous/self or heterologous/viral antigens induced comparable effector CD8+ T-cell responses by DNA vaccination. Vaccine. 2020;38:3711-3719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 54. | Zhang R, Tang L, Tian Y, Ji X, Hu Q, Zhou B, Zhenyu D, Heng X, Yang L. Cholesterol-modified DP7 enhances the effect of individualized cancer immunotherapy based on neoantigens. Biomaterials. 2020;241:119852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 55. | Loveland BE, Zhao A, White S, Gan H, Hamilton K, Xing PX, Pietersz GA, Apostolopoulos V, Vaughan H, Karanikas V, Kyriakou P, McKenzie IF, Mitchell PL. Mannan-MUC1-pulsed dendritic cell immunotherapy: a phase I trial in patients with adenocarcinoma. Clin Cancer Res. 2006;12:869-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 124] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 56. | Ni Q, Zhang F, Liu Y, Wang Z, Yu G, Liang B, Niu G, Su T, Zhu G, Lu G, Zhang L, Chen X. A bi-adjuvant nanovaccine that potentiates immunogenicity of neoantigen for combination immunotherapy of colorectal cancer. Sci Adv. 2020;6:eaaw6071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 186] [Article Influence: 37.2] [Reference Citation Analysis (0)] |