Copyright
©The Author(s) 2025.
World J Gastrointest Oncol. Apr 15, 2025; 17(4): 103048
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103048
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103048
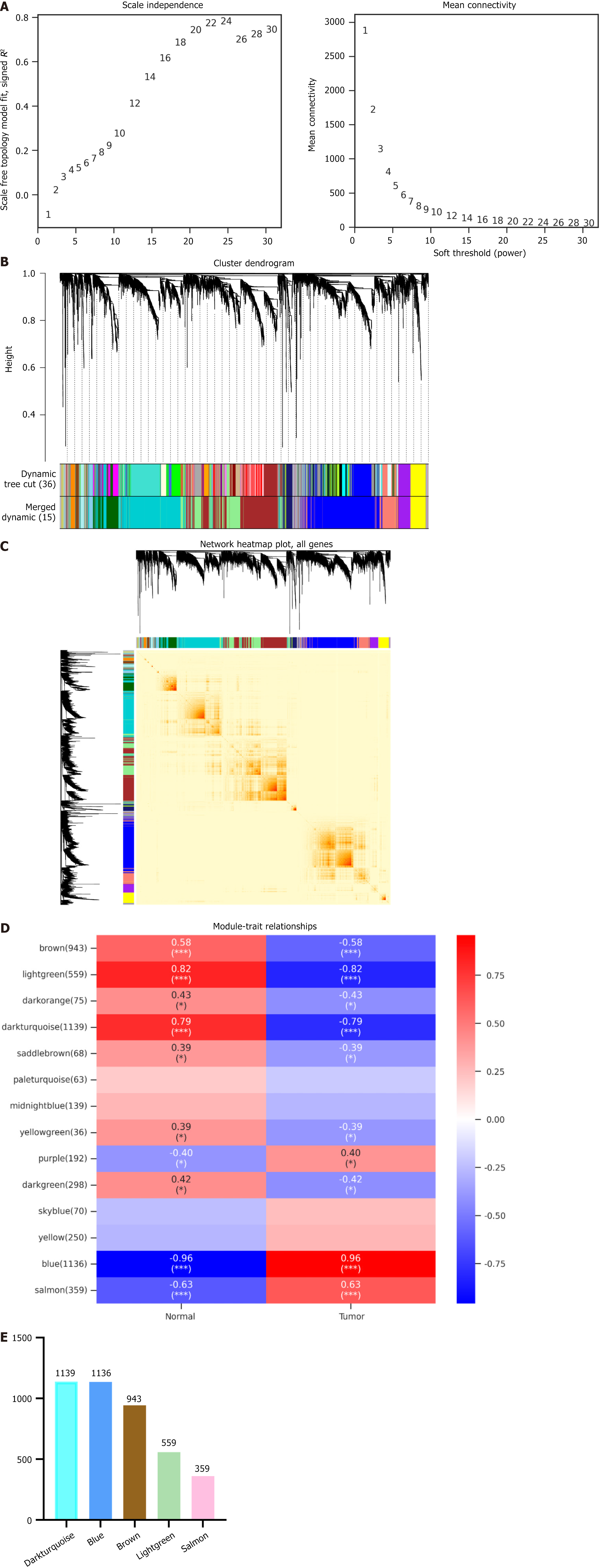
Figure 1 Construction of weighted gene co-expression network analysis.
A: Scale-free fitting index of the network topology obtained by the soft-threshold power analysis method; B: Cluster dendrogram of co-expression network modules from weighted gene co-expression network analysis depending on a dissimilarity measure; C: Genes from the network, using a heat map to describe the topological overlap matrix between genes in the analysis. On a linear scale, the depth of red is positively correlated with the correlation strength between the pair of modules; D: Module-trait relationships between the normal group and the tumor group; E: Number of genes in the first five modules.
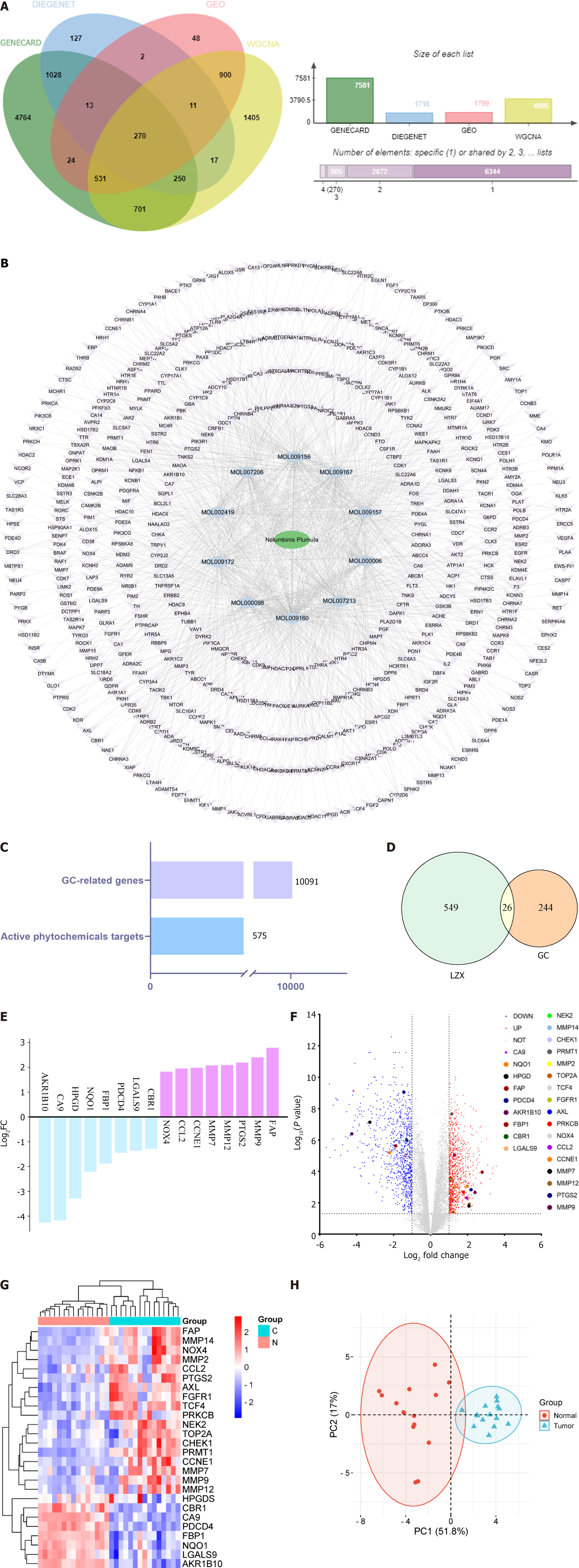
Figure 2 Targets of LZX active ingredients and differentially expressed genes in gastric cancer.
A: 279 intersection genes of GeneCards, GENENET, weighted gene co-expression network analysis and differential expressed genes (DEGs); B: 575 genes of LZX; C: All genes of gastric cancer and the number of targets of LZX active ingredients; D: 26 intersection genes of gastric cancer and LZX; E: Log2 fold change values of the top 10 up- and down-regulated genes in the intersection targets, where blue and purple represent up- and down-regulated genes, respectively; F: Volcano plot of DEGs in GSE118916; G: Heatmap of DEGs in GSE118916: Comparing the normal group (N) with the tumor group (C); H: Principal co-ordinates analysis plot of the samples of the intersecting targets. Red dots represent normal group samples and green dots represent tumor group samples. LZX: Lotus plumule; GEO: Gene Expression Omnibus; WGCNA: Weighted gene co-expression network analysis; GC: Gastric cancer; PC: Principal component.
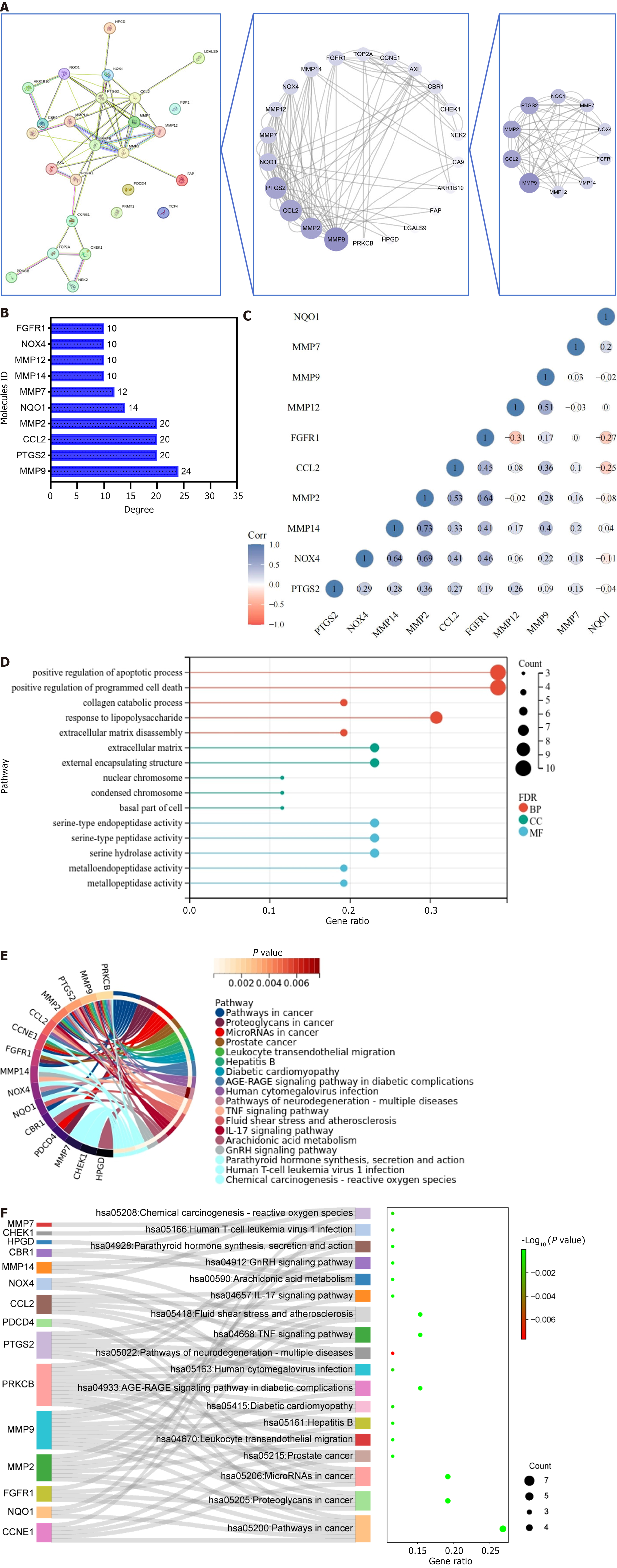
Figure 3 Screening and enrichment analysis of core targets.
A: Topology screening process of protein-protein interaction network. The larger the circle and the redder the color, the more important the target is in the network; B: Degree value of core targets; C: Correlation heatmap of the core targets. Blue represents positive correlation, red represents negative correlation, and darker color indicates stronger correlation; D: Gene ontology lollipop plot illustrating the intersecting targets. The colors red, blue, and green denote biological processes, cellular components, and molecular functions, respectively. The size of each circle corresponds to the number of enriched targets, with larger circles indicating a greater number of enriched targets; E: Kyoto encyclopedia of genes and genomes (KEGG) circle diagram of intersecting targets. The outermost left side of the circle represents the intersection target, the outermost right-side color represents the pathway, and the color of the innermost right side of the circle represents the P value of the enriched pathway. The lighter the color, the more significant the enriched pathway; F: Ranking of the front KEGG Sankey diagram. The left side represents the intersection target, the middle side represents the pathway, the curve indicates the correlation between the two, and the rightmost bubble graph represents the result of KEGG. The larger the circle and the redder the color, the more significant the enriched pathway. FDR: False discovery rate; BP: Biological processes; MF: Molecular functions; CC: Cellular components.
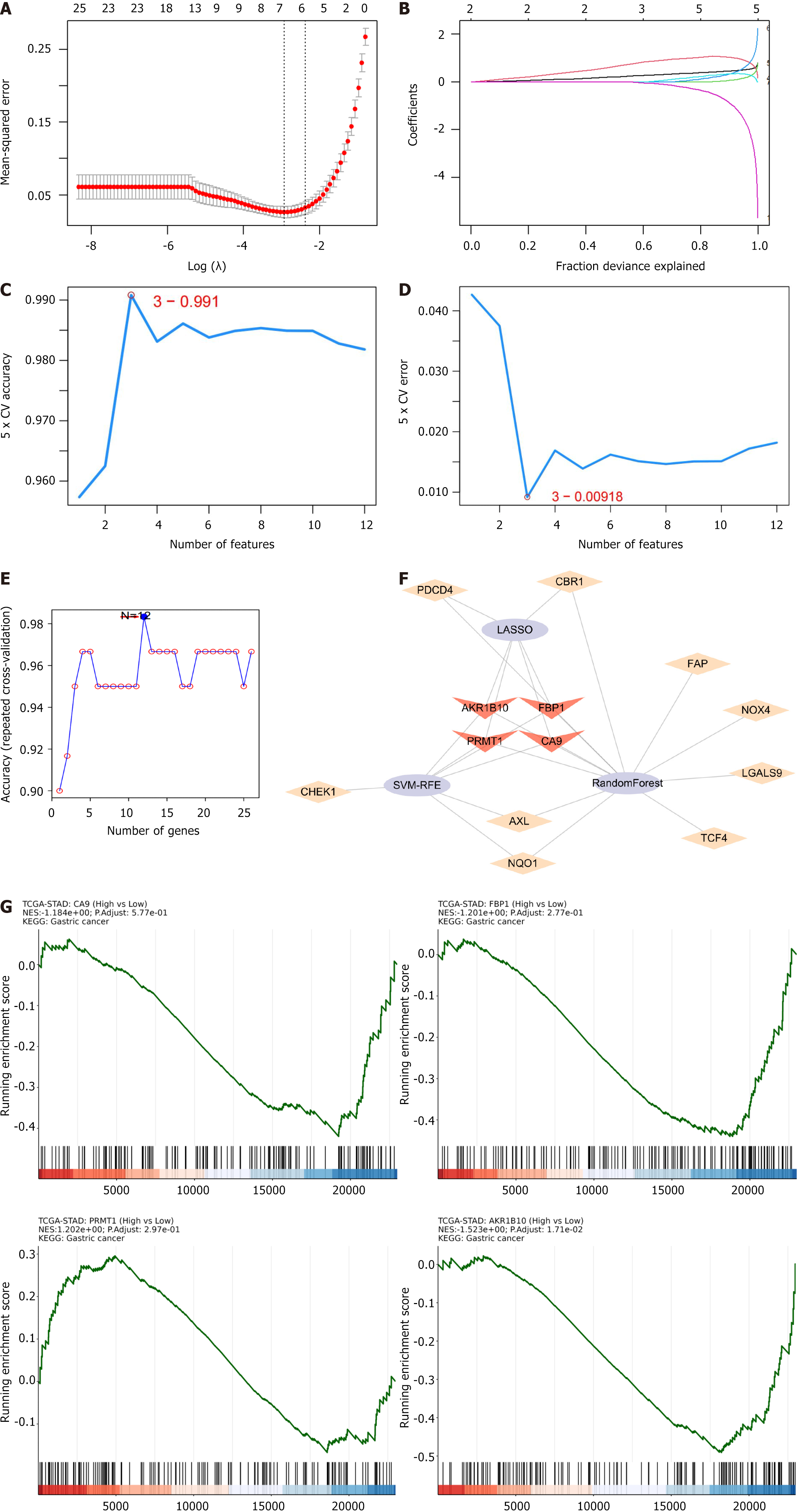
Figure 4 Application of machine learning for screening hub genes.
A and B: Six genes were identified through the LASSO regression algorithm; C and D: Support vector machines-recursive feature elimination algorithm to extract 3 genes; E: Random forest algorithm to select 12 genes; F: Venn network to intersect 4 gene subsets; G: Gene set enrichment analysis graph of hub genes.
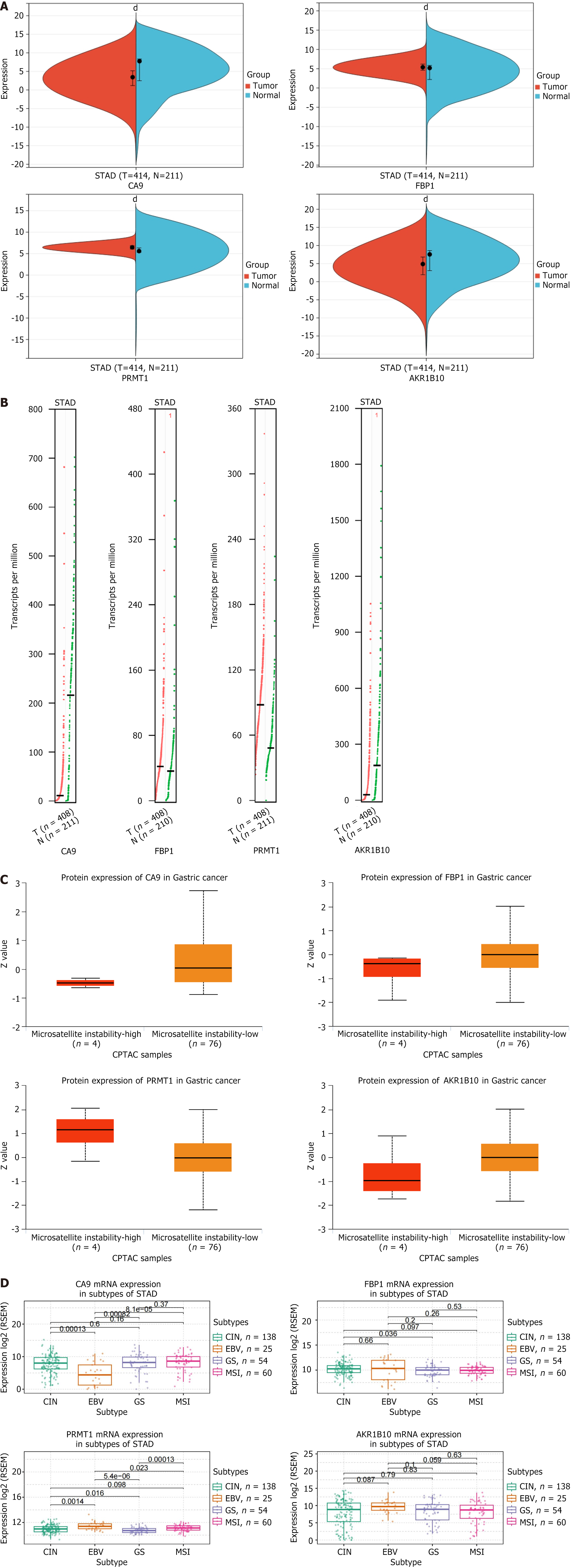
Figure 5 Clinical correlation analysis of hub genes.
A: Expression levels of hub gene mRNA. Red represents the tumor group and blue represents the normal group; B: Expression levels of hub gene copy number. Red represents the tumor group and green represents the normal group; C: Expression level of hub gene protein. Red represents the microsatellite instability group and orange represents the microsatellite stability group; D: Expression levels of hub genes in gastric cancer subtypes. dP < 0.0001. CA9: Carbonic anhydrase 9; FBP1: Fructose-bisphosphatase 1; PRMT1: Protein arginine methyltransferase 1; AKR1B10: Aldo-keto reductase family 1 member B10; CIN: Chromosomal instability; EBV: EB virus; GS: Genome stability; MSI: Microsatellite instability.
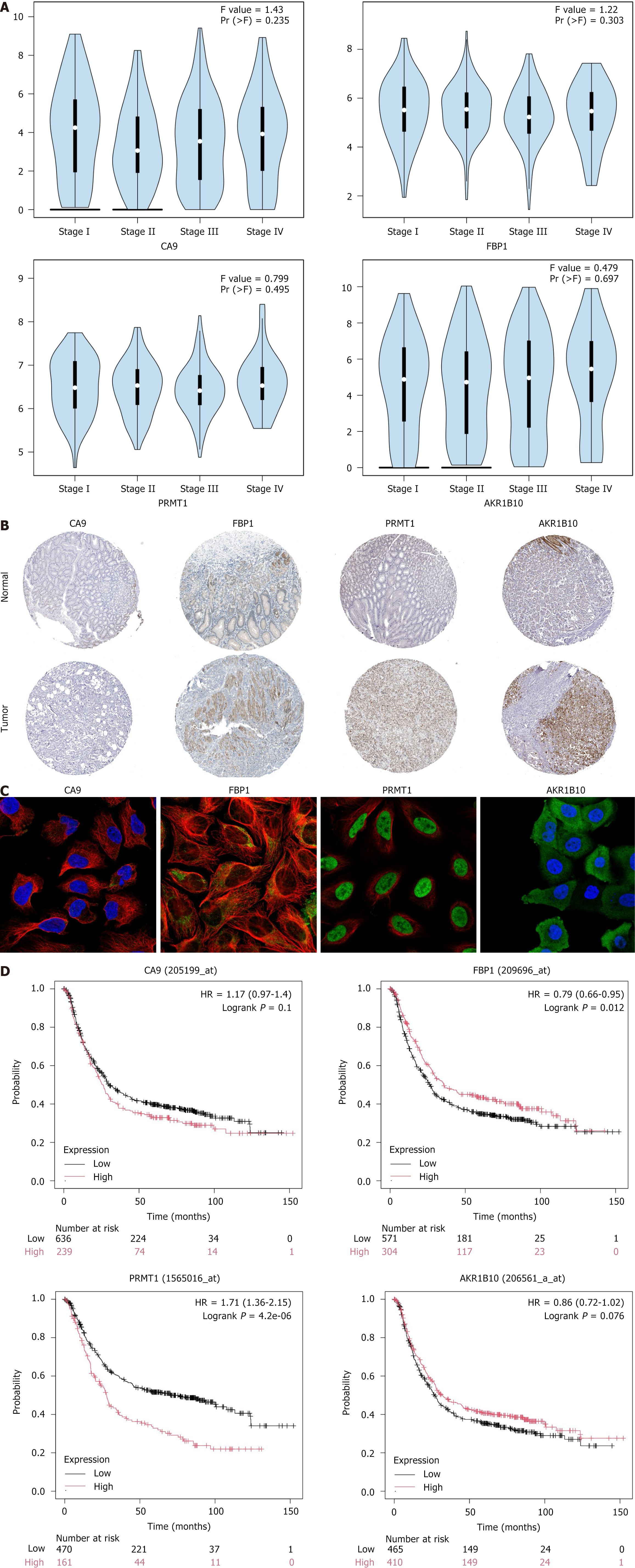
Figure 6 Expression and prognostic analysis of hub genes.
A. Correlation between hub genes and clinical stage of gastric cancer; B: Immuno
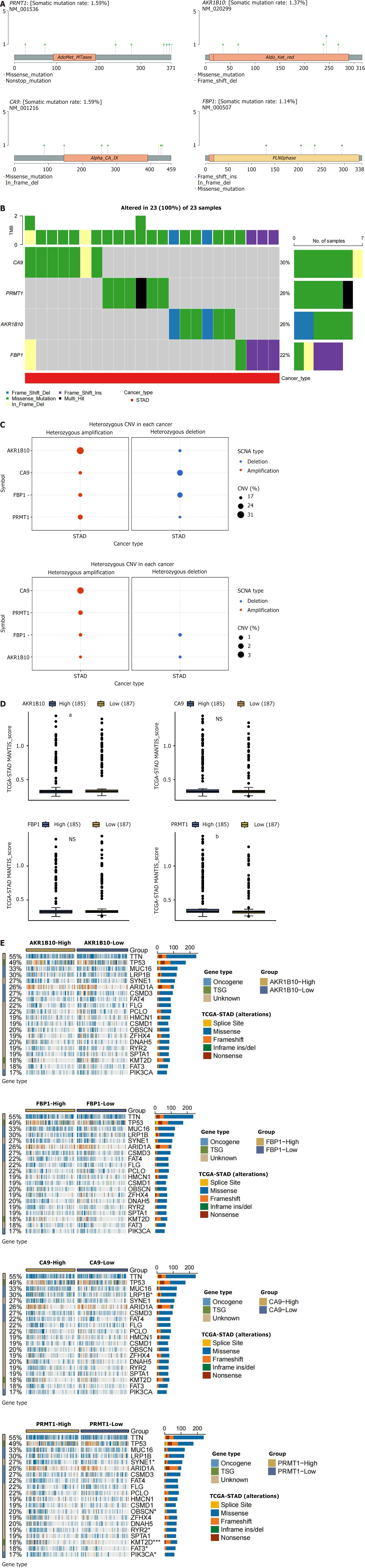
Figure 7 Effect of mutations in hub genes on gastric cancer.
A: Single nucleotide variation mutation sites and types of hub genes. The circle color represents the mutation type and the line length represents the mutation frequency; B: Waterfall plot of single nucleotide variation mutation frequency of hub genes. The upper bar indicates the proportion of hub gene mutations in the 23 samples; C: Bubble plots of heterozygous and pure heterozygous copy number variation mutations in the hub genes. The larger the bubble, the higher the proportion of mutations; D: Mutational associations of driver genes with the hub genes; E: Box plot of microsatellite instability expression levels of hub genes. CA9: Carbonic anhydrase 9; FBP1: Fructose-bisphosphatase 1; PRMT1: Protein arginine methyltransferase 1; AKR1B10: Aldo-keto reductase family 1 member B10; CNV: Copy number variation; SCNA: Somatic copy number alterations; TCGA: The Cancer Genome Atlas.
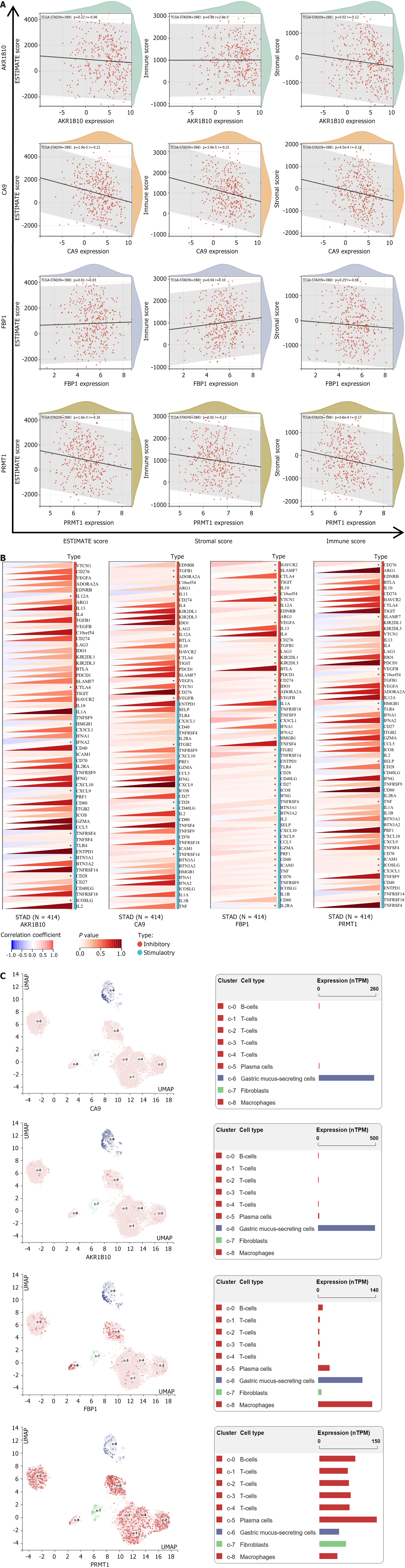
Figure 8 Relationship between hub genes and immune infiltration.
A: Scatter plot of correlation of StromalScore, ImmuneScore, and ESTIMATEScore with hub genes; B: Heatmap of correlation between hub genes and immune checkpoints. Blue color represents negative correlation and red color represents positive correlation; C: Single-cell sequencing and single-cell annotation map of hub genes. Different colors represent different sets of cells. CA9: Carbonic anhydrase 9; FBP1: Fructose-bisphosphatase 1; PRMT1: Protein arginine methyltransferase 1; AKR1B10: Aldo-keto reductase family 1 member B10.
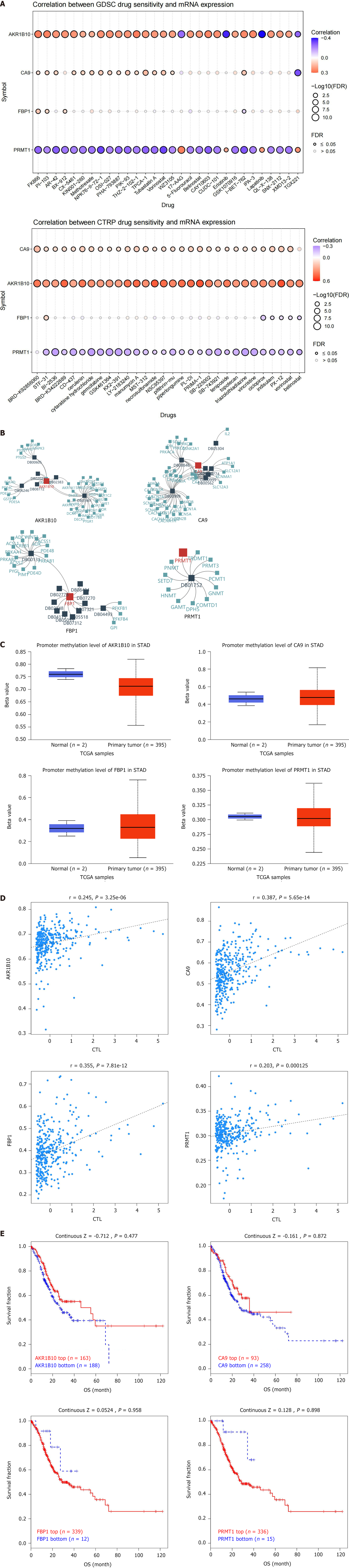
Figure 9 Hub genes are involved in epigenetic regulation and repair of damaged genes.
A: Correlation of hub genes with immunotherapy; B: Network diagram of hub genes and immunotherapy; C: Expression level map of hub gene methylation. Blue and red represent the normal and tumor groups, respectively; D: Correlation between hub gene methylation levels and cytotoxic T lymphocyte markers; E: Survival curves of hypermethylated and hypomethylated subgroups of hub genes. CTRP: Cancer Therapeutics Response Portal; CA9: Carbonic anhydrase 9; FBP1: Fructose-bisphosphatase 1; PRMT1: Protein arginine methyltransferase 1; AKR1B10: Aldo-keto reductase family 1 member B10; TCGA: The Cancer Genome Atlas; CTL: Cytotoxic T lymphocyte; OS: Overall survival.
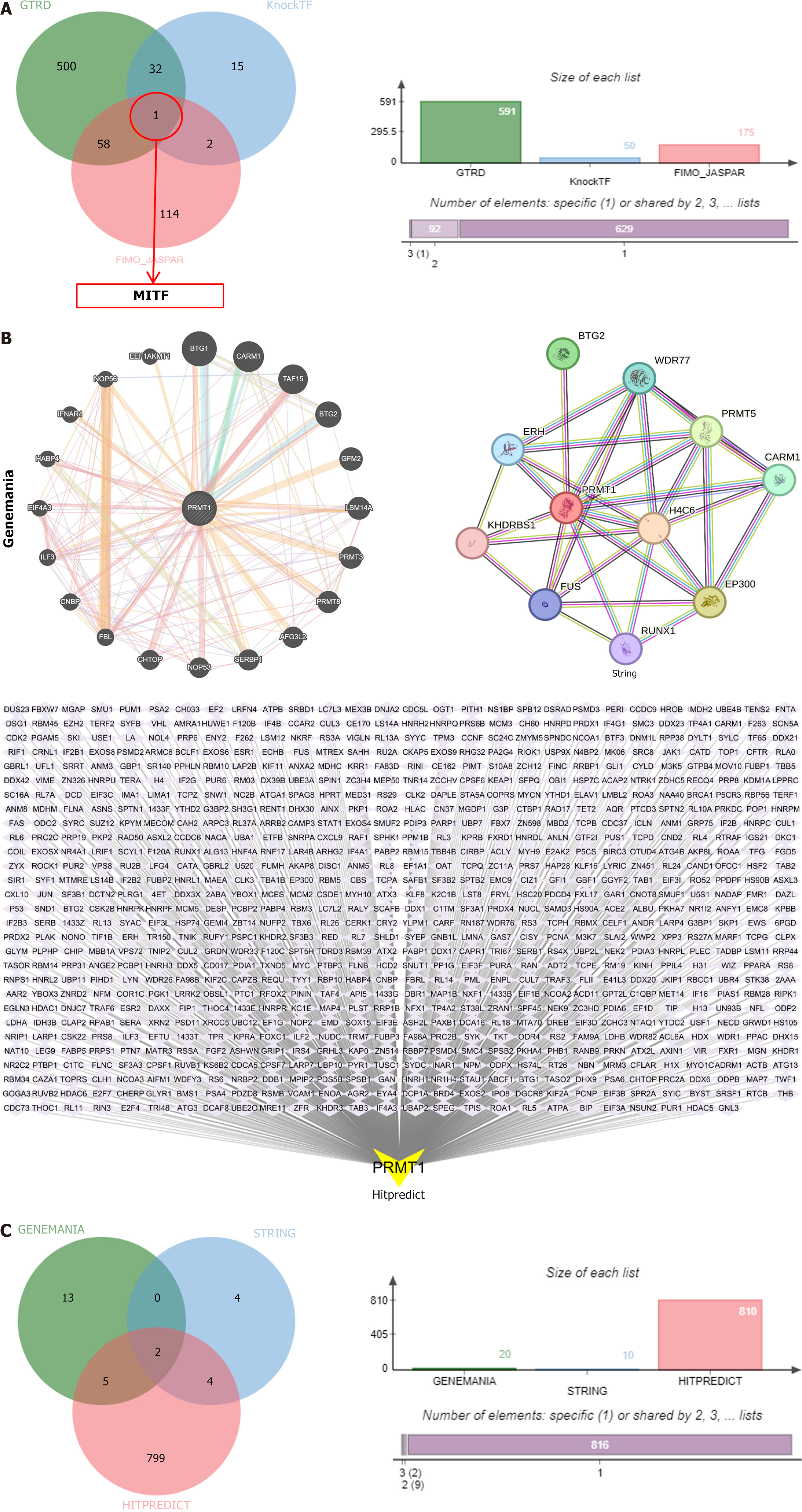
Figure 10 Single-cell sequencing and association of hub genes with antitumor drug therapy.
A. One intersection transcription factor of protein arginine methyltransferase 1 (PRMT1) from Gene Transcription Regulation Database, knockTF, and FIMO_JASPAR; B: Targets of PRMT1 from geneMANIA, STRING, and hitPredict; C: Two intersection targets of PRMT1 from geneMANIA, STRING, and hitPredict. MITF: Melanocyte inducing transcription factor.
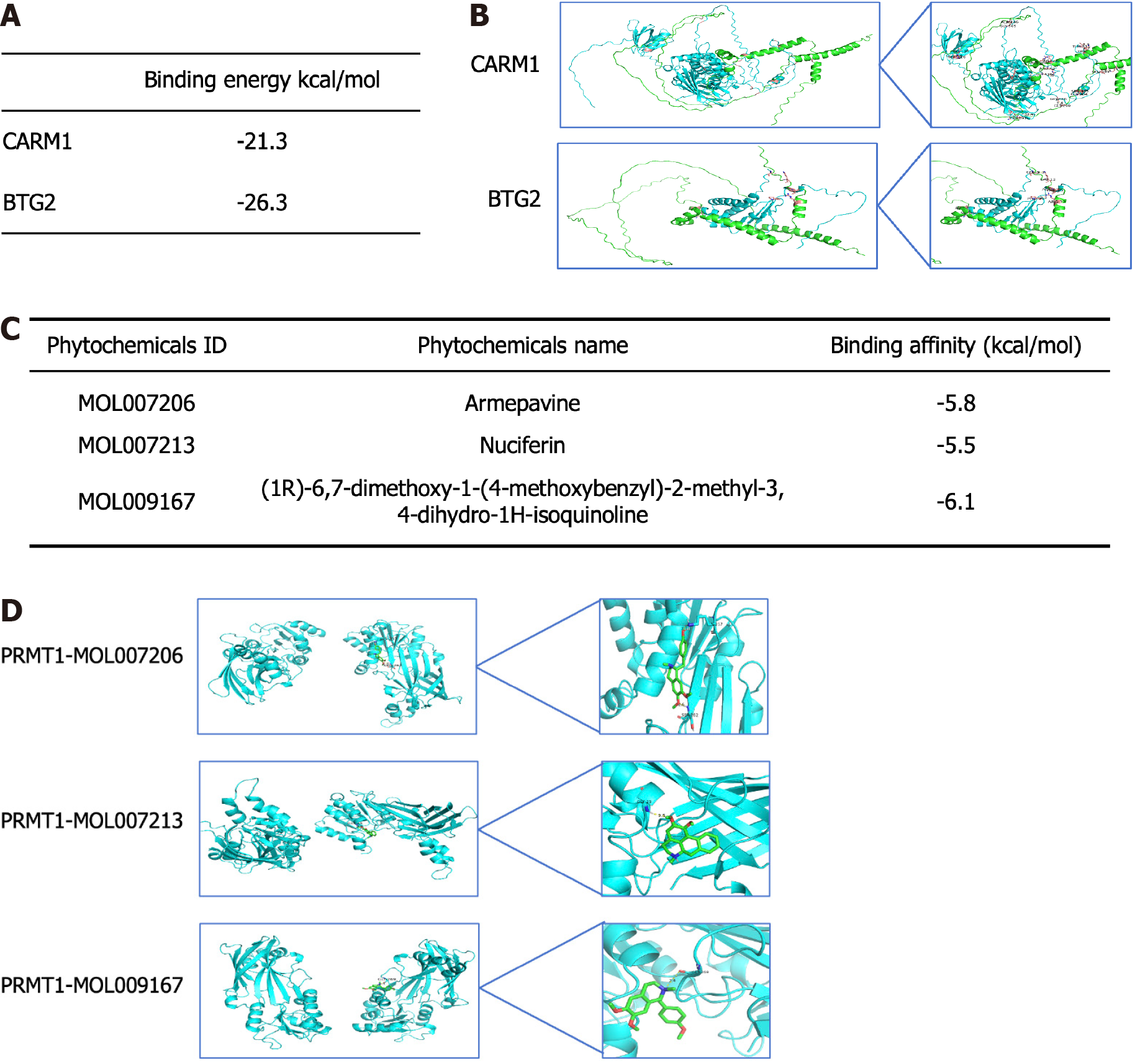
Figure 11 Molecular docking.
A: Protein-protein docking binding energy of protein arginine methyltransferase 1 and targets; B: Visual demonstration of protein-protein docking; C: Molecular docking binding energy of protein arginine methyltransferase 1 and core active ingredients; D: Visual demonstration of molecular docking. CARM1: Coactivator associated arginine methyltransferase 1; BTG2: B-cell translocation gene 2.
- Citation: Meng FD, Jia SM, Ma YB, Du YH, Liu WJ, Yang Y, Yuan L, Nan Y. Identification of key hub genes associated with anti-gastric cancer effects of lotus plumule based on machine learning algorithms. World J Gastrointest Oncol 2025; 17(4): 103048
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/103048.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.103048