Copyright
©The Author(s) 2022.
World J Gastrointest Oncol. Apr 15, 2022; 14(4): 935-946
Published online Apr 15, 2022. doi: 10.4251/wjgo.v14.i4.935
Published online Apr 15, 2022. doi: 10.4251/wjgo.v14.i4.935
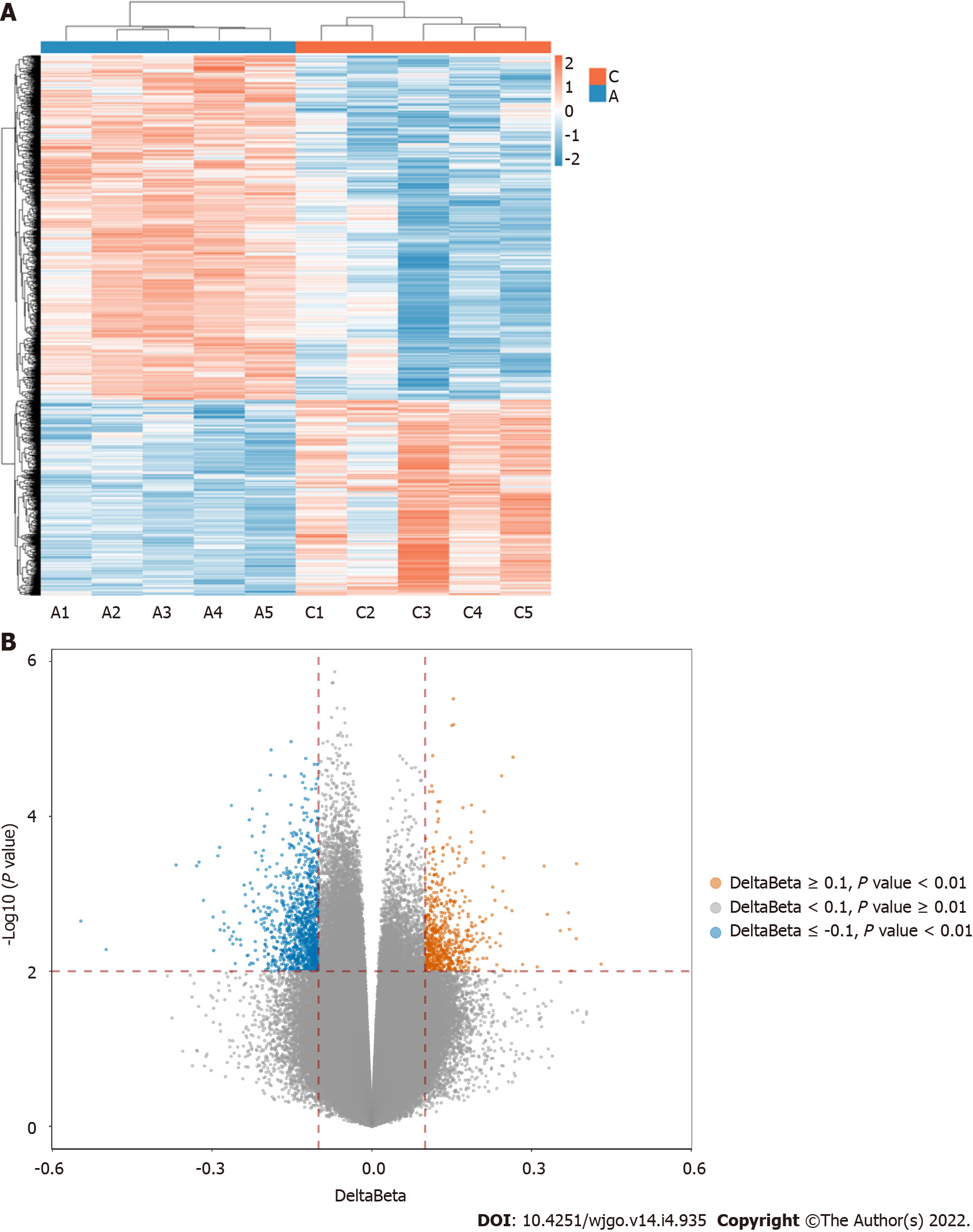
Figure 1 The heatmap and volcano of the differentially methylated sites.
A: Adenoma group; B: Delta β data. Ca: Cancer group.
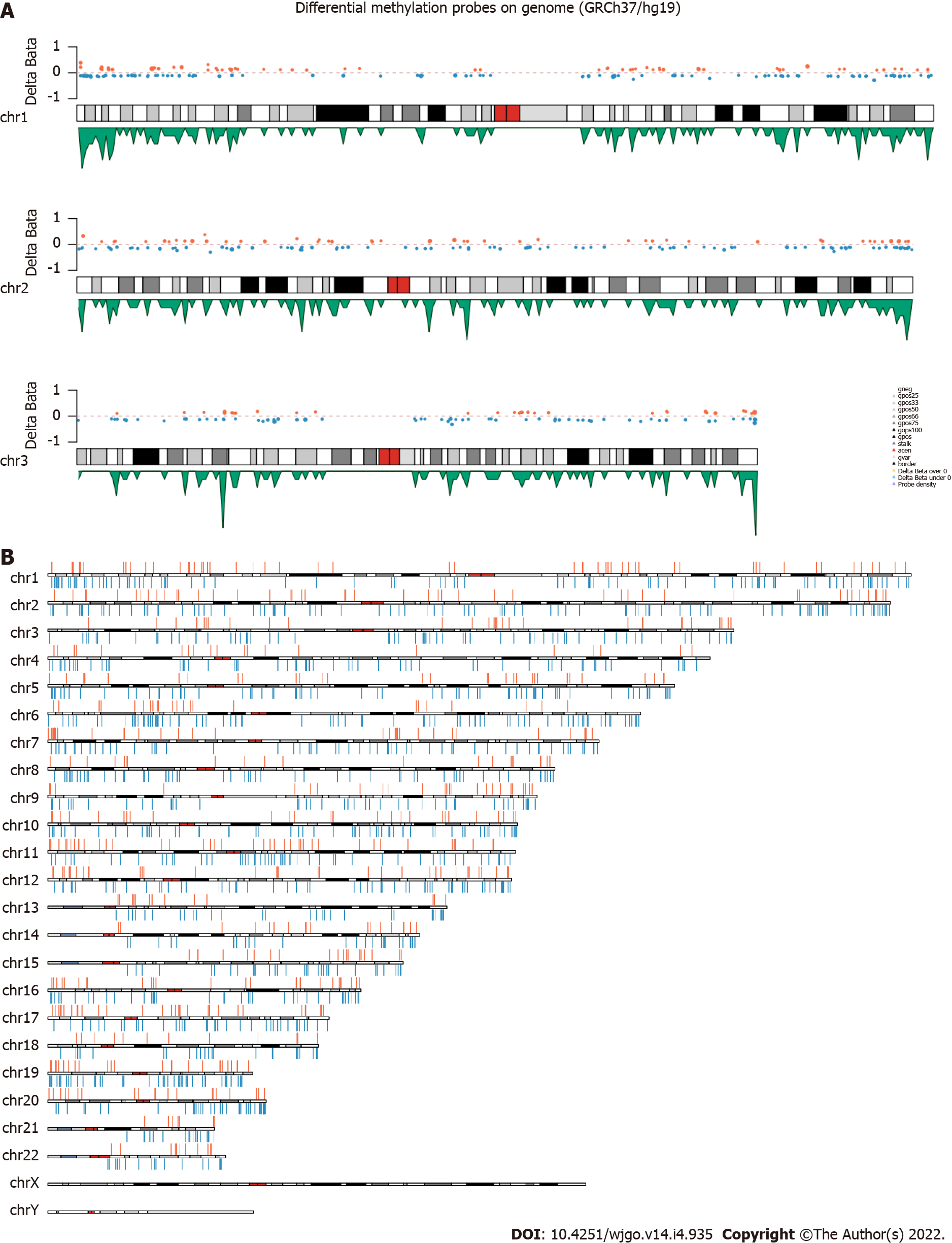
Figure 2 The distribution of differentially methylated sites.
A: Examples of the distribution of differentially methylated sites (DMSs) on chromosome 1, 2, and 3. The orange and blue dots represent the CpGs which are Δβ > 0 and Δβ < 0 respectively, the green peak represents the density of probes; B: Schematic map of the genomic DMSs, where red and blue represent the CpGs which are Δβ > 0 and Δβ < 0 respectively.
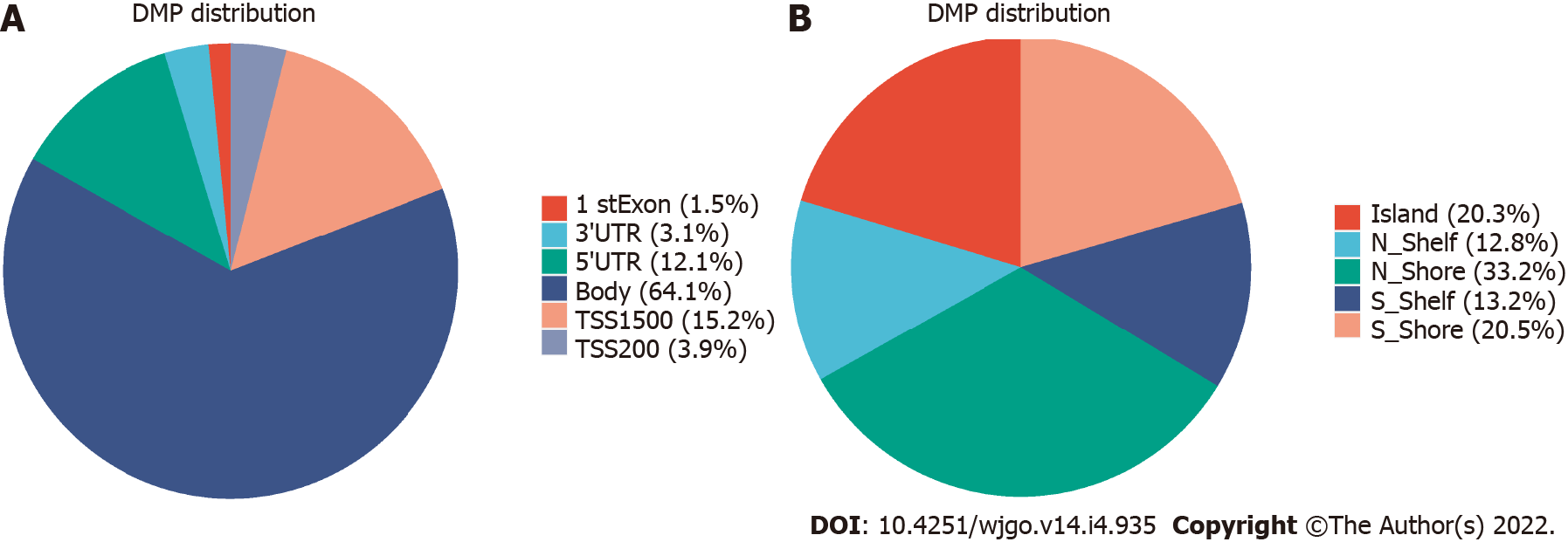
Figure 3 Distribution of differentially methylated sites.
A: Distribution of differentially methylated sites (DMSs) in gene structural elements; B: Distribution of DMSs in and around CpG island.
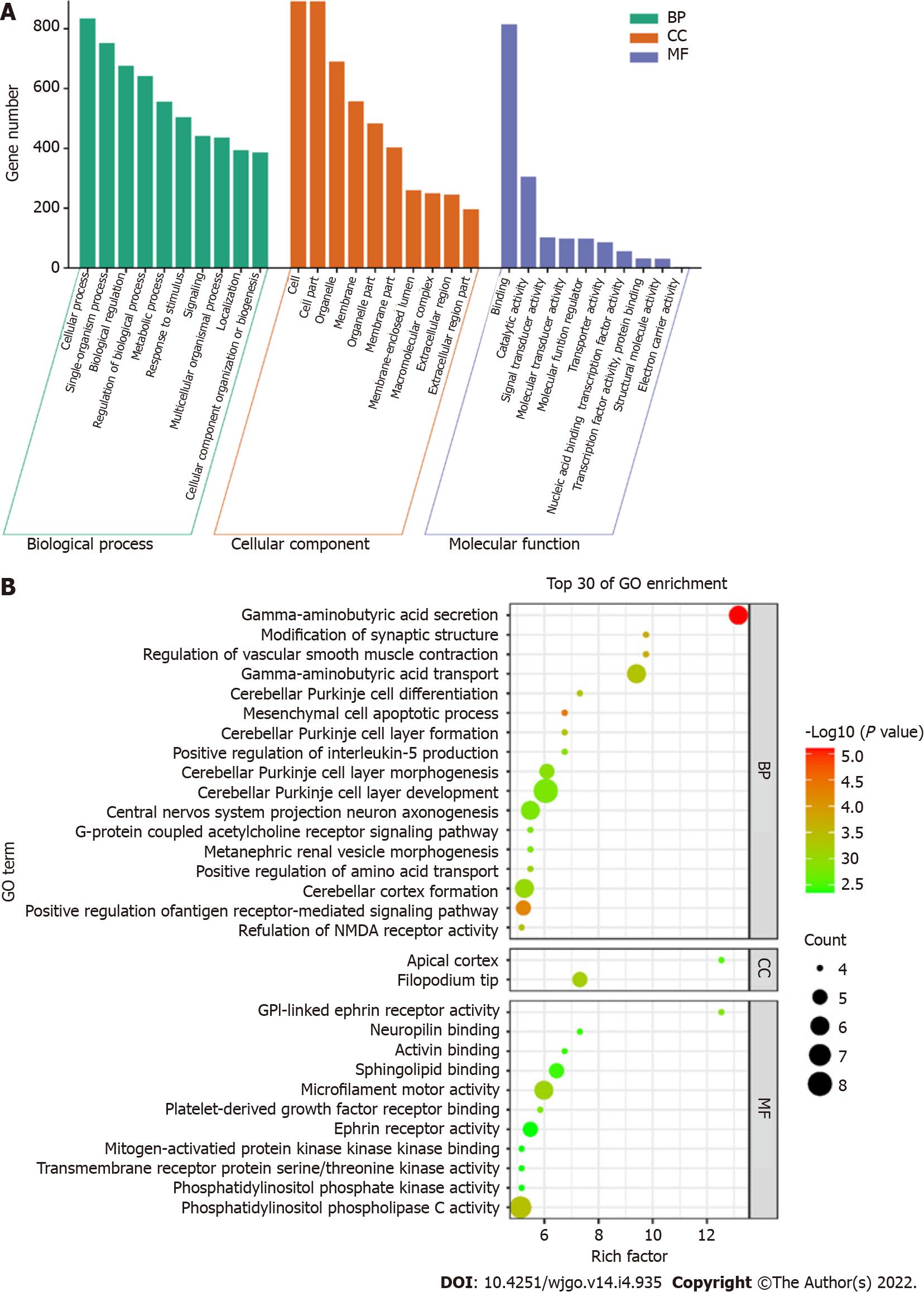
Figure 4 Gene ontology analysis.
A: Gene ontology (GO) function distribution of differentially methylated sites; B: Bubble diagram of GO enrichment results. GO: Gene ontology; BP: Biological process; CC: Cellular component; MF: Molecular function.
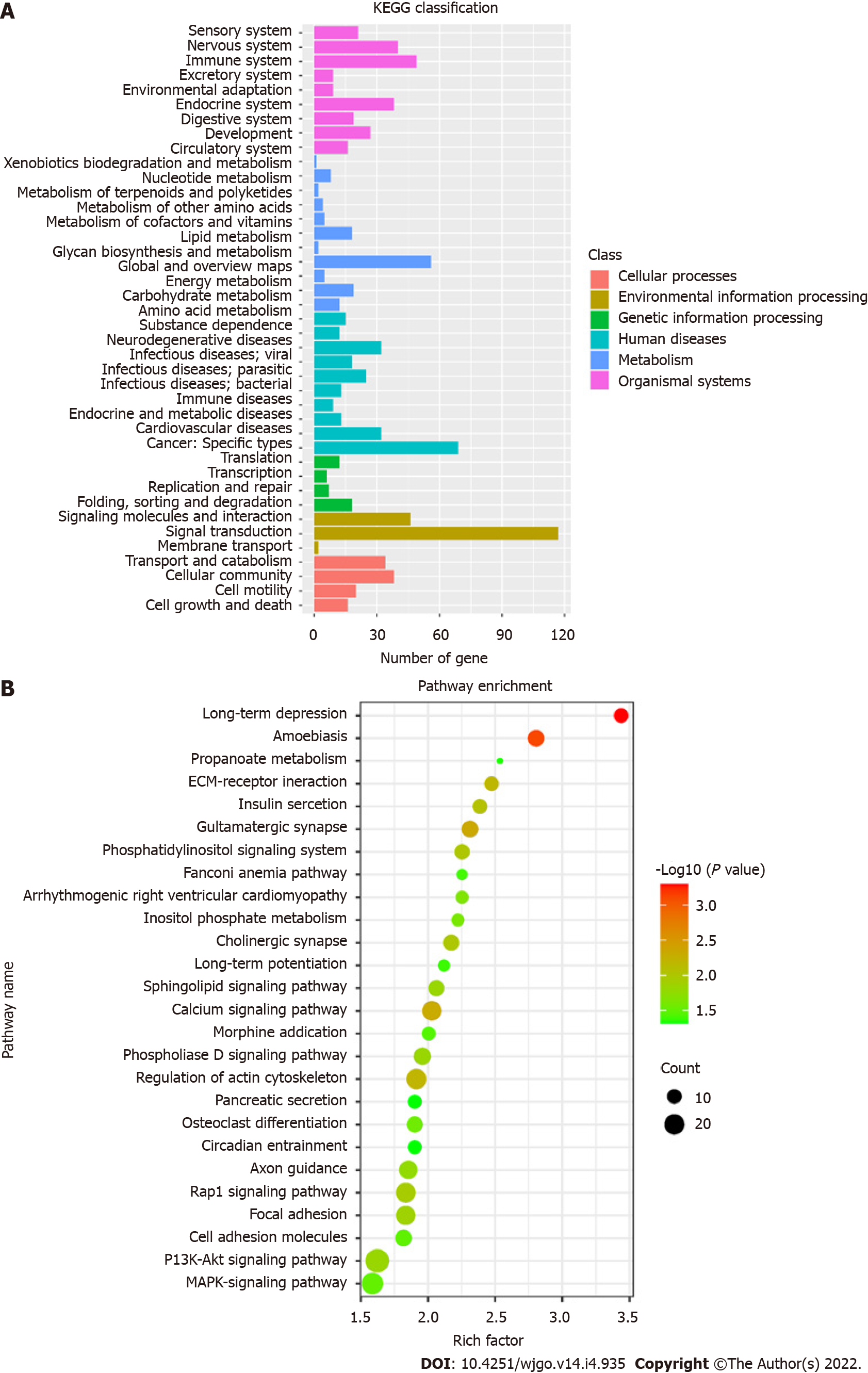
Figure 5 Kyoto Encyclopedia of Genes and Genomes analysis.
A: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification map; B: Bubble diagram of KEGG enrichment pathways. KEGG: Kyoto Encyclopedia of Genes and Genomes.
- Citation: Wu YL, Jiang T, Huang W, Wu XY, Zhang PJ, Tian YP. Genome-wide methylation profiling of early colorectal cancer using an Illumina Infinium Methylation EPIC BeadChip. World J Gastrointest Oncol 2022; 14(4): 935-946
- URL: https://www.wjgnet.com/1948-5204/full/v14/i4/935.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i4.935