Copyright
©The Author(s) 2019.
World J Gastrointest Oncol. Dec 15, 2019; 11(12): 1101-1114
Published online Dec 15, 2019. doi: 10.4251/wjgo.v11.i12.1101
Published online Dec 15, 2019. doi: 10.4251/wjgo.v11.i12.1101
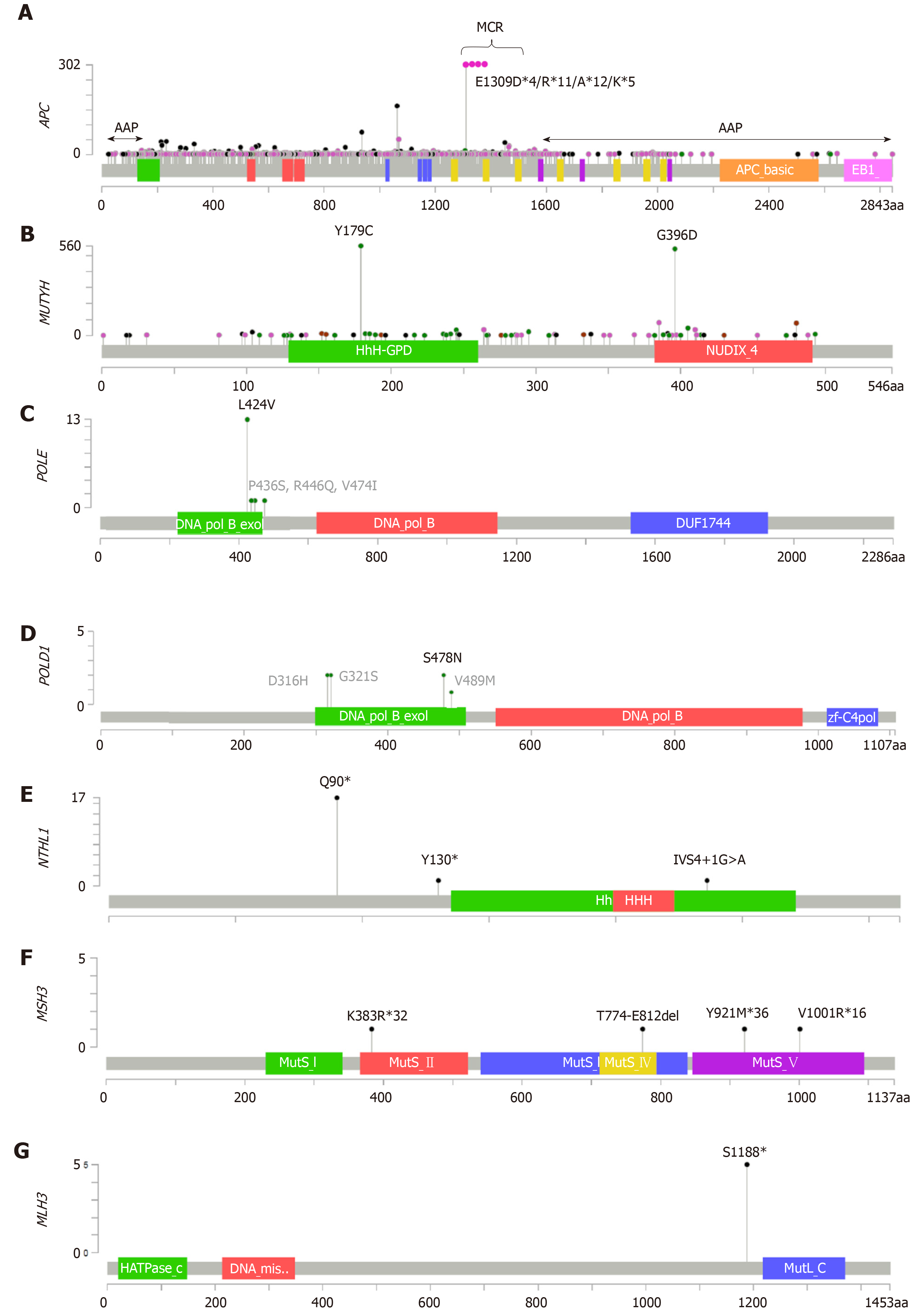
Figure 1 Distribution of germline mutations in attenuated adenomatous polyposis predisposition genes across protein domains.
A: APC likely pathogenic and pathogenic variants described in the LOVD database[115]. Most of the mutations are truncating variants. Mutations associated with AAP are located at both the 3’-end and 5’-end of the gene (indicated with arrows); B: MUTYH likely pathogenic and pathogenic mutations described in the LOVD database[115]. The two most prevalent mutations in Caucasians are shown; C-G: POLE, POLD1, NTHL1, MSH3 and MLH3 likely pathogenic and pathogenic mutations described in the literature and associated with AAP. Unclassified variants in the polymerase proofreading POLE and POLD1 domains are in gray. All lolliplots were designed with The cBio Cancer Genomics Portal[116,117]. Mutation types are coded as follows: black dots for nonsense variants; pink dots for frameshift and splicing variants; green dots for missense mutations; brown dots for in-frame indels. Reference sequences: APC: NM_000038, NP_000029; MUTYH: NM_001128425, NP_036354; POLE: NM_006231, NP_006222; POLD1: NM_001256849, NP_001121897; NTHL1: NM_002528, NP_002519; MSH3: NM_002439, NP_002430; MLH3: NM_001040108, NP_001035197.
- Citation: Lorca V, Garre P. Current status of the genetic susceptibility in attenuated adenomatous polyposis. World J Gastrointest Oncol 2019; 11(12): 1101-1114
- URL: https://www.wjgnet.com/1948-5204/full/v11/i12/1101.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v11.i12.1101