Published online Nov 15, 2019. doi: 10.4251/wjgo.v11.i11.971
Peer-review started: March 8, 2019
First decision: April 15, 2019
Revised: July 27, 2019
Accepted: September 12, 2019
Article in press: September 12, 2019
Published online: November 15, 2019
Processing time: 255 Days and 15.3 Hours
Calponin 3 (CNN3) is an actin-binding protein expressed in smooth muscle and non-smooth muscle cells. It is required for cytoskeletal rearrangement and wound healing.
To dissect the role of CNN3 in carcinogenesis with a focus on colon cancer.
A total of 20 cancer cell lines (8 breast, 11 colon, and HeLa cervical cancer cell as a positive control for mesenchymal phenotype) and 57 formalin-fixed, paraffin-embedded sections from archived sporadic colorectal carcinomas were included in this study. CNN3 expression analysis by western blot or immunohistochemistry was followed by functional analyses. The CNN3 gene was silenced by specific small interfering RNA (commonly known as siRNA), followed by confirmation of the silencing efficiency by western blotting. Then, the silenced cells and control siRNA-transfected cells were analyzed for changes in epithelial and mesenchymal markers, invasion, and response to 5-fluoruracil treatment. We also performed proteomics analysis using a phospho-kinase array-based panel of 45 proteins.
CNN3 showed positive expression in 6/8 breast and 9/11 colon cancer lines and in HeLa cells. Interestingly, the colorectal adenocarcinoma line SW480 was negative, while the cell line developed from its matching lymph node metastasis (SW620) was positive for CNN3. CNN3 expression was fairly consistent with the metastatic phenotype in colon cancer because it was absent in one other colon cell line from a primary site and expressed in all others. We selected SW620 for subsequent functional analyses. CNN3-silenced SW620 cells showed a reduction in collagen invasion and loss of mesenchymal markers. CNN3 silencing caused an increase in the SW620 colon cancer cell sensitivity to 5-fluorouracil. Phospho-kinase array-based proteomics analysis showed that CNN3 silencing in SW620 reduced extracellular signal-regulated kinase, β-Catenin, mutant p53, c-Jun, and heat shock protein 60 activities but increased that of checkpoint kinase 2. CNN3 was expressed in 20/57 (35%) colon cancer cases as shown by immunohistochemistry. CNN3 was associated with a decrease in overall survival in colon cancer in silico.
These results show the involvement of CNN3 in lymph node metastasis and resistance to chemotherapy in colon cancer and suggest that significant oncogenic pathways are involved in these CNN3-related actions.
Core tip: We hypothesized that calponin 3 (CNN3) may play a role in carcinogenesis based upon its known biological functions. We showed that it is expressed in colon and breast cancer cells and is associated with the metastatic phenotype in colon cancer via upregulating mesenchymal markers. CNN3 also improves the sensitivity to chemotherapy in these tumors. We also showed that it is linked to other carcinogenic pathways such as extracellular signal-regulated kinase 1/2, β-Catenin, mutant p53, c-Jun, and heat shock protein 60 in colorectal cancer. Thus, CNN3 is a promising biomarker in colon cancer.
- Citation: Nair VA, Al-khayyal NA, Sivaperumal S, Abdel-Rahman WM. Calponin 3 promotes invasion and drug resistance of colon cancer cells. World J Gastrointest Oncol 2019; 11(11): 971-982
- URL: https://www.wjgnet.com/1948-5204/full/v11/i11/971.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i11.971
The calponin family of actin-binding proteins consists of three isoforms: (1) Calponin-1 (CNN1; h1 or basic CNN); (2) CNN2 (h2 or neutral CNN); and (3) CNN3 (h3 or acidic CNN). All of them are generally involved in various forms of cell motility[1-3]. CNN1 is mainly expressed in smooth muscle cells in which it regulates smooth muscle contractions. It inhibits actin-activated myosin ATPase activity and thus inhibits smooth muscle contractility; it is also considered a troponin-like molecular switch[1]. CNN2 and CNN3 are expressed in smooth muscle and non-smooth muscle cells[3,4]. An accumulating body of evidence has shown that CNN3 has an important role in wound healing and cellular contractility and migration regulation. Appel et al[5] showed that CNN3 plays a role in fibroblast migration during wound healing; moreover, they proposed a model in which CNN3 can induce fibroblast migration through activation of extracellular signal-regulated kinase (ERK)1/2 and its direct, target l-caldesmon. CNN3 co-translocates with both ERK1/2 and protein kinase C-α to the cell cortex and podosome-like structures in a fibroblast cell stimulated by a phorbol ester[5]. These findings were corroborated by Daimon et al[6] who showed that CNN3 expression is induced by mechanical tension and is required for stress fiber formation in skin fibroblast after a wound occurs. When CNN3 was knocked out in these fibroblasts, the cells were not able to form the strong stress fibers necessary to generate the mechanical tension required for wound closure and contraction. Overall, CNN3 knockout resulted in a phenotype of decreased cellular dynamics[6]. CNN3 is mainly controlled by post-transcriptional modifications as evidenced by no changes in mRNA levels before and after a wound occurs in spite of a rise in its protein level at the time of wounding[6]. Mitogen-activated protein kinase kinase 1 (MEKK1 or MAP3K1), which is necessary for contractility and directs migration in many cell types, can phosphorylate CNN3 at Thr288 to increase the traction stress of the cell. Together, MEKK1 and CNN3 form an important hub in the positive feedback mechanism that promotes cell contraction and migration[7].
Cancer is a major devastating health problem worldwide, and colorectal cancer, in particular, is a notorious disease. Colorectal cancer is the third most commonly occurring cancer in men and the second most commonly occurring cancer in women. Over 1.8 million new cases were diagnosed in 2018[8]. More than half of the patients of colorectal cancer are doomed to die from this disease, particularly in the less developed regions of the world in which the disease outcome is influenced by adverse environmental factors and/or different genetic factors[8,9]. Generally, the course of cancer progression is essentially due to the capability of the cancer cells to invade, metastasize, and destroy normal tissues. Cancer cells, which undergo this complex process, simply trick the body into activating the biological wound-healing program to support their invasion. Rather than inventing a new mechanism to interact with their microenvironment during invasion and metastasis, cancer cells simply exploit their surrounding microenvironment that could have been created by the preexisting program of wound healing. Thus, tumors were long considered to be similar to “wounds that do not heal”[10,11]. The epithelial to mesenchymal transition (EMT) is an important step in both wound healing and epithelial cancer metastasis during which the epithelial cells shed their adhesive epithelial markers and acquire a set of mesenchymal markers in order to facilitate their movement. The EMT program is orchestrated by a complex network of signaling[12-16]. Not surprisingly, the EMT was found to be the most dominant program in a comprehensive, large-scale analysis of microarray data. The EMT is the first principal component of colorectal cancer, which is a well-known heterogeneous disease, and the tight correlation of EMT with recurrence and poor prognosis[12,13,16] highlights the significance of EMT-related genes in cancer development and progression.
It is expected that CNN3 plays a role in the EMT during carcinogenesis and tumor progression due to the similarity between wound healing and metastasis. The few available data on CNN3 in cancer are intriguing. In a murine model of ovarian carcinoma, CNN3 was on the list of genes associated with in vivo carcinogenicity[17]. The CNN3 locus on chromosome 1 is involved in a translocation that has been identified in mucosa-associated lymphoid tissue lymphoma[18]. CNN3 is expressed in mammary cells in response to erythroblastic oncogene B2 overexpression[19] and is associated with Duke’s stage C and lymph node metastasis of colon cancer[20]. Thus, the available data suggest that CNN3 could play a role in cancer invasion and progression.
In this study, we confirmed that CNN3 is differentially expressed in colorectal and breast carcinoma and showed that it is associated with EMT features, increased cancer cell invasion, and resistance to chemotherapeutic agents. We also demonstrated that CNN3 can upregulate multiple oncogenic pathways such as β-Catenin, ERK1/2, c-Jun, heat shock protein 60 (HSP60), and mutant p53.
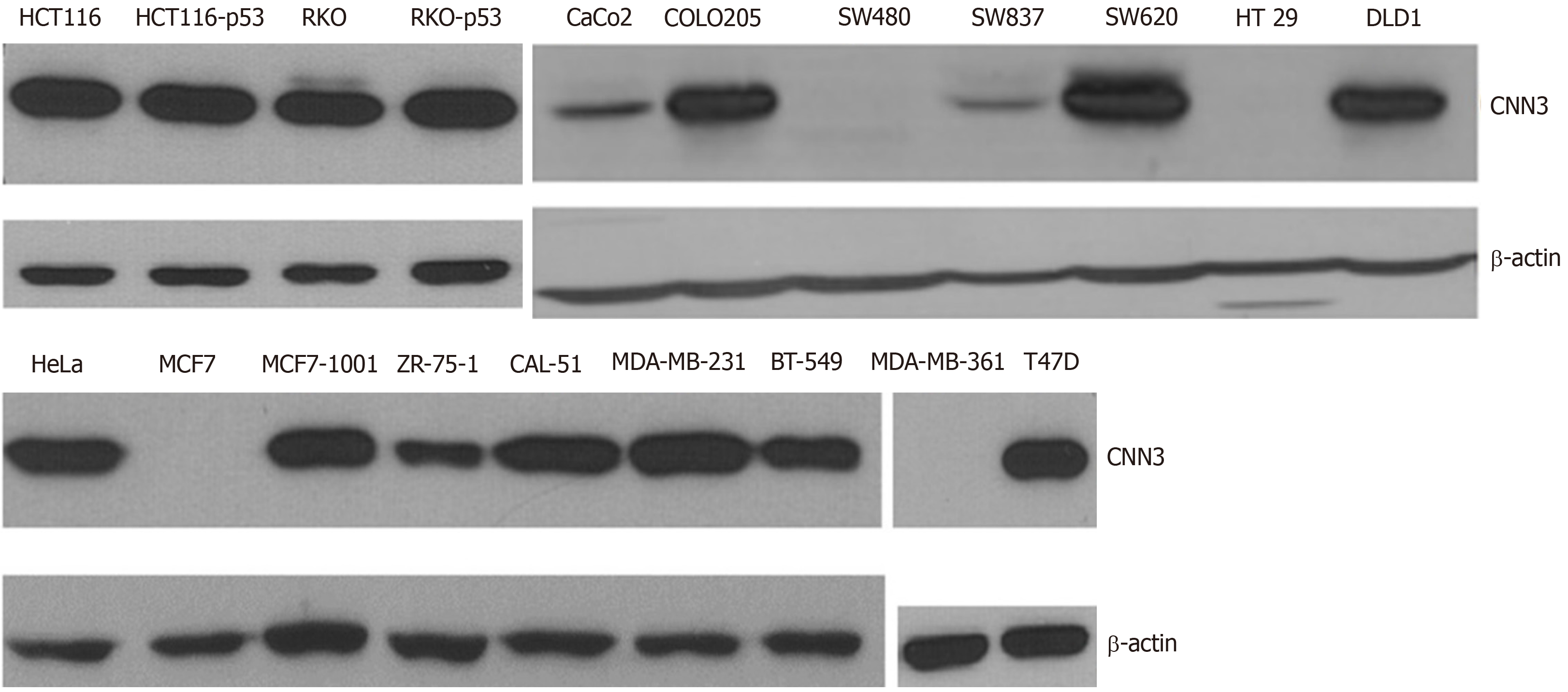
A total of 20 cell lines were used. The breast cancer cell lines were MCF7, 1001 (a tumor necrosis factor-resistant/TP53 mutant clone from the MCF7 parental cell line), CAL-51, MDA-MB-231, MDA-MB-361, ZR-75-1, T47D, and BT-549. Colon cancer cell lines included HCT116, HCT116.p53 mutant, RKO, RKO.P53-/-, SW480, SW620, CaCo2, COLO205, SW837, HT29, and DLD1. HeLa, a cervical cancer cell line, was used as a positive control for EMT. These cell lines were available from previous work[14,15] or purchased (Sigma Aldrich, St. Louis, MO, United States) for the purpose of this study. The cell lines were cultured in their corresponding growth media and specific conditions as previously described[14,15].
Cell lysates were prepared from cultured cells, washed twice with ice cold 1× phosphate-buffered saline, and the resulting pellets were then lysed using pre-chilled Triton lysis buffer (25 mM Tris HCL, pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% Triton) to which 50 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 2% protease inhibitor were added. Then, the lysed pellets were incubated on ice for 5 min and centrifuged at a speed of 14000 rpm for 10 min at 4°C in order to remove the cell debris. Protein concentration was quantified using the bicinchoninic acid protein assay (Thermo Fisher Scientific, Waltham, MA, United States). Approximately 30 µg total protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis consisting of a 7.5% resolving gel and 4% stacking gel. Subsequent steps were as previously described[14,15]. The primary antibodies were against CNN3 (clone H-55), rabbit polyclonal antibody [later replaced by clone A-2 mouse monoclonal (mAb) antibody due to discontinuation of the first (both from Santa Cruz, Dallas, TX, United States)], β-Actin (clone 13E5), rabbit mAb antibody (Cell Signaling, Danvers, MA, United States). EMT antibodies included vimentin (D21H3) rabbit mAb, N-cadherin (D4R1H) rabbit mAb, Claudin-1 (D5H1D) rabbit mAb, β-Catenin (D10A8), rabbit mAb, ZO-1 (D7D12) rabbit mAb, Snail (C15D3) rabbit mAb, Slug (C19G7) rabbit mAb, TCF8/ZEB1 (D80D3) rabbit mAb, and E-cadherin (24E10) rabbit mAb (all from Cell Signaling). All primary antibodies were used at a 1:1000 dilution.
We used siRNA for CNN3 protein (siRNA ID: s3256; Thermo Fisher Scientific). We transfected the SW620 cell line with a specific CNN3 siRNA along with control siRNA under the same conditions according to the Life Technologies protocol using the reverse transfection protocol available at https://http://www.lifetechnologies.com/.
For the invasion assay, we used QCMTM High Sensitivity Non-cross-linked Collagen Invasion Assay kit (Millipore, Burlington, MA, United States) according to the manufacturer’s protocol. Briefly, the assay was performed using a modified chamber with filter inserts (pore size 8 μm) coated with Matrigel in 24-well dishes. Approximately 0.5 million cells were prepared in serum-free media (RPMI 1640). Then 250 μL aliquots of cell suspension were added to each of the inserts (top chamber), and 500 μL of 15% fetal bovine serum-containing media was added to the bottom chamber. After a 48-h incubation, cells remaining in the top chamber were removed, and 400 μL cell stain was applied to the invasion chamber insert for 15 min. After several washes with water, the inserts were dried, viewed under the microscope, and photographed. Inserts were then transferred to 200 μL extraction buffer and allowed to incubate for 15 min at room temperature. The dye mixture was assessed by a plate reader at a wavelength of 630 nm.
The IC50 concentration of drugs used in the experiments were prepared by serial dilution in Dulbecco’s modified Eagles medium prior to performing the experiment. The IC50 concentrations were determined, and subsequent steps were as previously described[14,15].
For comprehensive proteomic analysis of proteins and pathways targeted by CNN3, we used a human phospho-kinase array ready-made kit that concurrently detects the relative phosphorylation levels of 43 kinase sites and two related total proteins using carefully selected specific capture antibodies. The experimental technique and analyses were according to manufacturer’s protocols and recommendations as detailed in the supplied booklet, which also contains a list of the target proteins and their phosphorylation sites (Proteome Profiler; R and D Systems, Minneapolis, MN, United States). This analysis was performed on SW620, SW620 transfected with siRNA control, and SW620-CNN3 silenced, repeated in duplicate, and two different exposures were taken. The average signal of the four analyses was finally used to generate graphs.
The primary antibody used for immunohistochemistry was anti-CNN3 (clone H-55), rabbit polyclonal antibody, from Santa Cruz. Immunohistochemistry was performed on 57 formalin-fixed, paraffin-embedded (FFPE) samples derived from a bigger series of tissues according to availability. The FFPE blocks were from surgical resection specimens of colorectal carcinoma submitted to pathology laboratories. All tumors were from the colon and/or rectum, generally diagnosed as adenocarcinoma, with no evidence of hereditary colon cancer syndrome. The histological variants of the 57 tumors included 44 colorectal adenocarcinomas, 9 mucinous adenocarcinomas, 1 signet ring carcinoma, 1 medullary carcinoma, 1 squamous cell carcinoma, and 1 undifferentiated carcinoma. Additional features of this series are included in Table 1. Molecular characterization of these tumors was described in our previous work[21,22]. We used the Envision+DAB system and immunohistochemistry staining protocol as previously described[23]. The work was done according to ethical standards of the Helsinki declaration and under the approval of the Ethics and Research Committee at the University of Sharjah. Five 200× power fields from different corners of the specimens were counted, and the percentage of stained cells was calculated. A tumor was graded as positive if it showed strong to moderate expression in 30% or more of the total counted tumor cells.
| Clinical and pathological features | Total | CNN3 expression | P value | ||
| Negative, n (%) | Positive, n (%) | ||||
| All cases | 57 | 37 (65) | 20 (35) | ||
| Gender | male | 32 | 21 (56) | 11 (34) | NS |
| female | 25 | 16 (64) | 9 (36) | ||
| Age in yr | ≤ 50 | 35 | 23 (66) | 12 (34) | NS |
| > 50 | 22 | 14 (64) | 8 (36) | ||
| Histological variant | adenocarcinoma | 44 | 27 (61) | 17 (39) | NS |
| mucinous adenocarcinoma | 9 | 8 (89) | 1 (11) | ||
| others1 | 4 | 2 (50) | 2 (50) | ||
| Differentiation/grade2 | well/moderate | 18 | 12 (67) | 6 (33) | NS |
| poor | 33 | 22 (67) | 11 (33) | ||
| Stage2 | early (I and II) | 32 | 20 (63.5) | 12 (37.5) | NS |
| late (III and IV) | 23 | 15 (65) | 8 (35) | ||
| Extracellular mucin | present | 21 | 13 (62) | 8 (38) | NS |
| absent | 36 | 24 (67) | 12 (33) | ||
| MSI status2 | MSI | 15 | 8 (53) | 7 (47) | NS |
| MSS | 38 | 25 (66) | 13 (34) | ||
| p53 | negative | 28 | 19 (68) | 9 (32) | NS |
| stabilized | 29 | 18 (62) | 11 (38) | ||
When appropriate, the Fisher's exact probability test, χ2 test, or Student t-test was used to evaluate differences between groups. Pearson’s correlation analysis was performed to test relationships between variables. Analyses were performed using MS Excel and/or VassarStats Web-based statistical program found at http://faculty.vassar. edu/lowry/VassarStats.html. All reported P values were two-tailed and P values < 0.05 were considered significant.
Overall, CNN3 was expressed in 9 of 11 (high expression in seven and low expression in two) colon cancer cells. SW480, which was developed from primary colorectal adenocarcinoma, was negative for CNN3, but the cell line derived from its lymph node metastasis, SW620, was strongly positive. This finding led us to focus on this cell line model (SW480/SW620) and to choose the SW620 to perform functional CNN3 analyses. CNN3 expression was not related to p53 status in the isogenic pairs of cells HCT116, HCT116.p53 mutant, or RKO, RKO.p53-/- (Figure 1). Six out of the eight breast cancer cell lines were positive for CNN3 regardless of p53 status. CNN3 was negative in the parental p53 wild type MCF7 while it was expressed in the MCF7 clone (MCF7-1001) that was a p53 mutant, but there was no relationship with p53 status in all of the other selected breast cancer cell lines. HeLa, used as EMT positive control, was positive for CNN3 as expected (Figure 1).
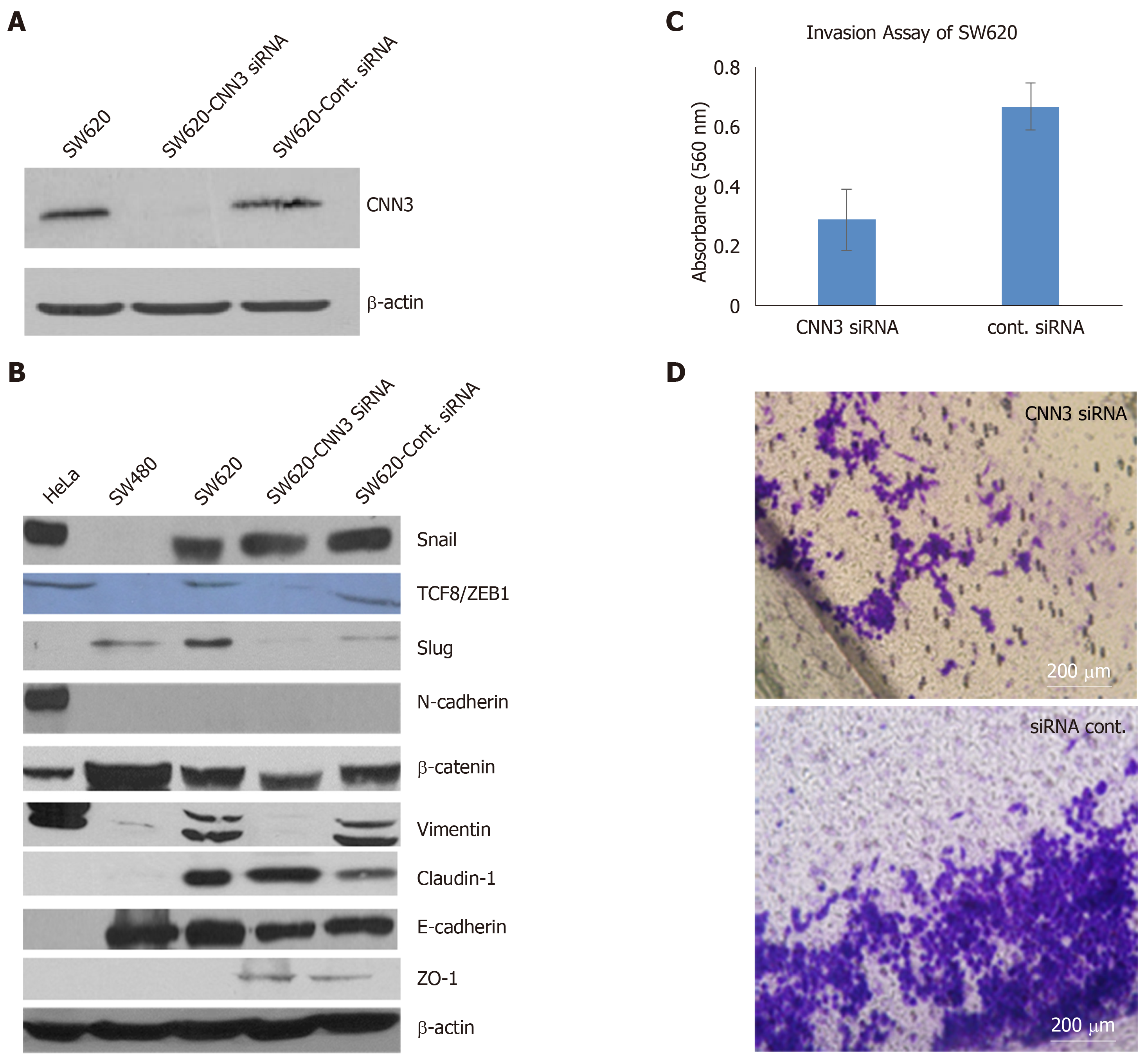
We chose SW620, which was developed from metastatic colon carcinoma, to silence CNN3 and carry out further functional analyses as this cell line shows high expression of CNN3 in contrast to its matching cell line from the primary tumor, SW480. Transfection of SW620 with CNN3-specific siRNA was highly efficient as evidenced by western blotting results and was consistent with data from the siRNA supplier (Figure 2A). Overall, the EMT marker set presented interesting findings. The positive control cell line for EMT (HeLa) confirmed our EMT marker set’s validity, showing positive expression of the mesenchymal markers Snail, TCF8/ZEB1, vimentin, N-cadherin, and β-Catenin and negative expression of the epithelial markers claudin-1, E-cadherin, and ZO-1. Comparison of SW480 with SW620 showed that the metastatic phenotype of SW620 was associated with gain of the mesenchymal markers Snail, TCF8/ZEB1, Slug, and vimentin, whereas the epithelial markers E-cadherin and ZO-1 showed no change. In contrast, claudin-1 was somewhat increased in SW620 (Figure 2B). CNN3 silencing resulted in total loss or reduction of the mesenchymal markers TCF8/ZB1, Slug, vimentin, and β-Catenin (Figure 2B). Collectively, these data demonstrated that CNN3 silencing predisposed metastatic cancer cells toward losing their mesenchymal markers.
The collagen invasion assay showed considerable reduction in SW620 invasiveness after siRNA silencing of CNN3 compared to its matching negative control transfected with control siRNA (Figure 2C and D).
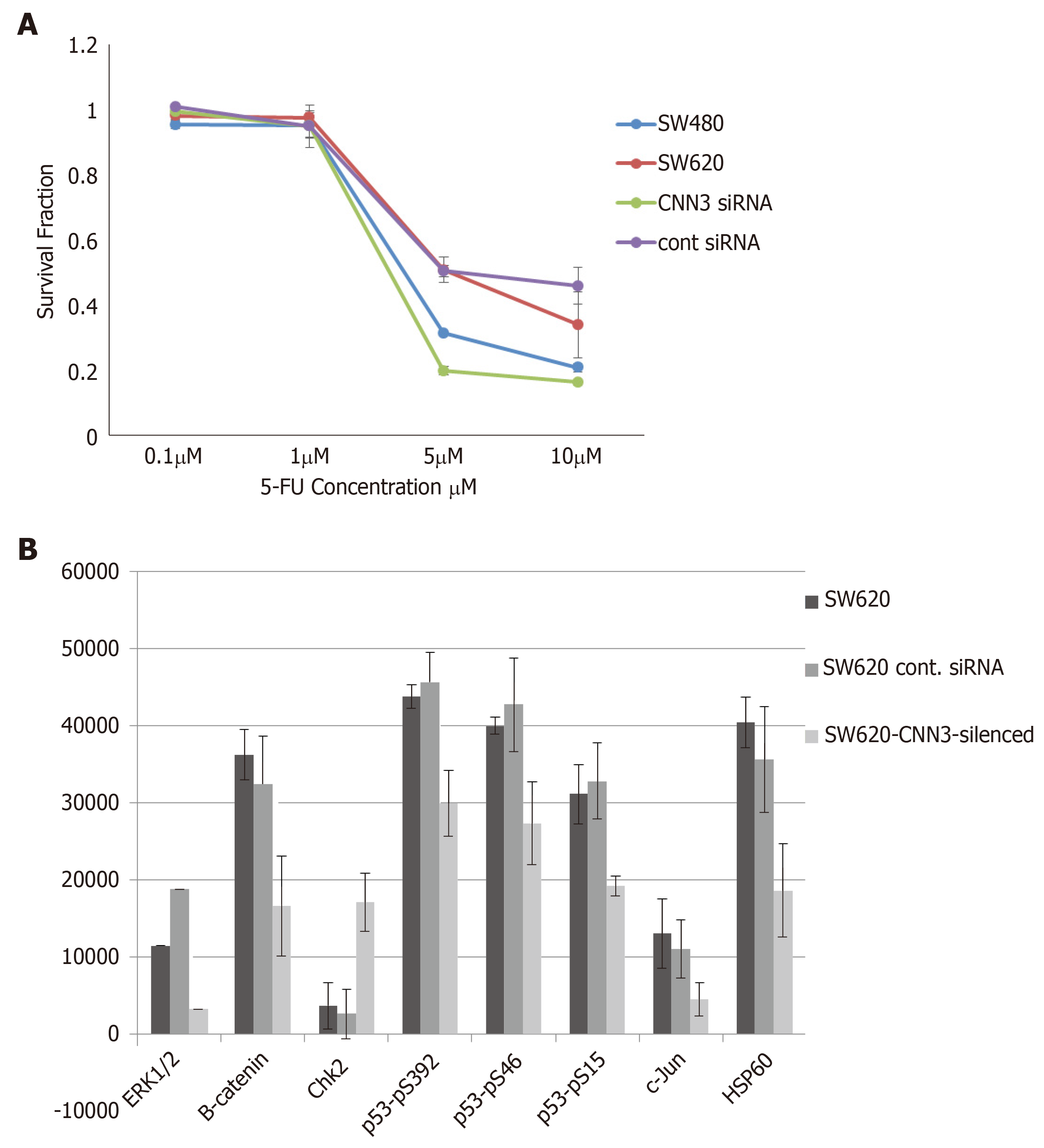
The SW620 cell line was more resistant to the standard chemotherapeutic agent 5-fluorouracil compared to its matching SW480 cell line. Interestingly, CNN3 silencing increased the sensitivity of SW620 to 5-fluorouracil compared to the matching cells transfected with control siRNA (Figure 3A).
To identify proteins associated with CNN3 silencing we compared SW620, SW620 transfected with control siRNA, and SW620-CNN3 silenced. CNN3 silencing in SW620 caused a reduction in ERK1/2, β-Catenin, mutant p53, c-Jun, and HSP60 but brought about an increase in checkpoint kinase 2 (Figure 3B). Changes in β-Catenin were noted on the western blot (Figure 2B).
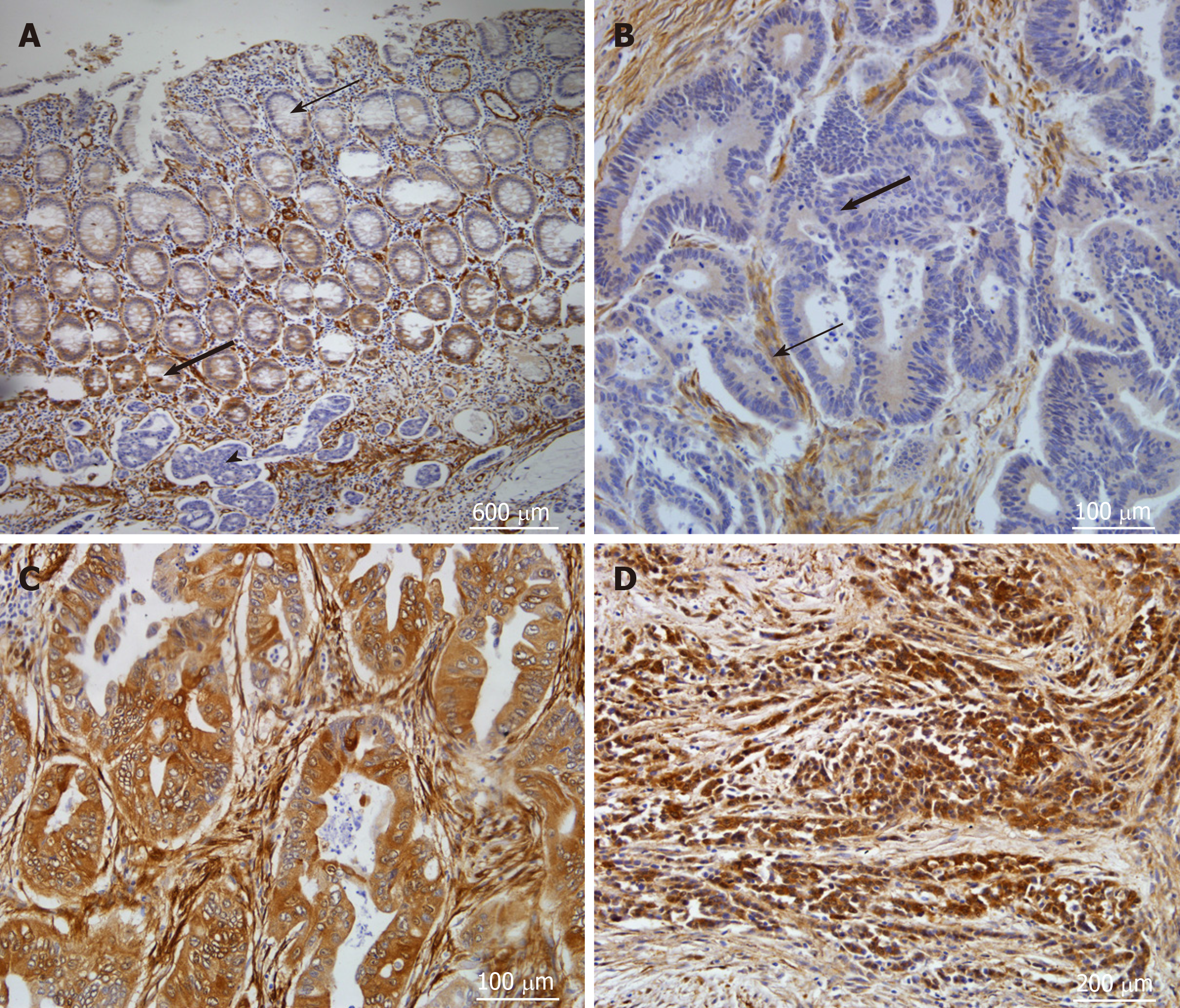
Immunohistochemical analyses for detection of CNN3 in uncultured colorectal cancer specimens were performed on 57 FFPE tissue sections derived from a series described in previous publications[21,22]. CNN3 expression was detected as cytoplasmic expression in smooth muscle and endothelial cells, stromal fibroblasts, and germinal centers of lymphoid follicles. Normal colonic mucosa showed positive cytoplasmic staining at the base of the crypt, which faded away until it was undetectable superficially (Figure 4A). CNN3 showed positive expression in 20/57 (35%) of tumors (Figure 4B-D), and it was not related to p53 stabilization in uncultured tumors consistent with the cell line findings. Furthermore, CNN3 did not show a significant relationship to any of the patient characteristics, pathological features, or other molecular characteristics in this tumor series (Table 1).
Here, we set out to obtain insights into the role of CNN3 in cancer development and metastasis. We showed that CNN3 is highly expressed in colon, breast, and cervical cancer cell lines, suggesting that it has widespread action in carcinomas of various lineages. This is consistent with some available literature data showing that CNN3 is expressed or altered in human colon, breast, and gastric cancers, mucosa-associated lymphoid tissue lymphoma, and murine ovarian carcinoma[17-20]. We extended our analysis in silico. The Cancer Cell Line Encyclopedia (https://portals.broadinstitute. org/ccle) showed that CNN3 mRNA was highly expressed in the majority of cell lines derived from various lineages in this database with the exception of hematological malignancies (lymphomas and leukemia). The highest expression of CNN3 was observed in osteosarcoma and glioma followed by various types of epithelial carcinomas.
We hypothesized that CNN3 plays a role in the EMT during invasion and lymph node metastasis based on the well-established biological role of CNN3 in wound healing and the finding that it was differentially and selectively expressed in a colon cancer cell line from lymph node metastasis compared to its matching line from a primary colon adenocarcinoma. We analyzed a good panel of EMT markers in the SW480/SW620 metastatic model. As expected, the metastatic cell line, SW620, showed evidence of mesenchymal transition compared to its non-metastatic version, SW480. Moreover, CNN3 silencing downregulated the majority of these mesenchymal markers suggesting that it is an important mediator of mesenchymal transition in carcinomas and is required to maintain this phenotype. The data from one of our markers, claudin-1, was apparently opposite to the general assumption that it is an epithelial phenotype marker. Originally, low claudin-1 levels were found in highly invasive breast cancers associated with high levels of Snail and Slug. Claudin-1 levels were efficiently downregulated by these transcription factors[24]. However, we found that claudin-1 was selectively upregulated in the metastatic colon cancer cells with concurrent upregulation of Slug, but it was not affected by the status of CNN3. It is becoming more acceptable that alterations in claudin-1 expression are lineage-dependent and may be regulated in different tissues in both normal and pathological situations under different conditions[25]. Claudin-1 expression and distribution were found to be associated with cell dissociation rather than cell adhesiveness in pancreatic cancer, and its expression was somewhat increased during metastasis of many cancer cells (reviewed in Jiang et al[25]). Colorectal cancers showed an increase in claudin-1 expression, in both primary and metastatic samples and in the cell lines derived from primary and metastatic tumors compared to their normal counterparts[26]; these results supported our findings.
The current data concerning CNN3 add significant molecules to the complex network that regulates EMT, particularly in colorectal metastasis. Our observation and that of many others suggests that EMT is a dominant phenotype in colon cancer and is controlled by a complex intricate network which includes also microRNA and epigenetic alterations[14-16]. Recently, miR-1 was shown to inhibit gastric and breast cancer growth and metastasis by targeting six genes that control cell cycle and EMT with CNN3 being one of them[27]. In addition to the role of EMT in cancer invasion and metastasis, many EMT markers also regulate tumor cell response to therapy[12,13]. In agreement with this, our data showed that CNN3 silencing increased the sensitivity of the metastatic cancer cell to standard therapy and provides a finding that could be exploited in targeted/personalized therapy. To date, we could not find detailed dedicated reports concerning CNN3 and cancer therapy, but we noted that the CNN3 gene name was mentioned in the middle of a list of 13 genes that were differentially expressed in the colon cancer cell line, HCT15, after all-trans retinoic acid treatment[28]. As suggested above, colorectal cancer is a heterogeneous disease with poor prognosis in some geographical locations or ethnicities in which it is characterized by advanced stages at diagnosis and poor outcomes as compared to the Western version of the disease[9]. Identification and exploitation of these molecules, such as CNN3 that play a role in later stages of this disease, could be useful in treatment of these aggressive tumor subsets.
We performed a comprehensive proteomic analysis of our chosen cell line models using a phosphokinase array. Silencing of CNN3 downregulated few highly important oncogenic proteins, namely ERK1/2, β-Catenin, mutant p53, c-Jun, and HSP60. β-Catenin downregulation was also noted on a western blot. β-Catenin, in particular, is a very significant finding as β-Catenin activation is the most common early change in the majority of colorectal carcinomas, suggesting that it has a major role in colorectal cancer development, together with its role in stemness[9]. The ERK1/2 finding is also interesting because it confirms the previous finding in which ERK1/2 is constitutively associated with CNN3[5]. The mutant p53 protein in SW480/SW620 is known to have a gain-of-function, oncogenic mutation that increases cell proliferation and resistance to DNA damage treatment; hence, its upregulation by CNN3 is expected[29].
Finally, we showed that CNN3 was overexpressed in a significant fraction of resected colon cancer cases, and it seems that it is an independent marker, but this requires further examination in a bigger series. Interestingly, CNN3 showed an expression gradient in the normal colonic mucosa with higher expression in the crypt bases, which is in agreement with its role in stemness and proliferation that is consistent with the status of β-Catenin activation at the base of the crypt. We interrogated the ProgGeneV2 database for the effect of CNN3 on survival and found that it was associated with low overall survival in 7/12 cohorts (one cohort was excluded because of ambiguity) and low relapse free survival in 4/7 cohorts. We found that CNN3 was associated with a decrease in metastasis-free survival in the single informative cohort (one cohort was excluded because of ambiguity).
In conclusion, we showed that CNN3 is widely expressed in many cancer types, and it is particularly associated with the lymph node metastasis in colon cancer cells. CNN3 appears to be an EMT trigger and exerts significant effects on tumor invasion and response to therapy in colorectal cancer cells.
Cancer is a major health problem and colorectal cancer, in particular, is a devastating disease with high mortality rates mainly because of metastasis. Hence there is a need to understand the molecular basis of invasion and metastasis of colorectal cancer cells and identify novel biomarkers of this process.
Calponin 3 (CNN3) is an actin-binding protein expressed in smooth muscle and non-smooth muscle cells. It is required for cytoskeletal rearrangement and wound healing. The cancer cells, which undergo the complex process of metastasis, simply trick the body into activating the biological wound healing program to support their invasion. Thus, CNN3 could play a role in cancer cell invasion and metastasis.
The objectives of this study were to find out the expression status of CNN3 in carcinomas and dissect the potential role of CNN3 in carcinogenesis with a focus on its role in colorectal cancer invasion and response to therapy.
We initially examined CNN3 expression in colon and breast cancer cell lines as well as formalin-fixed, paraffin-embedded sections from archived sporadic colorectal carcinomas, by western blot and immunohistochemistry, respectively. The CNN3 gene was silenced by specific siRNA, in metastatic colon cancer cell line and we confirmed the silencing efficiency by western blot. We then analyzed the silenced cells, along with control siRNA-transfected cells, for changes in epithelial and mesenchymal markers, invasion, and response to 5-fluoruracil treatment. We also performed a proteomic analysis using phospho-kinase array-based panel of 45 proteins.
CNN3 showed positive expression in 6/8 breast and 9/11 colon cancer lines and in HeLa. Interestingly, the colorectal adenocarcinoma line SW480 was negative, while the cell line developed from its matching lymph node metastasis (SW620) was positive for CNN3. The CNN3 expression was fairly consistent with the metastatic phenotype in colon cancer cells. We selected SW620 for further functional analyses. The CNN3-silenced SW620 showed a reduction in collagen invasion and loss of mesenchymal markers. CNN3 silencing caused an increase in the SW620 colon cancer cell sensitivity to 5-fluorouracil. Phospho-kinase array-based proteomic analysis revealed some potential pathways of CNN3 action. CNN3 was expressed in 20/57 (35%) colon cancer cases as shown by immunohistochemistry. CNN3 was associated with a decrease in overall survival in colon cancer in silico.
These results show the involvement of CNN3 in metastasis and resistance to chemotherapy in colon cancer and suggest that significant oncogenic pathways are involved in these CNN3 actions.
These data warrant further research into the role of CNN3 in colorectal carcinogenesis. The CNN3 protein interaction can be explored by various biochemical and proteomic methods to identify the mechanism of CNN3 action. The expression of CNN3 in larger series of various carcinomas and the association with response to various therapies can be investigated with the purpose to explore the potential application of CNN3 as a biomarker to predict metastasis and response to therapy.
We thank Dr. Maha Saber-Ayad for advice of SRB analysis, Ms. Harini Kumar for technical assistance, and Dr. Entesar Dalah for reading the manuscript. We thank the University of Sharjah for supplying and maintaining the research facility: The Sharjah Institute for Medical Research and the Environment and Cancer Research Group.
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country of origin: United Arab Emirates
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chung YH, Perse M S-Editor: Yan JP L-Editor: Filipodia E-Editor: Qi LL
| 1. | Rozenblum GT, Gimona M. Calponins: adaptable modular regulators of the actin cytoskeleton. Int J Biochem Cell Biol. 2008;40:1990-1995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 2. | Wu KC, Jin JP. Calponin in non-muscle cells. Cell Biochem Biophys. 2008;52:139-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 3. | Jin JP, Wu D, Gao J, Nigam R, Kwong S. Expression and purification of the h1 and h2 isoforms of calponin. Protein Expr Purif. 2003;31:231-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Applegate D, Feng W, Green RS, Taubman MB. Cloning and expression of a novel acidic calponin isoform from rat aortic vascular smooth muscle. J Biol Chem. 1994;269:10683-10690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 82] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 5. | Appel S, Allen PG, Vetterkind S, Jin JP, Morgan KG. h3/Acidic calponin: an actin-binding protein that controls extracellular signal-regulated kinase 1/2 activity in nonmuscle cells. Mol Biol Cell. 2010;21:1409-1422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Daimon E, Shibukawa Y, Wada Y. Calponin 3 regulates stress fiber formation in dermal fibroblasts during wound healing. Arch Dermatol Res. 2013;305:571-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Hirata H, Ku WC, Yip AK, Ursekar CP, Kawauchi K, Roy A, Guo AK, Vedula SR, Harada I, Chiam KH, Ishihama Y, Lim CT, Sawada Y, Sokabe M. MEKK1-dependent phosphorylation of calponin-3 tunes cell contractility. J Cell Sci. 2016;129:3574-3582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55814] [Article Influence: 7973.4] [Reference Citation Analysis (132)] |
| 9. | Abdel-Rahman WM, Faris ME, Peltomaki P. Molecular Determinants of Colon Cancer Susceptibility in the East and West. Curr Mol Med. 2017;17:34-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 10. | Dvorak HF. Tumors: wounds that do not heal-redux. Cancer Immunol Res. 2015;3:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 430] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 11. | Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3033] [Cited by in RCA: 3098] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 12. | Cai Z, Cao Y, Luo Y, Hu H, Ling H. Signalling mechanism(s) of epithelial-mesenchymal transition and cancer stem cells in tumour therapeutic resistance. Clin Chim Acta. 2018;483:156-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 13. | Boesch M, Spizzo G, Seeber A. Concise Review: Aggressive Colorectal Cancer: Role of Epithelial Cell Adhesion Molecule in Cancer Stem Cells and Epithelial-to-Mesenchymal Transition. Stem Cells Transl Med. 2018;7:495-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 14. | Abdel-Rahman WM, Al-Khayyal NA, Nair VA, Aravind SR, Saber-Ayad M. Role of AXL in invasion and drug resistance of colon and breast cancer cells and its association with p53 alterations. World J Gastroenterol. 2017;23:3440-3448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Alam F, Mezhal F, El Hasasna H, Nair VA, Aravind SR, Saber Ayad M, El-Serafi A, Abdel-Rahman WM. The role of p53-microRNA 200-Moesin axis in invasion and drug resistance of breast cancer cells. Tumour Biol. 2017;39:1010428317714634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Loboda A, Nebozhyn MV, Watters JW, Buser CA, Shaw PM, Huang PS, Van't Veer L, Tollenaar RA, Jackson DB, Agrawal D, Dai H, Yeatman TJ. EMT is the dominant program in human colon cancer. BMC Med Genomics. 2011;4:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 228] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 17. | Urzúa U, Roby KF, Gangi LM, Cherry JM, Powell JI, Munroe DJ. Transcriptomic analysis of an in vitro murine model of ovarian carcinoma: functional similarity to the human disease and identification of prospective tumoral markers and targets. J Cell Physiol. 2006;206:594-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Vinatzer U, Gollinger M, Müllauer L, Raderer M, Chott A, Streubel B. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 2008;14:6426-6431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 19. | Worthington J, Bertani M, Chan HL, Gerrits B, Timms JF. Transcriptional profiling of ErbB signalling in mammary luminal epithelial cells--interplay of ErbB and IGF1 signalling through IGFBP3 regulation. BMC Cancer. 2010;10:490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 20. | Nakarai C, Osawa K, Akiyama M, Matsubara N, Ikeuchi H, Yamano T, Hirota S, Tomita N, Usami M, Kido Y. Expression of AKR1C3 and CNN3 as markers for detection of lymph node metastases in colorectal cancer. Clin Exp Med. 2015;15:333-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 21. | Nieminen TT, Shoman S, Eissa S, Peltomäki P, Abdel-Rahman WM. Distinct genetic and epigenetic signatures of colorectal cancers according to ethnic origin. Cancer Epidemiol Biomarkers Prev. 2012;21:202-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Abdel-Rahman WM, Nieminen TT, Shoman S, Eissa S, Peltomaki P. Loss of p15INK⁴b expression in colorectal cancer is linked to ethnic origin. Asian Pac J Cancer Prev. 2014;15:2083-2087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Abdel-Rahman WM, Ruosaari S, Knuutila S, Peltomäki P. Differential roles of EPS8 in carcinogenesis: loss of protein expression in a subset of colorectal carcinoma and adenoma. World J Gastroenterol. 2012;18:3896-3903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Martínez-Estrada OM, Cullerés A, Soriano FX, Peinado H, Bolós V, Martínez FO, Reina M, Cano A, Fabre M, Vilaró S. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J. 2006;394:449-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 231] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 25. | Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, Thompson SK, Zollo M, Spano D, Dhawan P, Sliva D, Subbarayan PR, Sarkar M, Honoki K, Fujii H, Georgakilas AG, Amedei A, Niccolai E, Amin A, Ashraf SS, Ye L, Helferich WG, Yang X, Boosani CS, Guha G, Ciriolo MR, Aquilano K, Chen S, Azmi AS, Keith WN, Bilsland A, Bhakta D, Halicka D, Nowsheen S, Pantano F, Santini D. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol. 2015;35 Suppl:S244-S275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 353] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 26. | Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, Neff J, Washington MK, Beauchamp RD. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765-1776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 438] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 27. | Liu C, Zhang S, Wang Q, Zhang X. Tumor suppressor miR-1 inhibits tumor growth and metastasis by simultaneously targeting multiple genes. Oncotarget. 2017;8:42043-42060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 28. | Zhao J, Wen G, Ding M, Pan JY, Yu ML, Zhao F, Weng XL, Du JL. Comparative proteomic analysis of colon cancer cell HCT-15 in response to all-trans retinoic acid treatment. Protein Pept Lett. 2012;19:1272-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Yan W, Zhang Y, Zhang J, Liu S, Cho SJ, Chen X. Mutant p53 protein is targeted by arsenic for degradation and plays a role in arsenic-mediated growth suppression. J Biol Chem. 2011;286:17478-17486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |