Published online Nov 15, 2019. doi: 10.4251/wjgo.v11.i11.983
Peer-review started: March 28, 2019
First decision: April 15, 2019
Revised: July 26, 2019
Accepted: September 12, 2019
Article in press: September 12, 2019
Published online: November 15, 2019
Processing time: 233 Days and 22.6 Hours
Hepatitis B virus, together with hepatitis C virus, has been recognized as the leading causes of hepatocellular carcinoma (HCC). Long non-coding RNAs (lncRNAs) have been suggested in increasing studies to be the potential prognostic factors for HCC. However, the role of combined application of lncRNAs in estimating overall survival (OS) for hepatitis virus positive HCC (VHCC) is uncertain.
To construct an lncRNA signature related to the OS of VHCC patients to enhance the accuracy of prognosis prediction.
The expression patterns of lncRNAs, as well as related clinical data were collected from 149 VHCC patients from The Cancer Genome Atlas database. The R package was adopted to obtain the differentially expressed lncRNAs (DElncRNAs). LncRNAs significantly associated with OS were screened by means of univariate Cox regression analysis, so as to construct a least absolute shrinkage and selection operator (LASSO) model. Subsequently, the constructed lncRNA signature was developed and validated. Afterwards, the prognostic nomogram was established, which combined the as-established lncRNA signature as well as the clinical features. Meanwhile, subgroup analysis stratified by the virus type was also performed. Finally, the above-mentioned lncRNAs were enriched to corresponding pathways according to the markedly co-expressed genes.
A total of 1420 DElncRNAs were identified, among which 406 were significant in univariate Cox regression analysis. LASSO regression confirmed 8 out of the 406 lncRNAs, including AC005722.2, AC107959.3, AL353803.1, AL589182.1, AP000844.2, AP002478.1, FLJ36000, and NPSR1-AS1. Then, the prognostic risk score was calculated. Our results displayed a significant association between the risk model and the OS of VHCC [hazard ratio = 1.94, 95% confidence interval (CI): 1.61-2.34, log-rank P = 2e-10]. The inference tree suggested that the established lncRNA signature was useful in the risk stratification of VHCC. Furthermore, a nomogram was plotted, and the concordance index of internal validation was 0.763 (95%CI: 0.700-0.826). Moreover, the subgroup analysis regarding etiology confirmed this risk model. In addition, the Wnt signaling pathway, angiogenesis, the p53 pathway, and the PI3 kinase pathway were the remarkably enriched pathways.
An eight-lncRNA signature has been established to predict the prognosis for VHCC, which contributes to providing a novel foundation for the targeted therapy of VHCC.
Core tip: The existing long non-coding RNA (lncRNA) signatures for prognosis prediction are still controversial and need to be optimized for hepatocellular carcinoma (HCC). Typically, there is less knowledge concerning the lncRNA signature for hepatitis virus positive HCC (VHCC). As a result, this study was carried out to construct a robust lncRNA signature to predict the prognosis for VHCC. Typically, the eight-lncRNA signature constructed in this study displayed a potent potential to predict the prognosis of VHCC, which might contribute to risk stratification, and provide the individualized clinical suggestion for an individual case.
- Citation: Huang ZL, Li W, Chen QF, Wu PH, Shen LJ. Eight key long non-coding RNAs predict hepatitis virus positive hepatocellular carcinoma as prognostic targets. World J Gastrointest Oncol 2019; 11(11): 983-997
- URL: https://www.wjgnet.com/1948-5204/full/v11/i11/983.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i11.983
Hepatocellular carcinoma (HCC), one of the most frequent liver cancer types, is a severe worldwide health problem[1]. Chronic hepatitis B virus (HBV) infection and/or hepatitis C virus (HCV) infection have been identified as the leading causes of HCC[2]. Specifically, the prevalence of HCV infection is predominant in European countries, which accounts for about 60% of all HCC cases in Italy and Spain; by contrast, HBV infection is more commonly seen in eastern Asian countries such as China, with an incidence rate of over 2/3[3]. Specifically, hepatitis viruses have been epidemiologically associated with the development of HCC[4,5].
It has been extensively determined that hepatitis viruses are associated with HCC etiologically; nonetheless, it is still not completely clear about the molecular mechanism of hepatitis viruses in inducing liver cancer. An increasing number of mutation genes, like TP53, CTNNB1, and mTOR[6], are found to participate in HCC genesis as well as progression. Nonetheless, HCC is a highly heterogeneous disease, which can determine the altered biological progression of HCC and add to the difficulty in predicting prognosis. As a result, it is urgently needed to search for the efficient prognostic biomarkers for HCC.
Long non-coding RNAs (LncRNAs) have been identified to be the superior prognostic biomarkers over other cancer hallmarks, which can be ascribed to their unique advantages, as shown below. First, lncRNA expression is greatly distinct among different tissues, in various diseases, as well as at different disease progression stages; as a result, they can better represent the disease characteristics[7]. Second, lncRNAs have been found to regulate the expression of genes at the transcriptional, post-transcriptional, and epigenetic levels[8,9]; on this account, their levels as well as functions show a close association with cancer progression. As a result, increasing studies have been carried out to illustrate the clinical value of lncRNAs among cancers, such as HCC[10].
In this study, hepatitis virus positive HCC (VHCC) patients were appropriately selected to explore the difference in the lncRNA expression profiles, so as to identify the candidate lncRNA biomarkers based on The Cancer Genome Atlas (TCGA) database. Moreover, the key lncRNAs were determined using the least absolute shrinkage and selection operator (LASSO) algorithm; thereafter, the risk score system for VHCC was established. This study aimed to establish a potent lncRNA signature based on the expression profiles through thorough genomic data analysis, thus improving the accuracy in predicting the prognosis of VHCC.
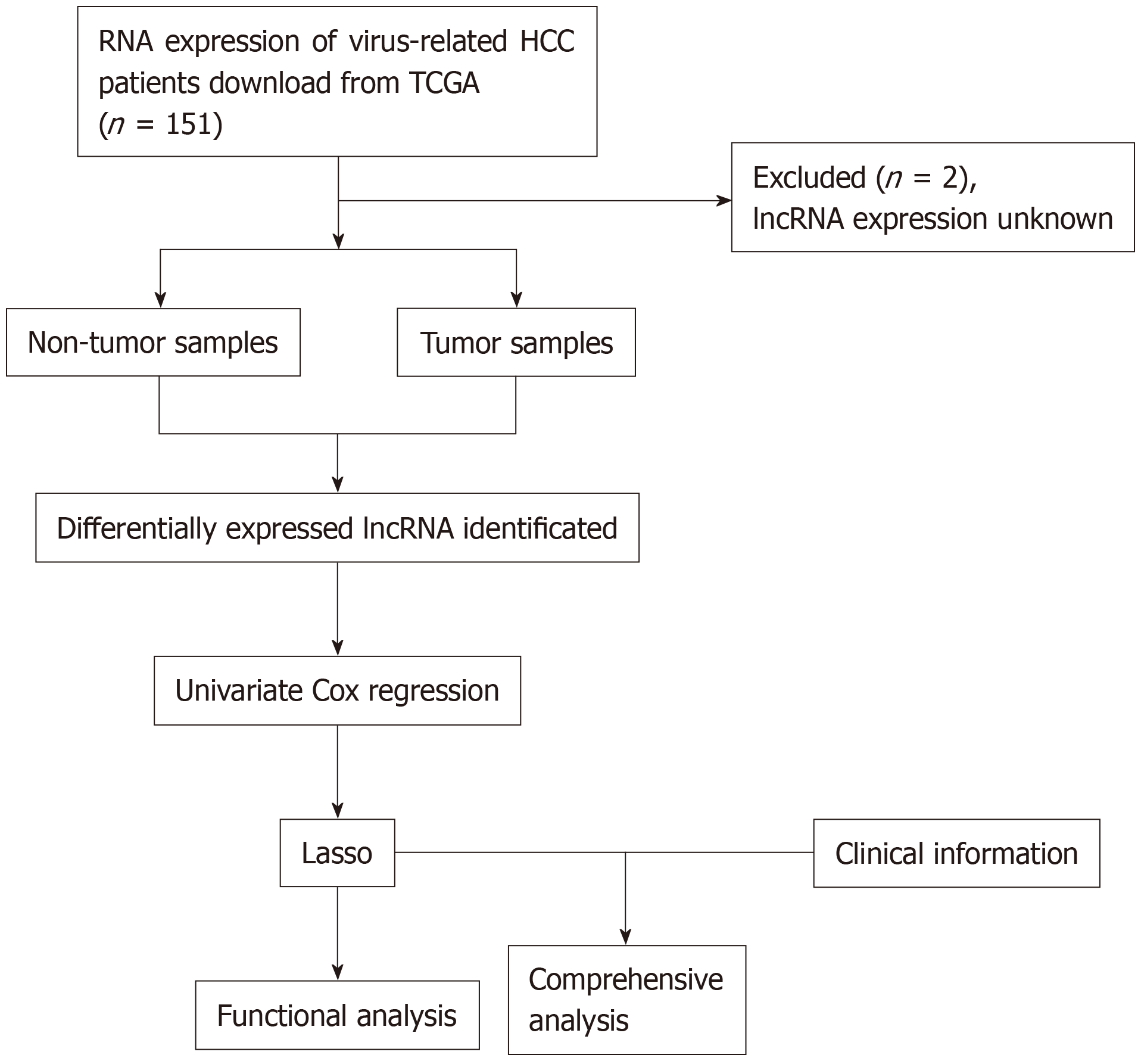
The data were downloaded from TCGA database. Gene expression, together with the clinical information of HCC patients (https://tcga-data.nci.nih.gov/), was downloaded using the Data Transfer Tool (provided by GDC Apps). Among them, 151 patients were virus-related. Patients with unknown lncRNA expression were eliminated from this study (n = 2); as a result, a total of 149 HCC cases were finally enrolled for analysis. The flowchart of analysis is shown in Figure 1. A total of 149 virus-induced HCC cases were enrolled for comprehensive integrated analysis. The data were publicly available, so approval was exempted from the Ethics Committee. Data were processed in accordance with the data access policies as well as the NIH TCGA human subject protection policies (http://cancergenome.nih.gov/publications/publicationguidelines).
LncRNA expression in HCC patients was derived from Illumina HiSeq RNASeq platform, which was then standardized based on TCGA. Then, the R software “edgeR” package was employed to detect the differentially expressed RNAs, so as to identify the differentially expressed lncRNAs (DElncRNAs), and the thresholds were set at log2 fold change > 2.0 as well as adjusted P value < 0.05.
The relationship of each lncRNA expression with the overall survival (OS) of patients was calculated using the univariate Cox model. Typically, lncRNAs would be deemed as statistically significant at P < 0.05 upon univariate Cox analysis. Subsequently, the chosen lncRNAs were screened and verified through LASSO regression with the R software “glmnet” package. Finally, the lncRNA-based prognostic risk score was constructed by linearly combining the products of expression level with the regression model (β) according to the formula below: Risk_score = βlncRNA1 × lncRNA1 expression + βlncRNA2 × lncRNA2 expression + · ···· + βlncRNAn × lncRNAn expression.
Patients together with their survival information were distributed according to the risk_score. Furthermore, patients were classified into a high-risk group and low-risk group according to the median risk_score. Then, the Kaplan-Meier survival curves were plotted, which could predict the high or low risk of patients. Thereafter, the univariate Cox proportional hazards regression analysis was performed. Then, the sensitivity and specificity regrading survival prediction were compared by the risk_score, and the receiver operating characteristic (ROC) curves with time dependence was then employed to evaluate the accuracy in predicting the 5-year prognosis. Moreover, multivariate Cox regression was also performed to examine whether lncRNA risk_score was predicted independently from other clinical factors. The R-package “party” “ctree” function was used to construct a conditional inference tree for better illustration. Besides, a nomogram was also established with the R project “rms” package. In addition, the predictive performance of the nomogram was verified for discrimination and calibration. Subsequently, subgroup analysis based on virus type was carried out. All the two-sided P < 0.05 would be deemed to be statistically significant. The R software (version 3.4.1, R Foundation) was employed for all analyses.
Pearson correlation analysis was carried out for the screened lncRNAs as well as the protein coding genes in the TCGA database according to their different expression quantities. Typically, a coefficient of correlation > 0.4 and P < 0.001 would be deemed to show a significant correlation. Enrichment analysis was performed with the DAVID Bioinformatics Tool (version 6.8, https://david-d.ncifcrf.gov/).
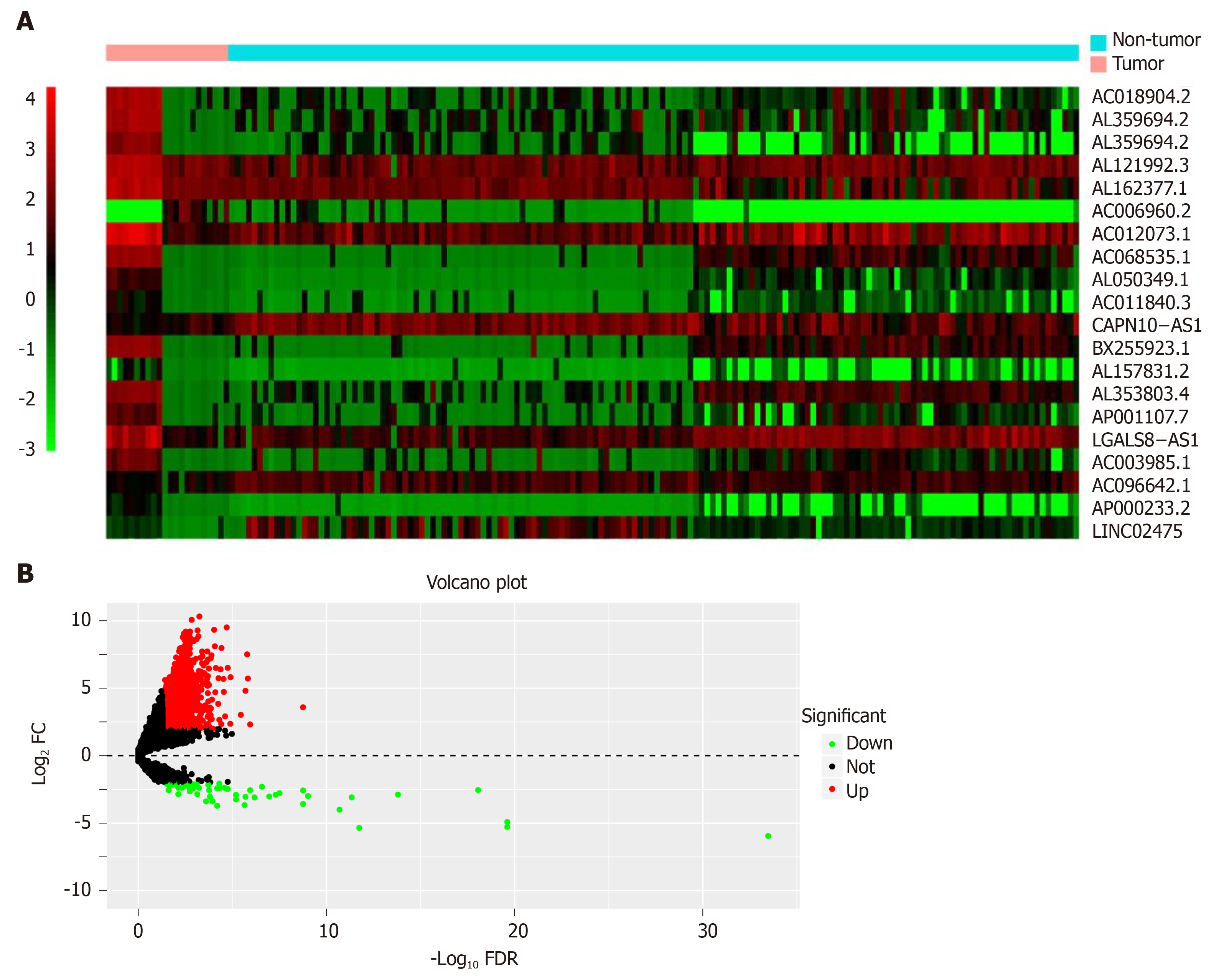
The significant DElncRNAs were identified by comparing the tumor samples with non-tumor samples. Altogether, 1420 DElncRNAs (including 1358 up-regulated and 62 down-regulated ones) were recognized using the R software “edgeR” package. Afterwards, the heat map (Figure 2A) and volcano of top 20 DElncRNAs (Figure 2B) were built.
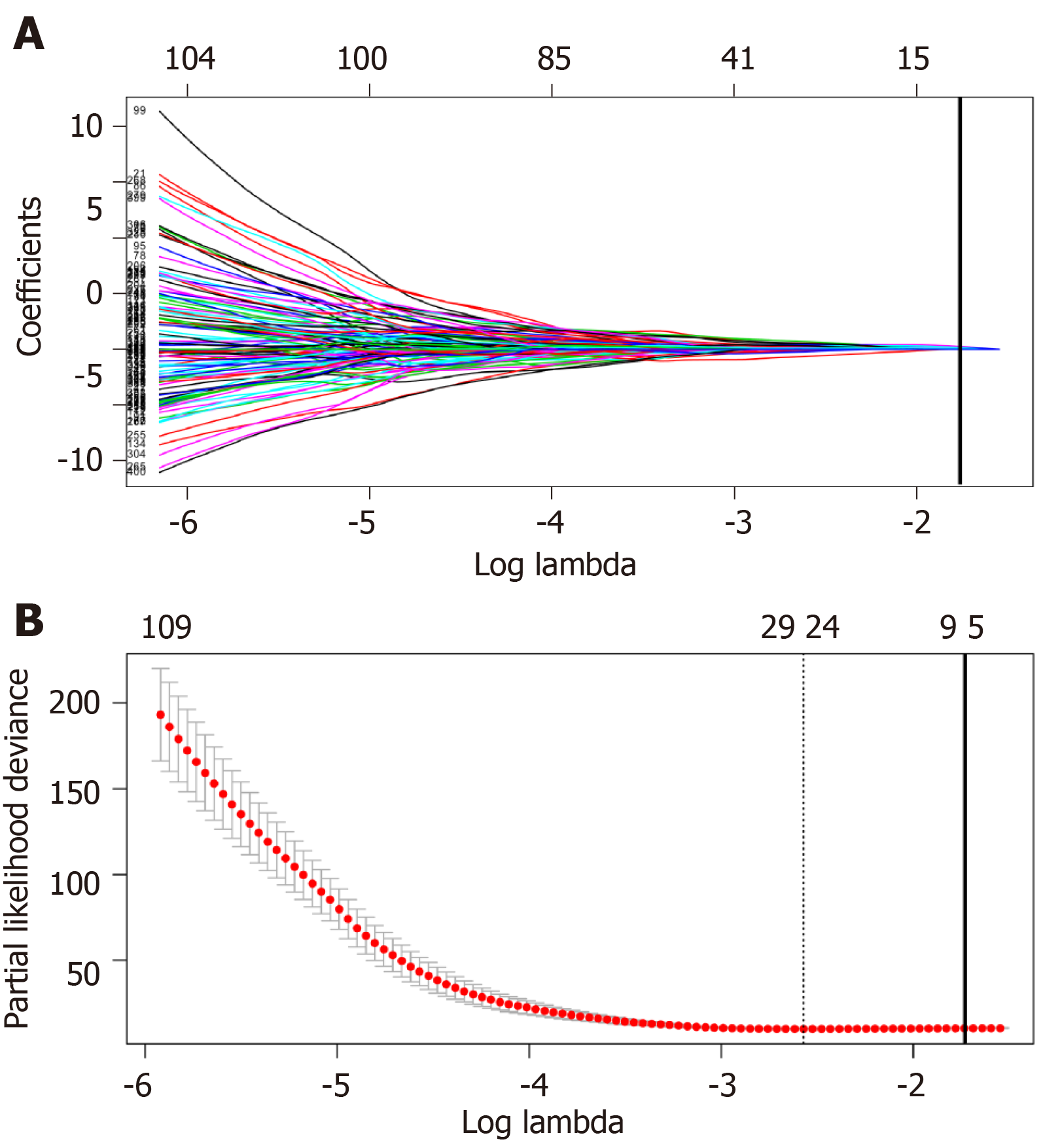
Univariate Cox regression analysis was performed between DElncRNAs and patients’ survival; as suggested by the results, altogether 406 lncRNAs were significantly associated with OS (P < 0.05). Next, LASSO regression was employed for verifying further variables (Figure 3). Eight lncRNAs were generated in this process, including AC005722.2, AC107959.3, AL353803.1, AL589182.1, AP000844.2, AP002478.1, FLJ36000, and NPSR1-AS1. Then, the prognostic risk_score was imputed as follows: (0.15103 *AC005722.2 expression) + (0.07857* AC107959.3 expression) + (0.09283 * AL353803.1 expression) + (0.17744* AL589182.1 expression) + (0.07857 * AP000844.2 expression) + (0.09888* AP002478.1 expression) + (0.13460 * FLJ36000 expression) + (0.06309* NPSR1-AS1 expression).
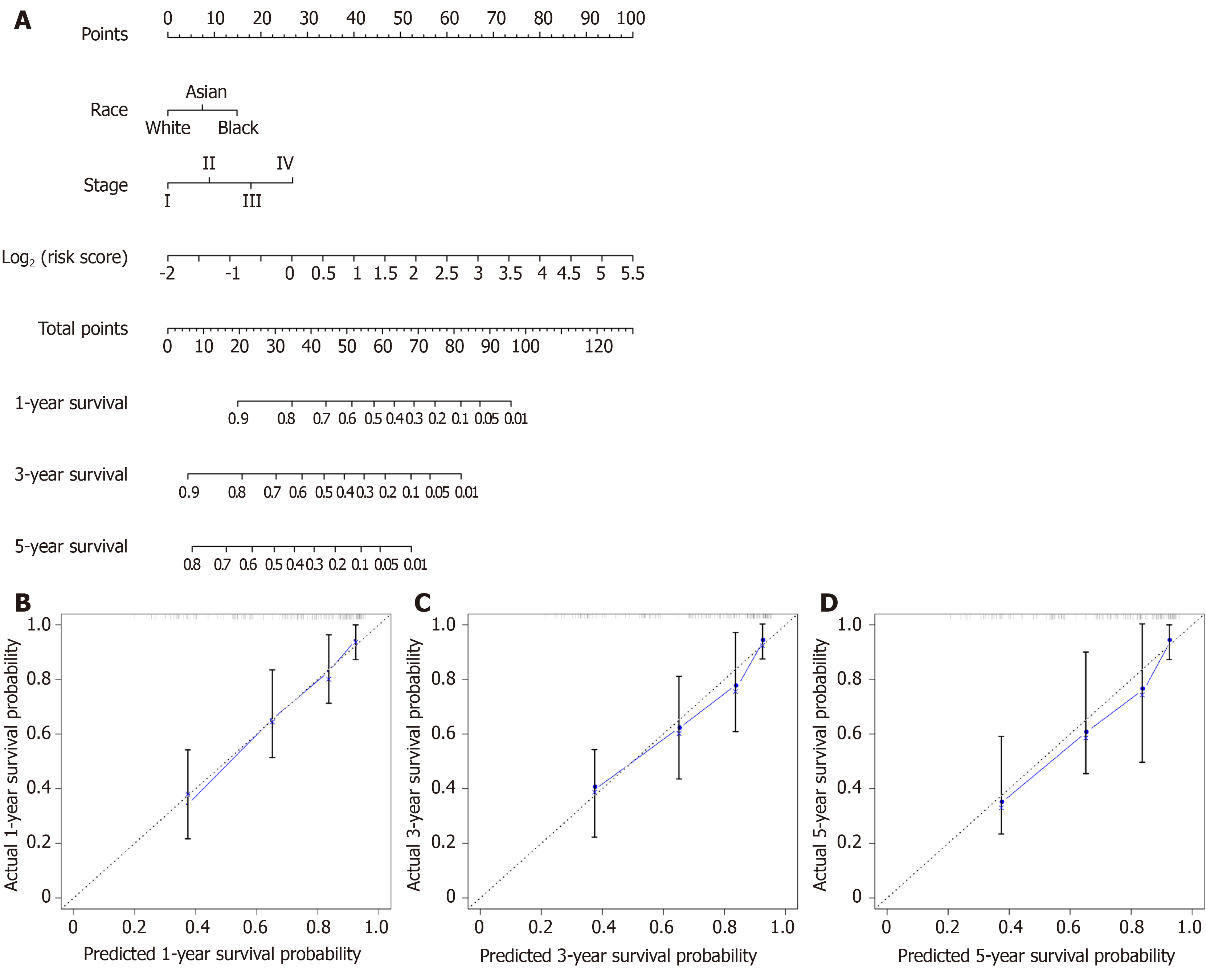
The risk score of every case was calculated; then, patients were classified into a high-risk group or low-risk group based on the median threshold (Figure 4A and B). The Kaplan-Meier curves for the high-risk as well as low-risk groups are presented in Figure 4C. Patients in the high-risk group were associated with a shorter OS compared with those in the low-risk group (P = 2e-10). Meanwhile, the risk score hazard ratio (HR) upon univariate Cox proportional hazards regression was 5.360 [95% confidence interval (CI): 3.012-9.538] (Table 1). Besides, multivariate Cox proportional hazards regression with adjusted clinical covariate also came to the same results (HR = 1.94, 95%CI: 1.61-2.34) (Table 2). Subsequently, the ROC curves with time dependency were adopted to assess the prognostic value of the eight-lncRNA signature. In addition, the areas under curves (AUCs) of the prognostic model constructed based on the eight-lncRNA signature were 0.73, 0.758, and 0.788 for 1-, 3-, and 5-year OS, respectively (Figure 4D).
| Characteristic | Overall survival | ||
| High-risk/low-risk | HR (95%CI) | P value | |
| Overall | 75/74 | 5.360 (3.012-9.538) | <0.001 |
| Age (yr) | |||
| ≤65 | 45/50 | 4.962 (2.361-10.428) | <0.001 |
| >65 | 29/25 | 5.512 (2.174-13.980) | <0.001 |
| Gender | |||
| Male | 51/49 | 5.324 (2.628-10.789) | <0.001 |
| Female | 23/26 | 5.509 (2.023-15.003) | 0.001 |
| Race | |||
| White | 38/47 | 7.473 (3.259-17.137) | <0.001 |
| Asian | 30/28 | 4.032 (1.774-9.164) | 0.001 |
| Grade | |||
| I/II | 45/57 | 4.959 (2.492-9.869) | <0.001 |
| III/IV | 29/18 | 5.033 (1.721-14.717) | 0.003 |
| Stage | |||
| I/II | 45/47 | 11.154 (3.882-32.049) | <0.001 |
| III/IV | 29/28 | 3.398 (1.652-6.988) | 0.001 |
| Variable | Multivariate analysis | ||
| HR | 95%CI | P value | |
| Risk_score | 1.944 | 1.613-2.343 | 2.94e-12 |
| Age | 1.004 | 0.986-1.023 | 0.662 |
| Gender | 0.832 | 0.480-1.442 | 0.512 |
| Race | 1.531 | 1.005-2.330 | 0.047 |
| Grade | 1.018 | 0.702-1.477 | 0.925 |
| Stage | 1.584 | 1.167-2.151 | 0.003 |
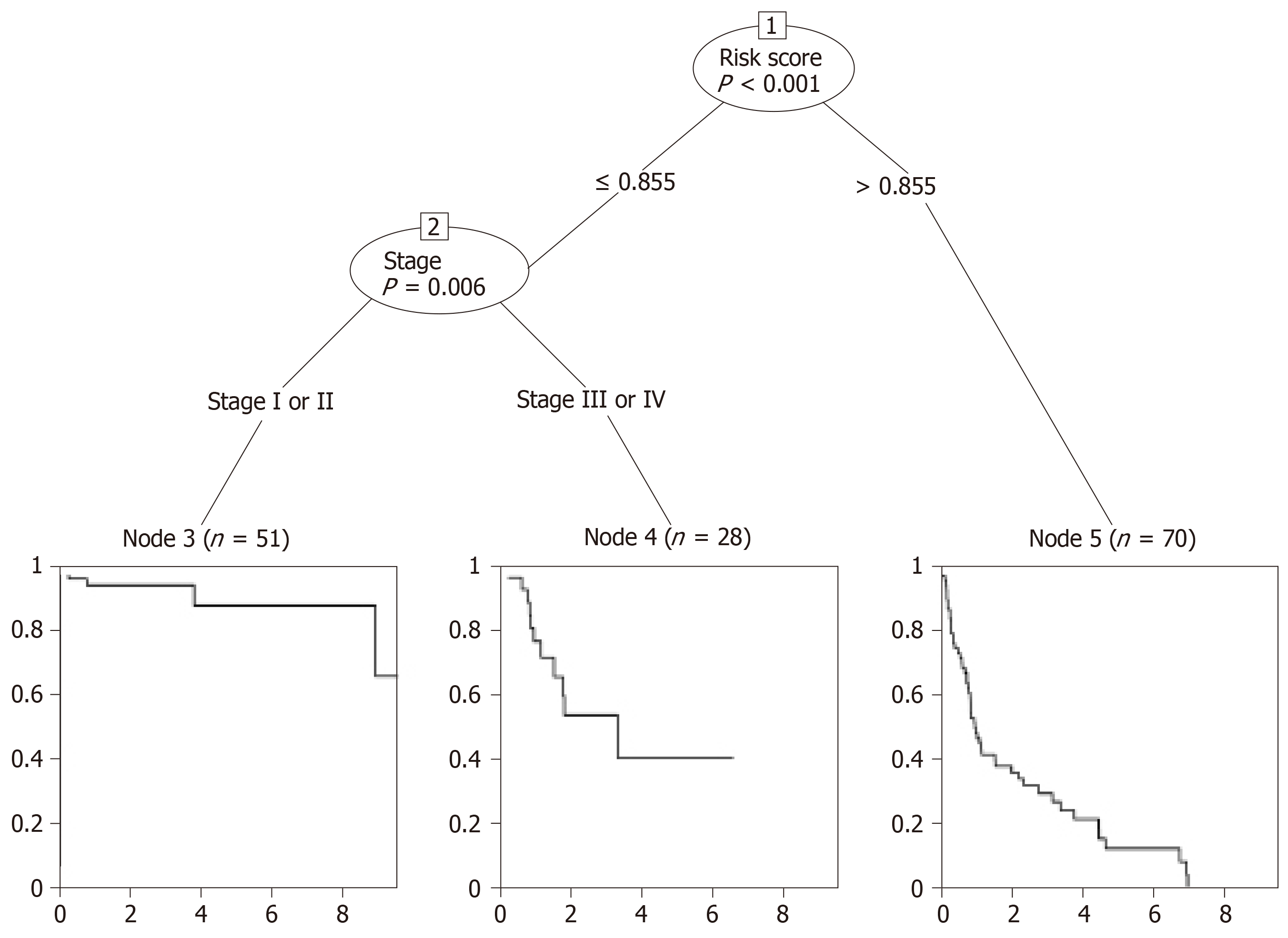
The first node of the conditional inference tree divided the data according to the risk_score, and the lower risk_score (≤0.855) was related to a better OS (P < 0.001) (Figure 5). For the lower risk_score (≤0.855), the branch was further divided depending on the stage (P = 0.006), and it was obvious that the risk_score ≤ 0.855 in stage I or II patients was associated with the best survival, while patients with the risk_score > 0.855 had the worst survival.
A nomogram for prognosis was constructed based on the above-mentioned independent factors for prognosis for the sake of visualization as well as prediction, so as to construct an integrated predicting factor for HCC. According to the multivariate Cox regression analysis, the eight-lncRNA risk_score, TNM stage, and race were recognized as independent risk factors (P < 0.05) (Table 2). Specifically, the nomogram was constituted by three factors (Figure 6A), with a C-index of 0.763 (95%CI: 0.700-0.826). Moreover, the calibrated curves for the risk of 1-, 3-, and 5-year recurrence were highly consistent between the nomogram predicted results and the observed results (Figure 6B-D).
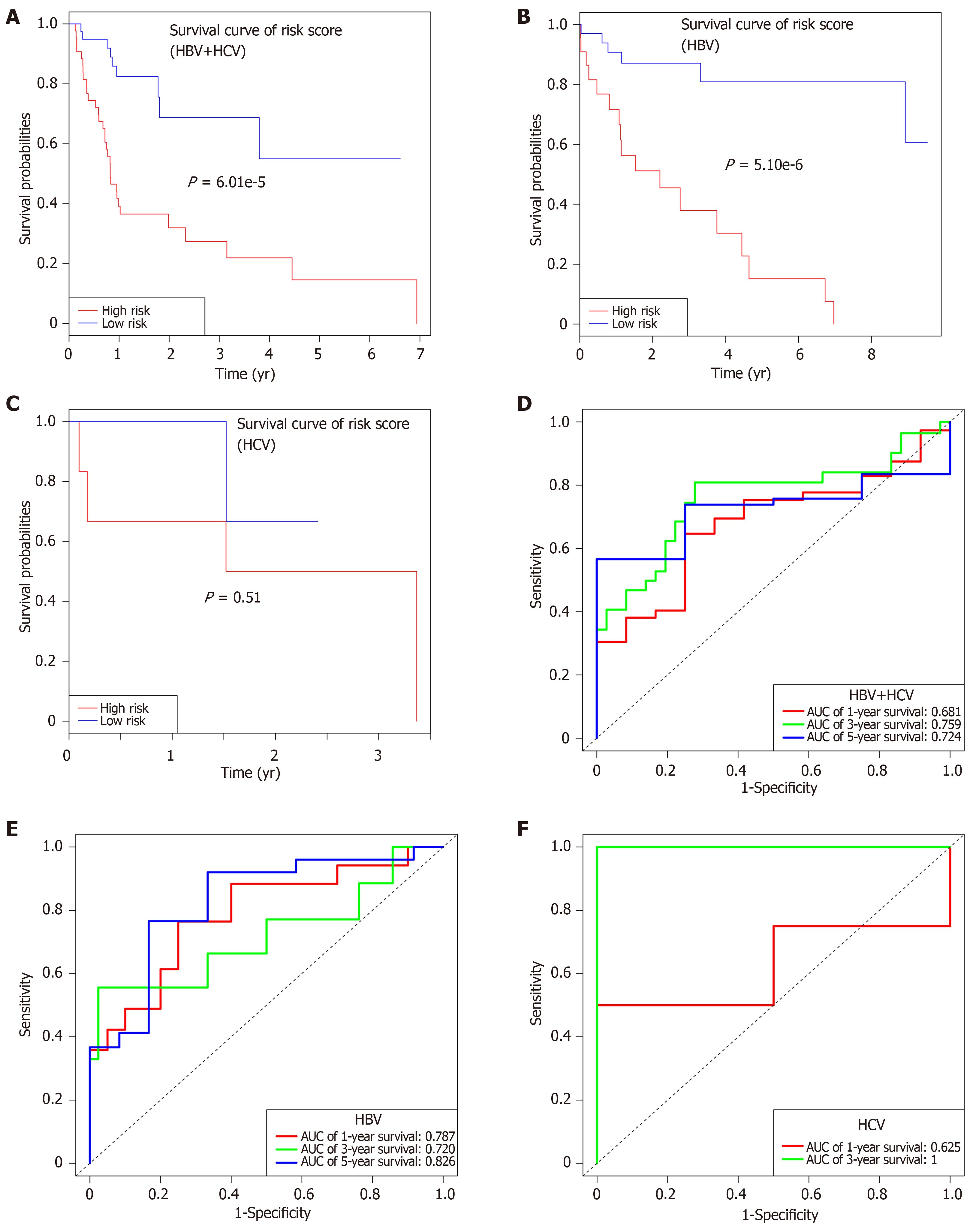
Subsequently, the patients were further classified into three subgroups, namely, HBV + HCV (n = 82), HBV alone (n = 58), and HCV alone (n = 9) groups. Kaplan-Meier curves for the high-risk and low-risk groups are displayed in Figure 7. The OS of patients benefited from the low-risk score in the HBV + HCV group (P = 6e-5, Figure 7A) and HBV alone group (P = 5e-6, Figure 7B). At the same time, the AUCs regarding the 1-, 3-, and 5-year OS in the HBV + HCV model were 0.681, 0.759, and 0.724, respectively (Figure 7D), while those in the HBV model were 0.787, 0.720, and 0.826, respectively (Figure 7E). Curves and corresponding P values for HCV patients are also shown in Figure 7C and F.
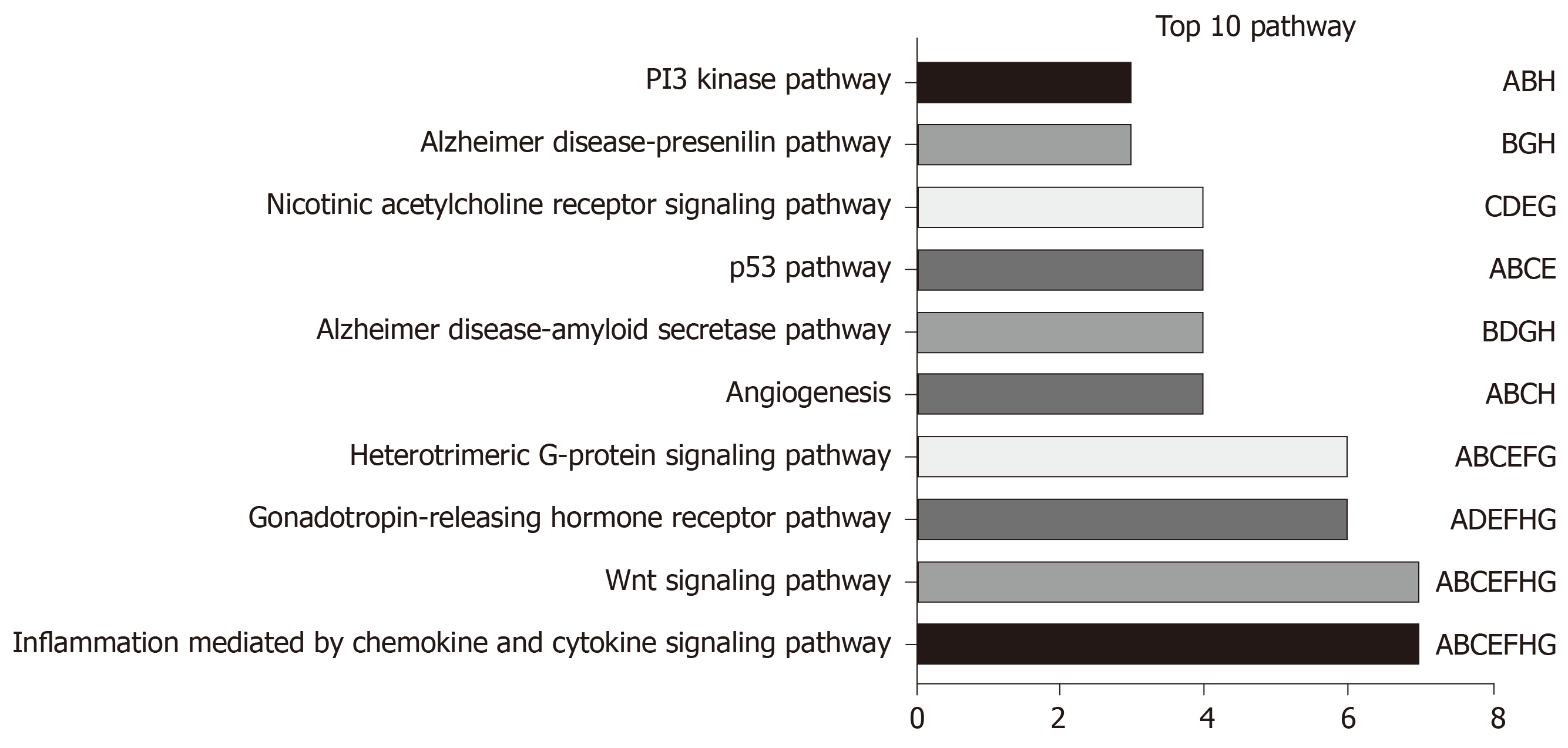
LncRNA-related mRNAs were first filtered out by Pearson correlation analysis and then applied into online tool DAVID for pathway enrichment, so as to examine the possible biological functions of the above-mentioned identified lncRNAs. The top 10 pathways with the highest enrichment are presented in Figure 8. Among those, the Wnt signaling pathway, angiogenesis, the p53 pathway, and the PI3 kinase pathway were recognized as the canonical pathways in the initiation as well as progression of HCC.
Few HCC prognostic models have been constructed on the basis of lncRNAs up to now. To our knowledge, little is known about lncRNA signature for hepatitis virus infection associated HCC patients. Meanwhile, the majority of studies analyzed virus combined with non-virus related HCC at the same time[11,12]. Different from previous studies that carried out analysis regardless of etiology, we conducted the current research in virus positive patients only. The DElncRNAs with marked correlation with OS were comprehensively screened by applying biostatistical methods and univariate Cox analysis. Then, the LASSO regression was applied to analyze the lncRNA data from the carefully chosen patients from TCGA, and eight lncRNAs were identified, which were recognized to be the independent factors for the prognosis of VHCC. Afterwards, the identified significant lncRNAs were employed for constructing a prognostic model, and its prognostic value was verified through Kaplan-Meier analysis, time-dependent ROC curve, and Cox regression analysis. A nomogram was developed to present the constructed risk_score system, and it displayed a favorable accuracy in OS prediction for VHCC. Further functional analysis uncovered that several pathways play crucial roles in the effects of the eight lncRNAs on the progression of HCC.
Our research displayed obvious advantages when compared with other research on the prognostic value of lncRNAs in HCC. First, patient homogeneity was markedly enhanced through careful patient selection before analysis, since the ignorance of patient features might result in false relationships, thus invaliding the conclusions. Existing treatments can treat chronic viral hepatitis, and some research has been performed previously to domenstrate the influence of antiviral treatment on disease progression as well as HCC development[13]. Moreover, many studies suggest that the survival of virus-related HCC patients can be prolonged through curative hepatectomy and/or local tumor ablation combined with adjuvant antiviral therapy[14-16]. On the other hand, virus and non-virus HCCs have been recognized to have apparently different outcomes, and different therapeutic strategies were recommended for them. Therefore, the prognostic models should also be developed in a distinct way. All the 149 patients were virus positive in our study. Second, with regard to the methodology, the use of LASSO penalized regression could boost the accuracy of bioinformatic analysis. Different from the conventional stepwise regression applied in prior research, the LASSO algorithm contributed to analyzing all independent factors at the same time, which tended to screen the most significant variables[17]. Then, the coefficients of the less significant variables would become zero following introduction into the penalty after regularization[18]. Consequently, such formulation approach would display a higher accuracy than stepwise regression using the multivariate Cox model, especially in the presence of huge datasets, such as genomics[19]. Third, an easy-to-use nomogram model was constructed on the basis of Cox analysis among the carefully screened patients with clinicopathological features, which would facilitate the prediction for individual patients.
As hepatitis viruses vary in different countries[3], our cohort was also divided into three subgroups based on the virus type (namely, HBV alone, HCV alone, and combined). Subgroup survival analysis showed that the as-constructed eight-lncRNA signature performed well in patients with combined virus infection (5-year AUC = 0.724), especially for those with HBV infection (5-year AUC = 0.826). No obvious OS heterogeneity was detected among the HCV patients (P = 0.51), which could be resulted from the small sample size (n = 9).
No study had investigated the biological functions for the eight lncRNAs identified in HCC; however, they could affect the genesis as well as the progression of HCC by the Wnt signaling pathway[20-24], angiogenesis[25,26], p53 pathway[27,28], and PI3 kinase pathways[29-31], as suggested by the pathway enrichment results. More importantly, this research could provide new evidence that lncRNAs, whose biological functions had not been reported before, might potentially serve as the potent markers to predict the prognosis of HCC. However, such findings should be further validated in future studies, and the molecular characteristics should also be investigated.
To our knowledge, this is the first study that constructs an lncRNA nomogram to predict the prognosis of VHCC. Nonetheless, some limitations should be noted in this study. First, no experimental study in vitro or in vivo was performed to verify the effect of the eight-lncRNA signature, which was constructed based on online datasets by the bioinformatic approach, in predicting the prognosis of HCCs. Second, data regarding more valuable clinical features are not available, including viral hepatitis duration, state of virus, Milan criteria, HCC treatment, and anti-viral treatment. Moreover, our results should be further validated in future studies.
To sum up, a novel lncRNA signature has been uncovered in this study based on comprehensive bioinformatic analysis combined with the expression patterns as well as the clinical information from the carefully selected cohort. Specifically, this risk model can be used as a biomarker to independently predict the prognosis of VHCC. Nonetheless, our results should be validated in studies that investigate the precise mechanisms regarding VHCC progression, as well as those that explore the functions of the eight identified lncRNAs.
Hepatocellular carcinoma (HCC), one of the most frequent liver cancer subtype, has posed a serious health issue in the world. Hepatitis virus is recognized to be a major factor leading to HCC. Recently, a verity of genetic markers as well as prediction models are proposed to improve HCC treatment. At the same time, numerous statistical techniques are also utilized to mine data in numerous large-scale public databases. Thanks to the well-developed clinical approaches, prognosis models with a higher accuracy and robustness can be established for HCC.
This study aimed to establish a prognosis model on the basis of HCC molecular biomarkers. Typically, long non-coding RNAs (lncRNAs) have been recognized as new predictive factors. Great efforts have been made to establish lncRNA-based HCC models, however, the lncRNA features for hepatitis virus positive HCC (VHCC) are not available at present.
This study aimed to develop a prognostic lncRNA feature for VHCC based on candidate lncRNAs through analyzing data collected from The Cancer Genome Atlas (TCGA) database.
Specifically, the least absolute shrinkage and selection operator (LASSO), the most advanced statistic algorithm, was used to establish the prediction model. This approach was carried out on the basis of typical lncRNAs selected according to the expression profiles of lncRNAs collected from the TCGA database. The as-established lncRNA feature was validated, and its clinical applicability was also examined.
Using LASSO, a risk score system was established to predict the overall survival (OS) for VHCC, which incorporated eight lncRNAs (including AC005722.2, AC107959.3, AL353803.1, AL589182.1, AP000844.2, AP002478.1, FLJ36000, and NPSR1-AS1). Notably, the as-constructed lncRNA feature was helpful in stratifying the risk for VHCC. To extend the model application range, a nomogram was also constructed, which involved both the lncRNA feature and other clinical features. Our results also suggested that the lncRNAs were markedly enriched in the Wnt signaling pathway, angiogenesis, the p53 pathway, and the PI3 kinase pathway.
The as-constructed signature based on eight lncRNAs displayed a favorable capacity in predicting the prognosis for VHCC patients, which can also contribute to risk stratification and may provide more useful clinical advice for individual patients.
Our results further support that lncRNAs can serve as possible functional regulating factors for the progression of VHCC. It is the future direction to search for the effective molecular biomarkers and predictive factors for the prognosis of HCC patients.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gougelet J, Jarcuska P S-Editor: Yan JP L-Editor: Wang TQ E-Editor: Qi LL
| 1. | Chen QF, Jia ZY, Yang ZQ, Fan WL, Shi HB. Transarterial Chemoembolization Monotherapy Versus Combined Transarterial Chemoembolization-Microwave Ablation Therapy for Hepatocellular Carcinoma Tumors ≤5 cm: A Propensity Analysis at a Single Center. Cardiovasc Intervent Radiol. 2017;40:1748-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2507] [Article Influence: 192.8] [Reference Citation Analysis (2)] |
| 3. | de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190-1200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 382] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 4. | Zhou HY, Luo Y, Chen WD, Gong GZ. Hepatitis B virus mutation may play a role in hepatocellular carcinoma recurrence: A systematic review and meta-regression analysis. J Gastroenterol Hepatol. 2015;30:977-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdom Radiol (NY). 2018;43:13-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 302] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 6. | Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1933] [Cited by in RCA: 1872] [Article Influence: 208.0] [Reference Citation Analysis (4)] |
| 7. | Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan CX, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 531] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 8. | Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1840] [Cited by in RCA: 2121] [Article Influence: 176.8] [Reference Citation Analysis (0)] |
| 9. | Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2002] [Cited by in RCA: 2315] [Article Influence: 192.9] [Reference Citation Analysis (0)] |
| 10. | Lanzafame M, Bianco G, Terracciano LM, Ng CKY, Piscuoglio S. The Role of Long Non-Coding RNAs in Hepatocarcinogenesis. Int J Mol Sci. 2018;19:pii: E682. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 11. | Gu JX, Zhang X, Miao RC, Xiang XH, Fu YN, Zhang JY, Liu C, Qu K. Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma. World J Gastroenterol. 2019;25:220-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 81] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 12. | Wu Y, Wang PS, Wang BG, Xu L, Fang WX, Che XF, Qu XJ, Liu YP, Li Z. Genomewide identification of a novel six-LncRNA signature to improve prognosis prediction in resectable hepatocellular carcinoma. Cancer Med. 2018;7:6219-6233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 13. | Hosaka T, Suzuki F, Kobayashi M, Seko Y, Kawamura Y, Sezaki H, Akuta N, Suzuki Y, Saitoh S, Arase Y, Ikeda K, Kobayashi M, Kumada H. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology. 2013;58:98-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 542] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 14. | Ogawa E, Furusyo N, Nomura H, Dohmen K, Higashi N, Takahashi K, Kawano A, Azuma K, Satoh T, Nakamuta M, Koyanagi T, Kato M, Shimoda S, Kajiwara E, Hayashi J; Kyushu University Liver Disease Study (KULDS) Group. Short-term risk of hepatocellular carcinoma after hepatitis C virus eradication following direct-acting anti-viral treatment. Aliment Pharmacol Ther. 2018;47:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 15. | Liu GM, Huang XY, Shen SL, Hu WJ, Peng BG. Adjuvant antiviral therapy for hepatitis B virus-related hepatocellular carcinoma after curative treatment: A systematic review and meta-analysis. Hepatol Res. 2016;46:100-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Shinn BJ, Martin A, Coben RM, Conn MI, Prieto J, Kroop H, DiMarino AJ, Hann HW. Persistent risk for new, subsequent new and recurrent hepatocellular carcinoma despite successful anti-hepatitis B virus therapy and tumor ablation: The need for hepatitis B virus cure. World J Hepatol. 2019;11:65-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Gao J, Kwan PW, Shi D. Sparse kernel learning with LASSO and Bayesian inference algorithm. Neural Netw. 2010;23:257-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 19. | McNeish DM. Using Lasso for Predictor Selection and to Assuage Overfitting: A Method Long Overlooked in Behavioral Sciences. Multivariate Behav Res. 2015;50:471-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 205] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 20. | Dai B, Ma Y, Yang T, Fan M, Yu R, Su Q, Wang H, Liu F, Yang C, Zhang Y. Synergistic effect of berberine and HMQ1611 impairs cell proliferation and migration by regulating Wnt signaling pathway in hepatocellular carcinoma. Phytother Res. 2019;33:745-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Li B, Cao Y, Meng G, Qian L, Xu T, Yan C, Luo O, Wang S, Wei J, Ding Y, Yu D. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine. 2019;39:239-254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 22. | Hu P, Ke C, Guo X, Ren P, Tong Y, Luo S, He Y, Wei Z, Cheng B, Li R, Luo J, Meng Z. Both glypican-3/Wnt/β-catenin signaling pathway and autophagy contributed to the inhibitory effect of curcumin on hepatocellular carcinoma. Dig Liver Dis. 2019;51:120-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 23. | Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch Biochem Biophys. 2019;661:196-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 24. | Huynh H, Ong R, Goh KY, Lee LY, Puehler F, Scholz A, Politz O, Mumberg D, Ziegelbauer K. Sorafenib/MEK inhibitor combination inhibits tumor growth and the Wnt/β‑catenin pathway in xenograft models of hepatocellular carcinoma. Int J Oncol. 2019;54:1123-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Miura K, Ohnishi H, Morimoto N, Minami S, Ishioka M, Watanabe S, Tsukui M, Takaoka Y, Nomoto H, Isoda N, Yamamoto H. Ezetimibe suppresses development of liver tumors by inhibiting angiogenesis in mice fed a high-fat diet. Cancer Sci. 2019;110:771-783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 26. | Morse MA, Sun W, Kim R, He AR, Abada PB, Mynderse M, Finn RS. The Role of Angiogenesis in Hepatocellular Carcinoma. Clin Cancer Res. 2019;25:912-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 387] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 27. | Meng X, Franklin DA, Dong J, Zhang Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res. 2014;74:7161-7167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 172] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 28. | Kong Y, Zhang L, Huang Y, He T, Zhang L, Zhao X, Zhou X, Zhou D, Yan Y, Zhou J, Xie H, Zhou L, Zheng S, Wang W. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017;407:76-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 29. | Jiao M, Nan KJ. Activation of PI3 kinase/Akt/HIF-1α pathway contributes to hypoxia-induced epithelial-mesenchymal transition and chemoresistance in hepatocellular carcinoma. Int J Oncol. 2012;40:461-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Liu W, Guo TF, Jing ZT, Yang Z, Liu L, Yang YP, Lin X, Tong QY. Hepatitis B virus core protein promotes hepatocarcinogenesis by enhancing Src expression and activating the Src/PI3K/Akt pathway. FASEB J. 2018;32:3033-3046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 31. | Zhang Y, Zhang T, Wu C, Xia Q, Xu D. ASIC1a mediates the drug resistance of human hepatocellular carcinoma via the Ca2+/PI3-kinase/AKT signaling pathway. Lab Invest. 2017;97:53-69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |