Published online Mar 15, 2019. doi: 10.4251/wjgo.v11.i3.181
Peer-review started: November 15, 2018
First decision: December 7, 2018
Revised: December 17, 2018
Accepted: December 23, 2018
Article in press: December 24, 2018
Published online: March 15, 2019
Processing time: 120 Days and 15.1 Hours
Based on the breakthrough of genomics analysis, The Cancer Genome Atlas Research Group recently proposed an integrative genomic analysis, dividing gastric cancer (GC) into four subtypes, characterized by the chromosomal instability (CIN) status. However, the CIN status of GC is still vaguely characterized and lacking the valuable easy-to-use CIN markers to diagnosis in molecular and histological detection.
To explore the associations of CIN with downstream lipidomics profiles.
We collected cancerous and noncancerous tissue samples from 18 patients with GC; the samples were divided into CIN and non-CIN types based on the system of The Cancer Genome Atlas Research Group and 409 sequenced oncogenes and tumor suppressor genes. We identified the lipidomics profiles of the GC samples and samples of their adjacent noncancerous tissues by using liquid chromatography–mass spectrometry. Furthermore, we selected leading metabolites based on variable importance in projection scores of > 1.0 and P < 0.05.
Twelve men and six women participated in this study; the participants had a median age of 67.5 years (range, 52–87 years) and were divided into CIN (n = 9) and non-CIN (n = 9) groups. The GC samples exhibited distinct profiles of lysophosphocholine, phosphocholine, phosphatidylethanolamine, phosphatidylinositol, phosphoserine, sphingomyelin, ceramide, and triglycerides compared with their adjacent noncancerous tissues. The glycerophospholipid levels (phosphocholine, phosphatidylethanolamine, and phosphatidylinositol) were 1.4- to 2.3-times higher in the CIN group compared with the non-CIN group (P < 0.05). Alterations in the glycerolipid and glycerophospholipid pathways indicated progression of GC toward CIN.
The lipidomics profiles of GC samples were distinct from those of their adjacent noncancerous tissues. CIN status of GC is primarily associated with downstream lipidomics in the glycerophospholipid pathway.
Core tip: We investigated the correlation between comprehensive lipidomic profiles of gastric cancer and the tumor’s chromosomal instability (CIN) status. In this disease landscape study, which involved no pre-specified hypotheses, we combined a gene molecule classification method with a lipidomic method to discover metabolic information for accurate tumor classification. CIN-status-based lipidomics profiling demonstrated translational potential for biomarker discovery and development of novel therapeutic strategies.
- Citation: Hung CY, Yeh TS, Tsai CK, Wu RC, Lai YC, Chiang MH, Lu KY, Lin CN, Cheng ML, Lin G. Glycerophospholipids pathways and chromosomal instability in gastric cancer: Global lipidomics analysis. World J Gastrointest Oncol 2019; 11(3): 181-194
- URL: https://www.wjgnet.com/1948-5204/full/v11/i3/181.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i3.181
Gastric cancer (GC) is traditionally subdivided into intestinal, diffuse, and mixed types according to Lauren classification based on histopathology[1,2]. Although widely used, the Lauren classification system does not provide precise information on treatments suitable for individual patients, and selecting a subtype-optimized therapeutic approach can be difficult[2]. Recently, The Cancer Genome Atlas (TCGA) Research Group proposed an integrative genomic analysis method, namely dividing GC into four subtypes—Epstein Barr Virus positive, microsatellite unstable, chromosomally instable, and genomically stable[3] - on the basis of gene expression profiling of exome sequences, copy-number alterations, gene expression, DNA methylation, and protein activity[2-4]. However, the chromosomal instability (CIN) status of GC is still characterized only vaguely and lacks valuable and user-friendly markers for diagnosis in molecular and histological detection[5].
Metabolomics—the study of results of interaction between the biosystem’s genome and its environment and the detection of end products of gene expression - offers opportunities to understand complex molecular mechanisms and identify the diagnostic biomarkers of human GC[4,6]. Previous metabolomics studies based on mass spectrometry (MS) and nuclear magnetic resonance systems have been limited to focusing on water-soluble compounds and volatile metabolites[7-10]. Lipid metabolites have several pivotal functions, including energy storage, modulation of cell membranes, the formation of “fat-soluble” vitamins, cellular massage, and hormonal regulation[11], and they thus warrant further research. Furthermore, increased de novo lipogenesis is frequently associated with the development of many cancer types[12]. For example, the lipid content of phospholipids could compromise membrane fluidity and signal transduction and in turn affect tumorigenesis and GC progression[13]. In addition, perturbation of lipid metabolism contributes to cancer progression through detection of dysregulated core enzyme activity in lipid pathways and global lipid metabolic alterations in cancer metastasis[14,15]. Global lipidomics analysis using liquid chromatography–MS (LC/MS) provides the most detailed detection and qualification of cellular lipids in systems biology. To the best of our knowledge, no prior studies have exploited the links between CIN and non-CIN status in GC and lipid alteration by using the lipidomics approach.
The present study hypothesized that lipidomic alternations reflect the CIN or non-CIN status of GC. Through global lipidomics profiling using LC/MS, we explored the correlation between lipidomic metabolites and the CIN status of GC.
The Institutional Review Board approved this prospective study (IRB103-7448B). Informed consent to screen patient enrollment was provided by a tertiary referral center with a GC-dedicated interdisciplinary team, and tissue samples were obtained from Chang Gung Memorial Hospital in Linkou, Taiwan. We screened a continuous cohort of patients with GC from May 2015 to April 2017. The inclusion criteria were (1) histologically confirmed GC with surgical resection; and (2) age of 20–80 years. The exclusion criteria were (1) receipt of neoadjuvant therapy before surgery; (2) tumor smaller than 1 cm in computed tomography images; (3) prior gastric surgery; (4) anti-Helicobacter pylori eradication therapy; and (5) receipt of nonsteroidal anti-inflammatory drugs within the 1 week prior to surgery[16]. We used 18 primary GC tissue samples for genomic analysis and re-evaluated the pathological diagnoses and histological Lauren classifications of all tumors, with samples from their adjacent noncancerous tissues as controls.
The tumor samples were divided into CIN or non-CIN by using TCGA system. We extracted genomic DNA from formalin-fixed paraffin-embedded tumor samples by using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) and quantified the DNA by using the Quant-iT dsDNA High-Sensitivity Assay Kit (Invitrogen, USA). In total, 409 leading oncogenes and tumor suppressor genes in GC tissue were sequenced; the protocol for TCGA analysis was detailed in our previous study[16]. The present study classified patients with GC based on high and low proportions of alteration genes.
Tumor tissue samples of similar weight were extracted from the organic layer through Folch extraction and analyzed using an LC/MS system for lipidomic analysis. A modified version of Folch’s method was employed[17]. In brief, we transferred approximately 50 mg of homogenized tissue into a glass tube and then added 6 mL of chloroform/methanol (2:1, v/v) solution and 1.5 mL of water. The sample was vortexed four times for 30 s each and then centrifuged at 8000 rpm for 30 min at 4 °C. The lower phase (hydrophobic phase and lipid layer) was transferred to new glass tubes and then dried using nitrogen gas. We stored the dried samples at −80 °C. Before analysis, the sample was dissolved in isopropanol/acetonitrile/water (2:1:1, V/V/V) through vortexing (four times for 30 s each) and centrifugation (12000 rpm for 20 min at 4 °C). Subsequently, the supernatant was transferred to vials for LC/MS analysis.
We performed liquid chromatographic separation on an ACQUITY CSH C18 column (2.1 × 100 mm, 1.7 μm; Waters Co.) at a constant temperature of 55 °C by using the ACQUITY UltraPerformance LC system (Waters MS Technologies, UK). For metabolite profiling, mobile phase A was acetonitrile/water (60:40, v/v) and mobile phase B was isopropanol/acetonitrile (90:10, v/v); both phases were solvents containing 10 mM ammonium formate and 0.1% formic acid. The flow rate was 0.4 mL/min with a time-resolved solvent gradient[18]. We performed MS analysis by using Waters time-of-flight (TOF)–MS (SYNAPT HDMS; Waters MS Technologies, UK) operated in electrospray ionization (ESI)-positive (ESI+) and ESI-negative (ESI−) ion modes. We set the capillary and cone voltage at 2700 V (2000 V in ESI− mode) and 35 V, respectively. The desolvation gas flow rate was 800 L/h, maintained at 25 L/h. The desolvation and source temperatures were 400 °C and 100 °C, respectively. We acquired MS data in centroid mode within 20 to 990 m/z at a rate of 10 scans/s. Leucine–enkephalin served as a reference compound. The LockSpray frequency was set at 0.5 s and averaged over 10 scans for correction. We performed three technical replicates for tissue samples in both ESI+ and ESI− modes.
We analyzed the lipidomic metabolites of the GC samples and their surrounding adjacent noncancerous tissues by using LC/TOF/MS with an untargeted metabolic approach to screen all potential biomarkers according to the application notes database (Waters, Milford, MA, USA)[19]. All MS data, namely retention times, m/z, and ion intensities, were extracted using MarkerLynx XS software (Waters) and then input to a matrix. Subsequently, the data were analyzed using orthogonal projections to latent structures discriminant analysis (OPLS-DA) run through SIMCA-P+ (version 13.0, Umetrics) with Pareto scaling. The variable importance in projection (VIP) score of each metabolite indicated a metabolite’s contribution to the model. In this analysis, VIP > 1.0 and P < 0.05 were considered significant. In addition, we evaluated diagnostic performance by analyzing receiver operating characteristic curves with 95% confidence intervals; the areas under these curves were calculated using MetaboAnalyst 4.0[20].
Lipids are composed of fats, oils, waxes, and sterols. As demonstrated by the LIPID MAPS classification system, lipids are broadly divided into eight categories: fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides[21]. Significant metabolites were sought in the Human Metabolome Database (http://www.hmdb.ca) and confirmed using in-house data (standards based on retention times and MS spectra). Candidates for LC/MS/MS analysis were confirmed according to chemical standards, the METLIN database[22], or LIPID MAPS database[21], depending on the m/z results for daughter fragments under chromatographic conditions identical to those of the profiling experiment. The sn- positions of fatty acids on the glycerol backbones of lipids were not identified in this study.
In total, 18 patients with GC enrolled in this study (median age, 67.5 years; range, 52–87 years) and were divided into CIN (n = 9) and non-CIN (n = 9) groups by using a 5% frequency of genetic variation as the demarcation point; no marked differences in demographics were observed (Table 1). In this study, 85.7% of the Lauren intestinal-type tumors (6/7) belonged to the CIN GC group and all of the Lauren diffuse-type tumors belonged to the non-CIN GC group. Lauren mixed-type tumors belonged to both the CIN (50%) and non-CIN (50%) groups. The intestinal-type tumors demonstrated a high alteration rate of 92.2% (377 genes), particularly those with copy-number changes; by contrast, the diffuse-type tumors exhibited a low alteration rate of 8.56% (35 genes).
| Term | TCGA system | |
| CIN | Non-CIN | |
| Number | 9 | 9 |
| Age (median yr, range) | 68.1 (56-79) | 66.9 (52-87) |
| Sex (male/female) | 7/2 | 5/4 |
| Size (cm) | 4.0 (1.8-6.9) | 5.4 (2.3-11.6) |
| Lauren's classification | ||
| Intestinal type | 6 | 1 |
| Diffuse type | 0 | 5 |
| Mixed type | 3 | 3 |
| Stage | ||
| I | 1 | 0 |
| II | 2 | 2 |
| III | 5 | 6 |
| IV | 1 | 1 |
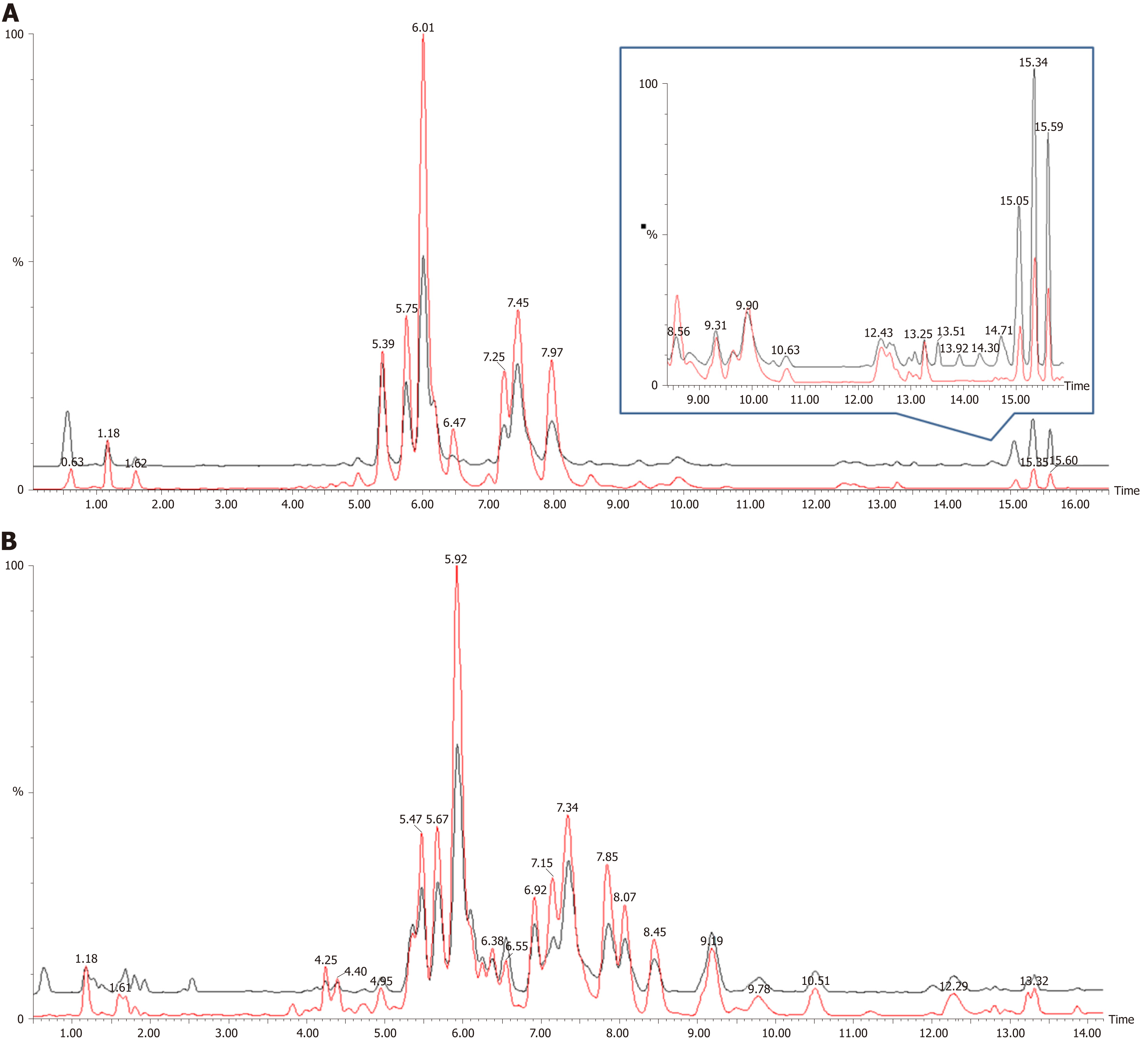
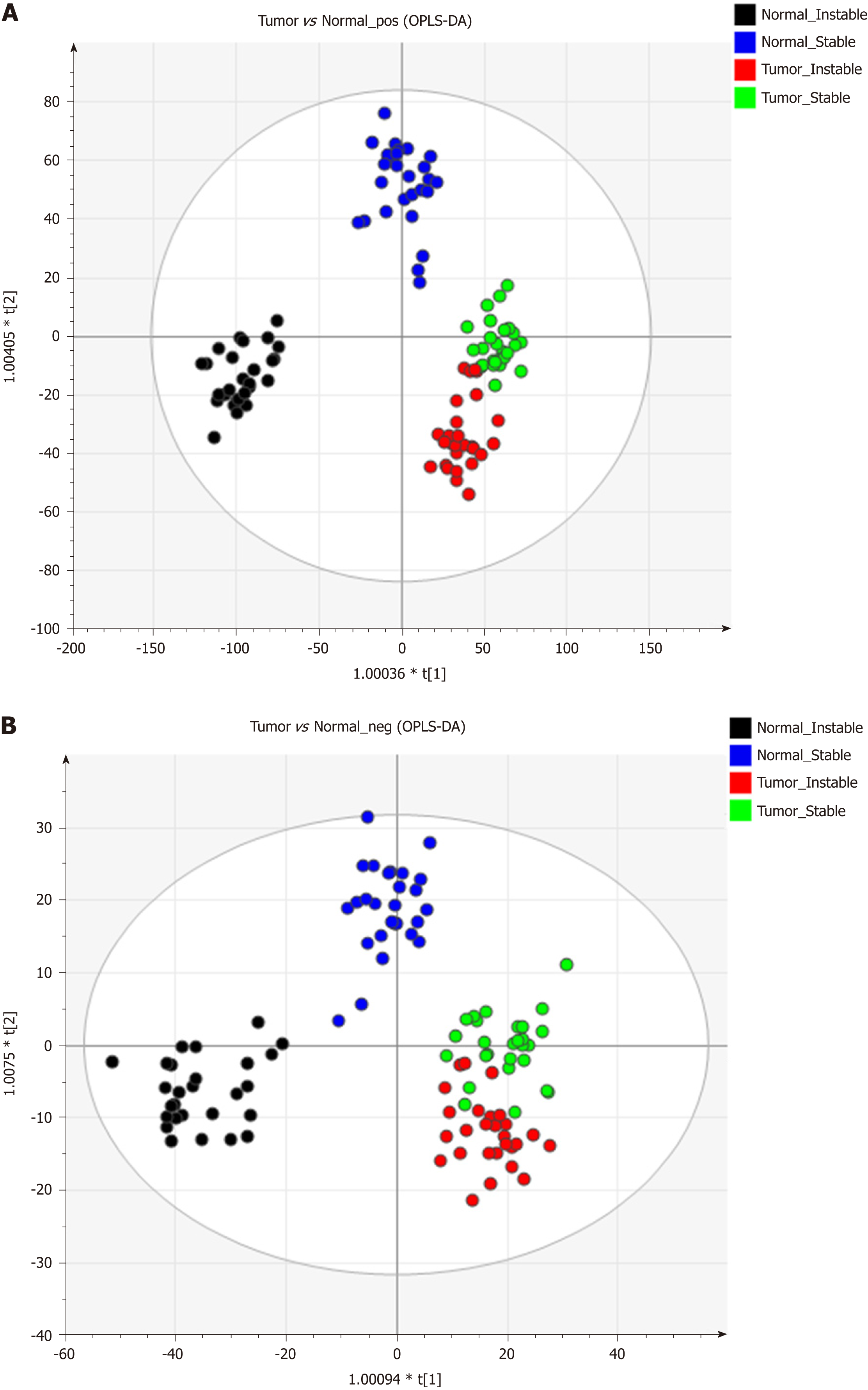
Figure 1 shows the representative MS spectra for both ESI modes. We observed significant changes in the lysoglycerophospholipid, GP, and triglyceride (TG) regions in the ESI+ mode and in the lysoglycerophospholipid, GP, and SP regions in the ESI− mode. After calculating data matrices by using MarkerLynx XS and exporting them to SIMCA-P+ software, we obtained 1374 variables (loadings) in the ESI+ mode and 539 variables in the ESI− mode. Four significant clusters between tumors and their adjacent noncancerous tissues were detected in both modes by using OPLS-DA (R2X = 0.844, R2Y = 0.89, and Q2 = 0.747 in ESI+ mode; R2X = 0.815, R2Y = 0.841, and Q2 = 0.603 in ESI- mode), as illustrated in Figure 2. These clusters were divided into tumor samples with CIN status, tumor samples with non-CIN status, adjacent noncancerous tissues with CIN status, and adjacent noncancerous tissues with non-CIN status.
Loading plots of the OPLS-DA and VIP scores were used to identify potential diagnostic markers in GC tissues. Significant metabolite differences between tumors and their adjacent noncancerous tissues were identified by VIP ≥ 1.0 and P < 0.05 and divided into lysophosphocholine (LysoPC), phosphocholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphoserine (PS), sphingomyelin (SM), ceramide, and TG in both ESI modes (Table 2). Compared with their adjacent noncancerous tissues, the GC samples exhibited higher levels of PC and SM but lower levels of PE and TG (all P < 0.05). We observed no lipid species that were present in only one group. All of the metabolites observed in this study exhibited dynamic differences between tumors and their adjacent noncancerous tissues.
| Catalog | Putative ID | RT (min) | m/z | Adduct ion | VIP | FC |
| TG | TG(54:3) | 15.6 | 902.8202 | [M+NH4]+ | 2.3 | 0.5 |
| TG(52:4) | 15.1 | 872.7729 | [M+NH4]+ | 3.2 | 0.4 | |
| TG(52:2) | 15.6 | 876.8048 | [M+NH4]+ | 3.2 | 0.5 | |
| TG(50:3) | 15.0 | 846.7568 | [M+NH4]+ | 2.9 | 0.3 | |
| TG(50:2) | 15.3 | 848.7722 | [M+NH4]+ | 3.3 | 0.4 | |
| SM | SM(d18:1/24:0) | 13.3 | 815.7015 | [M+H]+ | 1.6 | 1.3 |
| SM(d18:1/18:0) | 7.1 | 775.5980 | [M+FA-H]- | 1.7 | 0.7 | |
| SM(d18:0/22:0) | 9.9 | 811.6627 | [M+H]+ | 2.1 | 1.3 | |
| SM(d18:0/16:0) | 6.0 | 705.5912 | [M+H]+ | 1.0 | 1.4 | |
| PS | PS(P-18:0/22:6) | 9.2 | 818.5336 | [M-H]- | 1.0 | 0.7 |
| PI | PI(22:3/16:0) | 6.1 | 887.5682 | [M-H]- | 1.4 | 1.9 |
| PI(20:4/16:0) | 4.2 | 857.5202 | [M-H]- | 1.4 | 0.6 | |
| PI(16:0/18:2) | 4.4 | 833.520 | [M-H]- | 3.0 | 0.8 | |
| PE | PE(P-18:1/20:4) | 7.1 | 748.5279 | [M-H]- | 2.5 | 0.7 |
| PE(P-18:0/18:2) | 9.7 | 726.5437 | [M-H]- | 2.5 | 1.7 | |
| PE(P-16:0/18:2) | 7.3 | 698.5117 | [M-H]- | 4.0 | 1.9 | |
| PE(20:4/16:0) | 6.2 | 740.5237 | [M+H]+ | 3.6 | 0.4 | |
| PE(18:2/18:1) | 6.6 | 740.5221 | [M-H]- | 2.3 | 0.4 | |
| PE(18:2/16:0) | 6.4 | 714.5061 | [M-H]- | 2.2 | 0.4 | |
| PE(18:1/18:1) | 8.2 | 742.5385 | [M-H]- | 2.4 | 0.6 | |
| PC | PC(38:4) | 6.4 | 810.6034 | [M+H]+ | 2.6 | 1.7 |
| PC(34:3) | 4.8 | 756.5561 | [M+H]+ | 14.2 | 1.5 | |
| PC(30:0) | 5.5 | 706.5394 | [M+H]+ | 2.9 | 2.4 | |
| PC(18:2/16:0) | 6.0 | 758.5702 | [M+H]+ | 3.0 | 0.6 | |
| PC(18:0/20:3) | 8.6 | 812.6191 | [M+H]+ | 3.3 | 3.6 | |
| PC(18:0/18:1) | 9.8 | 832.6090 | [M+FA-H]- | 7.9 | 1.3 | |
| PC (16:1/16:0) | 5.6 | 776.5467 | [M+FA-H]- | 1.2 | 2.0 | |
| PC (16:0/16:0) | 7.3 | 734.5717 | [M+H]+ | 1.5 | 1.4 | |
| LysoPC | LysoPC (16:0) | 1.2 | 496.3395 | [M+H]+ | 3.1 | 0.7 |
| Ceramide | Cer (d18:1/24:0) | 13.9 | 694.6341 | [M+FA-H]- | 1.6 | 1.6 |
| carnitine | Linoleyl carnitine (C18:2) | 1.0 | 424.3408 | [M+H]+ | 1.2 | 0.6 |
| Oleoyl carnitine (C18:1) | 1.2 | 426.3560 | [M+H]+ | 1.1 | 0.8 |
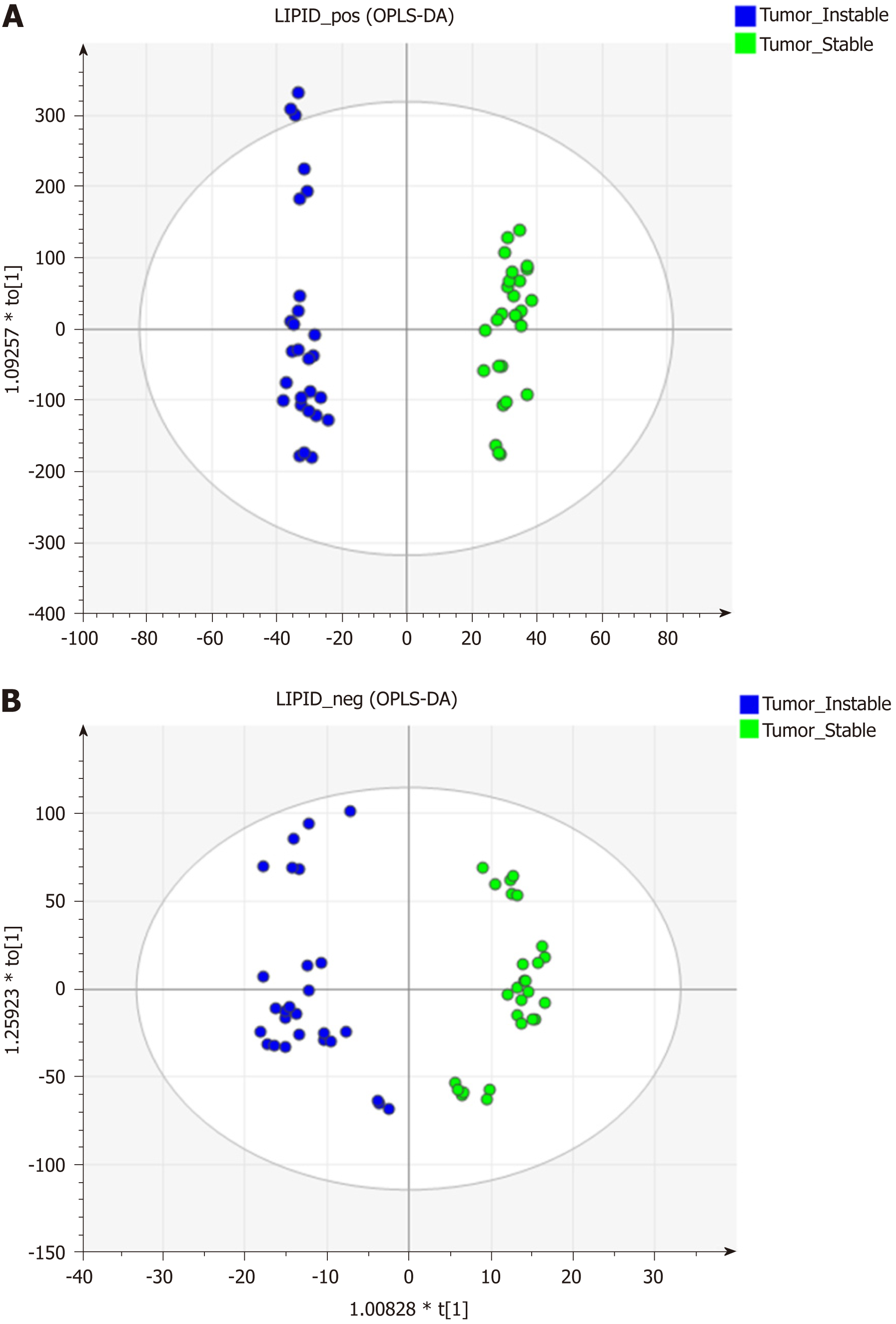
The data matrices were further exported for OPLS-DA in both ESI modes to show the lipid difference between CIN and non-CIN status within the GC samples. Two significant clusters are illustrated in Figure 3 (R2X = 0.79, R2Y = 0.988, and Q2 = 0.874 in ESI+ mode; R2X = 0.71, R2Y = 0.914, and Q2 = 0.694 in ESI− mode). This pattern suggests that the divergence of the OPLS-DA distribution was dependent on the CIN status with goodness of fit. Based on the loading plots of OPLS-DA, significant differences between the CIN and non-CIN GC samples were filtered by VIP ≥ 1.0 and P < 0.05 and divided into PC, PE, PI, SM, and diglycerides (DG) in both ESI modes (Table 3). No lipid species were present in only one group. The levels of almost all lipid species were different in the CIN tumors and exhibited higher intensity in the CIN tumors than in the non-CIN tumors, except for DG (38:4) and SM (d18:1/18:0) (all P < 0.05). Compared with the non-CIN group, GP levels (PC, PE, and PI) demonstrated were 1.4- to 2.3-times higher in the CIN group (P < 0.05). We observed alteration of the lipid metabolism for both GC status and CIN status in the GL, GP, and SL pathways. We also observed changes in lipid species in the GL and GP pathways in the CIN analysis only; these findings are shown in Figure 4.
| Catalog | Putative ID | RT (min) | m/z | Adduct ion | VIP | FC |
| SM | SM(d18:1/18:0) | 7.2 | 731.6077 | [M+H]+ | 5.8 | 0.49 |
| PI | PI(22:6/18:0) | 5.1 | 909.554 | [M-H]- | 1.6 | 1.53 |
| PI(22:3/16:0) | 6.1 | 887.568 | [M-H]- | 3.0 | 1.75 | |
| PE | PE(P-18:0/18:2) | 9.7 | 726.544 | [M-H]- | 2.8 | 1.92 |
| PE(P-16:0/20:5) | 5.8 | 720.499 | [M-H]- | 2.8 | 2.15 | |
| PE(37:4) | 5.8 | 754.5381 | [M+H]+ | 2.9 | 1.77 | |
| PE(22:6/18:1) | 5.8 | 788.523 | [M-H]- | 1.9 | 1.43 | |
| PE(18:2/16:0) | 6.5 | 716.5233 | [M+H]+ | 2.7 | 1.52 | |
| PC | PC(38:6) | 6.5 | 806.571 | [M+H]+ | 3.2 | 2.06 |
| PC(38:4) | 6.7 | 810.6041 | [M+H]+ | 5.9 | 2.34 | |
| PC(36:5) | 4.8 | 780.5556 | [M+H]+ | 5.7 | 2.30 | |
| PC(33:2) | 5.3 | 744.5562 | [M+H]+ | 3.2 | 1.73 | |
| PC(33:1) | 6.5 | 746.5715 | [M+H]+ | 3.2 | 1.59 | |
| PC(30:0) | 5.5 | 706.5394 | [M+H]+ | 4.6 | 1.89 | |
| PC(16:1/16:1) | 4.6 | 730.5399 | [M+H]+ | 4.1 | 1.98 | |
| PC(16:1/16:0) | 5.6 | 776.547 | [M+FA-H]- | 4.3 | 1.46 | |
| DG | DG(38:4) | 12.6 | 667.5287 | [M+Na]+ | 1.6 | 0.66 |
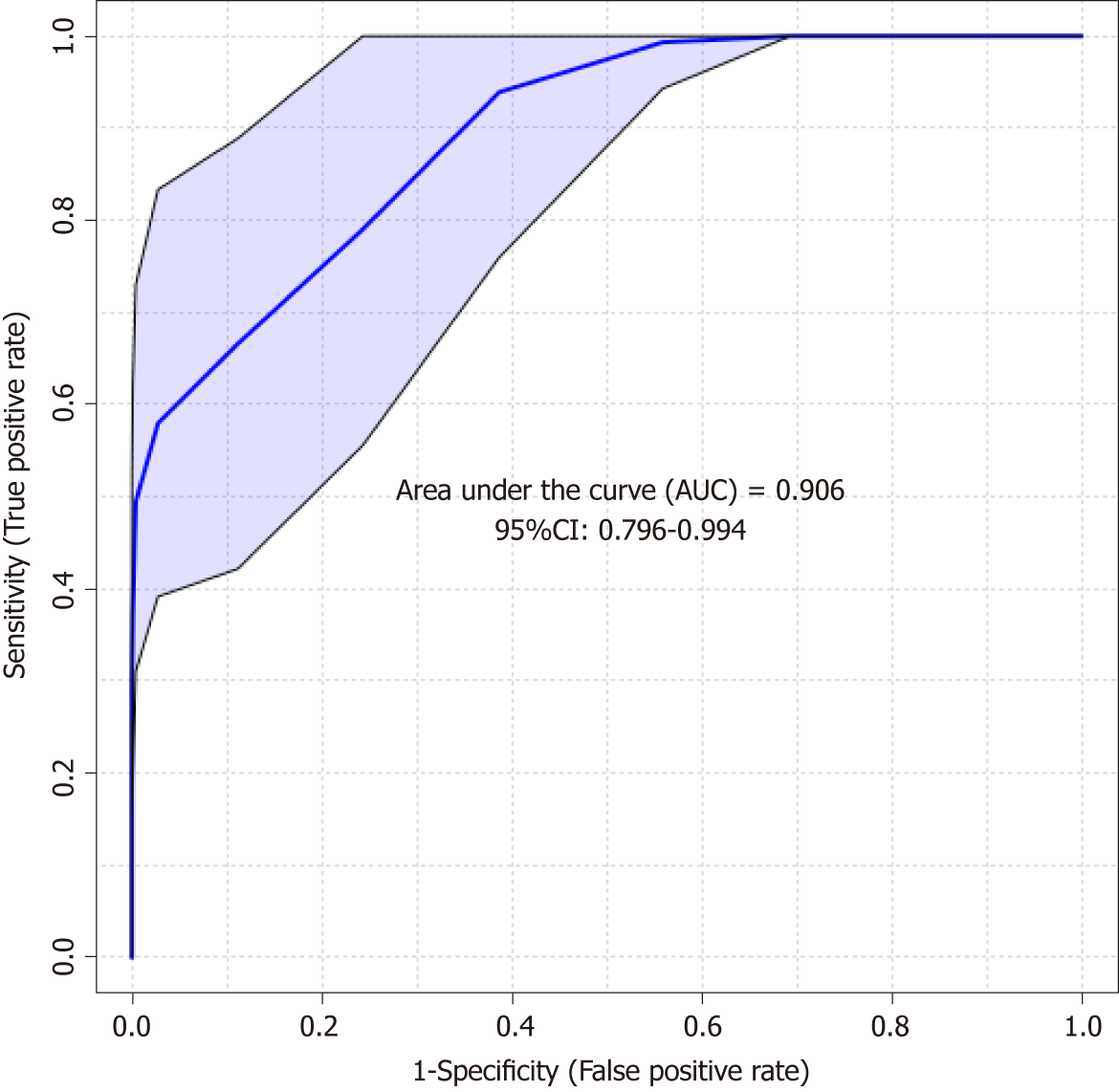
The predictive PLS-DA model based on the significant candidates (Table 3) demonstrated good differentiation between the CIN and non-CIN groups, with sensitivity of 0.852, specificity of 0.703, and an area under the curve of 0.906 (Figure 5).
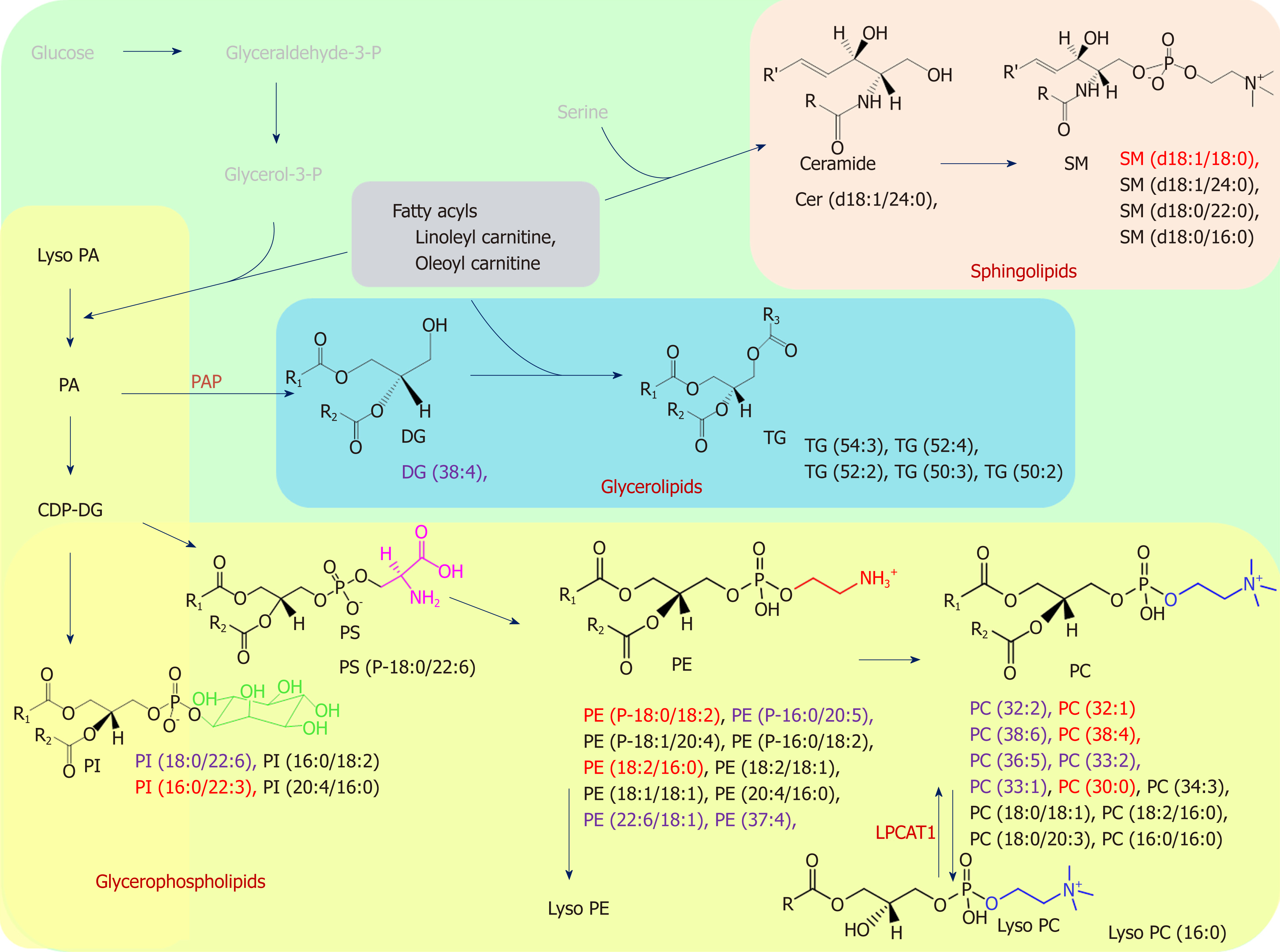
We found that several lipid species primarily affected the grouping of the GC samples and their adjacent noncancerous tissues; markedly higher levels of PC and SM and lower levels of PE and TG were detected in the GC samples, as shown in Figure 4. Alterations in lipid species discovered in the GL, GP, and SL pathways of the GC samples are marked in black. Few studies have examined the differing roles of lipid metabolomics in cancerous and noncancerous samples[6,23,24]. Abbassi-Ghadi et al[24] reviewed several metabolites of glycolysis, the tricarboxylic acid cycle, and lipid metabolism and suggested them to be biomarkers of esophagogastric cancers. Our findings on alterations in TG are supported by the higher prevalence of an olefinic group in noncancerous gastric spheroids at 5.29 ppm, detected using 1H nuclear magnetic resonance, compared with cancerous gastric spheroids[25]. Huang et al[26] reported the products of SL metabolism, including SM and ceramide, which act as bioactive molecules regulating cell survival and proliferation in apoptosis. In the present study, we observed dynamic differences in several SM species between tumors and their adjacent noncancerous tissues. The elevated PC level in cancerous tissue might have been related to overexpression of lysophosphatidylcholine acyltransferase 1[13]. Moreover, the lower level of LysoPC (16:0) observed in this study resulted from conversion of LysoPC into PC due to lysophosphatidylcholine acyltransferase 1 protein activity[13].
We further identified the undisclosed correlation between lipidomic profiling of GC and CIN status. We classified lipid alterations between the CIN and non-CIN GC samples into PC, PE, PI, SM (d18:1/18:0), and DG (38:4). Significant differences in CIN status were observed in the GP (PC, PE, and PI) category alongside various fatty acyl chain lengths and the degree of saturation in the fatty acyl chain in our findings. The features of CIN status are common p53 mutation and frequent activation of genomic amplification, which encodes the receptor tyrosine kinase pathway[5]. Mitogenic signaling conducted by growth factors regulates aberrant cell growth and proliferation, which are involved in the activation of numerous lipid-metabolism-related enzymes[26]. Genetic alterations and enzyme activity in lipid perturbation accumulate over time, resulting in severe changes in lipid metabolism and ultimately leading to tumor formation in CIN tissues[27]. Dysregulation of GP metabolism has previously been described in various cancers[15,28]. Luo et al[15] reviewed the emerging role of lipid metabolism in cancer metastasis and revealed higher levels of PS, PI and PC in metastatic groups than in noncancerous cells. Several core enzymes involved in the GP pathway might directly or indirectly regulate downstream biochemical alterations. Furthermore, Tsai et al[16] reported higher levels of PC in CIN samples after hydrophilic analysis. In our findings, CIN tumors contained significantly higher levels of PC (i.e., PC-containing lipids) than did non-CIN tumors; this finding facilitated discrimination between CIN and non-CIN status in lipidomic profiling, and this supports their results. Lipidomics analysis can provide further insight into other lipid classes. We provided evidence of the difference in the DG (38:4) level of CIN status, which could be affected by the activity of phosphatidic acid phosphatase—which is encoded by a family of genes named lipins—and dephosphorylate of phosphatidic acid to form diglycerides[15].
From the perspective of molecular biology, identification of genetic and epigenetic prognostic biomarkers in various cancers contributes to identification of potential therapeutic targets by upregulating genes in cancer tissues[29]. Potential roles of lipidomics identified by TCGA classification of genomic analysis facilitate diagnosis and surveillance of GC[3,23]. Metabolic phenotypes result from a combination of genomic, transcriptomic, and proteomic conditions and their interactions with the environment[30]. Our preliminary results have potential clinical implications. First, rapid lipidomics profiling could be used to identify patients at high risk of GC at various stages. We combined TCGA classification of genomic analysis with a lipidomics method to determine the distribution of lipid species for accurate diagnosis of GC and identify potential biomarkers for translational discovery and novel therapeutic strategies. Analyzing changes in GP levels (especially PC, PE, and PI) can not only provide insight into GC pathology and diagnosis but also determine novel biomarkers of CIN status in GC. Full molecular classification of GC advances the knowledge of the biology of GC, and identification of biomarkers for early diagnosis may improve effective treatment through precision medicine[8]. However, these preliminary results must be interpreted with caution until they are validated using an independent dataset because the small sample size relative to the number of features extracted may have resulted in model overfitting.
This study had some limitations. First, the sample size was small. Our objective of analyzing genomics and metabolomics data inadvertently limited the number of participants willing to contribute tissue samples in each category of this study. Therefore, more extensive research is warranted to further validate the utility of the analyzed biomarkers, and translation into clinical settings should follow. Second, the methodology of this study could be improved for development of a more comprehensive lipid extraction method for identifying more lipid species such as free fatty acids and cholesteryl ester and its derivatives. Third, potential classes were missing from this exploratory experiment. Although Helicobacter pylori plays a crucial role in gastric carcinogenesis, we aim to the CIN status influences on the outcome of gastric cancer, and tried to exclude the other possible factors including microbiota in gastrointestinal in this study. To further identify potential biomarkers, determining absolute concentrations in multiple biological organs is necessary. Therefore, further investigation that establishes a database of potential biomarkers - including their relative concentrations in multiple organs - for application in precision medicine is warranted.
In conclusion, CIN status of GC was primarily associated with downstream lipidomics in the GP pathway, namely PC, PE, and PI. These findings based on TCGA classification reflected regulation of the cellular signal pathway of apoptosis in CIN tumors. We employed a genomic classification method to obtain lipidomic information correlated with CIN status.
Gastric cancer (GC) leads to worldwide cancer mortality, especially in developing countries. Recently, The Cancer Genome Atlas (TCGA) Research Group proposed an integrative genomic analysis, dividing gastric cancer into four subtypes—Epstein Barr Virus positive, microsatellite unstable, chromosomally instable (CIN), and genomically stable, based on gene expression profiling of the exome sequences, copy-number alterations, gene expression, DNA methylation, and protein activities. However, the CIN status of GC is still vaguely characterized and lacking the valuable easy-to-use CIN markers to diagnosis in molecular and histological detection. Metabolomics, which study the result of the interaction of the biosystem’s genome with its environment and detect the end product of gene expression, offers the opportunity to understand the complex molecular mechanisms and to identify the diagnostic biomarkers of human GC. Although mass spectrometry (MS) and nuclear magnetic resonance system have been used widely to investigate metabolic changes in biological processes, most of those findings were limited to focus on water-soluble compounds, and volatile metabolites. Perturbation of lipid metabolism would also contribute to observing in the cancer progression by detecting the activity of the dysregulated core enzymes in lipid pathways and the global lipid metabolic alterations in cancer metastasis. Global lipidomics provides the most details detection and qualification of the cellular lipids in systems biology. The background, present status, and significance of the study should be described in detail.
In our previous study, metabolomic profiles of GC tumors and the adjacent healthy tissue are distinct, and altered pathways involving amino acid metabolism, glyoxylate and dicarboxylate metabolism. In this study, we hypothesize that lipidomic alternations reflect the CIN or non-CIN status of GC to provide the exploration of the correlation the lipidomic metabolites of GC with its CIN status.
The main objectives aimed to discover the numerous biomarkers from lipidomic studies and explore the associations of CIN with its downstream lipidomics profiles.
Tumor samples were categorized as CIN or non-CIN type by the TCGA system. We extracted the genomic DNA, and quantified them for genomic analysis. In total 409 leading oncogenes and tumor suppressor genes in the GC tumor tissue were sequenced. For lipidomic metabolite research, tissue extraction through Folch method and performed profiling using an LC/MS system. Data processing and statistical analysis for lipidomic analysis to discover the potential metabolites using MarkerLynx XS software, SIMCA-P+ and MetaboAnalyst 4.0.
This study demonstrated the Lipidomic profiling of GC tumors showed distinct profiles in glycerolipid, glycerophospholipid and sphingolipid compared with adjacent non-cancerous tissues. The glycerophospholipid levels (phosphocholine, phosphatidylethanolamine, and phosphatidylinositol) demonstrated a 1.4- to 2.3-fold increase in the CIN group, compared with the non-CIN group (P < 0.05). Alteration of the glycerolipid and glycerophospholipid pathways involved throughout the evolutions of GC formation toward chromosomal instability.
Lipidomics profiles of GC tumors were distinct against the adjacent non-cancerous tissue. The CIN status of GC primarily associated with the downstream lipidomics in glycerophospholipid pathway.
Our study provided the genomic classification method and discovered lipidomic information to correlate with its CIN status. To validate our initial findings, more sample collections with longer follow up times will be considered.
The authors thank all the members of the Cancer Centre, Chang Gung Memorial Hospital. LC-MS was carried out with the help from the Metabolomics Core Laboratory, Healthy Aging Research Center, Chang Gung University and Clinical Metabolomics Core Laboratory, Chang Gung Memorial Hospital.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: Taiwan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Pellicano R S- Editor: Wang JL L- Editor: A E- Editor: Song H
| 1. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4011] [Cited by in RCA: 4323] [Article Influence: 149.1] [Reference Citation Analysis (0)] |
| 2. | Grabsch HI, Tan P. Gastric cancer pathology and underlying molecular mechanisms. Dig Surg. 2013;30:150-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 3. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4854] [Article Influence: 441.3] [Reference Citation Analysis (2)] |
| 4. | Rochfort S. Metabolomics reviewed: a new "omics" platform technology for systems biology and implications for natural products research. J Nat Prod. 2005;68:1813-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 280] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 5. | Strand MS, Lockhart AC, Fields RC. Genetics of Gastric Cancer. Surg Clin North Am. 2017;97:345-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Jayavelu ND, Bar NS. Metabolomic studies of human gastric cancer: review. World J Gastroenterol. 2014;20:8092-8101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 7. | Chan AW, Mercier P, Schiller D, Bailey R, Robbins S, Eurich DT, Sawyer MB, Broadhurst D. (1)H-NMR urinary metabolomic profiling for diagnosis of gastric cancer. Br J Cancer. 2016;114:59-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 8. | Liang Q, Wang C, Li B. Metabolomic Analysis Using Liquid Chromatography/Mass Spectrometry for Gastric Cancer. Appl Biochem Biotechnol. 2015;176:2170-2184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 9. | Jung J, Jung Y, Bang EJ, Cho SI, Jang YJ, Kwak JM, Ryu DH, Park S, Hwang GS. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Ann Surg Oncol. 2014;21 Suppl 4:S736-S742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 10. | Yu L, Aa J, Xu J, Sun M, Qian S, Cheng L, Yang S, Shi R. Metabolomic phenotype of gastric cancer and precancerous stages based on gas chromatography time-of-flight mass spectrometry. J Gastroenterol Hepatol. 2011;26:1290-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 901] [Cited by in RCA: 987] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 12. | Guo S, Wang Y, Zhou D, Li Z. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci Rep. 2014;4:5959. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 13. | Uehara T, Kikuchi H, Miyazaki S, Iino I, Setoguchi T, Hiramatsu Y, Ohta M, Kamiya K, Morita Y, Tanaka H, Baba S, Hayasaka T, Setou M, Konno H. Overexpression of Lysophosphatidylcholine Acyltransferase 1 and Concomitant Lipid Alterations in Gastric Cancer. Ann Surg Oncol. 2016;23 Suppl 2:S206-S213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 14. | Lamaziere A, Wolf C, Quinn PJ. Perturbations of lipid metabolism indexed by lipidomic biomarkers. Metabolites. 2012;2:1-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 15. | Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, Cao Y. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. 2017;16:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 258] [Cited by in RCA: 471] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 16. | Tsai CK, Yeh TS, Wu RC, Lai YC, Chiang MH, Lu KY, Hung CY, Ho HY, Cheng ML, Lin G. Metabolomic alterations and chromosomal instability status in gastric cancer. World J Gastroenterol. 2018;24:3760-3769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 17. | Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497-509. [PubMed] |
| 18. | Cajka T, Fiehn O. Increasing lipidomic coverage by selecting optimal mobile-phase modifiers in LC-MS of blood plasma. Metabolomics. 2016;12:34-44. [DOI] [Full Text] |
| 19. | Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, Holmes E, Nicholson JK. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5:1005-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 808] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 20. | Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486-W494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2433] [Cited by in RCA: 2709] [Article Influence: 451.5] [Reference Citation Analysis (0)] |
| 21. | Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50 Suppl:S9-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1395] [Cited by in RCA: 1171] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 22. | Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1637] [Cited by in RCA: 1720] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 23. | Chan AW, Gill RS, Schiller D, Sawyer MB. Potential role of metabolomics in diagnosis and surveillance of gastric cancer. World J Gastroenterol. 2014;20:12874-12882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 24. | Abbassi-Ghadi N, Kumar S, Huang J, Goldin R, Takats Z, Hanna GB. Metabolomic profiling of oesophago-gastric cancer: a systematic review. Eur J Cancer. 2013;49:3625-3637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Ramachandran GK, Yong WP, Yeow CH. Identification of Gastric Cancer Biomarkers Using 1H Nuclear Magnetic Resonance Spectrometry. PLoS One. 2016;11:e0162222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Huang C, Freter C. Lipid metabolism, apoptosis and cancer therapy. Int J Mol Sci. 2015;16:924-949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 311] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 27. | Riquelme I, Saavedra K, Espinoza JA, Weber H, García P, Nervi B, Garrido M, Corvalán AH, Roa JC, Bizama C. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750-24779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 104] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 28. | Perrotti F, Rosa C, Cicalini I, Sacchetta P, Del Boccio P, Genovesi D, Pieragostino D. Advances in Lipidomics for Cancer Biomarkers Discovery. Int J Mol Sci. 2016;17:E1992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 29. | Shimizu D, Kanda M, Kodera Y. Review of recent molecular landscape knowledge of gastric cancer. Histol Histopathol. 2018;33:11-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 30. | Lario S, Ramírez-Lázaro MJ, Sanjuan-Herráez D, Brunet-Vega A, Pericay C, Gombau L, Junquera F, Quintás G, Calvet X. Plasma sample based analysis of gastric cancer progression using targeted metabolomics. Sci Rep. 2017;7:17774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |