Published online Apr 15, 2020. doi: 10.4251/wjgo.v12.i4.383
Peer-review started: December 22, 2019
First decision: January 19, 2020
Revised: February 6, 2020
Accepted: March 23, 2020
Article in press: March 23, 2020
Published online: April 15, 2020
Processing time: 115 Days and 0.6 Hours
In hepatocellular carcinoma (HCC), abnormal expression of multiple microRNAs (miRNAs) has been shown to be involved in the malignant biological behavior of liver cancer. The vast majority of liver cancer cases in China are closely related to hepatitis B virus (HBV) infection, but there are few studies on the changes of miRNA expression in the progression from HBV infection to hepatoma.
To explore the role of miRNAs in the progression of HBV infection to cirrhosis and even to liver cancer.
We screened differentially expressed miRNAs in 40 HBV cirrhosis, 40 normal and 15 HCC tissues by using a TaqMan Low Density Array and real time quantitative polymerase chain reaction. To evaluate the power of the selected miRNAs to predict disease, we calculated the area under the receiver-operating-characteristic curves. The overall survival of HBV cirrhosis patients was analyzed via Kaplan-Meier analysis.
The levels of miR-375, miR-122 and miR-143 were significantly lower in HBV cirrhosis tissues, while miR-224 was significantly higher than in the controls (P < 0.0001). The area under the curves of the receiver-operating-characteristic curve for the 4-miRNA panel was 0.991 (95%CI: 0.974-1). Patients with a lower expression level of miR-224 or higher expression levels of miR-375, miR-122 and miR-143 had longer overall survival.
The four miRNAs (miR-375, miR-122, miR-143 and miR-224) may be helpful for early diagnosis of HBV infection, HBV cirrhosis, and prediction of its overall survival.
Core tip: Abnormal expression of microRNAs (miRNAs) may lead to an abnormal physiological state and disease, such as, kinds of cancers. We detect the levels of miR-375, miR-122, miR-143 and miR-224. By combination of the miRNA panels, the area under the curves of the receiver-operating-characteristic curve was 0.991. In addition, the four miRNAs (miR-375, miR-122, miR-143 and miR-224) may be helpful for early detection and prognosis of hepatocellular carcinoma.
- Citation: Zhang Q, Xu HF, Song WY, Zhang PJ, Song YB. Potential microRNA panel for the diagnosis and prediction of overall survival of hepatocellular carcinoma with hepatitis B virus infection. World J Gastrointest Oncol 2020; 12(4): 383-393
- URL: https://www.wjgnet.com/1948-5204/full/v12/i4/383.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i4.383
Hepatitis B virus (HBV) is known as the smallest double-stranded DNA virus that infects humans, and HBV infection has become a global problem. The World Health Organization reported that an estimated 257 million people are living with HBV infection, defined as hepatitis B surface (HBsAg) antigen positivity, and it resulted in 887000 deaths in 2015, mostly from complications [including cirrhosis and hepatocellular carcinoma (HCC)][1]. Despite the availability of effective vaccines, the virus causes approximately 780000 deaths every year. It is estimated that approximately 15% to 30% of HBV carriers have a risk of developing cirrhosis[2]. For diagnosis of hepatitis B, the incubation period is long, and the disease cannot be diagnosed in the incubation period, which is from the invasion of the hepatitis virus to the initial clinical symptoms and can last 75 d on average but can vary from 30 to 180 d[1]. HBsAg, which is the main marker of HBV infection, can be detected in serum 2 to 6 wk before alanine aminotransferase elevation. DNA detection of HBV is sensitive to low-level HBV virus in vivo by amplifying viral nucleic acid. This assay is commonly used to assess viral replication, indicating HBV replication and contagiousness.
MicroRNAs (miRNAs) are a class of noncoding single-stranded RNA molecules of approximately 22 nt in size. They regulate the expression of the mRNA and protein of other genes by binding to target mRNAs to participate in various physiological processes, such as growth and development, inflammation, tumors and physiological and pathological processes[3-8]. Abnormal expression of miRNAs may lead to an abnormal physiological state and disease. In many cancers, the expression levels of miRNAs will change significantly, which may affect proto-oncogenes and tumor suppressor genes[9-12]. In HCC, abnormal expression of multiple miRNAs has been shown to be involved in the malignant biological behavior of liver cancer[13-17]. The vast majority of liver cancer cases in China are closely related to HBV infection, but there are few studies on the changes of miRNA expression in the progression from HBV infection to hepatoma.
In this study, TaqMan Low Density Array (TLDA) and real time quantitative polymerase chain reaction (RT-qPCR) were used to characterize the profile of miRNAs in chronic hepatitis B, HCC and normal control tissues to explore the role of miRNAs in the development of chronic hepatitis B to liver cancer.
Liver tissues from patients undergoing liver cancer resection or liver biopsy from July 2011 to January 2013 were collected, including 40 HBV cirrhosis, 40 normal, and 15 HCC tissues. HBsAg and/or HBV-DNA positivity for more than 6 months was considered to be chronic HBV infection, while the normal liver tissue was from the determination of no liver disease during the past and during hospitalization. All samples had no other basic liver disease and were confirmed by pathology. After the tissue was placed in liquid nitrogen for the first time, it was stored at -80 °C for later use.
The patients were followed up for 5 years, and their overall survival was recorded. For patients who survived 5 years later, their overall survival time was considered to be 5 years. All subjects had signed informed consent, and the study was approved by the Ethics Committee of Beijing Cancer Hospital.
Total RNA from 5 μg of normal liver and HBV cirrhosis samples was extracted according to the steps of the TRIzol reagent manual. The absorbance values of A260 and A280 of total RNA were determined by a UV spectrophotometer to calculate the concentration of RNA. The A260 value was used to detect the purity of RNA, and the value of A260/A280 was calculated to further test the quality of total RNA.
The TLDA (TaqMan Array Human MicroRNA A+B Cards Set v3.0, Life Technologies) was used to profile the 754 different human miRNAs described in previous literature[18]. To increase the sensitivity of the TLDA, we performed a pre-amplification using the QuantStudio 7 Flex RT-PCR System (Applied Biosystems)[19]. The threshold cycle (Cq) values showed the concentrations of miRNAs, which were normalized to an internal control. The fold changes of miRNA expression were calculated by the equation 2-ΔΔCq.
According to the manufacturer’s instructions (QuantStudio 7 Flex RT PCR System; Applied Biosystems) with slight modifications, hydrolysis probe–based qRT-PCR was performed. Reverse transcription was carried out as previously described[18]. All experiments were carried out in triplicate. An endogenous control, the combination of let-7d, let-7g and let-7i (let-7d/g/i) in this experiment, is important for normalizing qRT-PCR data[18,20]. Relative levels of miRNAs were normalized to let-7d/g/i and were calculated using the 2-ΔΔCq method[18,21].
Statistical analyses were performed with the Statistical Analysis System software SPSS 16.0, and data are presented as the mean ± SE for miRNAs or mean ± SD for other variables. With Student’s t-test and two-sided χ2 test, we compared the differences between the two groups, and the P value must be < 0.05, which will be considered statistically significant. The receiver operating characteristic (ROC) curves and the area under the ROC curves (AUC) were calculated to evaluate the predictive power of the selected miRNAs. Furthermore, risk score analysis was performed to evaluate the associations between miRNAs and HBV cirrhosis as previously described[20,22]. To indicate miRNAs’ contribution to the risk score function, the regression coefficient was used for the risk score as the weight[23,24]. Samples were divided into a high-risk group, predicting HBV cases, and a low-risk group, predicting control individuals, according to their risk score function, and then, we found the appropriate cutoff point. Analysis of patient survival was performed by Kaplan-Meier analysis.
A multiphase case control study was designed to investigate the differences in miRNA expression profiles between normal liver and HBV cirrhosis (Figure 1, Table 1). Using TLDA, we analyzed miRNA expression in three random pairs of samples of normal liver and HBV cirrhosis. The Cq values of the miRNAs were all < 25, and the concentrations of the miRNAs all showed > 2-fold differences between normal liver and HBV cirrhosis, which was defined as differential expression. Nine miRNAs, including miR-125b, miR-602, miR-210, miR-224, miR-129, miR-99a, miR-141, miR-342 and miR-145, were upregulated, while 14 miRNAs, including miR-122, miR-143, miR-199a, miR-375, miR-27a, miR-34b, miR-130a, miR-625, miR-142, miR-193a, miR-140, miR-100, miR-342 and miR-29c, were downregulated among the 754 miRNAs in HBV cirrhosis patients compared to normal liver tissue.
To verify the accuracy of differentially expressed miRNAs in the above TLDA results, we performed RT-qPCR analysis at the individual sample level.
In the training set, miRNAs were measured in a separate set of individual tissue samples from 25 HBV cirrhosis patients and 25 normal liver controls, and only miRNAs with a mean change of 2-fold and a P value of 0.001 were selected for further analysis. We used these criteria to generate a list of 4 miRNAs (miR-224, miR-375, miR-122 and miR-143), which showed significantly different miRNA patterns between HBV cirrhosis patients and normal controls.
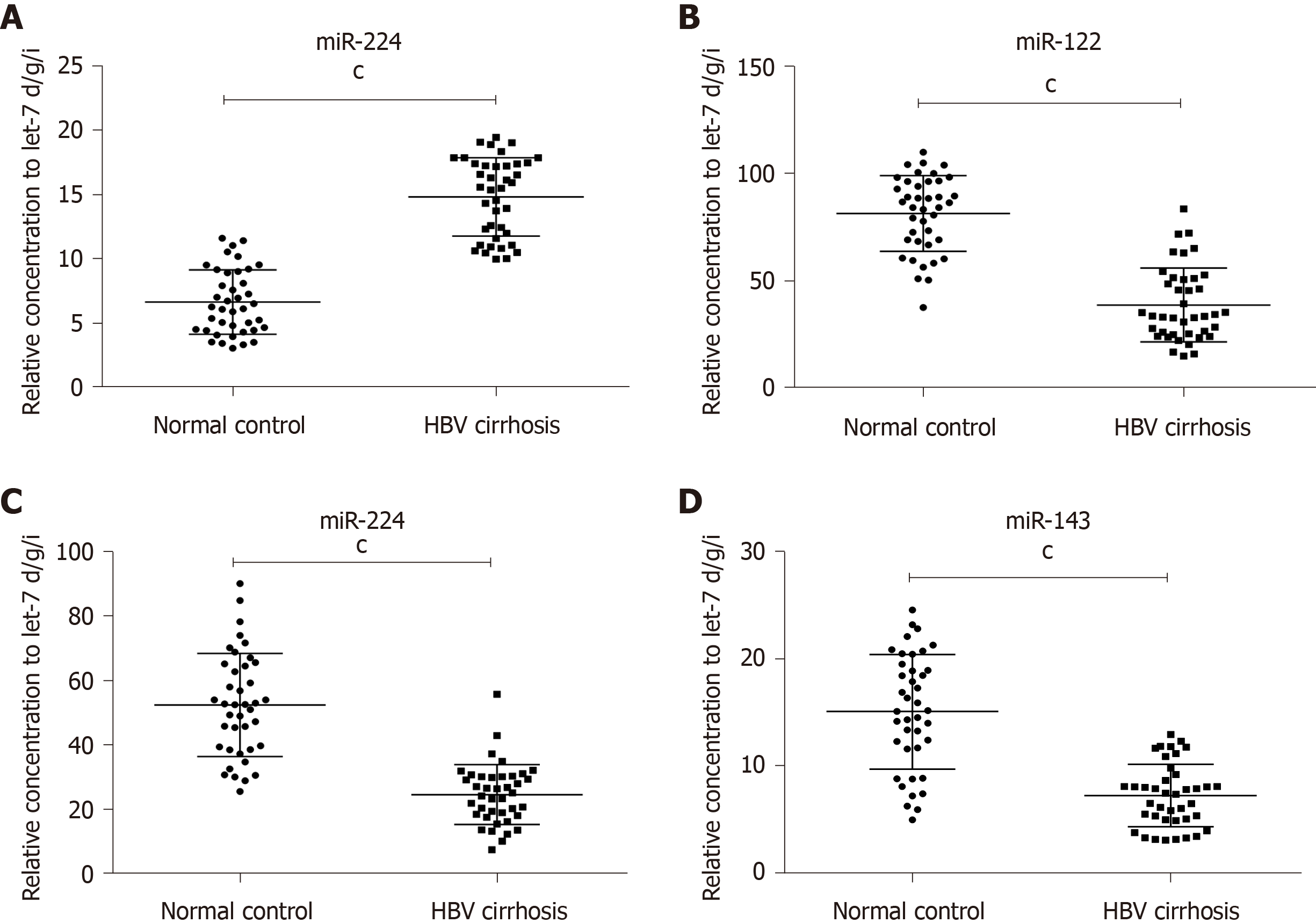
Furthermore, RT-qPCR was performed to verify the expression of the 4 miRNAs chosen above with another 15 HBV cirrhosis patients and 15 normal liver controls. The results showed that miR-224 was increased, while miR-375, miR-122 and miR-143 were significantly decreased in the tissues of HBV cirrhosis patients compared with the controls (at least P < 0.005), which was the same as the former cohort (Table 2, Figure 2A-D).
| miRNA | HBV cirrhosis | Normal control | Fold change2 | P value |
| miR-224 | 14.85 | 6.67 | 2.23 | < 0.0001 |
| miR-122 | 38.87 | 81.44 | 0.48 | < 0.0001 |
| miR-375 | 24.75 | 52.53 | 0.47 | < 0.0001 |
| miR-143 | 7.32 | 15.10 | 0.48 | < 0.0001 |
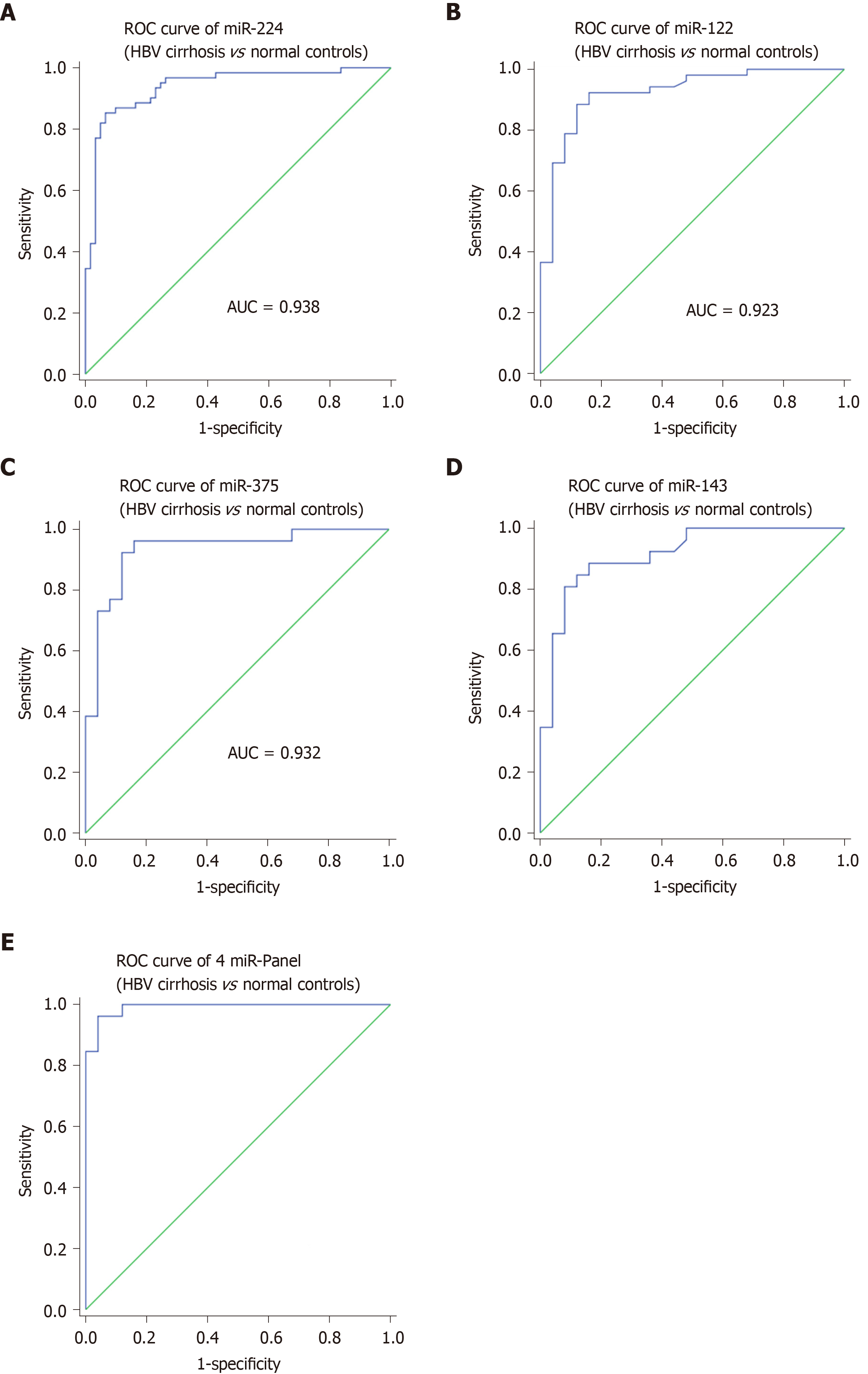
Subsequently, to evaluate the ability of the selected four tissue miRNAs to distinguish HBV cirrhosis from normal controls, we performed ROC curve analysis for each miRNA. For 40 cases of HBV cirrhosis and 40 normal control tissue samples, the AUC values of miR-224, miR-375, miR-122 and miR-143 were 0.938, 0.932, 0.923 and 0.915, respectively (Figure 3A-D). To further assess the diagnostic value of miRNAs in distinguishing between HBV cirrhosis and normal controls, we performed a risk score analysis of the dataset and used this risk scoring method to predict HBV cirrhosis and normal controls. The results showed that the best cutoff value (in this cutoff value, sensitivity + specificity is the largest) was 2.016, and 6 normal controls showed a risk score > 2.016, while 37 of the 40 HBV cirrhosis patients exhibited a risk score > 2.016 (Table 3). Furthermore, we integrated the 4-miRNA signature into a single biomarker using the risk score functions and evaluated the diagnostic accuracy of the miRNA signatures as HBV cirrhosis fingerprints. As expected, we obtained an AUC value of 0.991 (95%CI: 0.974-1) by combining miR-224, miR-375, miR-122 and miR-143 to differentiate HBV cirrhosis patients from healthy controls (Figure 3E).
| Score | 0-2.016 | > 20.16 | PPV | NPV |
| Normal subject (n = 40) | 34 | 6 | 0.92 | |
| HBV cirrhosis (n = 40) | 3 | 37 | 0.86 | |
| Total | 37 | 43 |
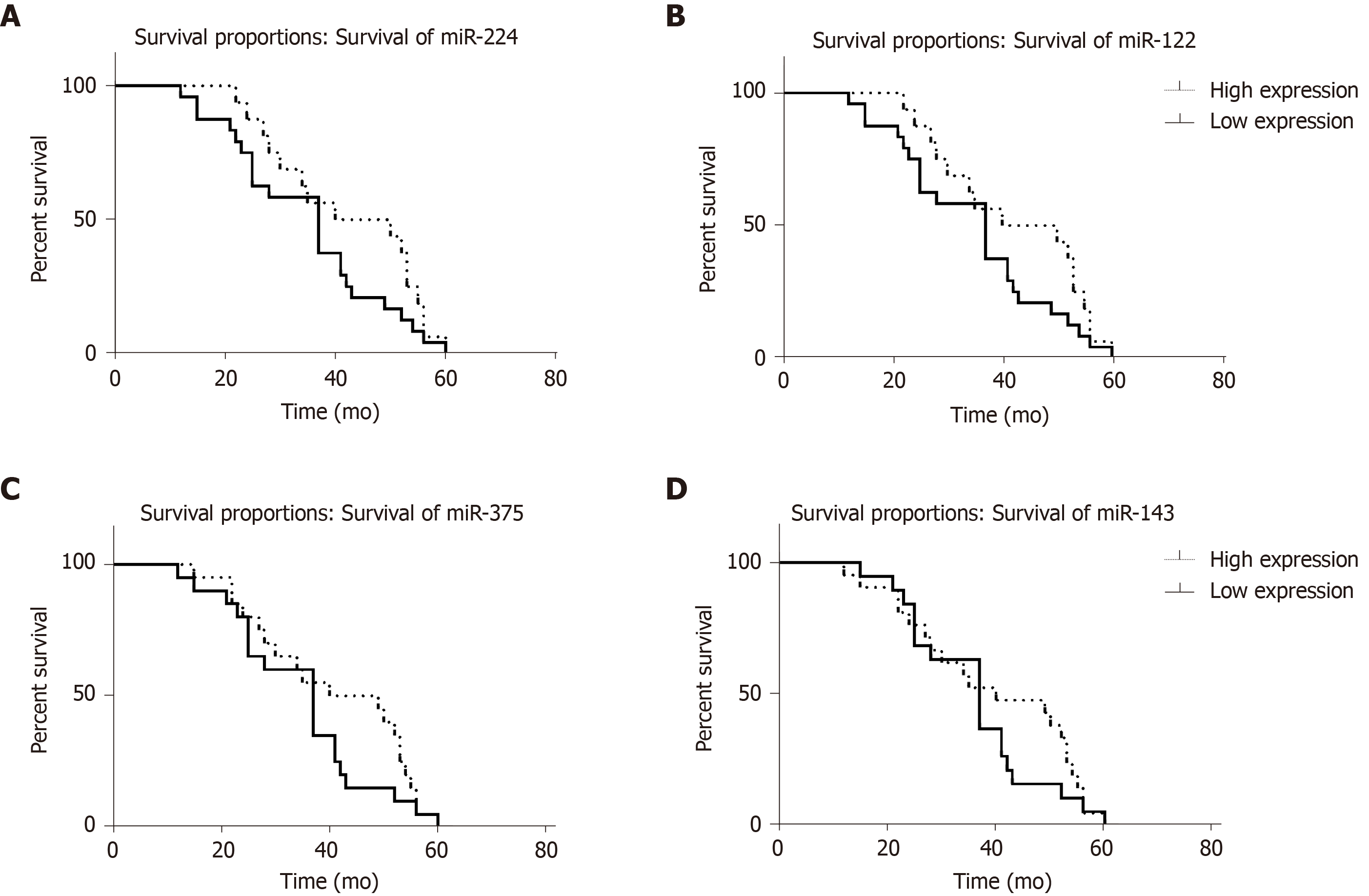
After statistical analysis of the follow-up of these 40 HBV cirrhosis patients, the overall survival of patients with different expression levels of the 4 miRNAs was determined and is shown in Table 4. Log rank analysis showed that patients with a lower expression level of miR-224 and higher expression levels of miR-375, miR-122 and miR-143 had longer overall survival (Figure 4) than those with the opposite expression pattern (P < 0.01). According to Cox analysis, miR-224, miR-375, miR-122 and miR-143 are important factors affecting overall survival.
| miR-224 | miR-122 | miR-375 | miR-143 | |
| Higher expression | 34.45 ± 13.09 | 42.19 ± 12.98 | 40.75 ± 14.12 | 39.38 ± 15.08 |
| Lower expression | 41.72 ± 13.42 | 34.75 ± 13.39 | 34.70 ± 12.60 | 35.89 ± 11.77 |
In this study, we used TLDA and RT-qPCR validation to systematically detect miRNA expression in HBV cirrhosis and found a new miR-panel (miR-224, miR-375, miR-122 and miR-143) that can effectively distinguish HBV cirrhosis patients from controls. Furthermore, we examined the expression of the 4 miRNAs in the tumor tissues of patients with HCC and found that the results were consistent with those in HBV cirrhosis. Compared with normal controls, in the tissue/serum of patients with HBV cirrhosis, miR-375, miR-92a, miR-10a, miR-223, miR-423, miR-23b/a, miR-342-3p, miR-150, let-7c, miR-99a, miR-125b, miR-22, miR-720, miR-1275, miR-486-3p, miR-1908, miR-675, and miR-1231 were significantly upregulated[25-29]. However, this 4-miRNA combination in our study has not been reported, and the combination has a high ROC curve AUC of 0.991, suggesting a strong ability to distinguish HBV cirrhosis from normal controls.
Simultaneously, we analyzed the relationship of each miRNA with overall survival and found that patients with lower abnormal miRNA expression will have a longer overall survival. MiR-224 can offset the effects on the reduction of tumor growth and cell proliferation of glycine N-methyltransferase (GNMT) by targeting GNMT, which is a tumor suppressor for HCC[30]. The receptor tyrosine-protein kinase erbB-2, a direct target gene of miR-375, was associated with human liver cancer growth, and the upregulation of miR-375 can inhibit human liver cancer cell growth by regulating its cell apoptosis[31]. The overexpression of GATA-binding factor 6, which is a downstream target of miR-143 in HCC, significantly increased cell proliferation and invasion rates in HCC, suggesting that miR-143 may suppress the malignancy of HCC by targeting GATA-binding factor 6[32]. Liver-specific miR-122, which is essential for metabolic homeostasis, suppresses glucose-6-phosphate-dehydrogenase (G6PD) expression by directly interacting with its 3'UTR to achieve its anti-HCC efficacy. G6PD is the rate-limiting enzyme of the pentose phosphate pathway, which is often activated in human malignancies to generate precursors for nucleotide and lipid synthesis[33]. MiR-122 overexpression inhibited the epithelial-mesenchymal transition by targeting Snail1 and Snail2 and regulated their expression levels to inhibit cell proliferation, colony formation and cell invasion in HCC cells[34]. In summary, the high expression of miRNA-224 and low expression of miR-375, miR-122, and miR143 in HBV cirrhosis tissue may promote the development of liver cirrhosis to liver cancer. This may be the reason that smaller miRNA expression differences result in longer survival.
When performing RT-qPCR, it is critical to use a stably expressed gene as an internal standard for standardization. To date, no consensus has been established for endogenous controls in the study of circulating miRNAs. There are many different internal controls, such as RNU6B, RNU44, RNU48 and miR-16[35,36], and as expected, the results are not the same. In this study, we chose the combination of let-7d, let-7g and let-7i as an internal reference, which is statistically superior and can better correct experimental changes[20]. In addition, some other levels detection method also have been combined with miRNA, such as, CRISPER/Cas 9 system[37,38], DNA methylation[39], proteomics[40], and metabolics, to involve the development of HCC.
Overall, we found a new miRNA group (miR-122, miR-375, miR-224 and miR-143) for the initial diagnosis of HBV cirrhosis and HBV infection, and compared with normal controls, patients with HBV cirrhosis had high expression of miR-224 and low expression of miR-375, miR-122 and miR-143. In addition, miR-224 low expression/miR-375, miR-122 and miR-143 high expression patients had a longer overall survival. In short, we identified four miRNAs as potential biomarkers for early diagnosis of HBV infection, HBV cirrhosis, and prediction of its overall survival.
The vast majority of liver cancer cases in China are closely related to hepatitis B virus (HBV) infection, but there are few studies on the changes of microRNAs (miRNA) expression in the progression from HBV infection to hepatoma.
In this study, TaqMan Low Density Array and real time quantitative polymerase chain reaction were used to characterize the profile of miRNAs in chronic hepatitis B, HCC and normal control tissues.
This study aimed to explore the role of miRNAs in the progression of HBV infection to cirrhosis and even to liver cancer.
The authors screened differentially expressed miRNAs in 40 HBV cirrhosis, 40 normal and 15 HCC tissues. Authors calculated the area under the receiver-operating-characteristic curves.
The levels of miR-375, miR-122 and miR-143 were significantly lower in HBV cirrhosis tissues, while miR-224 was significantly higher than in the controls. The area under the curves of the receiver-operating-characteristic curve for the 4-miRNA panel was 0.991. Patients with a lower expression level of miR-224 or higher expression levels of miR-375, miR-122 and miR-143 had longer overall survival.
The miR-375, miR-122, miR-143 and miR-224 may be helpful for early diagnosis of HBV infection, HBV cirrhosis, and prediction of its overall survival.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Abd el Moety HA, Campanale M, Sogabe I S-Editor: Wang JL L-Editor: A E-Editor: Qi LL
| 1. | World Health Organization. Global Hepatitis Report 2017. Geneva: World Health Organization, 2017. Available from: http://apps.who.int/iris/bitstream/handle/10665/255016/9789241565455-eng.pdf;jsessionid=7E29131834B6D97C84AC00E8742A0BFE?sequence=1. |
| 2. | Han Y, Zeng A, Liao H, Liu Y, Chen Y, Ding H. The efficacy and safety comparison between tenofovir and entecavir in treatment of chronic hepatitis B and HBV related cirrhosis: A systematic review and Meta-analysis. Int Immunopharmacol. 2017;42:168-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 3. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2930] [Cited by in RCA: 3055] [Article Influence: 152.8] [Reference Citation Analysis (0)] |
| 4. | He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075-19080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 890] [Cited by in RCA: 928] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 5. | Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029-6033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1865] [Cited by in RCA: 1897] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 6. | Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442-4452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 537] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 7. | Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 305] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 8. | Lei KJ, Lin YM, An GY. miR156 modulates rhizosphere acidification in response to phosphate limitation in Arabidopsis. J Plant Res. 2016;129:275-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Matullo G, Naccarati A, Pardini B. MicroRNA expression profiling in bladder cancer: the challenge of next-generation sequencing in tissues and biofluids. Int J Cancer. 2016;138:2334-2345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 10. | Xian X, Tang L, Wu C, Huang L. miR-23b-3p and miR-130a-5p affect cell growth, migration and invasion by targeting CB1R via the Wnt/β-catenin signaling pathway in gastric carcinoma. Onco Targets Ther. 2018;11:7503-7512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 11. | Jiménez-Wences H, Martínez-Carrillo DN, Peralta-Zaragoza O, Campos-Viguri GE, Hernández-Sotelo D, Jiménez-López MA, Muñoz-Camacho JG, Garzón-Barrientos VH, Illades-Aguiar B, Fernández-Tilapa G. Methylation and expression of miRNAs in precancerous lesions and cervical cancer with HPV16 infection. Oncol Rep. 2016;35:2297-2305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Bagheri A, Khorshid HRK, Tavallaie M, Mowla SJ, Sherafatian M, Rashidi M, Zargari M, Boroujeni ME, Hosseini SM. A panel of noncoding RNAs in non-small-cell lung cancer. J Cell Biochem. 2019;120:8280-8290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 13. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 876] [Cited by in RCA: 896] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 14. | Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419-427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 402] [Cited by in RCA: 422] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 15. | Huang YS, Dai Y, Yu XF, Bao SY, Yin YB, Tang M, Hu CX. Microarray analysis of microRNA expression in hepatocellular carcinoma and non-tumorous tissues without viral hepatitis. J Gastroenterol Hepatol. 2008;23:87-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 16. | Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, Tang ZY, Wang XW. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 568] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 17. | Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P, Lee CG. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205-13215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 299] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 18. | Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, Zhang Y, Chen X, Wang J, Zen K, Zhang CY, Zhang C. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9:e92292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 19. | Luo Y, Wang C, Chen X, Zhong T, Cai X, Chen S, Shi Y, Hu J, Guan X, Xia Z, Wang J, Zen K, Zhang CY, Zhang C. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin Chem. 2013;59:658-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 20. | Chen X, Liang H, Guan D, Wang C, Hu X, Cui L, Chen S, Zhang C, Zhang J, Zen K, Zhang CY. A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS One. 2013;8:e79652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 21. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133962] [Article Influence: 5581.8] [Reference Citation Analysis (1)] |
| 22. | Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, Wang J, Li L, Zhou X, Li N, Pan H, Zhang J, Zen K, Zhang CY, Zhang C. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132:116-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 23. | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [PubMed] |
| 24. | Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245-5250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5406] [Cited by in RCA: 5239] [Article Influence: 249.5] [Reference Citation Analysis (0)] |
| 25. | Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 26. | Kohno T, Tsuge M, Murakami E, Hiraga N, Abe H, Miki D, Imamura M, Ochi H, Hayes CN, Chayama K. Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J Viral Hepat. 2014;21:e89-e97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 27. | Yang X, Li H, Sun H, Fan H, Hu Y, Liu M, Li X, Tang H. Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 28. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 29. | Akamatsu S, Hayes CN, Tsuge M, Miki D, Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S, Aikata H, Kawaoka T, Kawakami Y, Ohishi W, Chayama K. Differences in serum microRNA profiles in hepatitis B and C virus infection. J Infect. 2015;70:273-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 30. | Hung JH, Li CH, Yeh CH, Huang PC, Fang CC, Chen YF, Lee KJ, Chou CH, Cheng HY, Huang HD, Chen M, Tsai TF, Lin AM, Yen CH, Tsou AP, Tyan YC, Chen YA. MicroRNA-224 down-regulates Glycine N-methyltransferase gene expression in Hepatocellular Carcinoma. Sci Rep. 2018;8:12284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Li L, Jia L, Ding Y. Upregulation of miR-375 inhibits human liver cancer cell growth by modulating cell proliferation and apoptosis via targeting ErbB2. Oncol Lett. 2018;16:3319-3326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 32. | Xue F, Yin J, Xu L, Wang B. MicroRNA-143 inhibits tumorigenesis in hepatocellular carcinoma by downregulating GATA6. Exp Ther Med. 2017;13:2667-2674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Barajas JM, Reyes R, Guerrero MJ, Jacob ST, Motiwala T, Ghoshal K. The role of miR-122 in the dysregulation of glucose-6-phosphate dehydrogenase (G6PD) expression in hepatocellular cancer. Sci Rep. 2018;8:9105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 34. | Jin Y, Wang J, Han J, Luo D, Sun Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/β-cadherin signaling pathway. Exp Cell Res. 2017;360:210-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 35. | Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 186] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 36. | Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50:298-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 867] [Cited by in RCA: 932] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 37. | Gao W, Long L, Tian X, Xu F, Liu J, Singh PK, Botella JR, Song C. Genome Editing in Cotton with the CRISPR/Cas9 System. Front Plant Sci. 2017;8:1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 38. | Guo J, Li K, Jin L, Xu R, Miao K, Yang F, Qi C, Zhang L, Botella JR, Wang R, Miao Y. A simple and cost-effective method for screening of CRISPR/Cas9-induced homozygous/biallelic mutants. Plant Methods. 2018;14:40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 39. | Sun Q, Qiao J, Zhang S, He S, Shi Y, Yuan Y, Zhang X, Cai Y. Changes in DNA methylation assessed by genomic bisulfite sequencing suggest a role for DNA methylation in cotton fruiting branch development. PeerJ. 2018;6:e4945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 40. | Yu J, Zhang Y, Liu J, Wang L, Liu P, Yin Z, Guo S, Ma J, Lu Z, Wang T, She Y, Miao Y, Ma L, Chen S, Li Y, Dai S. Proteomic discovery of H2O2 response in roots and functional characterization of PutGLP gene from alkaligrass. Planta. 2018;248:1079-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |