Published online Apr 15, 2020. doi: 10.4251/wjgo.v12.i4.492
Peer-review started: November 26, 2019
First decision: January 14, 2020
Revised: February 5, 2020
Accepted: March 12, 2020
Article in press: March 12, 2020
Published online: April 15, 2020
Processing time: 140 Days and 19.5 Hours
The relationship between microRNAs, such as miR-654-5p and miR-376b-3p, and the prognosis of colon cancer has not been studied until now.
To evaluate the expression levels of miR-654-5p and miR-376b-3p and their clinical significance in colon cancer.
RT-qPCR was performed to evaluate miR-654-5p and miR-376b-3p expression in 34 pairs of colon cancer and adjacent noncancerous tissues. Subsequently, the association of miR-654-5p and miR-376b-3p expression with clinical factors or the survival of patients suffering from colon cancer was determined by using The Cancer Genome Atlas.
miR-654-5p was upregulated and miR-376b-3p was downregulated in colon cancer tissues compared with adjacent noncancerous tissues (P < 0.001). Increased miR-654-5p and decreased miR-376b-3p expression levels were significantly associated with metastasis and clinical stage. Moreover, a univariate analysis demonstrated that colon cancer patients with high miR-654-5p or low miR-376b-3p expression (P = 0.044 and 0.007, respectively) had a poor overall survival rate. A multivariate analysis identified high miR-654-5p expression and low miR-376b-3p expression as independent predictors of poor survival in colon cancer patients.
Upregulated miR-654-5p and downregulated miR-376b-3p may be associated with tumour progression in colon cancer, and these microRNAs may serve as independent prognostic markers for colon cancer.
Core tip: This was a prospective study. The relationship between microRNAs, such as miR-654-5p and miR-376b-3p, and the prognosis of colon cancer has not been studied until now. In this present study, we found that upregulated miR-654-5p and downregulated miR-376b-3p may be associated with tumour progression in colon cancer, and these microRNAs may serve as independent prognostic markers for colon cancer.
- Citation: Li P, Cai JX, Han F, Wang J, Zhou JJ, Shen KW, Wang LH. Expression and significance of miR-654-5p and miR-376b-3p in patients with colon cancer. World J Gastrointest Oncol 2020; 12(4): 492-502
- URL: https://www.wjgnet.com/1948-5204/full/v12/i4/492.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i4.492
Colon cancer is the third most common malignancy and a major cause of cancer-related deaths worldwide[1]. Approximately 600000 patients die from colon cancer every year. Despite the availability of effective diagnostic and treatment modalities, most patients are at a risk of recurrence and progression of this cancer[2]. Extensive studies have revealed new diagnostic and prognostic markers based on novel molecular networks, but only a few of these tests are recommended for daily practice because of their limited diagnostic and prognostic performance[3]. The identification of more reliable and accurate prognostic mediators of tumour occurrence, development, invasion and metastasis is necessary to define the progression of patients with colon cancer and to improve postoperative treatment strategies.
MicroRNAs (miRNAs) regulate gene expression and are involved in numerous cellular and physiological processes, including tumour growth, proliferation, apoptosis, and angiogenesis; they have a high degree of sequence conservation among distantly related organisms and may participate in critical biological processes[4-6]. Some miRNAs are differentially expressed in colon cancer tissues, body fluids, serum and plasma and may be useful as biomarkers for diagnosis and as therapeutic targets and are clinically important[7-10]. However, few miRNA expression profiling studies have been conducted in colon cancer to reveal diagnostic and prognostic biomarkers.
miR-654-5p and miR-376b-3p play a major role in regulating various biological activities, including cancer progression, liver regeneration, angiogenesis, and cardioprotection. miR-654-5p has been implicated in breast cancer, prostate cancer, Hodgkin lymphoma, and oral squamous cell carcinoma[11-14]. miR-376b-3p plays major roles in cancer progression, liver regeneration, angiogenesis, and cardioprotection[15,16]. Some studies have suggested that miR-654-5p and miR-376b-3p are metastasis-associated miRNAs in colon cancer[17]. However, the diagnostic and prognostic role of miR-654-5p and miR-376b-3p in colon cancer remains largely unknown. In this study, our primary aim was to investigate the expression of miR-654-5p and miR-376b-3p in colon cancer. Consequently, we studied the association between the expression of miR-654-5p and miR-376b-3p and the clinical features of colon cancer patients, including survival, and biomarkers that could be used as factors to predict prognosis. We investigated mainly the miR-654-5p and miR-376b-3p expression levels in colon cancer tissues by using an open source database, The Cancer Genome Atlas (TCGA). Our findings indicate that miR-654-5p and miR-376b-3p might provide a novel prognostic approach for colon cancer.
Tumour and adjacent noncancerous tissue samples of 34 patients (68 samples in total) were obtained from the Department of General Surgery, Northern Jiangsu Province Hospital. All samples from patients were obtained by taking their informed consent, and this project was approved by the Clinical Research Ethics Committees of the participating institutions. We retrieved the registered data of colon cancer patients from the TCGA data portal (https://cancergenome.nih.gov/) in January 2019. Demographic information, such as age and sex, and cancer characteristics, such as cancer location and status, were collected. We followed the TNM Classification of Malignant Tumors (TNM) staging according to the AJCC 7th edition, and patients whose TNM stage and age were not described in the data registry were excluded from the analysis.
RNA isolation encapsulates all steps required for purifying total RNA, including small RNAs from tissues. RNA isolation was performed for the 34 patient samples using a miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. After RNA extraction, the total RNA samples were stored at -80 °C until required. RNA concentrations were measured using a NanoDrop spectrophotometer.
Real-time quantitative PCR (RT-qPCR) was performed using the SYBR detection system. A total of 68 samples were tested for the estimation of miRNAs. Primers used for the detection of gene expression (miR-654-5p and miR-376b-3p) were purchased from the QuantiTect Primer Collection (Qiagen, Germany). The lyophilized primers were dissolved in 10× Tris-EDTA buffer (TE buffer; pH 8.0), and RNU6 was selected as a housekeeping gene for miR-654-5p and miR-376b-3p. The PCR programme used for the detection of miRNAs was as follows: Segment 1 (1 cycle), 95 °C for 15 min; segment 2, 94 °C for 15 s, 55 °C for 30 s and 70 °C for 30 s; 40 cycles.
miRNA expression was quantified using the delta delta CT (∆∆Ct) method as previously described. Average crossing point (Ct) values were obtained for the housekeeping gene of the selected miRNAs under control and experimental conditions. The four sets of values included the housekeeping gene under the experimental condition, the housekeeping gene under the control condition, and the selected miRNA under both experimental and control conditions. The calculated differences between the control and experimental values signified the ∆Ct values under the experimental (∆CTE) and control (∆CTC) conditions. The difference between ∆CTE and ∆CTC was calculated to derive the ∆∆Ct value, and the value of (2^-∆∆Ct) was calculated to obtain fold changes in miRNA expression.
SPSS software (v.19.0; IBM SPSS, Chicago, IL, United States) was used to statistically analyse all the results. Analysis of variance and least significance difference post hoc tests were used to analyse miR-654-5p and miR-376b-3p expression. Student’s t-test, Pearson’s χ2 test, and Fisher’s exact test were used to compare measurement data. The Kaplan-Meier method was used to analyse overall survival, and the log-rank test was uses to evaluate differences in survival. Cox’s proportional hazards regression model was performed for an independent prognosis. P ≤ 0.05 indicated a statistically significant difference.
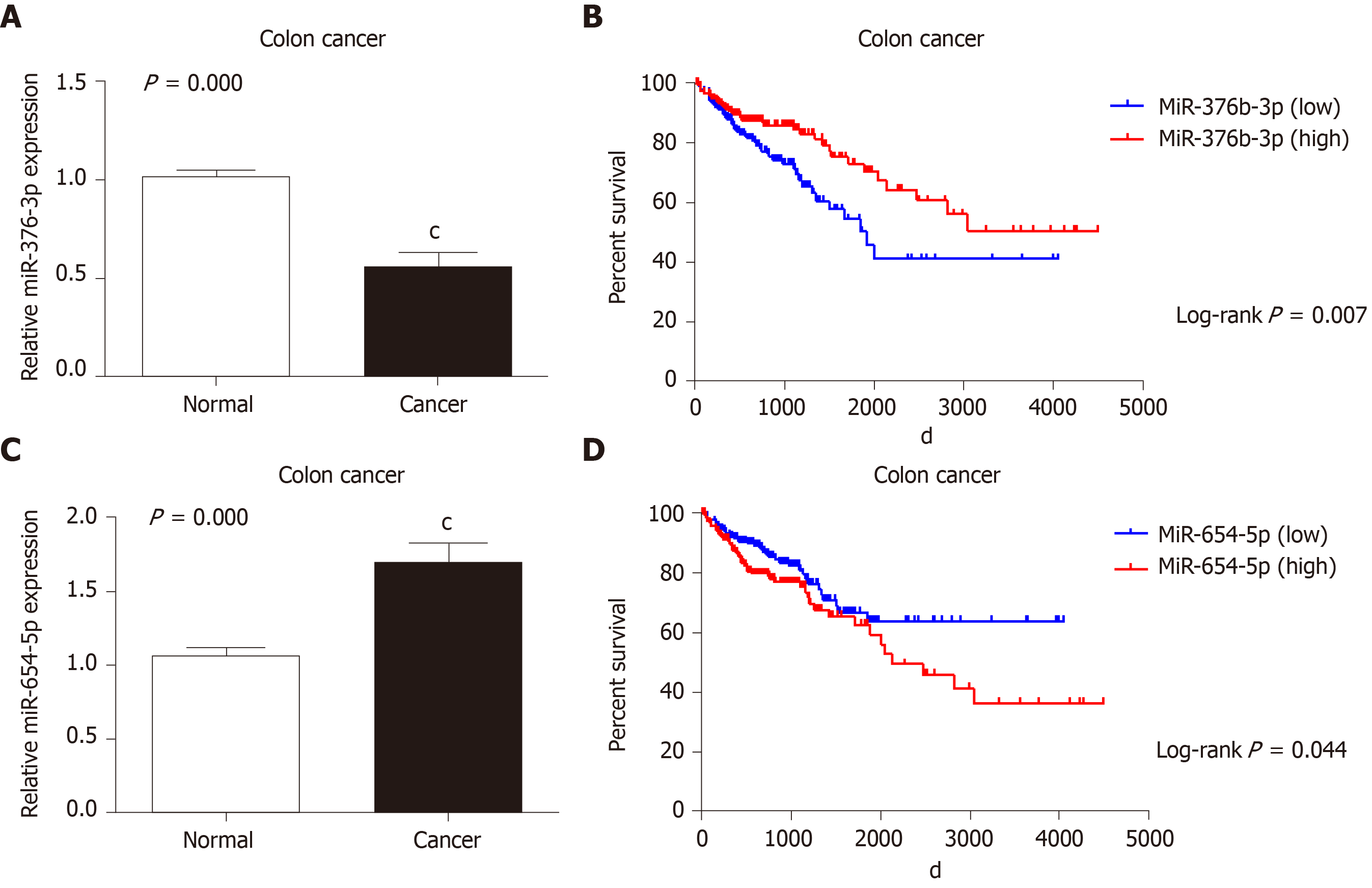
The expression levels of miR-654-5p and miR-376b-3p were detected by RT-qPCR. The expression level of miR-654-5p was significantly higher (1.698 ± 0.952) in colon cancer tissues than in adjacent noncancerous tissues (1.062 ± 0.329) (P < 0.001) (Figure 1A). However, the expression level of miR-376b-3p was lower (0.562 ± 0.564) in colon cancer tissues than in adjacent noncancerous tissues (1.02 ± 0.226) (P < 0.001) (Figure 1C).
To validate the outcomes of the RT-qPCR analysis, data from the TCGA were used to evaluate miR-654-5p and miR-376b-3p expression in 413 colon cancer tissues (Table 1 and Table 2). Patients with colon cancer were divided into 2 groups (low and high groups) according to miR-654-5p and miR-376b-3p expression levels, and the results were compared by analysis of variance and least significance difference tests. In the miR-654-5p group, the results revealed that the expression of miR-654-5p was upregulated in colon cancer tissues and positively associated with miR-376b-3p (P = 0.005), metastasis (P = 0.036), race (P = 0.016), organ in which the cancer was detected (P = 0.044), and TNM stage (P = 0.043). In the miR-376b-3p group, the results revealed that the expression of miR-376b-3p was downregulated in colon cancer tissues and positively associated with miR-552-5p, TNM stage, and metastasis (P = 0.001, 0.002, and 0.049, respectively).
| n | MiR-654-5p expression | ||||
| low | high | χ2 | P value | ||
| Age | 0.006 | 0.938 | |||
| ≤ 60 | 119 | 59 | 60 | ||
| > 60 | 294 | 147 | 147 | ||
| Gender | 0.414 | 0.520 | |||
| Female | 195 | 94 | 101 | ||
| Male | 218 | 112 | 106 | ||
| Race | 4.613 | 0.099 | |||
| White | 202 | 90 | 112 | ||
| African | 57 | 30 | 27 | ||
| Asian | 11 | 7 | 4 | ||
| Site | 0.144 | 0.931 | |||
| Ascending | 162 | 81 | 81 | ||
| Transverse | 36 | 17 | 19 | ||
| Descending | 122 | 62 | 60 | ||
| TNM stage | 9.759 | 0.002 | |||
| I-II | 229 | 130 | 99 | ||
| III-IV | 184 | 86 | 108 | ||
| Metastasis | 5.463 | 0.019 | |||
| M0 | 352 | 184 | 168 | ||
| M1 | 61 | 22 | 39 | ||
| MiR-376b-3p expression | 7.866 | 0.005 | |||
| Low | 206 | 117 | 89 | ||
| High | 207 | 89 | 118 | ||
| n | MiR-376b-3p expression | ||||
| low | high | χ2 | P value | ||
| Age | 0.087 | 0.768 | |||
| ≤ 60 | 119 | 58 | 61 | ||
| > 60 | 294 | 148 | 146 | ||
| Gender | 0.707 | 0.4 | |||
| Female | 195 | 93 | 102 | ||
| Male | 218 | 113 | 105 | ||
| Race | 8.274 | 0.016 | |||
| White | 202 | 91 | 111 | ||
| African | 57 | 20 | 37 | ||
| Asian | 11 | 9 | 2 | ||
| Site | 6.251 | 0.044 | |||
| Ascending | 162 | 84 | 78 | ||
| Transverse | 36 | 11 | 25 | ||
| Descending | 122 | 52 | 70 | ||
| TNM stage | 4.097 | 0.043 | |||
| I-II | 229 | 104 | 125 | ||
| III-IV | 184 | 102 | 82 | ||
| Metastasis | 4.413 | 0.036 | |||
| M0 | 352 | 168 | 184 | ||
| M1 | 61 | 38 | 23 | ||
| MiR-654-5p expression | 7.866 | 0.005 | |||
| Low | 206 | 117 | 89 | ||
| High | 207 | 89 | 118 | ||
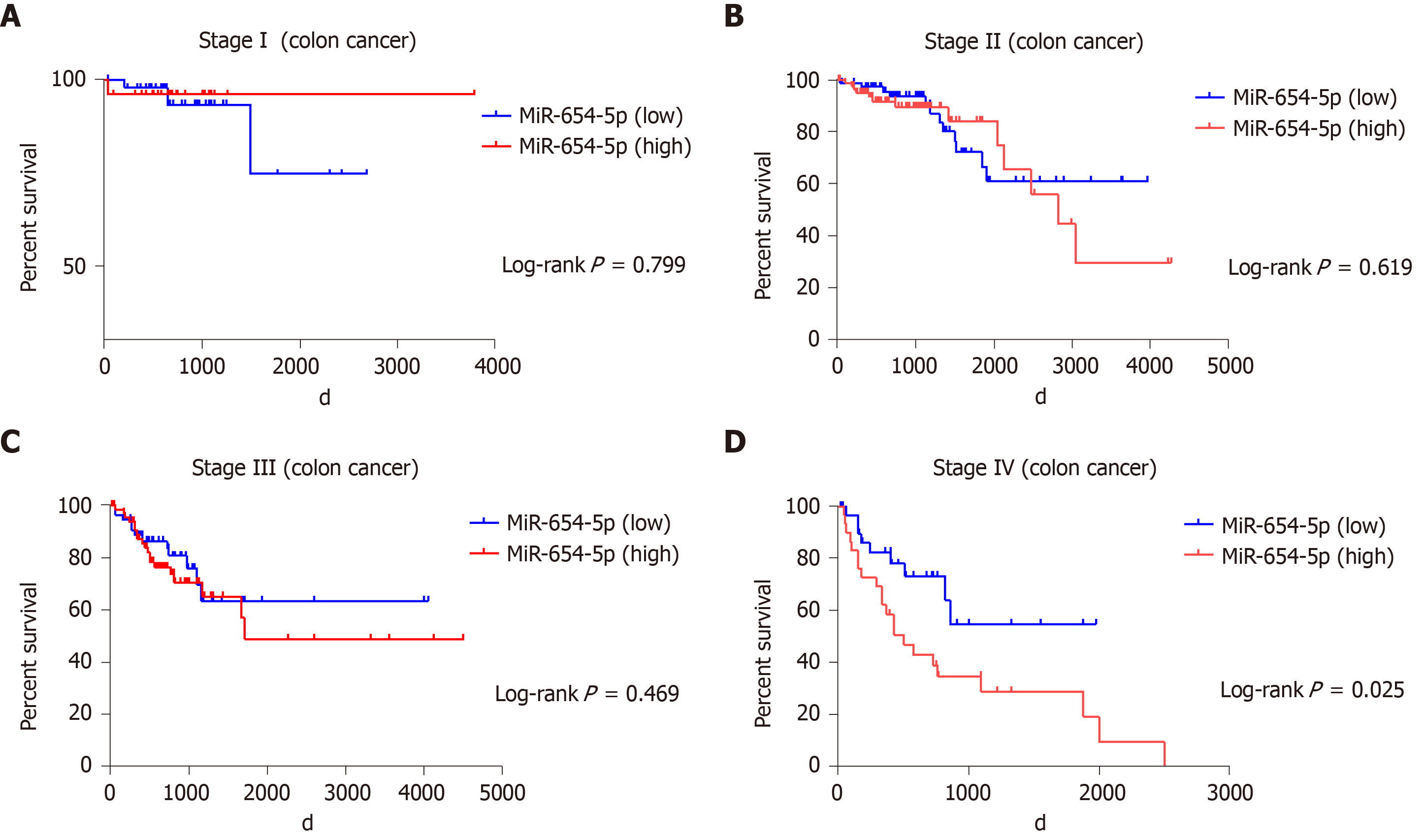
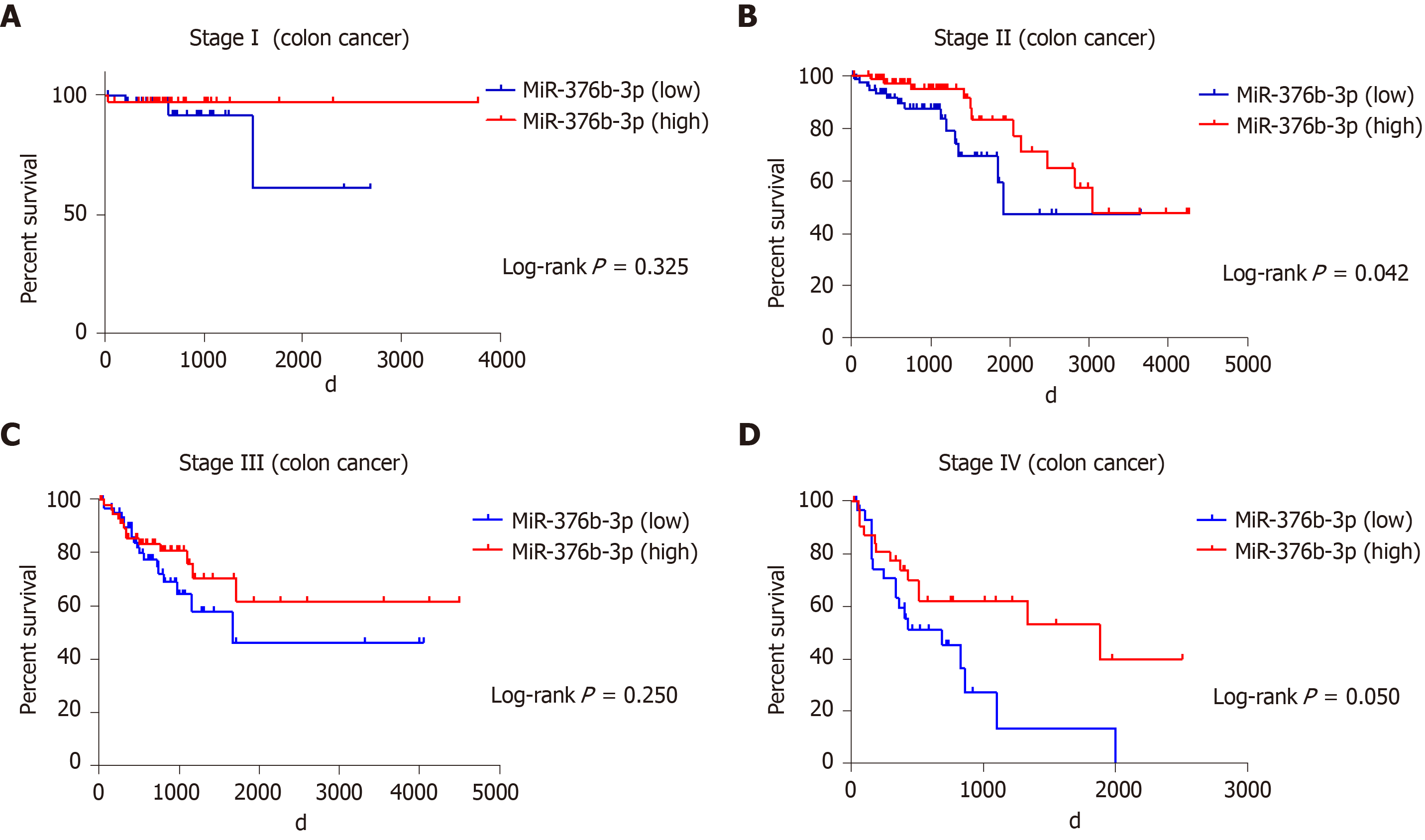
The prognostic value of miR-654-5p and miR-376b-3p relative to the survival time of colon cancer patients was evaluated by the Kaplan-Meier method. The overall survival rate of all colon cancer patients with high miR-654-5p expression was significantly lower than that of patients with low miR-654-5p expression (P = 0.044) (Figure 1B). Similarly, the overall survival rate of all colon cancer patients with high miR-376b-3p expression was significantly lower than that of patients with low miR-376b-3p expression (P = 0.007) (Figure 1D). A similar result was also obtained in the survival rate of patients with advanced colon cancer. At stage IV of colon cancer, patients with high miR-654-5p expression had low survival rates (P = 0.025) (Figure 2D). In contrast, at stages II and IV of colon cancer, patients with low miR-376b-3p expression had low survival rates (P = 0.042 and 0.05, respectively) (Figure 3B, Figure 3D). However, differences in miR-654-5p expression in patients with colon cancer were not significant at stages I, II, or III, and the differences in miR-376b-3p expression were not significant at stages I or III (Figure 2 and 3).
Cox’s proportional hazards model was used to decipher data obtained from the TCGA and determine whether miR-654-5p and miR-376b-3p could be used as independent prognostic markers for the survival rate of colon cancer patients. The univariate analysis revealed that miR-654-5p and miR-376b-3p expression levels were significant prognostic markers for the overall survival rate of patients with colon cancer: miR-654-5p (HR = 0.655; 95%CI: 0.434-0.989; P = 0.044); miR-376b-3p (HR = 1.782; 95%CI: 1.173-2.707; P = 0.007); TNM stage (HR = 0.305; 95%CI: 0.200-0.465; P = 0.000); metastasis (HR = 0.093; 95%CI: 0.048-0.180; P = 0.000). The multivariate analysis demonstrated that high miR-654-5p expression and low miR-376b-3p expression were significant independent prognostic markers in colon cancer: miR-654-5p (HR = 0.725; 95%CI: 0.421-1.372; P = 0.058); miR-376b-3p (HR = 0.518; 95%CI: 0.339-0.792; P = 0.012); TNM stage (HR = 2.199; 95%CI: 1.323-3.656; P = 0.002), metastasis (HR = 2.886; 95%CI: 1.739-4.790; P = 0.000) (Table 3).
| Univariate analysis | Multi variate analysis | |||
| P value | HR (95%CI) | P value | HR (95%CI) | |
| Age | 0.354 | 0.806 (0.511-1.272) | ||
| Gender | 0.575 | 0.889 (0.588-1.343) | ||
| Race | 0.671 | 0.911 (0.592-1.403) | ||
| TNM stage | 0.000 | 0.305 (0.200-0.465) | 0.002 | 2.199 (1.323-3.656) |
| Metastasis | 0.000 | 0.093 (0.048-0.180) | 0.000 | 2.886 (1.739-4.790) |
| MiR-376b-3p expression | 0.007 | 1.782 (1.173-2.707) | 0.012 | 0.518 (0.339-0.792) |
| MiR-654-5p expression | 0.044 | 0.655 (0.434-0.989) | 0.058 | 0.725 (0.421-1.372) |
With improvements in living standards and changes in the living environment, the incidence of colon cancer has significantly improved[18]. Multiple genetic mutations, multiple factors and multiple stages lead to a poor prognosis and an aggressive progression of colon cancer in which multiple tumour suppressor genes and oncogenes are involved. Alterations in the expression of miRNAs in cancer and correlations between miRNA expression profiles and clinicopathological features could be used in the diagnosis and therapies of different types of malignancies[19,20]. A number of deregulated miRNAs have been studied by expression analyses in colon cancer and compared with adjacent noncancerous tissues in patients with colon cancer and have indicated that such classes of miRNAs are closely associated with colon cancer.
However, no reports describing the biological role of miR-654-5p and miR-376b-3p in patients with colon cancer are available. Our study is the first of its kind to examine the expression of miR-654-5p and miR-376b-3p in patients with colon cancer. In this prospective study, we analysed the expression levels of miR-654-5p and miR-376b-3p using tissues from patients with colon cancer and adjacent noncancerous tissues. We performed experiments in which 68 tissue samples (cancer tissues and adjacent noncancerous tissues) were obtained from 34 cancer patients from the Department of General Surgery, Northern Jiangsu Province Hospital. Our findings reveal that miR-654-5p and miR-376b-3p are promising candidates that could be used as markers for colon cancer given that they demonstrated well-defined patterns of expression in the tested tissue samples.
Metastasis is a main cause of the majority of colon cancer-related deaths. Metastasis is a major prognostic factor that should be considered in patients with colon cancer[21,22]. Our research suggests that high miR-654-5p expression and low miR-376b-3p expression in tumours may impact tumour aggression and prognosis. When we tested the effects of both the upregulated and downregulated expression levels of miR-654-5p and miR-376b-3p, we observed that they are closely related to metastasis in tumours. We observed that miR-654-5p and miR-376b-3p were associated with the overall survival rate of colon cancer patients and were likely to have different levels of expression depending on the stage of cancer at diagnosis. When interpreting these results, several aspects of the study need to be considered. One of the drawbacks of this study was that the number of samples taken was small. Another factor was the difficulty encountered when carrying out follow-ups of cancer patients, and because such follow-ups are important, they were likely to have affected overall survival. Additionally, since we used an open source database, we had several limitations to our analysis. Incomplete information on cancer patients limited our understanding of the relationship between miR-654-5p, miR-376b-3p and colon cancer prognosis.
Transforming growth factor β (TGF-β) plays important pathological and physiological roles in regulating cell differentiation and proliferation, invasion, apoptosis, migration and modification of the microenvironment and cancer metastasis[23]. Interestingly, the TGF-β signalling pathway has a paradoxical effect on biology. It maintains proliferation, apoptosis and differentiation in normal cells and early-stage cancer cells but promotes cancer cell invasion, migration and metastasis in advanced-stage cancer cells. miR-654-5p plays a major role in regulating various biological activities, including cancer progression[11-14]. Similarly, miR-376b-3p is also implicated in regulating various biological activities, such as cancer progression, liver regeneration, angiogenesis, and cardioprotection[15,16]. To date, no report has demonstrated the roles of miR-654-5p and miR-376b-3p in colon diseases; however, our study revealed that miR-654-5p and miR-376b-3p participate in regulating the progression of colon cancer. To explain the relation between miR-654-5p and miR-376b-3p and metastasis, we used miRWalk (http://mirwalk.umm.uni-heidelberg.de/) to identify the targets of miR-654-5p and miR-376b-3p. We found that miR-654-5p is related to mothers against decapentaplegic homologue 2 (SMAD2), SMAD6, SMAD7, and SMAD9 and that miR-376b-3p is related to SMAD1, SMAD4, SMAD2, and SMAD7. SMADs are a crucial part of the TGF-β signalling pathway. Dysregulated miRNA expression targets SMAD7, which has been shown to be a TGF-β receptor type 1 antagonist. In contrast, the inhibition of SMAD7 led to an increase in the expression of the TGF-β pathway member TGF-β receptor type 1. This pattern of regulation indicates that the dysregulation of miRNAs may provide a positive feedback mechanism to the TGF-β pathway through SMAD7[24,25].
The results of this study showed that the expression levels of miR-654-5p and miR-376b-3p in colon cancer tissues were significantly higher and lower, respectively, than those in adjacent healthy tissues (P < 0.01). The expression levels of miR-654-5p and miR-376b-3p were associated with TNM stage and metastasis (P < 0.05). The survival rate of patients with high miR-654-5p expression was significantly low. The survival time of the group with high miR-376b-3p expression was long (P ≤ 0.05). In addition, the Cox multivariate regression analysis showed that TNM stage, metastasis, and miR-654-5p and miR-376b-3p expression were independent risk factors for a poor prognosis and the aggressive progression of colon cancer (Table 3). The expression of miR-654-5p was positively associated with the risk of disease progression and the degree of malignancy and negatively associated with the survival time of patients, while miR-376b-3p demonstrated the opposite effect. Thus, it is apparent that the expression of miR-654-5p and miR-376b-3p in colon cancer is an essential index that can be used determine the prognosis of colon cancer. The combination of miR-654-5p and miR-376b-3p expression and an immunohistochemical pathological examination may improve the diagnosis and prognosis of colon cancer and is worth pursuing further for clinical applications.
Our results have shown the association of the expression levels of miR-654-5p and miR-376b-3p with clinicopathologic features and prognosis in colon cancer patients. Nevertheless, we should further perform both in vitro analyses of molecular mediators and in vivo analyses in animal models of colon cancer to evaluate whether genes, inflammatory molecules, gastrointestinal tract microbiota, and the extracellular matrix could together contribute to carcinogenesis of the colon.
In conclusion, our study provides sensitive and accurate biomarkers that may aid in the prognosis of colon cancer. However, sustained effort is required to clarify the underlying molecular mechanisms by which miR-654-5p and miR-376b-3p may improve colon cancer metastasis.
Our study found that Upregulated miR-654-5p and downregulated miR-376b-3p may be associated with tumour progression in colon cancer. However, incomplete information on cancer patients limited our understanding of the relationship between miR-654-5p, miR-376b-3p and colon cancer prognosis. Sustained effort is required to clarify the underlying molecular mechanisms by which miR-654-5p and miR-376b-3p may improve colon cancer metastasis.
The relationship between microRNAs (miRNAs), such as miR-654-5p and miR-376b-3p, and the prognosis of colon cancer has not been studied until now. It is the first time that our study found that Upregulated miR-654-5p and downregulated miR-376b-3p may be associated with tumour progression in colon cancer.
Colon cancer is the third most common malignancy and a major cause of cancer-related deaths worldwide. Extensive studies have revealed new diagnostic and prognostic markers based on novel molecular networks, but only a few of these tests are recommended for daily practice because of their limited diagnostic and prognostic performance. This study evaluated the expression levels of miR-654-5p and miR-376b-3p and their clinical significance in colon cancer. In this present study, we found that upregulated miR-654-5p and downregulated miR-376b-3p may be associated with tumour progression in colon cancer, and these miRNAs may serve as independent prognostic markers for colon cancer.
Evaluated the expression levels of miR-654-5p and miR-376b-3p and their clinical significance in colon cancer.
Tumour and adjacent noncancerous tissue samples of 34 patients (68 samples in total) were obtained from the Department of General Surgery, Northern Jiangsu Province Hospital. We retrieved the registered data of colon cancer patients from the TCGA data portal (https://cancergenome.nih.gov/) in January 2019. Demographic information, such as age and sex, and cancer characteristics, such as cancer location and status, were collected. RT-qPCR was performed using the SYBR detection system. A total of 68 samples were tested for the estimation of miRNAs. miRNA expression was quantified using the delta delta CT (∆∆Ct) method. Average crossing point (Ct) values were obtained for the housekeeping gene of the selected miRNAs under control and experimental conditions. SPSS software (v.19.0; IBM SPSS, Chicago, IL, United States) was used to statistically analyse all the results. Analysis of variance and least significance difference post hoc tests were used to analyse miR-654-5p and miR-376b-3p expression. Student’s t-test, Pearson’s χ2 test, and Fisher’s exact test were used to compare measurement data. The Kaplan-Meier method was used to analyse overall survival, and the log-rank test was uses to evaluate differences in survival. Cox’s proportional hazards regression model was performed for an independent prognosis. P ≤ 0.05 indicated a statistically significant difference.
miR-654-5p and miR-376b-3p showed significant differences between tumour tissues and adjacent noncancerous tissues. Correlation of miR-654-5p and miR-376b-3p expression in colon cancer with clinicopathological features. High miR-654-5p expression and low miR-376b-3p expression are associated with a poor prognosis and the aggressive progression of colon cancer. miR-654-5p and miR-376b-3p serve as independent prognostic markers for the survival of patients with colon cancer.
Upregulated miR-654-5p and downregulated miR-376b-3p may be associated with tumour progression in colon cancer, and these miRNAs may serve as independent prognostic markers for colon cancer.
Our study provides sensitive and accurate biomarkers that may aid in the prognosis of colon cancer. However, sustained effort is required to clarify the underlying molecular mechanisms by which miR-654-5p and miR-376b-3p may improve colon cancer metastasis.
The authors thank Xiang-Ming Li for data analysis.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ahmed M, Aykan NF S-Editor: Dou Y L-Editor: A E-Editor: Qi LL
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20491] [Article Influence: 2049.1] [Reference Citation Analysis (20)] |
| 2. | Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, Su F, Yao H, Song E. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127-19137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 248] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 3. | Jung SB, Lee HI, Oh HK, Shin IH, Jeon CH. Clinico-pathologic Parameters for Prediction of Microsatellite Instability in Colorectal Cancer. Cancer Res Treat. 2012;44:179-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Amirkhah R, Schmitz U, Linnebacher M, Wolkenhauer O, Farazmand A. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosomes Cancer. 2015;54:129-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 5. | Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 240] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 6. | Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1353] [Cited by in RCA: 1268] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 7. | Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116:762-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 8. | Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10:197-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 9. | Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 520] [Cited by in RCA: 895] [Article Influence: 111.9] [Reference Citation Analysis (2)] |
| 10. | Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233:901-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 11. | Tan YY, Xu XY, Wang JF, Zhang CW, Zhang SC. MiR-654-5p attenuates breast cancer progression by targeting EPSTI1. Am J Cancer Res. 2016;6:522-532. [PubMed] |
| 12. | Östling P, Leivonen SK, Aakula A, Kohonen P, Mäkelä R, Hagman Z, Edsjö A, Kangaspeska S, Edgren H, Nicorici D, Bjartell A, Ceder Y, Perälä M, Kallioniemi O. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956-1967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 222] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 13. | Paydas S, Acikalin A, Ergin M, Celik H, Yavuz B, Tanriverdi K. Micro-RNA (miRNA) profile in Hodgkin lymphoma: association between clinical and pathological variables. Med Oncol. 2016;33:34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 14. | Lu M, Wang C, Chen W, Mao C, Wang J. miR-654-5p Targets GRAP to Promote Proliferation, Metastasis, and Chemoresistance of Oral Squamous Cell Carcinoma Through Ras/MAPK Signaling. DNA Cell Biol. 2018;37:381-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 15. | Huang Q, Wang C, Hou Z, Wang G, Lv J, Wang H, Yang J, Zhang Z, Zhang H. Serum microRNA-376 family as diagnostic and prognostic markers in human gliomas. Cancer Biomark. 2017;19:137-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 16. | Pan Z, Guo Y, Qi H, Fan K, Wang S, Zhao H, Fan Y, Xie J, Guo F, Hou Y, Wang N, Huo R, Zhang Y, Liu Y, Du Z. M3 subtype of muscarinic acetylcholine receptor promotes cardioprotection via the suppression of miR-376b-5p. PLoS One. 2012;7:e32571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Mudduluru G, Abba M, Batliner J, Patil N, Scharp M, Lunavat TR, Leupold JH, Oleksiuk O, Juraeva D, Thiele W, Rothley M, Benner A, Ben-Neriah Y, Sleeman J, Allgayer H. A Systematic Approach to Defining the microRNA Landscape in Metastasis. Cancer Res. 2015;75:3010-3019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Ji BC, Yu CC, Yang ST, Hsia TC, Yang JS, Lai KC, Ko YC, Lin JJ, Lai TY, Chung JG. Induction of DNA damage by deguelin is mediated through reducing DNA repair genes in human non-small cell lung cancer NCI-H460 cells. Oncol Rep. 2012;27:959-964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Masuda T, Hayashi N, Kuroda Y, Ito S, Eguchi H, Mimori K. MicroRNAs as Biomarkers in Colorectal Cancer. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 20. | Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233:901-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 21. | Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol Res Pract. 2015;211:557-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 289] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 22. | Tauriello DV, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11:97-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 23. | García-Vizcaíno EM, Liarte S, Alonso-Romero JL, Nicolás FJ. Sirt1 interaction with active Smad2 modulates transforming growth factor-β regulated transcription. Cell Commun Signal. 2017;15:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 24. | Yu J, Lei R, Zhuang X, Li X, Li G, Lev S, Segura MF, Zhang X, Hu G. MicroRNA-182 targets SMAD7 to potentiate TGFβ-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat Commun. 2016;7:13884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 104] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 25. | Luo M, Tan X, Mu L, Luo Y, Li R, Deng X, Chen N, Ren M, Li Y, Wang L, Wu J, Wan Q. MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci Rep. 2017;7:43427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |