Published online Jun 26, 2024. doi: 10.4252/wjsc.v16.i6.656
Revised: January 23, 2024
Accepted: April 12, 2024
Published online: June 26, 2024
Processing time: 214 Days and 21.2 Hours
Validation of the reference gene (RG) stability during experimental analyses is essential for correct quantitative real-time polymerase chain reaction (RT-qPCR) data normalisation. Commonly, in an unreliable way, several studies use genes involved in essential cellular functions [glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 18S rRNA, and β-actin] without paying attention to whether they are suitable for such experimental conditions or the reason for choosing such genes. Furthermore, such studies use only one gene when Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines recom
To verify the most stable RG during osteogenic differentiation of human dental pulp stem cells (DPSCs) by RT-qPCR.
We cultivated DPSCs under two conditions: Undifferentiated and osteogenic dif
All of the data sets from clonogenic and osteogenic samples were analysed using the RefFinder algorithm. The final ranking showed RPLP0/TBP as the two most stable RGs and TUB/B2M as the two least stable RGs. Either the ΔCt method or NormFinder analysis showed TBP/RPLP0 as the two most stable genes. However, geNorm analysis showed RPLP0/EF1α in the first place. These algorithms’ two least stable RGs were B2M/GAPDH. For BestKeeper, ALAS1 was ranked as the most stable RG, and SDHA as the least stable RG. The pair RPLP0/TBP was detected in most subgroups as the most stable RGs, following the RefFinfer ranking.
For the first time, we show that RPLP0/TBP are the most stable RGs, whereas TUB/B2M are unstable RGs for long-term osteogenic differentiation of human DPSCs in traditional monolayers.
Core Tip: Detecting the best reference genes (RGs) under specific conditions is a good practice to improve the understanding of gene expression. Stem cells have been largely studied during commitment to particular cell lineages for many applications, such as tissue engineering. In this way, dental pulp stem cells (DPSCs) are promising for craniofacial reconstruction. For the first time, we show that the best pair of RGs for the osteogenic differentiation of human DPSCs are ribosomal protein, large, P0/TATA-binding protein by quantitative real-time polymerase chain reaction through the four algorithms (ΔCt comparative method, geNorm, BestKeeper, and NormFinder) and ranked by RefFinder.
- Citation: Ferreira DB, Gasparoni LM, Bronzeri CF, Paiva KBS. RPLP0/TBP are the most stable reference genes for human dental pulp stem cells under osteogenic differentiation. World J Stem Cells 2024; 16(6): 656-669
- URL: https://www.wjgnet.com/1948-0210/full/v16/i6/656.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v16.i6.656
Quantitative real-time polymerase chain reaction (RT-qPCR) is a powerful messenger RNA expression analysis te
A systematic review highlighted that between 2010 and 2015, the most used RGs were actin beta (ACTB) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), even though only 15% of the papers have shown evidence that they have checked the stability condition of the genes used[2]. Currently, these genes are still the most used, and in particular, GAPDH has established itself in many fields as the standard gene used for normalisation. It is essential to highlight that there is no absolute RG, and it is always necessary to test the stability of these genes among all tested conditions to choose the gene with the best normalising potential. The stability can be evaluated by algorithms named ΔCt comparative method[3], geNorm[4], NormFinder[5], and BestKeeper[6]. These algorithms are accessible through the web-based RefFinder, an initiative that calculates these four algorithms and has its ranking method[7].
Stem cells from many sources have been studied extensively for regenerative medicine approaches. In tissue bone engineering, it is suitable for stem cells to be committed to the osteoblastic lineage to create a new bone. Among them, mesenchymal stem cells (MSCs) are largely investigated because they can be found in all tissues and organs, have many pro-regenerative properties (differentiation multipotential, immunomodulation, secretion of trophic factors, etc.), but can display different potentials for osteoblastic differentiation, according to their embryonic/tissue origin. MSC populations from dental tissues have the same embryonic origin as craniofacial bones, making them promising candidates for craniofacial reconstruction[8]. Dental pulp stem cells (DPSCs) were first isolated and characterised by Gronthos et al[9]. DPSCs are obtained in a non-invasive way from the permanent tooth and have a high proliferative rate, immunomodulatory properties, and multilineage differentiation capacity, especially for osteogenic ones. Together, they are an attractive source of cells for Bone Tissue Engineering and Regenerative Medicine applications[10].
However, few studies have addressed the most stable RG during osteogenic differentiation and no one in DPSCs. Thus, our study aimed to verify the most stable RG during osteogenic differentiation of human DPSCs by RT-qPCR. To validate, we chose the ten most used RG found during osteogenic differentiation in other MSCs [ribosomal protein, large, P0 (RPLP0), TATA-binding protein (TBP), GAPDH, ACTB, tubulin (TUB), aminolevulinic acid synthase 1 (ALAS1), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta (YWHAZ), eukaryotic translational elongation factor 1 alpha (EF1a), succinate dehydrogenase complex, subunit A, flavoprotein (SDHA), and beta-2-microglobulin (B2M)] through the most common and reliable methods (algorithms) ΔCt comparative method, geNorm, BestKeeper, and NormFinder and ranked by RefFinder.
Three human third molar teeth were extracted from 15- to 23-year-old healthy donors (University Hospital, University of São Paulo, Brazil). The analysis will designate them as donors #1, #2, and #3. Informed consent was obtained from donors (approval from the Human Ethics Committee - CAAE: 51097315.7.0000.5467 and CAAE: 51097315.7.3001.0076). An incision on the enamel-dentin junction was made, and the dental pulps were harvested. Then they were mechanically and enzymatically disaggregated (collagenase type I - 6 mg/mL and dispase - 8 mg/mL for 1 h at 37 °C), and single cells were obtained by filtration through a 70-μm mesh filter. Cells were seeded in T25 flasks and maintained in a clonogenic medium (α-MEM supplement with 2 mM glutamine + 10% BFS + 50 μg/mL ascorbic acid + 100 U/mL ampicillin + 100 + 100 μg/mL streptomycin). Cells were trypsinized when they reached 80%-90% subconfluence. The medium was refre
Undifferentiated DPSCs (#4) were seeded (5000 cells/cm2) in P35 dishes and induced for osteogenic (α-MEM supplement with 2 mM glutamine + 10% BFS + 50 μg/mL ascorbic acid + 1 μM dexamethasone + 10 mM β-glycerophosphate + 100 U/mL ampicillin + 100 μg/mL streptomycin) differentiation for 1, 7, 14, 21, 28, and 35 d. The medium was refreshed every 2-3 d, and cells were incubated under a humidity atmosphere at 37 °C and 5% CO2. The validation of osteogenic differentiation was performed by alizarin red staining.
Cells were seeded on P100 plates (5000 cells/cm2). Samples were collected and lysed in 1 mL TRIzol reagent (15596-026; Life Technologies, Carlsbad, CA, United States), following the manufacturer’s protocol. The purified total RNA was resuspended in 50 μL DEPC water. An aliquot of each sample was quantified in Nanodrop (2000c Spectrophotometer; Thermo Fisher Scientific, Waltham, MA, United States), and only RNA with an optical density A260/280 ratio between 1.9 and 2.1 was used for RT-qPCR analysis. The purified mRNA was stored at -80 °C until further use. Then total RNA (1 μg or 500 ng) was treated with 1 μL DNAse I (18068-015; Invitrogen), 1 μL buffer, and DEPC H2O to a final volume of 10 μL. It was left for 15 min at room temperature and 10 min at 65 °C. The 1 μL EDTA (25 mM) was added to inactivate the enzyme. Subsequently, complementary DNAs were synthesised by RT-PCR (18080-093; SuperScript™ III Reverse Transcriptase, Invitrogen). RT-PCR was performed in two steps: (1) Alignment (65 °C for 5 min): For a final volume of 13 μL, 11 μL RNA treated with DNase I, 1 μL oligo(dT) (18418-012; Invitrogen) and 1 μL of 10 mM dNTP (2.5 mM dATP, 2.5 mM dCTP, 2.5 mM dGTP, and 2.5 mM dTTP) (100 mM dNTP Set, PCR grade, 10297-018; Invitrogen); and (2) Reverse transcription (50 °C for 60 min and 70 °C for 15 min): After alignment, 4 μL of 5X Buffer, 1 μL DTT (0.1 M), 1 μL RNaseOUTTM (40 U/μL) (10777-019; Invitrogen) and 1 μL SuperScript III (200 U/μL), for a final volume of 20 μL. At the end of the second stage, the cDNA was obtained.
The synthesised cDNA was the template used for the reaction using the SYBR Green Dye I method and the evaluation of relative gene expression using Pfaffl[11]. Samples in osteogenic or clonogenic media in 1 d were used as calibrator samples. Candidate RGs and Runt-related transcription factor 2 (RUNX2) were used for osteogenic differentiation validation, which is described in Table 1. Samples synthesised from 1 μg RNA were diluted 1:1, and samples synthesised from 500 ng RNA were used neat. Reactions were performed in a total volume of 10 μL, containing 1 μL of the sample, 10 pM of each primer (400 nM), 5 μL SYBR Green Master Mix® (Applied Biosystems, Waltham, MA, United States) and water q.s.p. The reactions were performed on Applied Biosystems equipment (7500; Real-Time PCR System) at 60 °C in 40 cycles.
| Gene symbol | Function | NCBI access | Primers sequences (5’-3’) | Temperature (°C) |
| RPLP0 | Component of the 60S subunit | NM_001002 | F: AGCCCAGAACACTGGTCTC | 60 |
| R: ACTCAGGATTTCAATGGTGCC | ||||
| TBP | Transcription, metabolic pathways | NM_003194.5 | F: CACGAACCACGGCACTGATT | 62 |
| R: TTTTCTTGCTGCCAGTCTGGA | ||||
| YWHAZ | Signal transduction | NM_001135702 | F: TGATCCCCAATGCTTCACAAG | 61 |
| R: GCCAAGTAACGGTAGTAATCTCC | ||||
| RUNX2 | Master transcription factor for osteogenesis | NM_001015051 | F: TGGTTACTGTCATGGCGGGTA | 62 |
| R: TCTCAGATCGTTGAACCTTGCTA | ||||
| EF1α | Translation | NM_001402 | F: GAAGCTGGTATCTCCAAGAATGG | 61 |
| R: CGACAATTAGTTGTTTCACACCC | ||||
| GAPDH | Mitochondrial metabolism | NM_002046 | F: GCATCCTGGGCTACACTGA | 60 |
| R: CCACCACCCTGTTGCTGTA | ||||
| ACTB | Cytoskeleton | NM_001101 | F: CGACAGGATGCAGAAGGAG | 60 |
| R: TCCTGCTTGCTGATCCACAT | ||||
| B2M | Immune response | NM_004048 | F: GAGGCTATCCAGCGTACTCCA | 62 |
| R: CGGCAGGCATACTCATCTTTT | ||||
| ALAS1 | Mitochondrial metabolism | NM_199166 | F: AAATGAATGCCGTGAGGAAAGA | 60 |
| R: CCCTCCATCGGTTTTCACACTA | ||||
| TUB | Cytoskeleton | NM_001293212 | F: TCAACACCTTCTTCAGTGAAACG | 60 |
| R: AGTGCCAGTGCGAACTTCATC | ||||
| SDHA | Mitochondrial metabolism | NM_004168 | F: CAAACAGGAACCCGAGGTTTT | 60 |
| R: CAGCTTGGTAACACATGCTGTAT |
BestKeeper: BestKeeper algorithm performs for each candidate gene data analysis; the results are interpreted mainly by evaluating standard deviation (SD) coefficient of variation (CV) values and Pearson’s correlation coefficient - the smaller the SD and CV, the more stable the gene. Typically, genes are considered stable with an SD < 1. For Pearson’s correlation test, the higher the correlation value, the better, and P < 0.05. RefFinder, however, only uses SD for its ranking. It differs from other algorithms because it considers the intragene variation, not only intergene.
GeNorm: GeNorm considers that since a set of potentially stable genes was already selected, the Ct variation throughout the conditions must be similar for most genes, so it determines the two most stable genes that share a similar expression profile throughout all samples. It does so in a pairwise variation system where it calculates and compares the stability for all possible gene pair combinations, thus obtaining the gene stability value (M), defined as the arithmetic mean of the standard deviations of a gene. The smaller, the more stable. The geNorm performs several “cycles” of calculating M, always excluding the least stable one and recalculating it with the remaining data, thus finding the most stable pair. In the final result, as a criterion (threshold), only genes with M less than 1.5 are considered stable for this algorithm. In addition, we must calculate the value of Vn/Vn + 1 when its value is less than 0.15, using the n RGs with the lowest M value; the contribution of 1 or more genes will not significantly improve the data normalisation. We run geNorm using the RefFinder value and R ctrlGene library package version 1.0.1 (https://rdrr.io/cran/ctrlGene/) for Vn/Vn + 1 calculation.
NormFinder: It uses a pairwise variation system similar to the geNorm; its differential analyses are of intra- and inter-group variation. In this way, it is not as significantly influenced by mutually co-regulated genes. The lower its value, the more stable the genes. Furthermore, the criterion (threshold) is the same as geNorm; only genes with M less than 1.5 are considered stable for the algorithm. The number of samples affects the calculation and is more accurate as n increases (n ≥ 8).
ΔCt comparative method: It is based on mathematical principles similar to the pairwise comparison of the NormFinder and geNorm, with a more accessible calculation for researchers less articulated in mathematics. To do so, it performs the ΔCt method for each pair of genes within each sample, then evaluates the ΔCt mean for each pair and between every two groups. RGs are ranked by their associated arithmetic mean of standard deviation values, as the smaller the number, the more stable the gene. We will consider the two best-ranked genes as the best RG pair. This method bypasses the need to quantify the RNA, using ΔCt comparisons between genes accurately.
RefFinder: It is a user-friendly web-based tool containing the four main statistical gene stability algorithms (geNorm, NormFinder, BestKeeper, and the comparative ΔCt method). After calculation, it addresses a weight for each gene by their ranking position in each algorithm and calculates the geometric mean of their weights for the comprehensive final ranking. To date, it is available at https://blooge.cn/RefFinder/. A table for each group was made, with each column representing a gene and each row the original Ct value (not Ct mean) from a sample’s qRT-PCR. All three patients’ data were on all tables; thus, we calculated simultaneously.
Subsets: To better identify endogenous genes in the initial and final phases of differentiation, we divided the data into the early phase (from 1 to 21 d) and late phase (from 21 to 35 d) after induction of osteogenic differentiation.
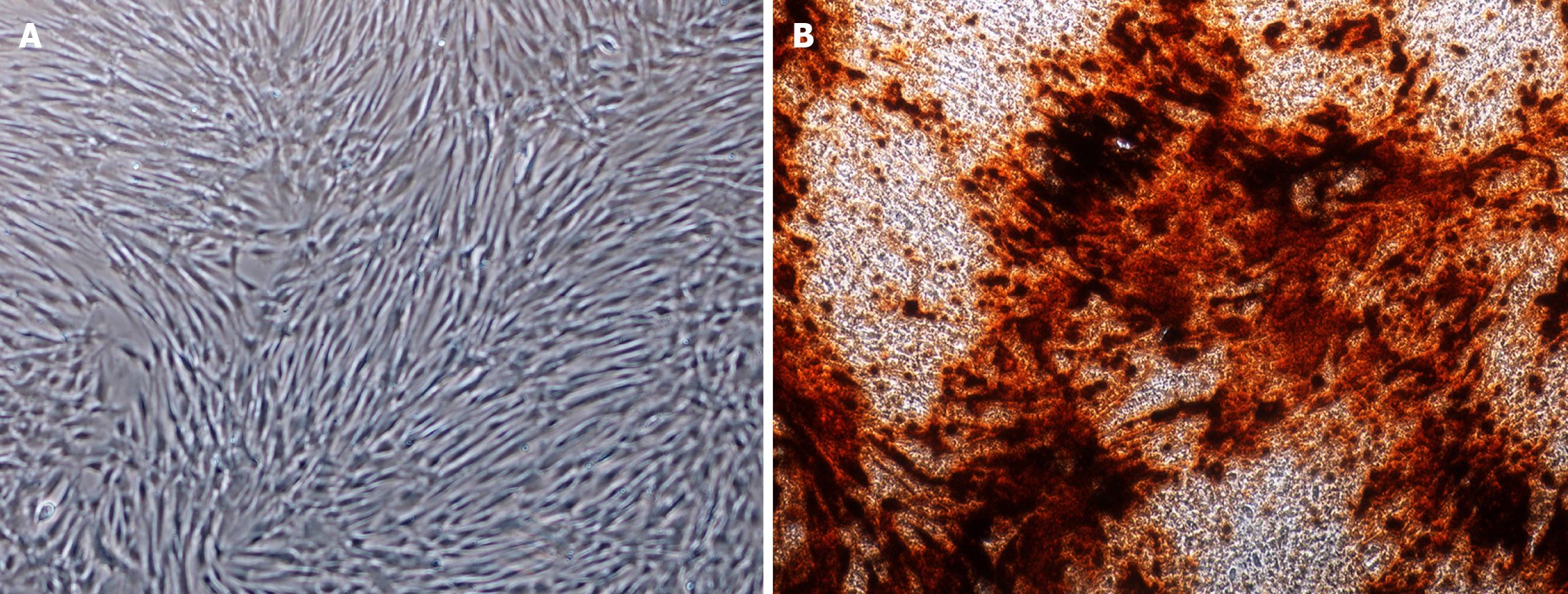
The DPSCs display a fibroblast-like morphology (Figure 1A) and differentiate into osteoblast-like cells under osteogenic induction, confirmed by the alizarin red staining (Figure 1B).
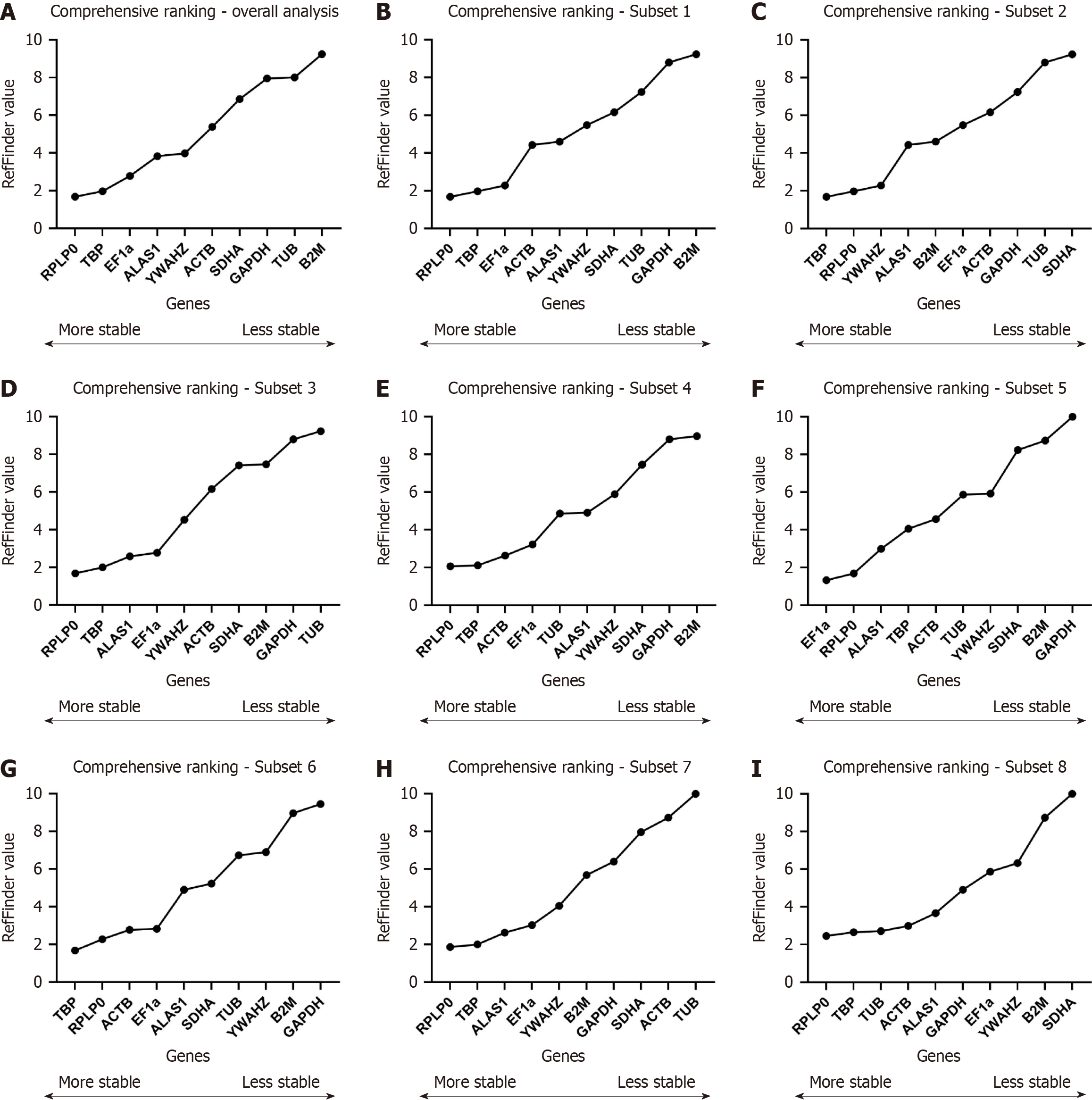
All of the data sets from clonogenic and osteogenic samples were analysed by the RefFinder algorithm; since Vn/Vn + 1 were lower than 1.5, we will consider the top two ranked genes. The final ranking showed RPLP0/TBP as the two most stable RGs and TUB/B2M as the two least stable RGs (Figure 2A).
For the ΔCt method, we will consider the top two ranked genes. NormFinder and geNorm, the best RGs were those with stability values less than 1.5. Either ΔCt method and NormFinder analysis showed TBP/RPLP0 as the two most stable genes. However, geNorm analysis showed RPLP0/EF1α in the first place. These algorithms’ two least stable RG were B2M/GAPDH (Table 2). For BestKeeper, the best-ranked RGs must be SD < 1. ALAS1 was ranked as the most stable RG, and SDHA as the least stable RG.
| Main group | Rank | Delta Ct | NormFinder | geNorm | BestKeeper | ||||||
| Gene | Stability value | Gene | Stability value | Gene | M value | Gene | Std Dev | R | P value | ||
| 1 to 35 d (clo + ost) | 1 | TBP | 2.13 | TBP | 1.078 | RPLP0/EFα1 | 0.889 | ALAS1 | 0.70 | 0.335 | 0.003 |
| 2 | RPLP0 | 2.27 | RPLP0 | 1.451 | - | - | RPLP0 | 1.33 | 0.726 | 0.001 | |
| 3 | YWAHZ | 2.37 | YWAHZ | 1.62 | TBP | 1.712 | EFα1 | 1.41 | 0.668 | 0.001 | |
| 4 | EFα1 | 2.43 | ACTB | 1.659 | YWAHZ | 1.923 | GAPDH | 1.78 | 0.571 | 0.001 | |
| 5 | ACTB | 2.43 | EFα1 | 1.707 | SDHA | 2.037 | TBP | 1.87 | 0.857 | 0.001 | |
| 6 | ALAS1 | 2.55 | ALAS1 | 1.84 | ALAS1 | 2.152 | ACTB | 2.00 | 0.785 | 0.001 | |
| 7 | SDHA | 2.57 | SDHA | 1.948 | ACTB | 2.261 | YWAHZ | 2.40 | 0.876 | ||
| 8 | TUB | 2.68 | TUB | 2.06 | B2M | 2.337 | TUB | 2.41 | 0.777 | ||
| 9 | B2M | 2.72 | B2M | 2.122 | TUB | 2.402 | SDHA | 2.59 | 0.847 | ||
| 10 | GAPDH | 2.93 | GAPDH | 2.385 | GAPDH | 2.508 | B2M | 2.63 | 0.799 | 0.001 | |
We analysed all samples from each group separately to identify differences between RG stability between clonogenic and osteogenic groups for long-time cultivation. By RefFinder ranking, TBP/RPLP0 were the most stable for Subset 1 and RPLP0/TBP for Subset 2, respectively. The least stable RG for Subset 1 were TUB/SDHA and GAPDH/B2M for Subset 2 (Figure 2B and C).
In both subsets, the two most stable genes by ΔCt method and NormFinder were equal to those by RefFinder (TBP/RPLP0). However, other genes demonstrated stability values less than 1.5 in NormFinder, such as YWAHZ (Subset 1) and EF1α/ACTB (Subset 2). For geNorm, while TBP/YWAHZ/B2M/SDHA are the most stable genes in Subset 1, the RPL0/EF1α/TBP were considered the best for Subset 2. Bestkeeper showed ALAS1 as the most stable in both subsets and SDHA and B2M as the least stable genes, respectively (Table 3).
| Subset | Rank | Delta Ct | NormFinder | geNorm | BestKeeper | ||||||
| Gene | Stability value | Gene | Stability value | Gene | M value | Gene | Std Dev | R | P value | ||
| 1: 1 to 35 d (clo) | 1 | TBP | 2.040 | TBP | 1.024 | TBP/YWAHZ | 0.996 | ALAS1 | 0.660 | 0.326 | 0.043 |
| 2 | RPLP0 | 2.220 | RPLP0 | 1.423 | - | - | RPLP0 | 1.200 | 0.700 | 0.001 | |
| 3 | YWAHZ | 2.220 | YWAHZ | 1.466 | B2M | 1.319 | EFα1 | 1.250 | 0.537 | 0.001 | |
| 4 | B2M | 2.430 | B2M | 1.790 | SDHA | 1.449 | GAPDH | 1.660 | 0.647 | 0.001 | |
| 5 | ACTB | 2.460 | ACTB | 1.818 | RPLP0 | 1.880 | TBP | 1.900 | 0.866 | 0.001 | |
| 6 | ALAS1 | 2.480 | ALAS1 | 1.821 | EFα1 | 2.068 | ACTB | 2.140 | 0.799 | 0.001 | |
| 7 | GAPDH | 2.500 | GAPDH | 1.824 | ALAS1 | 2.159 | YWAHZ | 2.240 | 0.850 | 0.001 | |
| 8 | EFα1 | 2.560 | EFα1 | 1.989 | GAPDH | 2.272 | TUB | 2.510 | 0.800 | 0.001 | |
| 9 | SDHA | 2.620 | SDHA | 2.099 | ACTB | 2.356 | B2M | 2.560 | 0.855 | 0.001 | |
| 10 | TUB | 2.680 | TUB | 2.161 | TUB | 2.421 | SDHA | 2.710 | 0.875 | 0.001 | |
| 2: 1 to 35 d (ost) | 1 | TBP | 2.140 | TBP | 1.087 | RPLP0/EFα1 | 0.515 | ALAS1 | 0.740 | 0.324 | 0.044 |
| 2 | RPLP0 | 2.170 | RPLP0 | 1.276 | - | - | RPLP0 | 1.460 | 0.786 | 0.001 | |
| 3 | EFα1 | 2.180 | EFα1 | 1.291 | TBP | 1.450 | EFα1 | 1.570 | 0.812 | 0.001 | |
| 4 | ACTB | 2.320 | ACTB | 1.467 | YWAHZ | 1.777 | ACTB | 1.720 | 0.767 | 0.001 | |
| 5 | YWAHZ | 2.450 | YWAHZ | 1.758 | SDHA | 1.860 | TBP | 1.850 | 0.846 | 0.001 | |
| 6 | SDHA | 2.480 | SDHA | 1.789 | ACTB | 2.010 | GAPDH | 1.920 | 0.519 | 0.001 | |
| 7 | TUB | 2.570 | ALAS1 | 1.853 | TUB | 2.108 | TUB | 2.190 | 0.744 | 0.001 | |
| 8 | ALAS1 | 2.570 | TUB | 1.882 | ALAS1 | 2.184 | SDHA | 2.460 | 0.818 | 0.001 | |
| 9 | B2M | 2.970 | B2M | 2.456 | B2M | 2.323 | YWAHZ | 2.530 | 0.901 | 0.001 | |
| 10 | GAPDH | 3.270 | GAPDH | 2.833 | GAPDH | 2.512 | B2M | 2.640 | 0.739 | 0.001 | |
Here, we analysed the early and late differentiation steps groups, considering clonogenic and osteogenic samples in the same group. By RefFinder ranking, RPLP0/TBP were again the two most stable for Subsets 3 and 4. The least stable RG for Subset 3 were TUB/GAPDH and B2M/GAPDH for Subset 4 (Figure 2D and E).
The most stable genes by ΔCt method were TBP/RPLP0 and TBP/ACTB for Subset 3 and 4, respectively. By Norm
| Subset | Rank | Delta Ct | Normfinder | geNorm | BestKeeper | ||||||
| Gene | Stability value | Gene | Stability value | Gene | M value | Gene | Std Dev | R | P value | ||
| 3: 1 to 21 d (clo + ost) | 1 | TBP | 2.160 | TBP | 1.202 | RPLP0/EFα1 | 0.594 | ALAS1 | 0.750 | 0.433 | 0.001 |
| 2 | RPLP0 | 2.230 | RPLP0 | 1.419 | - | - | RPLP0 | 1.100 | 0.614 | 0.001 | |
| 3 | YWAHZ | 2.320 | ALAS1 | 1.492 | ALAS1 | 1.207 | EFα1 | 1.100 | 0.535 | 0.001 | |
| 4 | EFα1 | 2.320 | YWAHZ | 1.550 | TBP | 1.639 | TBP | 1.770 | 0.801 | 0.001 | |
| 5 | ALAS1 | 2.330 | EEF1α1 | 1.591 | YWAHZ | 1.814 | ACTB | 1.940 | 0.703 | 0.001 | |
| 6 | ACTB | 2.580 | ACTB | 1.938 | SDHA | 1.989 | GAPDH | 2.010 | 0.683 | 0.001 | |
| 7 | SDHA | 2.640 | B2M | 1.979 | B2M | 2.104 | YWAHZ | 2.080 | 0.819 | 0.001 | |
| 8 | B2M | 2.640 | SDHA | 2.048 | ACTB | 2.275 | B2M | 2.210 | 0.709 | 0.001 | |
| 9 | TUB | 2.790 | TUB | 2.252 | TUB | 2.383 | SDHA | 2.290 | 0.785 | 0.001 | |
| 10 | GAPDH | 2.970 | GAPDH | 2.411 | GAPDH | 2.499 | TUB | 2.350 | 0.738 | 0.001 | |
| 4: 21 to 35 d (clo + ost) | 1 | TBP | 1.980 | TBP | 0.749 | RPLP0/EFα1 | 1.039 | ALAS1 | 0.630 | 0.345 | 0.039 |
| 2 | ACTB | 2.090 | ACTB | 1.075 | - | - | RPLP0 | 1.740 | 0.860 | 0.001 | |
| 3 | RPLP0 | 2.120 | RPLP0 | 1.155 | ACTB | 1.427 | EFα1 | 1.890 | 0.815 | 0.001 | |
| 4 | TUB | 2.280 | TUB | 1.447 | TBP | 1.636 | ACTB | 1.920 | 0.893 | 0.001 | |
| 5 | YWAHZ | 2.300 | YWAHZ | 1.604 | TUB | 1.738 | TBP | 1.940 | 0.915 | 0.001 | |
| 6 | EFα1 | 2.360 | EEF1α1 | 1.605 | YWAHZ | 1.866 | GAPDH | 1.970 | 0.456 | 0.005 | |
| 7 | SDHA | 2.540 | SDHA | 1.979 | SDHA | 1.980 | TUB | 2.120 | 0.829 | 0.001 | |
| 8 | ALAS1 | 2.670 | ALAS1 | 2.044 | B2M | 2.087 | YWAHZ | 2.620 | 0.899 | 0.001 | |
| 9 | B2M | 2.760 | B2M | 2.283 | ALAS1 | 2.210 | SDHA | 2.810 | 0.838 | 0.001 | |
| 10 | GAPDH | 3.420 | GAPDH | 3.075 | GAPDH | 2.452 | B2M | 2.930 | 0.830 | 0.001 | |
We now analysed only early and late osteogenic differentiation step groups under osteogenic differentiation. By RefFinder ranking, the two most stable genes for Subset 5 (the early stages) were EF1α/RPLP0, whereas TBP/RPLP0 for Subset 6 (the late stages). The two least stable RG for Subsets 5 and 6 were GAPDH/B2M (Figure 2F and G).
The most stable genes by ΔCt method were EF1α/RPLP0 for Subset 5 and TBP/ACTB for Subset 6. By NormFinder, the most stable genes were EF1α/RPLP0/TBP/ALAS1 for Subset 5 and TBP/ACTB/RPL0/EF1α/SDHA for Subset 6. By geNorm, the RPLP0/EF1α were the most stable in Subset 5 and RPLP0/EF1α/ACTB/TBP/SDHA for Subset 6. Even though those genes have been ranked differently between Subsets 5 and 6, they coincide. Bestkeeper showed ALAS1 as the most stable in both Subsets, similar to those found by Subsets 1, 2, 3, and 4, and GAPDH and B2M as the least stable genes, respectively (Table 5).
| Subset | Rank | Delta Ct | NormFinder | geNorm | BestKeeper | ||||||
| Gene | Stability value | Gene | Stability value | Gene | M value | Gene | Std Dev | R | P value | ||
| 5: 1 to 21 d (ost) | 1 | EFα1 | 2.200 | EFα1 | 1.255 | RPLP0/EFα1 | 0.463 | ALAS1 | 0.810 | 0.440 | 0.022 |
| 2 | RPLP0 | 2.210 | RPLP0 | 1.278 | - | - | RPLP0 | 1.330 | 0.731 | 0.001 | |
| 3 | TBP | 2.280 | TBP | 1.325 | ACTB | 1.514 | EFα1 | 1.330 | 0.755 | 0.001 | |
| 4 | ALAS1 | 2.380 | ALAS1 | 1.485 | TUB | 1.629 | ACTB | 1.540 | 0.555 | 0.003 | |
| 5 | YWAHZ | 2.450 | YWAHZ | 1.706 | ALAS1 | 1.742 | TBP | 1.860 | 0.787 | 0.001 | |
| 6 | ACTB | 2.480 | ACTB | 1.757 | TBP | 1.978 | TUB | 1.880 | 0.620 | 0.001 | |
| 7 | TUB | 2.600 | TUB | 1.948 | YWAHZ | 2.115 | YWAHZ | 2.170 | 0.845 | 0.001 | |
| 8 | SDHA | 2.660 | SDHA | 2.043 | SDHA | 2.222 | B2M | 2.240 | 0.609 | 0.001 | |
| 9 | B2M | 2.950 | B2M | 2.369 | B2M | 2.345 | SDHA | 2.290 | 0.752 | 0.001 | |
| 10 | GAPDH | 3.460 | GAPDH | 3.031 | GAPDH | 2.568 | GAPDH | 2.310 | 0.647 | 0.001 | |
| 6: 21 to 35 d (ost) | 1 | TBP | 1.820 | TBP | 0.578 | RPLP0/EFα1 | 0.495 | ALAS1 | 0.560 | 0.257 | 0.304 |
| 2 | ACTB | 1.900 | ACTB | 0.584 | - | - | TBP | 1.750 | 0.963 | 0.001 | |
| 3 | RPLP0 | 2.050 | RPLP0 | 1.096 | ACTB | 1.024 | RPLP0 | 1.810 | 0.878 | 0.001 | |
| 4 | EFα1 | 2.110 | EFα1 | 1.224 | TBP | 1.134 | EFα1 | 2.040 | 0.883 | 0.001 | |
| 5 | SDHA | 2.160 | SDHA | 1.368 | SDHA | 1.440 | ACTB | 2.070 | 0.963 | 0.001 | |
| 6 | YWAHZ | 2.300 | TUB | 1.551 | YWAHZ | 1.557 | SDHA | 2.250 | 0.843 | 0.001 | |
| 7 | TUB | 2.310 | YWAHZ | 1.648 | TUB | 1.655 | TUB | 2.400 | 0.856 | 0.001 | |
| 8 | ALAS1 | 2.830 | ALAS1 | 2.254 | B2M | 1.842 | GAPDH | 2.510 | 0.492 | 0.038 | |
| 9 | B2M | 2.840 | B2M | 2.401 | ALAS1 | 2.035 | YWAHZ | 2.760 | 0.927 | 0.001 | |
| 10 | GAPDH | 4.040 | GAPDH | 3.776 | GAPDH | 2.435 | B2M | 2.800 | 0.810 | 0.001 | |
We now analysed only early and late osteogenic differentiation step groups under clonogenic differentiation. By RefFinder ranking, the two most stable genes were RPLP0/TBP for Subsets 7 and 8. The least stable RG for Subset 5 were ACTB/TUB and B2M/SDHA for Subset 8 (Figure 2H and I).
The most stable genes by ΔCt method were TBP/RPLP0 for both Subsets 7 and 8. By and NormFinder, TBP/RPLP0/YWAHZ/ALAS1/B2M for Subset 7 and TBP/RPLP0/TUB/ACTB for Subset 8. By geNorm, the RPLP0/EF1
| Subset | Rank | Delta Ct | NormFinder | geNorm | BestKeeper | ||||||
| Gene | Stability value | Gene | Stability value | Gene | M value | Gene | Std Dev | R | P value | ||
| 7: 1 to 21 d (clo) | 1 | TBP | 2.010 | TBP | 1.091 | RPLP0/EFα1 | 0.661 | ALAS1 | 0.690 | 0.379 | 0.051 |
| 2 | RPLP0 | 2.120 | RPLP0 | 1.379 | - | - | EEF1α1 | 0.820 | 0.144 | 0.473 | |
| 3 | YWAHZ | 2.150 | YWAHZ | 1.398 | ALAS1 | 1.033 | RPLP0 | 0.870 | 0.516 | 0.006 | |
| 4 | ALAS1 | 2.150 | ALAS1 | 1.404 | TBP | 1.430 | TBP | 1.600 | 0.778 | 0.001 | |
| 5 | B2M | 2.210 | B2M | 1.442 | YWAHZ | 1.617 | GAPDH | 1.630 | 0.722 | 0.001 | |
| 6 | EFα1 | 2.370 | GAPDH | 1.615 | B2M | 1.718 | YWAHZ | 1.950 | 0.759 | 0.001 | |
| 7 | GAPDH | 2.370 | EFα1 | 1.839 | SDHA | 1.890 | B2M | 1.980 | 0.762 | 0.001 | |
| 8 | SDHA | 2.610 | SDHA | 2.089 | GAPDH | 2.032 | ACTB | 2.180 | 0.789 | 0.001 | |
| 9 | ACTB | 2.630 | ACTB | 2.133 | ACTB | 2.212 | SDHA | 2.320 | 0.800 | 0.001 | |
| 10 | TUB | 2.920 | TUB | 2.546 | TUB | 2.354 | TUB | 2.660 | 0.815 | 0.001 | |
| 8: 21 to 35 d (clo) | 1 | TBP | 2.070 | TBP | 1.063 | ACTB/TUB | 0.766 | ALAS1 | 0.670 | 0.480 | 0.044 |
| 2 | RPLP0 | 2.130 | RPLP0 | 1.247 | - | - | GAPDH | 1.470 | 0.455 | 0.058 | |
| 3 | TUB | 2.200 | TUB | 1.378 | RPLP0 | 1.417 | RPLP0 | 1.680 | 0.841 | 0.001 | |
| 4 | ACTB | 2.210 | ACTB | 1.430 | GAPDH | 1.621 | EEF1α1 | 1.780 | 0.752 | 0.001 | |
| 5 | YWAHZ | 2.280 | YWAHZ | 1.601 | ALAS1 | 1.757 | ACTB | 1.780 | 0.801 | 0.001 | |
| 6 | ALAS1 | 2.480 | ALAS1 | 1.813 | EEF1α1 | 1.932 | TUB | 1.850 | 0.799 | 0.001 | |
| 7 | EFα1 | 2.580 | EFα1 | 1.953 | TBP | 2.055 | TBP | 2.170 | 0.901 | 0.001 | |
| 8 | GAPDH | 2.680 | B2M | 2.231 | YWAHZ | 2.172 | YWAHZ | 2.520 | 0.862 | 0.001 | |
| 9 | B2M | 2.700 | GAPDH | 2.232 | B2M | 2.315 | B2M | 3.070 | 0.857 | 0.001 | |
| 10 | SDHA | 2.820 | SDHA | 2.388 | SDHA | 2.415 | SDHA | 3.240 | 0.884 | 0.001 | |
Comparing the results obtained for the two most stable genes from the overall ranking with each group, by RefFinder, the pair RPLP0/TBP or TBP/RPLP0 were observed in 7 over 8. By the ΔCt method, the pair RPLP0/TBP or TBP/RPLP0 was detected in 5 over 8. NormFinder detected RPLP0/TBP or TBP/RPLP0 in 7 over 8. By geNorm, RPLP0/EF1α was detected in 6 over 8. Finally, ALAS1 was the most stable gene in all subsets.
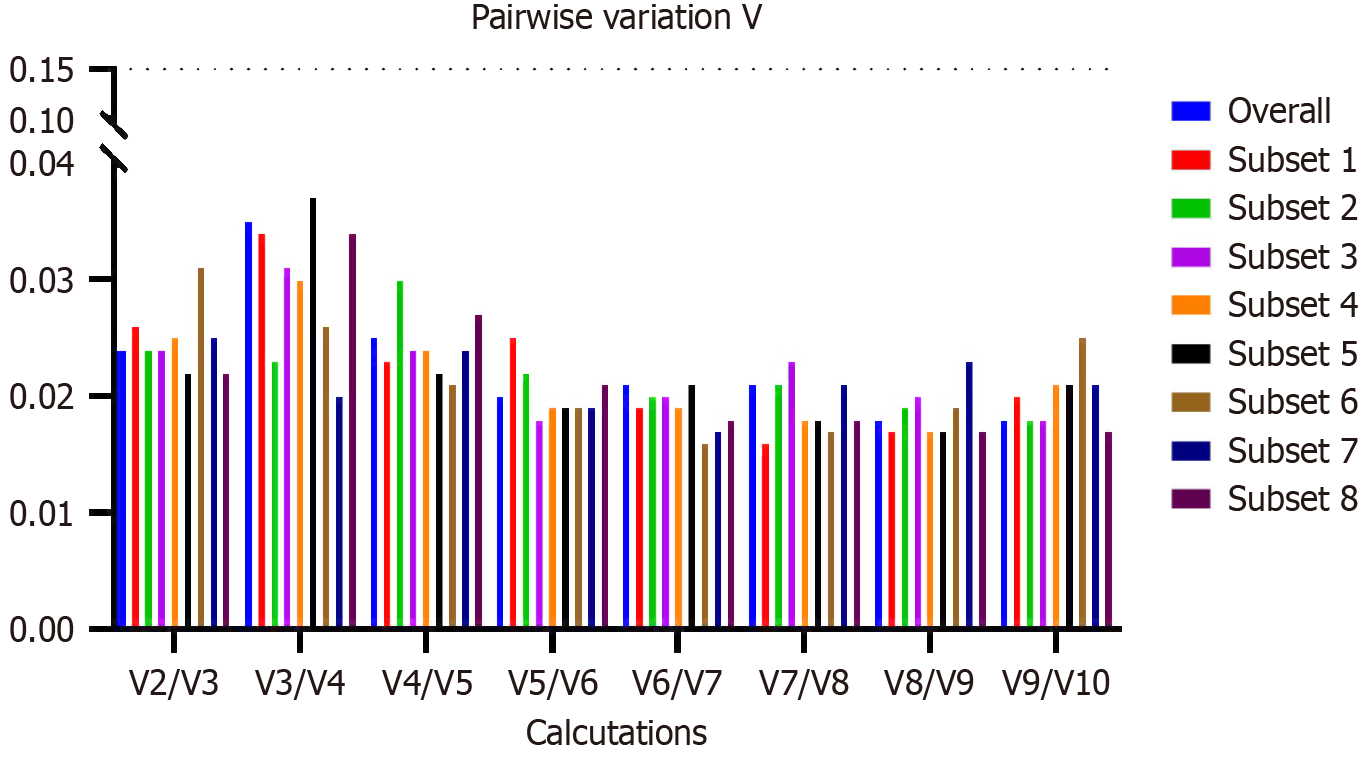
MIQE guideline highlights that more than one gene is preferable for data normalisation[1]. The geNorm algorithm can evaluate the recommended number of genes for normalisation by Vn/Vn + 1 calculation. This means that the optimal number of RGs is determined by calculating the pairwise variation (V) between a given number of RGs and including an additional gene. A cut-off value of 0.15 has been suggested, where the inclusion of an extra gene has little effect on the normalisation. For the first n value that shows a number less than 0.15 after Vn/Vn + 1 calculation, it is recommended to use the n most stable genes calculated. We evaluated the Vn/Vn + 1 for the leading group (all samples). The result was that V2/V3 was already lower than 0.15; therefore, we found that only the two most stable genes were necessary for data normalisation, TBP/RPLP0 (Figure 3). To do this normalisation, as Vandesompele et al[4] recommended, we simply take the geometric mean of the Ct of the two most stable genes and use it as a normalisation factor.
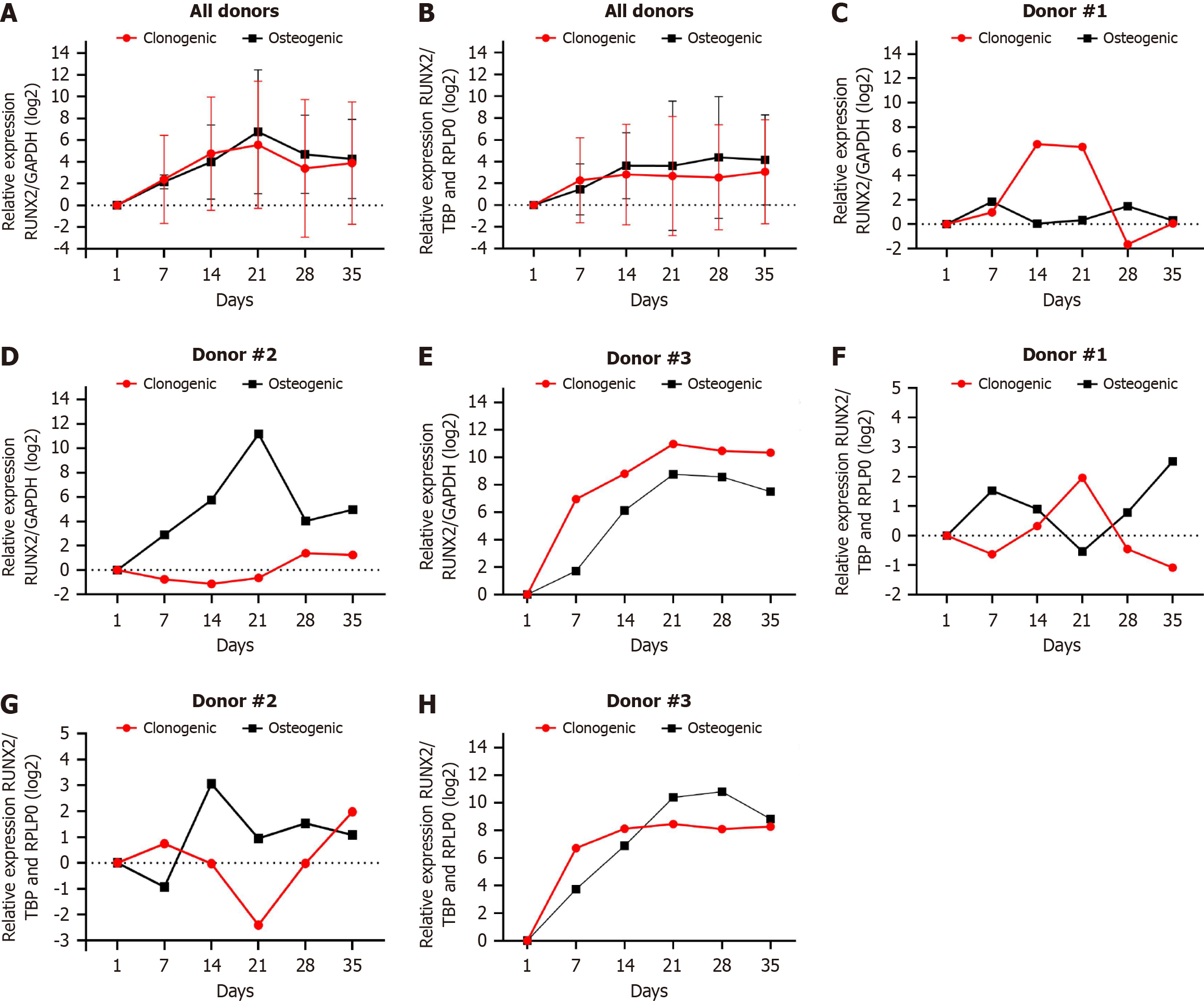
We demonstrated the individual gene expression of each donor and the results together. As we used DPSC primary cell cultures, we expected differences in gene expression between the donors because of individual factors. The alizarin red staining (Figure 1) from donor #1 to donor #3 increased at 7, 14, and 21 d, respectively. When we normalise RUNX2 with TBP/RPLP0, the gene expression profile is similar to those seen for alizarin red staining (Figure 4F-H). When the RUNX2 normalisation with GAPDH occurs, the gene expression is overestimated or underestimated in both clonogenic and osteogenic media (Figure 4C-E). When we compared the relative expression of RUNX2 normalised by GAPDH or TBP/RPLP0, we observed that GAPDH tends to super estimate or underestimated in both clonogenic or osteogenic media gene expression than TBP/RPLP0 (Figure 4A and B).
For the first time, we show that the best pair of RGs for osteogenic differentiation of human DPSCs are RPLP0/TBP and TUB/B2M are the two least stable RGs through the four algorithms (ΔCt method, geNorm, BestKeeper, and NormFinder) and ranked by RefFinder. The RT-qPCR technique is an excellent tool for detecting gene transcription levels. Good practices must be adopted to generate solid results supporting eventual interpretations and statements. An essential step in good RT-qPCR practices is checking the stability of the RGs adapted to normalise the results[1].
During induction of differentiation in MSCs in vitro, an intense change in gene expression profile is expected. For osteogenic differentiation, the first step is the cell commitment to the osteoblastic lineage. It generally happens around 1 to 7 d after the MSC induction with the osteogenic medium. The second step is the proliferation and differentiation of osteoprogenitor cells (7 to 14 d). The third step is differentiating these cells towards mature osteoblasts (14 to 21 d). Finally, from 21 d, matrix maturation and mineralisation are observed, as well as the early stages of osteoblast-osteocyte transition[12]. We cultivated DPSCs under clonogenic (negative control for differentiation) and osteogenic media for 35 d, and we analysed the gene expression every 7 d to understand the influence of cell status on RGs. Since we are evaluating long-term osteogenic differentiation, we separated the analyses into early (1 to 21 d) and late (21 to 35 d) stages of differentiation in DPSCs.
Quiroz et al[13] conducted the first study, which addressed the study of RGs during osteogenic differentiation in human bone marrow stem cells (BMSCs) from 14 to 20 d. They evaluated only ACTB, GAPDH, and RPL13a by two different mathematical approaches based on the ΔCt method[14], and RPL13a was the most stable gene, confirming that the most commonly employed RGs are not stable during differentiation. Even one decade later, many basic studies still ignore the stability of RG for specific MSC origins and differentiation conditions, such as cell culture methods (traditional monolayer or tridimensional techniques) and oxygen tension (normoxia and hypoxia), which will impact clinical studies. To explore the RGs in DPSCs, we selected ten putative genes previously explored to normalise gene expression data of osteogenic differentiation in traditional monolayer culture in different origins of human MSCs, such as fatty tissue[15-17], Wharton’s Jelly[16], cord blood[15], umbilical cord[18], bone marrow[13,15,16,18-22], gingiva[23], fetal tissue[21], and from induced pluripotent stem cells (iPSCs)[24]. Other human cell lineages have also been analysed[25].
For the overall group, we noticed that by RefFinder ranking, the pair TBP/RPLP0 is recommended for normalising undifferentiated DPSCs and undergoing osteogenic differentiation by 35 d. RPLP0 is a gene member of the ribosomal protein family. TBP is a transcription factor that binds to the TATA-box sequences in the DNA promoter region, which belongs to the RNA-polymerase II pre-initiation complex (transcription of all protein-encoded genes). Jacobi et al[20] showed RPLP0 as the most stable gene during BMSC osteogenic differentiation for 14 d and B2M as the most unstable RG calculated by geNorm. Ragni et al[15] showed TBP/YWHAZ/GUSB as the most stable gene of bone marrow, adipose and cord blood MSCs under mesodermal differentiation (osteogenic, chondrogenic, and adipogenic for 21 d) and B2M as the most unstable RG ranked by geNorm and NormFinder. Specifically by geNorm, RPLP0/EEF1α were the best genes for osteogenic differentiation. Ayanoğlu et al[17] showed EF1α/RPLP13a/RPLP0 as the most stable gene of adipose-derived stem cell (ADSC) under osteogenic differentiation for 21 d and B2M as the most unstable RG ranked by ΔCt method and NormFinder. Recently, Okamura et al[24] also showed that TBP/RPLP0 are the most stable genes during human iPSC cell line into osteogenic differentiation for 28 d and B2M as the most unstable RG calculated by ΔCt method, geNorm, NormFinder, and BestKeeper. However, these authors did not recommend the REfFinder ranking because the ranking method is unclear. Even though the ranking method is not explicit, it is still a user-friendly tool for researchers to verify RGs. It is easy to format and input data and gives a well-determined ranking, suitable for those unfamiliar with inter
When we analysed the overall data and Subgroups, EF1α is ranked in the third or fourth place as a stable gene by RefFinder. In all subgroups, except subgroup 8, it composes a second position with RPLP0, which is calculated by geNorm. Specifically, in Subgroup 5 (only osteogenic medium from 1 to 21 d - early stage), it was classified in the first position: EEF1α/RPLP0 (RefFinder), EEF1/RPLP0 (ΔCt method), EEF1/RPLP0/TBP/ALAS1 (NormFinder), and RPLP0/EEF1α (geNorm). Previous works have shown EF1α is a suitable RG[17] or fluctuates in the first positions in the algori
GAPDH and ACTB are widely used for the normalisation of RT-qPCR data. However, almost all studies to find the most stable RGs have identified that these genes are unsuitable for that, especially during osteogenic differentiation. In the overall analysis, B2M (RefFinder), B2M/GAPDH (ΔCt method and NormFinder), TUB/GAPDH (geNorm), and SDHA (BestKeeper) were the most unstable RGs. Our results show that B2M and GAPDH are the two least stable RGs in half of the Subgroups, calculated by ΔCt method, NormFinder and geNorm. SDHA and TUB also appear in combination with B2M and GAPDH. By BestKeeper, SDHA, B2M, and TUB are ranked as the least stable RGs in most of the Sub
When we compared the relative expression of RUNX2 normalised by GAPDH or TBP/RPLP0, we observe that GAPDH tends to overestimate gene expression more than TBP/RPLP0. This was reported in most of the studies cited before and generates a misinterpretation of gene expression during osteogenic differentiation.
For the first time, we show that RPLP0/TBP are the most stable RGs, while TUB/B2M are unstable RGs for long-term osteogenic differentiation of human DPSCs in traditional monolayers.
| 1. | Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, Penning LC, Toegel S. MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 2010;11:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 466] [Cited by in RCA: 495] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 2. | Chapman JR, Waldenström J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS One. 2015;10:e0141853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 224] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 3. | Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1345] [Cited by in RCA: 1177] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 4. | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12858] [Cited by in RCA: 14552] [Article Influence: 632.7] [Reference Citation Analysis (0)] |
| 5. | Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245-5250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5406] [Cited by in RCA: 5236] [Article Influence: 249.3] [Reference Citation Analysis (0)] |
| 6. | Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3334] [Cited by in RCA: 3577] [Article Influence: 170.3] [Reference Citation Analysis (0)] |
| 7. | Xie F, Wang J, Zhang B. RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct Integr Genomics. 2023;23:125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 209] [Reference Citation Analysis (0)] |
| 8. | Alarcón-Apablaza J, Prieto R, Rojas M, Fuentes R. Potential of Oral Cavity Stem Cells for Bone Regeneration: A Scoping Review. Cells. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97:13625-13630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3174] [Cited by in RCA: 3363] [Article Influence: 134.5] [Reference Citation Analysis (0)] |
| 10. | Moeenzade N, Naseri M, Osmani F, Emadian Razavi F. Dental pulp stem cells for reconstructing bone defects: A systematic review and meta-analysis. J Dent Res Dent Clin Dent Prospects. 2022;16:204-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24144] [Cited by in RCA: 26011] [Article Influence: 1083.8] [Reference Citation Analysis (0)] |
| 12. | Paiva KBS, Granjeiro JM. Matrix Metalloproteinases in Bone Resorption, Remodeling, and Repair. Prog Mol Biol Transl Sci. 2017;148:203-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 147] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 13. | Quiroz FG, Posada OM, Gallego-Perez D, Higuita-Castro N, Sarassa C, Hansford DJ, Agudelo-Florez P, López LE. Housekeeping gene stability influences the quantification of osteogenic markers during stem cell differentiation to the osteogenic lineage. Cytotechnology. 2010;62:109-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112-114, 116, 118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 718] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 15. | Ragni E, Viganò M, Rebulla P, Giordano R, Lazzari L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: how to choose the most reliable housekeeping genes. J Cell Mol Med. 2013;17:168-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 16. | Amable PR, Teixeira MV, Carias RB, Granjeiro JM, Borojevic R. Identification of appropriate reference genes for human mesenchymal cells during expansion and differentiation. PLoS One. 2013;8:e73792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Ayanoğlu FB, Elçin AE, Elçin YM. Evaluation of the stability of standard reference genes of adipose-derived mesenchymal stem cells during in vitro proliferation and differentiation. Mol Biol Rep. 2020;47:2109-2122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Jeon RH, Lee WJ, Son YB, Bharti D, Shivakumar SB, Lee SL, Rho GJ. PPIA, HPRT1, and YWHAZ Genes Are Suitable for Normalization of mRNA Expression in Long-Term Expanded Human Mesenchymal Stem Cells. Biomed Res Int. 2019;2019:3093545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Studer D, Lischer S, Jochum W, Ehrbar M, Zenobi-Wong M, Maniura-Weber K. Ribosomal protein l13a as a reference gene for human bone marrow-derived mesenchymal stromal cells during expansion, adipo-, chondro-, and osteogenesis. Tissue Eng Part C Methods. 2012;18:761-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Jacobi A, Rauh J, Bernstein P, Liebers C, Zou X, Stiehler M. Comparative analysis of reference gene stability in human mesenchymal stromal cells during osteogenic differentiation. Biotechnol Prog. 2013;29:1034-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Li X, Yang Q, Bai J, Yang Y, Zhong L, Wang Y. Identification of optimal reference genes for quantitative PCR studies on human mesenchymal stem cells. Mol Med Rep. 2015;11:1304-1311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Cagnan I, Aerts Kaya F, Çetinkaya D, Özcan A. Stably expressed reference genes during differentiation of bone marrow-derived mesenchymal stromal cells. Turkish J Biol. 2017;41:88-97. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Taïhi I, Nassif A, Berbar T, Isaac J, Berdal A, Gogly B, Fournier BP. Validation of Housekeeping Genes to Study Human Gingival Stem Cells and Their In Vitro Osteogenic Differentiation Using Real-Time RT-qPCR. Stem Cells Int. 2016;2016:6261490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Okamura K, Inagaki Y, Matsui TK, Matsubayashi M, Komeda T, Ogawa M, Mori E, Tanaka Y. RT-qPCR analyses on the osteogenic differentiation from human iPS cells: an investigation of reference genes. Sci Rep. 2020;10:11748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Franko N, Vrščaj LA, Zore T, Ostanek B, Marc J, Lojk J. TBP, PPIA, YWHAZ and EF1A1 Are the Most Stably Expressed Genes during Osteogenic Differentiation. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |