Published online Dec 15, 2019. doi: 10.4251/wjgo.v11.i12.1101
Peer-review started: April 18, 2019
First decision: July 31, 2019
Revised: August 18, 2019
Accepted: October 14, 2019
Article in press: October 14, 2019
Published online: December 15, 2019
Processing time: 237 Days and 9.3 Hours
Adenomatous polyposis (AP) is classified according to cumulative adenoma number in classical AP (CAP) and attenuated AP (AAP). Genetic susceptibility is the major risk factor in CAP due to mutations in the known high predisposition genes APC and MUTYH. However, the contribution of genetic susceptibility to AAP is lower and less understood. New predisposition genes have been recently proposed, and some of them have been validated, but their scarcity hinders accurate risk estimations and prevalence calculations. AAP is a heterogeneous condition in terms of severity, clinical features and heritability. Therefore, clinicians do not have strong discriminating criteria for the recommendation of the genetic study of known predisposition genes, and the detection rate is low. Elucidation and knowledge of new AAP high predisposition genes are of great importance to offer accurate genetic counseling to the patient and family members. This review aims to update the genetic knowledge of AAP, and to expound the difficulties involved in the genetic analysis of a highly heterogeneous condition such as AAP.
Core tip: Attenuated adenomatous polyposis (AAP) is a highly genetically and clinically heterogeneous condition in terms of severity, clinical features, heritability, and genetics. The major high predisposition genes APC and MUTYH explain a small fraction of AAP (10%-20%). Several predisposition genes have been recently proposed, and some of them have been validated, but studies addressing their global contribution to AAP genetic predisposition is scarce. Clinicians do not have strong discriminating criteria for the recommendation of genetic testing, and the detection rate is low. Therefore, multigene panel testing and a redefinition of strong clinical criteria could improve the outcome of AAP genetic testing.
- Citation: Lorca V, Garre P. Current status of the genetic susceptibility in attenuated adenomatous polyposis. World J Gastrointest Oncol 2019; 11(12): 1101-1114
- URL: https://www.wjgnet.com/1948-5204/full/v11/i12/1101.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i12.1101
Adenomatous polyposis (AP) can be defined as the tendency to develop adenomatous polyps (adenomas) along the large intestine and/or rectum. Although adenomas are benign growths, they are considered the precursor lesions of colorectal carcinoma (CRC)[1]; thus, AP is classified as a cancer risk syndrome with cumulative risks ranging from 40% to 100% depending on the severity of the polyposis (adenoma burden).
AP is usually classified according to the adenoma burden in two major groups: classical AP (CAP) and attenuated AP (AAP). Classical forms are characterized by the detection of hundreds or thousands of adenomas, and have a very low prevalence in the population (1/10000[2]), whereas attenuated forms are defined by the detection of between 10-100 adenomas, and are more prevalent in the adult population. CAP shows aggressive phenotypes, usually triggered during the second decade of life, and with a cumulative absolute cancer risk if adenomas are not removed. Extracolonic manifestations are frequent, and most of the cases show a dominant inheritance pattern[3]. By contrast, AAP is a much more heterogeneous group in terms of polyposis severity and family history[4-6]. Clinical features are distinctive from classical forms; adenoma detection is low or mild, ranging from ten synchronic or 20 cumulative to 100 adenomas, and the polyposis diagnosis age is significantly later than CAP. Cancer risk is also lower and later, ranging from 40% to 80% depending on the adenoma burden. Extracolonic manifestations are uncommon, and a family history of polyposis is frequently absent. AAP is sometimes accompanied by other types of polyps, such as hyperplastic or serrated polyps[3,7].
There are currently two clearly clinically-actionable genes that can lead to AP: APC (MIM#611731) and MUTYH (MIM#604933). Thus, prevalence and cancer risk estimations are well-defined, allowing accurate genetic counseling and effective high-risk monitoring programs for carriers. Heterozygous germline truncating mutations in the tumor suppressor gene APC mainly give rise to CAP, and sometimes to AAP, with dominant inheritance patterns. In contrast, germline biallelic mutations in the DNA repair gene MUTYH mainly lead to AAP and less frequently to CAP, with recessive inheritance patterns. In these cases, identification of APC or MUTYH carriers is important, not only to define the risks and follow-up strategies for the patient, but also to discriminate between high- and low-risk individuals among the family members who could benefit from high-risk follow-up or, on the contrary, avoid unnecessary and invasive monitoring. Ambiguously, even though both genes explain the vast majority of CAP, together they are only able to explain between 10%-20% of AAP.
AAP incidence is significantly increasing in hospital settings, mainly due to the improvement of imaging techniques and the implementation of CRC population screening programs. This increase translates into a problem in the Genetic Counseling Units due to the high heterogeneity of the disease. On the one hand, it is difficult to discriminate not only between sporadic multiple polyposis and real AP in patients with low adenoma burden, but also between attenuated and classical forms in patients with adenoma counts close to 100. On the other hand, family history is not a discriminant criterion for genetic studies due to the high rate of de novo mutations described in APC (10%-25%)[8,9] and the recessive inheritance pattern of MUTYH[10]. Furthermore, only a minority of AAP cases (< 20%) is explained by germline mutations in APC or MUTYH[11,12], leaving a substantial fraction of AAP cases unexplained. This means that undiscriminating and invasive follow-up programs will be recommended to all first-degree relatives of these patients.
Under this scenario, the elucidation of genetic susceptibility, which could explain the etiology of the disease and improve the accuracy of genetic counseling, has become a priority for scientists and clinicians. Thanks to the advance of sequencing technologies, new genes have been recently associated with primary predisposition to the development of adenomas by genome/exome sequencing studies in unexplained AP cohorts[13-16]. In the same way, other genetic alterations not detected by conventional coding germline DNA sequencing screening strategies have also been described in the APC gene, such as mutations in the promoter[17] or introns[18], large inversions[19] or mosaicism phenomes[20]. In addition, the use of wide gene panels for the genetic diagnosis of AP has incidentally revealed an overlap between different polyposis syndromes[21]. However, all these studies together are only able to explain the etiology of a very small fraction of AAP cases, and the unexplained cases are still a major group that needs to be clarified. Most likely, polygenic inheritance models in which the accumulation of multiple low penetrance alleles[22] and lifestyle risk factors such as smoking, alcohol, body mass index, diet and physical activity[23] play a major role in unexplained AAP cases.
Despite the low frequency of high predisposition genes in AAP, their knowledge is important for the detection of carriers, allowing the discrimination of high- and normal-risk individuals among family members, and leading to accurate and cost-effective monitoring programs.
The aim of this review is to describe the current knowledge of the genetic susceptibility of AAP, with emphasis on genes with a primary predisposition to AP that have been described so far, which are either already implemented in clinical practice, in process, or have recently been proposed.
Until recently, APC and MUTYH were the only known AP syndrome predisposition genes. With the advent of next-generation sequencing (NGS) technologies, new AP predisposition genes have emerged. There are currently three new validated genes [POLE (MIM#174762), POLD1 (MIM#174761), NTHL1 (MIM#602656)], and two more genes that have been described but not validated [MSH3 (MIM#600887), MLH3 (MIM#604395)]. The discovery of new AP predisposition genes has allowed for considerable advancement in the biology of AP development and, therefore, in colorectal carcinogenesis. However, the newly described genes are still poorly implemented in clinical practice, mainly because of their low frequency and the lack of accurate risk estimations. Thus, time is needed to increase the number of described cases that allow better prevalence and risk estimations to be obtained.
APC is a tumor suppressor gene closely involved in colorectal carcinogenesis; APC somatic mutations are the first event in the canonical CRC carcinogenesis model, which is followed by more than 80% of all CRCs[24]. The APC gene encodes a multifunctional protein that is mainly involved in signal transduction, cell adhesion and migration, microtubule assembly and chromosome segregation[25]. Its tumor-suppressing ability relies on its capacity to negatively regulate intracellular β-catenin levels, the main effector of the Wnt pathway. Therefore, inactivation of APC leads to increased β-catenin levels and overexpression of its different target genes involved in cell proliferation, differentiation, migration and apoptosis[26], which histologically correlates with adenoma formation.
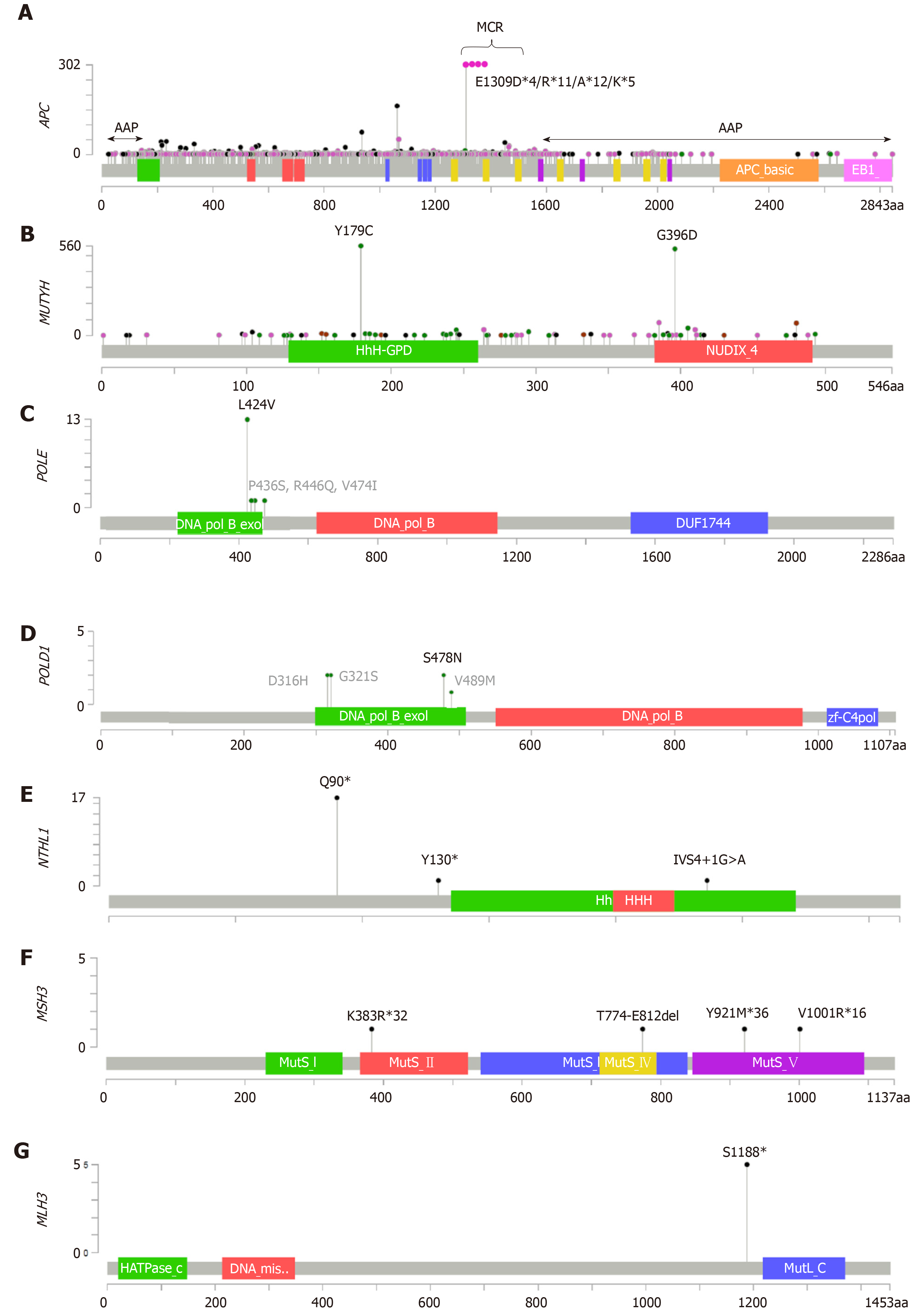
APC is located on the long arm of chromosome 5 (5q21), has 15 exons, and encodes a 2,843 amino acid protein[27]. Most of the somatic mutations lie in the mutation cluster region (MCR), which is located between amino acids 1286 and 1513 and overlaps with the β-catenin binding region[28]. Heterozygous APC germline mutations have been associated with AP predisposition in a gene location-dependent manner[29]. Most of the germline APC mutations are truncating variants lying between codons 178 and 1580, and give rise to stable mutant peptides that exert a dominant-negative effect on the wild-type protein[30,31]. These mutations lead to classical forms of the disease called familial AP (FAP), whereas germline mutations located at both the 5’ and 3’ ends of the transcript, as well as splicing mutations that lead to exon 9 skipping, give rise to attenuated forms of the disease called attenuated familial AP (AFAP) (Figure 1A). Germline mutations at the 3’ end give rise to stable proteins with a certain capability to regulate β-catenin levels[30], and 5’ end mutations upstream of codon 177 produce functional proteins by initiation of translation at codon 184[31,32]. This internal initiation of translation is relatively inefficient, leading to a haploinsufficient phenotype rather than a dominant-negative phenotype. Mutations at the splice donor site in intron 9 lead to inefficient exon skipping with some expression of normal transcript, and therefore with an attenuated form of the disease[33].
Both FAP and AFAP show autosomal dominant inheritance patterns. However, there are some exceptions without any family history. De novo mutations have been described in 10%-25% of APC carriers[8,9], and recent studies report APC mosaicism rates of 20%-50% in unexplained AP cases[20,34]. Whereas de novo mutations have been observed in both FAP and AFAP, it is noteworthy that mosaicism carriers present an attenuated form of the disease[20], likely due to the nonubiquitous distribution of the mutant allele.
MUTYH is a DNA repair gene involved in the base excision repair (BER) pathway[35]. It encodes a monofunctional DNA glycosylase responsible for the recognition and excision of the deoxyadenosine misincorporated with 8-hydroxy-2' -deoxyguanosine (8-OHdG) in the DNA molecule. 8-OHdG arises as a consequence of the oxidation of deoxyguanosine, which is a mutagenic base because it has the ability to pair indiscriminately with deoxycytosine or deoxyadenosine, leading to an increase in somatic G>T transversions[36]. Therefore, inactivation of MUTYH leads to an increase in the G>T mutation rate, which especially affects known cancer driver genes such as KRAS or APC[37], both of which are involved in adenoma formation.
MUTYH is located on the short arm of chromosome 1 (1p34.1) and is formed by 6 exons, encoding two major transcripts, which leads to 546 and 535 amino acid isoforms[35]. Biallelic MUTYH germline mutations have been associated with AP predisposition, leading to an autosomal recessive syndrome[10]. Because it is a recessive condition, there is no vertical transmission of the disease, and family history is often absent or is presented horizontally (siblings)[38]. MUTYH-associated polyposis (MAP) is characterized by the presence of 10–100 adenomatous polyps in the colon rectum resembling AFAP, but in some cases it may be accompanied by hyperplastic or serrated polyps[39]. A minor fraction of MAP presents classical forms of the disease with the detection of more than 100 adenomas. In contrast to APC, no relationship has been observed between the location of the mutation and the phenotype of the disease. Mutations located throughout the entire MUTYH have been described in MAP, but only two missense mutations, NM_001128425: c.1187G>A p.(Gly396Asp) and c.536A>G p.(Tyr179Cys), are the most prevalent in Caucasians. Other recurrent mutations have been described in more specific populations[40] (Figure 1B).
POLE and POLD1 encode the catalytic subunits of the polymerase enzyme complexes ε (Polε) and δ (Polδ), respectively, which are the principal leading- and lagging-strand DNA polymerases during S phase[41]. In addition, they also catalyze DNA synthesis in several DNA repair pathways, such as nucleotide excision repair (NER) or mismatch repair (MMR). Both POLE and POLD1 encompass not only a binding DNA region and polymerase domain, but also an exonuclease domain, which confers proofreading capability by the recognition and removal of misincorporated nucleotides during DNA replication[42]. Polymerase proofreading activity, together with high base selectivity and the MMR pathway, are the main cellular mechanisms responsible for minimizing errors during DNA replication[43]. Inactivating point mutations within the exonuclease domains lead to proteins with an active polymerase domain that lack proofreading activity, which causes high genetic instability during DNA replication. Indeed, somatic mutations within the exonuclease domains have been described in human cancer, leading to a high increase in mutational rates[44]. Tumor mutations in the POLE exonuclease domain have been identified in 1%-2% of sporadic CRC and in 7%-12% of endometrial cancers, as well as in tumors of the brain, pancreas, ovary, breast and stomach, showing ultramutated and microsatellite-stable tumors[45].
POLE is located on the long arm of chromosome 12 (12q24.33), consists of 49 exons, and encodes a 2,286 amino acid protein. Its exonuclease domain lies between codons 268 and 471[46]. POLD1 is located on the long arm of chromosome 19 (19q13.33) and consists of 27 exons, encoding an 1,133 amino acid protein. Its exonuclease domain is located between codons 304 and 517[47]. Heterozygous germline mutations within the exonuclease (proofreading) domains of both POLE and POLD1 were recently associated with AAP[13], leading to an autosomal dominant inheritance condition that is characterized by high-penetrance predisposition to multiple colorectal adenomas, large adenomas, early-onset CRC, or multiple CRCs, as well as other extracolonic tumors such as endometrial tumors[48].
Since the first association of POLE and POLD1 with AAP, several studies have validated the results and found new germline mutations in the exonuclease domains[49-55] (Figures 1C and 1D). However, due to the small number of families described so far, accurate risk estimations and the contribution of polymerases to AP are still not well-defined.
Similar to MUTYH, NTHL1 is a DNA repair gene involved in the BER pathway. It encodes a bifunctional N-glycosylase protein that recognizes and removes oxidized pyrimidines, such as 2′-deoxy-5-hydroxycytldine (5-OHdC) and ring-opened purines[56]. 5-OHdC arises as a consequence of the oxidation of deoxycytosine, and it has the ability to pair both deoxyguanosine and deoxyadenine, leading to an accumulation of somatic C>T transitions, which can affect important CRC driver genes such as APC, TP53 or KRAS, among others.
NTHL1 is located on the short arm of chromosome 16 (16p13.3), consists of 6 exons, and encodes a 312 amino acid protein[57]. NTHL1 homozygous or compound heterozygous germline mutations have been recently detected in AAP, delineating an autosomal recessive polyposis syndrome called NTHL1-associated polyposis (NAP)[14]. All NTHL1 biallelic carriers described so far showed AAP, and also frequently showed other extracolonic tumors such as endometrial or breast[58]. One nonsense mutation at codon 90 seems to be involved in nearly all the biallelic carriers described; however, novel pathogenic mutations are arising as new studies emerge[58-62] (Figure 1E).
Theoretical estimations of NAP suggest a prevalence of at least five times lower than that of MAP[62]. Due to the limited number of NAP families described until now, the phenotypic spectrum and cancer risk estimates have not been properly established.
MSH3 is one of the six MMR genes identified to date in eukaryotic cells[63]. It is involved in the detection of replication errors in microsatellite sequences together with MSH2 and MSH6. MSH3 encodes an alternative binding partner for MSH2, which is required for the specific detection of insertion or deletion loops of two or more nucleotides[64], as well as for double strand break repair[65]. MSH2 requires the binding of MSH6 or MSH3 to exercise its function. The MSH2-MSH6 dimer recognizes single substitutions and small indel mispairs, whereas MSH2-MSH3 recognizes errors in di- and larger nucleotide repeats[66]. Inactivation of MSH3 leads to a high microsatellite instability of di- and tetranucleotides (EMAST), which has been associated with a characteristic somatic APC mutation spectrum in colorectal adenoma from AAP patients[15].
The MSH3 gene is located in the long arm of chromosome 5 (5q14.1) and consists of 24 exons, encoding an 1,137 amino acid protein[67]. Biallelic truncating variants in MSH3 have been recently reported in two patients with AAP, suggesting an additional recessive subtype of colorectal AP[15] (Figure 1F).
Until now, no more studies have validated these results, so its association with AAP and phenotype estimations remain to be defined.
MLH3 is a member of the MutL homolog family of MMR proteins[63]. MLH3 dimerizes with MLH1, resulting in the MutLγ complex, which is primarily involved in meiotic recombination rather than in mitotic genetic stability[68].
The MLH3 gene is located in the long arm of chromosome 14 (14q24.3), consists of 13 exons, and encodes a 1,453 amino acid protein. The homozygous truncating germline variant S1188* was first detected in an unexplained Swedish AAP case[21], and more recently in one more AAP and two CAP subjects from Finland, suggesting a founder effect[16] (Figure 1G). Authors hypothesize the involvement of a defective DNA damage response and/or recombination-related processes in the pathogenesis of these cases[16].
Once again, research on additional cohorts is needed to reinforce the significance of MLH3 as an AP predisposition gene.
Other candidate genes, including AXIN2 (MIM#604025), FOCAD (MIM#614606), GALNT12 (MIM#610290) and BUB1 (MIM#602452) /BUB3 (MIM#603719), are involved in the AP predisposition. However, evidence for these genes is not as thorough as those previously discussed.
AXIN2 encodes the Wnt pathway component conductin; it is the scaffold protein of the β-catenin destruction complex and main negative regulator of the pathway[69]. Mutations in this gene have been described in CRC, and similar to APC, they increase β-catenin levels and activate β-catenin/T-cell factor signaling, thus promoting CRC development[70]. AXIN2 is located on the long arm of chromosome 17 (17q24.1), consists of 11 exons, and encodes two major transcripts, which leads to 843 and 778 amino acid isoforms[69]. Deleterious germline mutations have been reported in four families, showing a strong association with oligodontia as well as gastrointestinal neoplasias[71-73]. More recently, a novel missense variant has been described in an AAP family without signs of oligodontia or ectodermal dysplasia, suggesting the possibility of different phenotypes depending on the protein domain affected[74]. Two other works have screened mutations for AXIN2 in different CRC populations, both in polyposis and nonpolyposis, without any success[75,76]. Therefore, although there is a clear association between AXIN2 and oligodontia, further studies are needed to clarify its role in CRC syndromes, particularly with AAP.
FOCAD encodes a focal adhesion protein with a potential tumor suppressor function in glyomas[77]. The FOCAD gene is located on the short arm of chromosome 9 (9p21.3) and is formed by 46 exons encoding an 1,801 amino acid protein[77]. Two studies identified large deletions and truncating point mutations in a total of five CRC cases: 2/221 cases of unexplained AP[78] and 3/1232 early-onset and familial CRC cases[79]. Altogether, four cases had a diagnosis of AAP. Since FOCAD shows high expression levels in colonic epithelial cells and has been involved in cell survival and proliferation, the authors suggest a potential role of this gene in polyposis/CRC susceptibility[79]. Regardless, this association and its contribution to AAP predisposition requires further clarification.
GALNT12 encodes a hexosyltransferase involved in the initial steps of the mucin-type O-glycosylation process[80]. Alterations in this process lead to aberrant glycosylation, which has been associated with alterations in cell growth, differentiation, transformation, adhesion, metastasis and immune surveillance in cancers[81]. GALNT12 is highly expressed in the digestive tract, and is frequently downregulated in CRC[82]. The GALNT12 gene is located on the long arm of chromosome 9 (9q22.33), has 10 exons, and encodes a 581 amino acid protein[80]. Evidence for the association between GALNT12 and CRC has been reported[83], but its association with familial CRC, particularly AP, remains a controversial issue. Partially inactivating variants have been detected in familial CRC along with a mild polyp burden, suggesting the involvement of this gene in CRC predisposition[84]. However, later studies do not support its involvement in nonpolyposis and polyposis CRC predisposition[85,86].
BUB1 and BUB3 (mitotic checkpoint serine/threonine kinases)
BUB1 and BUB3 encode components of the spindle assembly checkpoint complex, which controls chromosome biorientation on the mitotic spindle, delaying the anaphase transition until all kinetochores are properly attached[87]. Alterations in the activity of this complex lead to alterations in chromosome copy number, i.e. aneuploidies[88]. The BUB1 gene is located on the long arm of chromosome 2 (2q13), consists of 25 exons, and encodes a 1,085 amino acid protein, whereas BUB3 is located on the long arm of chromosome 10 (10q26.13), has 8 exons, and encodes a 328 amino acid protein[89]. Deleterious germline mutations in both genes have been associated not only with increased levels of constitutive aneuploidy, but also with gastrointestinal neoplasms, including adenocarcinomas and adenomas[90,91]. Furthermore, aneuploidy caused by Bub1 insufficiency has been proven to drive colorectal adenoma formation in mice through APC loss of heterozygosity (LOH)[92]. Screening of the BUB1 and BUB3 genes in familial and AP CRC cohorts has shown functionally relevant germline mutations in a low fraction of patients with CRC who also presented increased levels of constitutive aneuploidy[93,94]. However, the causality of these mutations in CRC/adenoma susceptibility remains unproven.
Although phenotypes for related CRC risk syndromes are generally well-defined, there are some overlapping features that can lead to confusion in the clinical suspicion and subsequent misdirection of the genetic testing approach. The cancer risk syndromes prone to phenotypically overlap with AAP are described below.
Lynch syndrome is the main hereditary nonpolyposis colorectal cancer syndrome caused by heterozygous deleterious mutations in MMR genes (MSH2, MLH1, MSH6 and PMS2) that can be accompanied by early-onset adenomas[95]. Usually, the adenoma burden does not exceed 10, but it can sometimes mimic AAP.
Constitutional MMR deficiency is due to loss-of-function biallelic germline mutations in the main MMR genes. It is an aggressive recessive cancer predisposition syndrome with a wide tumor spectrum, very early age of onset and poor outcome[96]. In addition, nearly 36% of affected subjects develop colorectal AP ranging from a few up to 100 adenomas[97].
Hereditary mixed polyposis syndrome is characterized by multiple colon polyps of mixed pathologic subtypes and an increased risk for CRC[98]. It is caused by large duplications in the 5' regulatory region of GREM1 (MIM 603054), leading to an excess of coding protein expression[99]. GREM1 is an antagonist of bone morphogenic protein (BMP), so its overexpression can lead to inactivation of the BMP pathway and subsequent hyperproliferation of colonic epithelium[100].
The pathogenesis of polyps in hereditary mixed polyposis syndrome likely overlaps with that of juvenile polyposis syndrome (JPS), which is caused by inactivating mutations in other genes of the BMP pathway, including BMPR1A (MIM 601299), SMAD4 (MIM 600993), ENG (MIM 131195) and BMP4 (MIM 112262)[101-104]. JPS is a hamartomatous polyposis syndrome with an increased risk of CRC as well as other digestive cancers. Cancer risk arises from adenomatous components present in the juvenile polyps, which can sometimes lead to misinterpretations[105].
Germline alterations in genes involved in the PTEN/PI3K/AKT pathway are also associated with hamartomatous polyposis syndromes. Cowden syndrome is caused by heterozygous PTEN (MIM 601728) germline mutations, and is characterized by the development of hamartomatous and neoplastic lesions of the skin, mucous membranes, thyroid, breast, endometrium, and brain[106]. Although hamartomatous polyps are the most characteristic gastrointestinal lesions in Cowden syndrome, adenomatous polyps in the colon have been detected in 30% of affected individuals[107].
In contrast, germline heterozygous mutations in STK11 (MIM 602216) lead to Peutz-Jeghers syndrome (PJS), which is characterized by mucocutaneous pigmentation and diffuse gastrointestinal hamartomas[108]. Similar to other hamartomatous syndromes, polyps with large adenomatous transformation areas and adenomatous polyps have been described in PJS[109].
Currently, thanks to NGS technology and the widespread use of multigene panels for hereditary cancer testing, the detection of overlapping phenotypes between different CRC syndromes is greatly increasing, improving the diagnosis and follow-up of these patients[12,21,110].
AAP is a highly heterogeneous disease, covering both moderate and mild forms of AP, as well as hereditary and sporadic forms, recessive and dominant conditions, and the presence or absence of other gastrointestinal or extracolonic manifestations. Thus, the genetic heterogeneity of the syndrome, where several high predisposition genes are involved in the polyposis predisposition of a minor subset of AAP, is not surprising. Two previous studies have investigated the prevalence of pathogenic mutations in large cohorts of AP, detecting approximately 6%-15% of pathogenic mutations in either the APC or MUTYH genes when analyzing patients with an adenoma burden between 10 and 99[11,12]. These detection rates were decreased (2%-9%, respectively) when only patients between 10 and 19 adenomas were considered, showing that adenoma burden and the likelihood of detecting pathogenic mutations in APC and MUTYH are directly proportional in AAP. Regarding the prevalence of the new AP predisposition genes, Stanich and collaborators included the analysis of POLE and POLD1 in their cohorts, but the contribution of these genes was scarce (one detection in 2,979 AAP cases), and it did not alter the overall mutation detection rate[12]. The NTHL1 contribution to AAP has been recently estimated to be five times less prevalent than that of MUTYH[62]. Therefore, it seems that the heritability of AAP lies in different predisposition genes, each of which explains a small fraction of the total. Recently, other newly associated genes have been described, but the contribution of genetics to the etiology of the disease, as well as its heritability, are difficult to estimate. The high clinical and genetic heterogeneity, as well as the low prevalence of pathogenic mutations in the described genes, reflects the necessity of multigene panel testing for the effective genetic diagnosis of AAP.
To increase diagnostic sensitivity in such a heterogeneous syndrome, clinical guidelines have been developed with broad criteria, recommending genetic testing in patients with more than 10 adenomas, even in those patients with oligopolyposis (< 10 adenomas) or early CRC[111-113]. These criteria increase the genetic testing requests in diagnostic laboratories, thus decreasing the mutation detection rate, which makes genetic studies not cost-effective, even if they are performed by multigene panel testing. Furthermore, most of the genetic testing results are not informative, and the probability of unclassified variant detection with multigene panel testing is high, which leads to a major group of patients with anxiety and confusion. Therefore, more stringent clinical criteria, especially in the cumulative number of adenomas, should be redefined to ascertain those patients who are most likely to harbor a hereditary polyposis syndrome. The stricter the recommendation criteria for the genetic study is, the greater the mutation detection rate and lower the ambiguous results. We are in agreement with the last guideline of the American Society of Colon and Rectal Surgeons (ASCRS) that a cutoff of 20 cumulative adenomas should be used to prompt genetic counseling and testing[114].
In conclusion, the contribution of genetics to the etiology of the disease and its heritability are difficult to estimate. The high clinical and genetic heterogeneity, as well as the low prevalence of each AP predisposing gene, reflects the necessity of multigene panel testing for an effective diagnosis of AAP. Nevertheless, the decline in diagnosis rates that comes with the decrease in adenoma burden shows the necessity of stricter clinical criteria when genetic testing is recommended for AAP predisposition genes.
The authors wish to thank Dra. Trinidad Caldés and Dra. Vanesa Barberán for their critical revision.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kadiyska T, Jiang L S-Editor: Ma YJ L-Editor: Filipodia E-Editor: Ma YJ
| 1. | Shinya H, Wolff WI. Morphology, anatomic distribution and cancer potential of colonic polyps. Ann Surg. 1979;190:679-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 353] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 2. | Vasen HF, Möslein G, Alonso A, Aretz S, Bernstein I, Bertario L, Blanco I, Bülow S, Burn J, Capella G, Colas C, Engel C, Frayling I, Friedl W, Hes FJ, Hodgson S, Järvinen H, Mecklin JP, Møller P, Myrhøi T, Nagengast FM, Parc Y, Phillips R, Clark SK, de Leon MP, Renkonen-Sinisalo L, Sampson JR, Stormorken A, Tejpar S, Thomas HJ, Wijnen J. Guidelines for the clinical management of familial adenomatous polyposis (FAP). Gut. 2008;57:704-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 471] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 3. | Lucci-Cordisco E, Risio M, Venesio T, Genuardi M. The growing complexity of the intestinal polyposis syndromes. Am J Med Genet A. 2013;161A:2777-2787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Knudsen AL, Bisgaard ML, Bülow S. Attenuated familial adenomatous polyposis (AFAP). A review of the literature. Fam Cancer. 2003;2:43-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 181] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 5. | Mongin C, Coulet F, Lefevre JH, Colas C, Svrcek M, Eyries M, Lahely Y, Fléjou JF, Soubrier F, Parc Y. Unexplained polyposis: a challenge for geneticists, pathologists and gastroenterologists. Clin Genet. 2012;81:38-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | de Leon MP, Pedroni M, Roncucci L, Domati F, Rossi G, Magnani G, Pezzi A, Fante R, Bonetti LR. Attenuated polyposis of the large bowel: a morphologic and molecular approach. Fam Cancer. 2017;16:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044-2058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 945] [Cited by in RCA: 857] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 8. | Aretz S, Uhlhaas S, Caspari R, Mangold E, Pagenstecher C, Propping P, Friedl W. Frequency and parental origin of de novo APC mutations in familial adenomatous polyposis. Eur J Hum Genet. 2004;12:52-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Rustin RB, Jagelman DG, McGannon E, Fazio VW, Lavery IC, Weakley FL. Spontaneous mutation in familial adenomatous polyposis. Dis Colon Rectum. 1990;33:52-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 960] [Cited by in RCA: 930] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 11. | Grover S, Kastrinos F, Steyerberg EW, Cook EF, Dewanwala A, Burbidge LA, Wenstrup RJ, Syngal S. Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. JAMA. 2012;308:485-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 12. | Stanich PP, Pearlman R, Hinton A, Gutierrez S, LaDuca H, Hampel H, Jasperson K. Prevalence of Germline Mutations in Polyposis and Colorectal Cancer-Associated Genes in Patients With Multiple Colorectal Polyps. Clin Gastroenterol Hepatol. 2019;17:2008-2015.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 13. | Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S; CORGI Consortium; WGS500 Consortium, Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJ, McVean G, Houlston RS, Tomlinson I. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136-144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 766] [Cited by in RCA: 756] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 14. | Weren RD, Ligtenberg MJ, Kets CM, de Voer RM, Verwiel ET, Spruijt L, van Zelst-Stams WA, Jongmans MC, Gilissen C, Hehir-Kwa JY, Hoischen A, Shendure J, Boyle EA, Kamping EJ, Nagtegaal ID, Tops BB, Nagengast FM, Geurts van Kessel A, van Krieken JH, Kuiper RP, Hoogerbrugge N. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet. 2015;47:668-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 279] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 15. | Adam R, Spier I, Zhao B, Kloth M, Marquez J, Hinrichsen I, Kirfel J, Tafazzoli A, Horpaopan S, Uhlhaas S, Stienen D, Friedrichs N, Altmüller J, Laner A, Holzapfel S, Peters S, Kayser K, Thiele H, Holinski-Feder E, Marra G, Kristiansen G, Nöthen MM, Büttner R, Möslein G, Betz RC, Brieger A, Lifton RP, Aretz S. Exome Sequencing Identifies Biallelic MSH3 Germline Mutations as a Recessive Subtype of Colorectal Adenomatous Polyposis. Am J Hum Genet. 2016;99:337-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 173] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 16. | Olkinuora A, Nieminen TT, Mårtensson E, Rohlin A, Ristimäki A, Koskenvuo L, Lepistö A; Swedish Extended Genetic Analysis of Colorectal Neoplasia (SWEN) Study Group, Gebre-Medhin S, Nordling M, Peltomäki P. Biallelic germline nonsense variant of MLH3 underlies polyposis predisposition. Genet Med. 2019;21:1868-1873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Rohlin A, Engwall Y, Fritzell K, Göransson K, Bergsten A, Einbeigi Z, Nilbert M, Karlsson P, Björk J, Nordling M. Inactivation of promoter 1B of APC causes partial gene silencing: evidence for a significant role of the promoter in regulation and causative of familial adenomatous polyposis. Oncogene. 2011;30:4977-4989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Nieminen TT, Pavicic W, Porkka N, Kankainen M, Järvinen HJ, Lepistö A, Peltomäki P. Pseudoexons provide a mechanism for allele-specific expression of APC in familial adenomatous polyposis. Oncotarget. 2016;7:70685-70698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Shirts BH, Salipante SJ, Casadei S, Ryan S, Martin J, Jacobson A, Vlaskin T, Koehler K, Livingston RJ, King MC, Walsh T, Pritchard CC. Deep sequencing with intronic capture enables identification of an APC exon 10 inversion in a patient with polyposis. Genet Med. 2014;16:783-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Spier I, Drichel D, Kerick M, Kirfel J, Horpaopan S, Laner A, Holzapfel S, Peters S, Adam R, Zhao B, Becker T, Lifton RP, Perner S, Hoffmann P, Kristiansen G, Timmermann B, Nöthen MM, Holinski-Feder E, Schweiger MR, Aretz S. Low-level APC mutational mosaicism is the underlying cause in a substantial fraction of unexplained colorectal adenomatous polyposis cases. J Med Genet. 2016;53:172-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 21. | Rohlin A, Rambech E, Kvist A, Törngren T, Eiengård F, Lundstam U, Zagoras T, Gebre-Medhin S, Borg Å, Björk J, Nilbert M, Nordling M. Expanding the genotype-phenotype spectrum in hereditary colorectal cancer by gene panel testing. Fam Cancer. 2017;16:195-203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Cheng TH, Gorman M, Martin L, Barclay E, Casey G, Colon Cancer Family Registry, CGEMS, Saunders B, Thomas H, Clark S, Tomlinson I. Common colorectal cancer risk alleles contribute to the multiple colorectal adenoma phenotype, but do not influence colonic polyposis in FAP. Eur J Hum Genet. 2015;23:260-263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Øines M, Helsingen LM, Bretthauer M, Emilsson L. Epidemiology and risk factors of colorectal polyps. Best Pract Res Clin Gastroenterol. 2017;31:419-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 24. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8005] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 25. | Hankey W, Frankel WL, Groden J. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: implications for therapeutic targeting. Cancer Metastasis Rev. 2018;37:159-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 135] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 26. | Fodde R. The APC gene in colorectal cancer. Eur J Cancer. 2002;38:867-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 283] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 27. | Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1920] [Cited by in RCA: 1863] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 28. | Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 631] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 29. | Nagase H, Miyoshi Y, Horii A, Aoki T, Ogawa M, Utsunomiya J, Baba S, Sasazuki T, Nakamura Y. Correlation between the location of germ-line mutations in the APC gene and the number of colorectal polyps in familial adenomatous polyposis patients. Cancer Res. 1992;52:4055-4057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 30. | Dihlmann S, Gebert J, Siermann A, Herfarth C, von Knebel Doeberitz M. Dominant negative effect of the APC1309 mutation: a possible explanation for genotype-phenotype correlations in familial adenomatous polyposis. Cancer Res. 1999;59:1857-1860. [PubMed] |
| 31. | Sieber OM, Segditsas S, Knudsen AL, Zhang J, Luz J, Rowan AJ, Spain SL, Thirlwell C, Howarth KM, Jaeger EE, Robinson J, Volikos E, Silver A, Kelly G, Aretz S, Frayling I, Hutter P, Dunlop M, Guenther T, Neale K, Phillips R, Heinimann K, Tomlinson IP. Disease severity and genetic pathways in attenuated familial adenomatous polyposis vary greatly but depend on the site of the germline mutation. Gut. 2006;55:1440-1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Heppner Goss K, Trzepacz C, Tuohy TM, Groden J. Attenuated APC alleles produce functional protein from internal translation initiation. Proc Natl Acad Sci U S A. 2002;99:8161-8166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 44] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Varesco L, Gismondi V, Presciuttini S, Groden J, Spirio L, Sala P, Rossetti C, De Benedetti L, Bafico A, Heouaine A. Mutation in a splice-donor site of the APC gene in a family with polyposis and late age of colonic cancer death. Hum Genet. 1994;93:281-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Jansen AM, Crobach S, Geurts-Giele WR, van den Akker BE, Garcia MV, Ruano D, Nielsen M, Tops CM, Wijnen JT, Hes FJ, van Wezel T, Dinjens WN, Morreau H. Distinct Patterns of Somatic Mosaicism in the APC Gene in Neoplasms From Patients With Unexplained Adenomatous Polyposis. Gastroenterology. 2017;152:546-549.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 35. | Slupska MM, Baikalov C, Luther WM, Chiang JH, Wei YF, Miller JH. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178:3885-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 286] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 36. | Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1634] [Cited by in RCA: 1683] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 37. | Lipton L, Halford SE, Johnson V, Novelli MR, Jones A, Cummings C, Barclay E, Sieber O, Sadat A, Bisgaard ML, Hodgson SV, Aaltonen LA, Thomas HJ, Tomlinson IP. Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res. 2003;63:7595-7599. [PubMed] |
| 38. | Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evans DG, Frayling I, Jordan S, Maher ER, Mak T, Maynard J, Pigatto F, Shaw J, Cheadle JP. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet. 2003;362:39-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 289] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 39. | Boparai KS, Dekker E, Van Eeden S, Polak MM, Bartelsman JF, Mathus-Vliegen EM, Keller JJ, van Noesel CJ. Hyperplastic polyps and sessile serrated adenomas as a phenotypic expression of MYH-associated polyposis. Gastroenterology. 2008;135:2014-2018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 129] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 40. | Poulsen ML, Bisgaard ML. MUTYH Associated Polyposis (MAP). Curr Genomics. 2008;9:420-435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 41. | Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20:281-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 42. | Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 43. | Henninger EE, Pursell ZF. DNA polymerase ε and its roles in genome stability. IUBMB Life. 2014;66:339-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 44. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6673] [Article Influence: 513.3] [Reference Citation Analysis (0)] |
| 45. | Church DN, Stelloo E, Nout RA, Valtcheva N, Depreeuw J, ter Haar N, Noske A, Amant F, Tomlinson IP, Wild PJ, Lambrechts D, Jürgenliemk-Schulz IM, Jobsen JJ, Smit VT, Creutzberg CL, Bosse T. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst. 2014;107:402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 230] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 46. | Kesti T, Frantti H, Syväoja JE. Molecular cloning of the cDNA for the catalytic subunit of human DNA polymerase epsilon. J Biol Chem. 1993;268:10238-10245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 47. | Yang CL, Chang LS, Zhang P, Hao H, Zhu L, Toomey NL, Lee MY. Molecular cloning of the cDNA for the catalytic subunit of human DNA polymerase delta. Nucleic Acids Res. 1992;20:735-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 48. | Briggs S, Tomlinson I. Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol. 2013;230:148-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 230] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 49. | Spier I, Holzapfel S, Altmüller J, Zhao B, Horpaopan S, Vogt S, Chen S, Morak M, Raeder S, Kayser K, Stienen D, Adam R, Nürnberg P, Plotz G, Holinski-Feder E, Lifton RP, Thiele H, Hoffmann P, Steinke V, Aretz S. Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int J Cancer. 2015;137:320-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 107] [Article Influence: 10.7] [Reference Citation Analysis (1)] |
| 50. | Bellido F, Pineda M, Aiza G, Valdés-Mas R, Navarro M, Puente DA, Pons T, González S, Iglesias S, Darder E, Piñol V, Soto JL, Valencia A, Blanco I, Urioste M, Brunet J, Lázaro C, Capellá G, Puente XS, Valle L. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med. 2016;18:325-332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 193] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 51. | Esteban-Jurado C, Giménez-Zaragoza D, Muñoz J, Franch-Expósito S, Álvarez-Barona M, Ocaña T, Cuatrecasas M, Carballal S, López-Cerón M, Marti-Solano M, Díaz-Gay M, van Wezel T, Castells A, Bujanda L, Balmaña J, Gonzalo V, Llort G, Ruiz-Ponte C, Cubiella J, Balaguer F, Aligué R, Castellví-Bel S. POLE and POLD1 screening in 155 patients with multiple polyps and early-onset colorectal cancer. Oncotarget. 2017;8:26732-26743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 52. | Rohlin A, Eiengård F, Lundstam U, Zagoras T, Nilsson S, Edsjö A, Pedersen J, Svensson J, Skullman S, Karlsson BG, Björk J, Nordling M. GREM1 and POLE variants in hereditary colorectal cancer syndromes. Genes Chromosomes Cancer. 2016;55:95-106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 53. | Wimmer K, Beilken A, Nustede R, Ripperger T, Lamottke B, Ure B, Steinmann D, Reineke-Plaass T, Lehmann U, Zschocke J, Valle L, Fauth C, Kratz CP. A novel germline POLE mutation causes an early onset cancer prone syndrome mimicking constitutional mismatch repair deficiency. Fam Cancer. 2017;16:67-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 54. | Hansen MF, Johansen J, Bjørnevoll I, Sylvander AE, Steinsbekk KS, Sætrom P, Sandvik AK, Drabløs F, Sjursen W. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam Cancer. 2015;14:437-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 55. | Lorca V, Rueda D, Martín-Morales L, Fernández-Aceñero MJ, Grolleman J, Poves C, Llovet P, Tapial S, García-Barberán V, Sanz J, Pérez-Segura P, de Voer RM, Díaz-Rubio E, de la Hoya M, Caldés T, Garre P. Contribution of New Adenomatous Polyposis Predisposition Genes in an Unexplained Attenuated Spanish Cohort by Multigene Panel Testing. Sci Rep. 2019;9:9814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 56. | Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, Teebor GW. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst). 2009;8:786-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 93] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 57. | Imai K, Sarker AH, Akiyama K, Ikeda S, Yao M, Tsutsui K, Shohmori T, Seki S. Genomic structure and sequence of a human homologue (NTHL1/NTH1) of Escherichia coli endonuclease III with those of the adjacent parts of TSC2 and SLC9A3R2 genes. Gene. 1998;222:287-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 58. | Grolleman JE, de Voer RM, Elsayed FA, Nielsen M, Weren RDA, Palles C, Ligtenberg MJL, Vos JR, Ten Broeke SW, de Miranda NFCC, Kuiper RA, Kamping EJ, Jansen EAM, Vink-Börger ME, Popp I, Lang A, Spier I, Hüneburg R, James PA, Li N, Staninova M, Lindsay H, Cockburn D, Spasic-Boskovic O, Clendenning M, Sweet K, Capellá G, Sjursen W, Høberg-Vetti H, Jongmans MC, Neveling K, Geurts van Kessel A, Morreau H, Hes FJ, Sijmons RH, Schackert HK, Ruiz-Ponte C, Dymerska D, Lubinski J, Rivera B, Foulkes WD, Tomlinson IP, Valle L, Buchanan DD, Kenwrick S, Adlard J, Dimovski AJ, Campbell IG, Aretz S, Schindler D, van Wezel T, Hoogerbrugge N, Kuiper RP. Mutational Signature Analysis Reveals NTHL1 Deficiency to Cause a Multi-tumor Phenotype. Cancer Cell. 2019;35:256-266.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 123] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 59. | Belhadj S, Mur P, Navarro M, González S, Moreno V, Capellá G, Valle L. Delineating the Phenotypic Spectrum of the NTHL1-Associated Polyposis. Clin Gastroenterol Hepatol. 2017;15:461-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 60. | Chubb D, Broderick P, Dobbins SE, Frampton M, Kinnersley B, Penegar S, Price A, Ma YP, Sherborne AL, Palles C, Timofeeva MN, Bishop DT, Dunlop MG, Tomlinson I, Houlston RS. Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat Commun. 2016;7:11883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 61. | Rivera B, Castellsagué E, Bah I, van Kempen LC, Foulkes WD. Biallelic NTHL1 Mutations in a Woman with Multiple Primary Tumors. N Engl J Med. 2015;373:1985-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 62. | Weren RD, Ligtenberg MJ, Geurts van Kessel A, De Voer RM, Hoogerbrugge N, Kuiper RP. NTHL1 and MUTYH polyposis syndromes: two sides of the same coin? J Pathol. 2018;244:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 63. | Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 614] [Cited by in RCA: 619] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 64. | Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629-13634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 405] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 65. | Burdova K, Mihaljevic B, Sturzenegger A, Chappidi N, Janscak P. The Mismatch-Binding Factor MutSβ Can Mediate ATR Activation in Response to DNA Double-Strand Breaks. Mol Cell. 2015;59:603-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 66. | Haugen AC, Goel A, Yamada K, Marra G, Nguyen TP, Nagasaka T, Kanazawa S, Koike J, Kikuchi Y, Zhong X, Arita M, Shibuya K, Oshimura M, Hemmi H, Boland CR, Koi M. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res. 2008;68:8465-8472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 125] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 67. | Fujii H, Shimada T. Isolation and characterization of cDNA clones derived from the divergently transcribed gene in the region upstream from the human dihydrofolate reductase gene. J Biol Chem. 1989;264:10057-10064. [PubMed] [DOI] [Full Text] |
| 68. | Wang TF, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci U S A. 1999;96:13914-13919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 220] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 69. | Dong X, Seelan RS, Qian C, Mai M, Liu W. Genomic structure, chromosome mapping and expression analysis of the human AXIN2 gene. Cytogenet Cell Genet. 2001;93:26-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 70. | Liu W, Dong X, Mai M, Seelan RS, Taniguchi K, Krishnadath KK, Halling KC, Cunningham JM, Boardman LA, Qian C, Christensen E, Schmidt SS, Roche PC, Smith DI, Thibodeau SN. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat Genet. 2000;26:146-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 390] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 71. | Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, Pirinen S, Nieminen P. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 481] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 72. | Marvin ML, Mazzoni SM, Herron CM, Edwards S, Gruber SB, Petty EM. AXIN2-associated autosomal dominant ectodermal dysplasia and neoplastic syndrome. Am J Med Genet A. 2011;155A:898-902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 73. | Renkonen ET, Nieminen P, Abdel-Rahman WM, Moisio AL, Järvelä I, Arte S, Järvinen HJ, Peltomäki P. Adenomatous polyposis families that screen APC mutation-negative by conventional methods are genetically heterogeneous. J Clin Oncol. 2005;23:5651-5659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 74. | Rivera B, Perea J, Sánchez E, Villapún M, Sánchez-Tomé E, Mercadillo F, Robledo M, Benítez J, Urioste M. A novel AXIN2 germline variant associated with attenuated FAP without signs of oligondontia or ectodermal dysplasia. Eur J Hum Genet. 2014;22:423-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 75. | Peterlongo P, Howe LR, Radice P, Sala P, Hong YJ, Hong SI, Mitra N, Offit K, Ellis NA. Germline mutations of AXIN2 are not associated with nonsyndromic colorectal cancer. Hum Mutat. 2005;25:498-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 76. | Lipton L, Sieber OM, Thomas HJ, Hodgson SV, Tomlinson IP, Woodford-Richens K. Germline mutations in the TGF-beta and Wnt signalling pathways are a rare cause of the "multiple" adenoma phenotype. J Med Genet. 2003;40:e35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 77. | Brockschmidt A, Trost D, Peterziel H, Zimmermann K, Ehrler M, Grassmann H, Pfenning PN, Waha A, Wohlleber D, Brockschmidt FF, Jugold M, Hoischen A, Kalla C, Waha A, Seifert G, Knolle PA, Latz E, Hans VH, Wick W, Pfeifer A, Angel P, Weber RG. KIAA1797/FOCAD encodes a novel focal adhesion protein with tumour suppressor function in gliomas. Brain. 2012;135:1027-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 78. | Horpaopan S, Spier I, Zink AM, Altmüller J, Holzapfel S, Laner A, Vogt S, Uhlhaas S, Heilmann S, Stienen D, Pasternack SM, Keppler K, Adam R, Kayser K, Moebus S, Draaken M, Degenhardt F, Engels H, Hofmann A, Nöthen MM, Steinke V, Perez-Bouza A, Herms S, Holinski-Feder E, Fröhlich H, Thiele H, Hoffmann P, Aretz S. Genome-wide CNV analysis in 221 unrelated patients and targeted high-throughput sequencing reveal novel causative candidate genes for colorectal adenomatous polyposis. Int J Cancer. 2015;136:E578-E589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 79. | Weren RD, Venkatachalam R, Cazier JB, Farin HF, Kets CM, de Voer RM, Vreede L, Verwiel ET, van Asseldonk M, Kamping EJ, Kiemeney LA, Neveling K, Aben KK, Carvajal-Carmona L, Nagtegaal ID, Schackert HK, Clevers H, van de Wetering M, Tomlinson IP, Ligtenberg MJ, Hoogerbrugge N, Geurts van Kessel A, Kuiper RP. Germline deletions in the tumour suppressor gene FOCAD are associated with polyposis and colorectal cancer development. J Pathol. 2015;236:155-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 80. | Guo JM, Zhang Y, Cheng L, Iwasaki H, Wang H, Kubota T, Tachibana K, Narimatsu H. Molecular cloning and characterization of a novel member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, pp-GalNAc-T12. FEBS Lett. 2002;524:211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 81. | Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 2006;7:599-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 409] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 82. | Guo JM, Chen HL, Wang GM, Zhang YK, Narimatsu H. Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-12 in gastric and colonic cancer cell lines and in human colorectal cancer. Oncology. 2004;67:271-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 83. | Guda K, Moinova H, He J, Jamison O, Ravi L, Natale L, Lutterbaugh J, Lawrence E, Lewis S, Willson JK, Lowe JB, Wiesner GL, Parmigiani G, Barnholtz-Sloan J, Dawson DW, Velculescu VE, Kinzler KW, Papadopoulos N, Vogelstein B, Willis J, Gerken TA, Markowitz SD. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci U S A. 2009;106:12921-12925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 118] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 84. | Clarke E, Green RC, Green JS, Mahoney K, Parfrey PS, Younghusband HB, Woods MO. Inherited deleterious variants in GALNT12 are associated with CRC susceptibility. Hum Mutat. 2012;33:1056-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 85. | Seguí N, Pineda M, Navarro M, Lázaro C, Brunet J, Infante M, Durán M, Soto JL, Blanco I, Capellá G, Valle L. GALNT12 is not a major contributor of familial colorectal cancer type X. Hum Mutat. 2014;35:50-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 86. | Lorca V, Rueda D, Martín-Morales L, Poves C, Fernández-Aceñero MJ, Ruiz-Ponte C, Llovet P, Marrupe D, García-Barberán V, García-Paredes B, Pérez-Segura P, de la Hoya M, Díaz-Rubio E, Caldés T, Garre P. Role of GALNT12 in the genetic predisposition to attenuated adenomatous polyposis syndrome. PLoS One. 2017;12:e0187312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 87. | Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr Biol. 2015;25:R1002-R1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 524] [Cited by in RCA: 581] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 88. | Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 402] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 89. | Cahill DP, da Costa LT, Carson-Walter EB, Kinzler KW, Vogelstein B, Lengauer C. Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics. 1999;58:181-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 148] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 90. | Rio Frio T, Lavoie J, Hamel N, Geyer FC, Kushner YB, Novak DJ, Wark L, Capelli C, Reis-Filho JS, Mai S, Pastinen T, Tischkowitz MD, Marcus VA, Foulkes WD. Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N Engl J Med. 2010;363:2628-2637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 91. | de Voer RM, Hoogerbrugge N, Kuiper RP. Spindle-assembly checkpoint and gastrointestinal cancer. N Engl J Med. 2011;364:1279-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 92. | Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 181] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 93. | de Voer RM, Geurts van Kessel A, Weren RD, Ligtenberg MJ, Smeets D, Fu L, Vreede L, Kamping EJ, Verwiel ET, Hahn MM, Ariaans M, Spruijt L, van Essen T, Houge G, Schackert HK, Sheng JQ, Venselaar H, van Ravenswaaij-Arts CM, van Krieken JH, Hoogerbrugge N, Kuiper RP. Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are risk factors for colorectal cancer. Gastroenterology. 2013;145:544-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 94. | Mur P, De Voer RM, Olivera-Salguero R, Rodríguez-Perales S, Pons T, Setién F, Aiza G, Valdés-Mas R, Bertini A, Pineda M, Vreede L, Navarro M, Iglesias S, González S, Brunet J, Valencia A, Esteller M, Lázaro C, Kops GJPL, Urioste M, Puente XS, Capellá G, Valle L. Germline mutations in the spindle assembly checkpoint genes BUB1 and BUB3 are infrequent in familial colorectal cancer and polyposis. Mol Cancer. 2018;17:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 95. | Jass JR, Smyrk TC, Stewart SM, Lane MR, Lanspa SJ, Lynch HT. Pathology of hereditary non-polyposis colorectal cancer. Anticancer Res. 1994;14:1631-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 96. | Abedalthagafi M. Constitutional mismatch repair-deficiency: current problems and emerging therapeutic strategies. Oncotarget. 2018;9:35458-35469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 97. | Wimmer K, Kratz CP, Vasen HF, Caron O, Colas C, Entz-Werle N, Gerdes AM, Goldberg Y, Ilencikova D, Muleris M, Duval A, Lavoine N, Ruiz-Ponte C, Slavc I, Burkhardt B, Brugieres L; EU-Consortium Care for CMMRD (C4CMMRD). Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium 'care for CMMRD' (C4CMMRD). J Med Genet. 2014;51:355-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 334] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 98. | Whitelaw SC, Murday VA, Tomlinson IP, Thomas HJ, Cottrell S, Ginsberg A, Bukofzer S, Hodgson SV, Skudowitz RB, Jass JR, Talbot IC, Northover JM, Bodmer WF, Solomon E. Clinical and molecular features of the hereditary mixed polyposis syndrome. Gastroenterology. 1997;112:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 92] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 99. | Jaeger E, Leedham S, Lewis A, Segditsas S, Becker M, Cuadrado PR, Davis H, Kaur K, Heinimann K, Howarth K; HMPS Collaboration, East J, Taylor J, Thomas H, Tomlinson I. Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet. 2012;44:699-703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 100. | Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, Walther A, Spain S, Pittman A, Kemp Z, Sullivan K, Heinimann K, Lubbe S, Domingo E, Barclay E, Martin L, Gorman M, Chandler I, Vijayakrishnan J, Wood W, Papaemmanuil E, Penegar S, Qureshi M; CORGI Consortium, Farrington S, Tenesa A, Cazier JB, Kerr D, Gray R, Peto J, Dunlop M, Campbell H, Thomas H, Houlston R, Tomlinson I. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 233] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 101. | Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001;28:184-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 442] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 102. | Howe JR, Roth S, Ringold JC, Summers RW, Järvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 658] [Cited by in RCA: 604] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 103. | Howe JR, Haidle JL, Lal G, Bair J, Song C, Pechman B, Chinnathambi S, Lynch HT. ENG mutations in MADH4/BMPR1A mutation negative patients with juvenile polyposis. Clin Genet. 2007;71:91-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 30] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 104. | Lubbe SJ, Pittman AM, Matijssen C, Twiss P, Olver B, Lloyd A, Qureshi M, Brown N, Nye E, Stamp G, Blagg J, Houlston RS. Evaluation of germline BMP4 mutation as a cause of colorectal cancer. Hum Mutat. 2011;32:E1928-E1938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 105. | Schreibman IR, Baker M, Amos C, McGarrity TJ. The hamartomatous polyposis syndromes: a clinical and molecular review. Am J Gastroenterol. 2005;100:476-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 206] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 106. | Starink TM, van der Veen JP, Arwert F, de Waal LP, de Lange GG, Gille JJ, Eriksson AW. The Cowden syndrome: a clinical and genetic study in 21 patients. Clin Genet. 1986;29:222-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 329] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 107. | Levi Z, Baris HN, Kedar I, Niv Y, Geller A, Gal E, Gingold R, Morgenstern S, Baruch Y, Leach BH, Bronner MP, Eng C. Upper and Lower Gastrointestinal Findings in PTEN Mutation-Positive Cowden Syndrome Patients Participating in an Active Surveillance Program. Clin Transl Gastroenterol. 2011;2:e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 108. | Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Höglund P, Järvinen H, Kristo P, Pelin K, Ridanpää M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1136] [Cited by in RCA: 1083] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 109. | McGarrity TJ, Kulin HE, Zaino RJ. Peutz-Jeghers syndrome. Am J Gastroenterol. 2000;95:596-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 154] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 110. | Stoffel EM, Koeppe E, Everett J, Ulintz P, Kiel M, Osborne J, Williams L, Hanson K, Gruber SB, Rozek LS. Germline Genetic Features of Young Individuals With Colorectal Cancer. Gastroenterology. 2018;154:897-905.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 246] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 111. | Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL; Guideline Development Group, American College of Medical Genetics and Genomics Professional Practice and Guidelines Committee and National Society of Genetic Counselors Practice Guidelines Committee. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17:70-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 397] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 112. | Hegde M, Ferber M, Mao R, Samowitz W, Ganguly A; Working Group of the American College of Medical Genetics and Genomics (ACMG) Laboratory Quality Assurance Committee. ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis). Genet Med. 2014;16:101-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 113. | Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW; American College of Gastroenterology. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110:223-62; quiz 263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 1090] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 114. | Herzig D, Hardiman K, Weiser M, You N, Paquette I, Feingold DL, Steele SR. The American Society of Colon and Rectal Surgeons Clinical Practice Guidelines for the Management of Inherited Polyposis Syndromes. Dis Colon Rectum. 2017;60:881-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |