Published online Jul 26, 2023. doi: 10.4252/wjsc.v15.i7.701
Peer-review started: December 29, 2022
First decision: April 27, 2023
Revised: May 18, 2023
Accepted: June 25, 2023
Article in press: June 25, 2023
Published online: July 26, 2023
Processing time: 208 Days and 3.2 Hours
Mesenchymal stromal cells (MSCs) are multipotent cell populations obtained from fetal and adult tissues. They share some characteristics with limb bud meso
To evaluate the potential of MSCs to differentiate into skeletal lineages and gene
We used the experimental system of RLs from dissociated-reaggregated human placenta (PL) and umbilical cord blood (UCB) MSCs. After being harvested and reaggregated in a pellet, cultured cells were introduced into an ectodermal cover obtained from an early chicken limb bud. Next, this filled ectoderm was grafted into the back of a donor chick embryo. Under these conditions, the cells received and responded to the ectoderm’s embryonic signals in a spatiotemporal manner to differentiate and pattern into skeletal elements. Their response to differentiation and morphogenetic signals was evaluated by quantitative poly
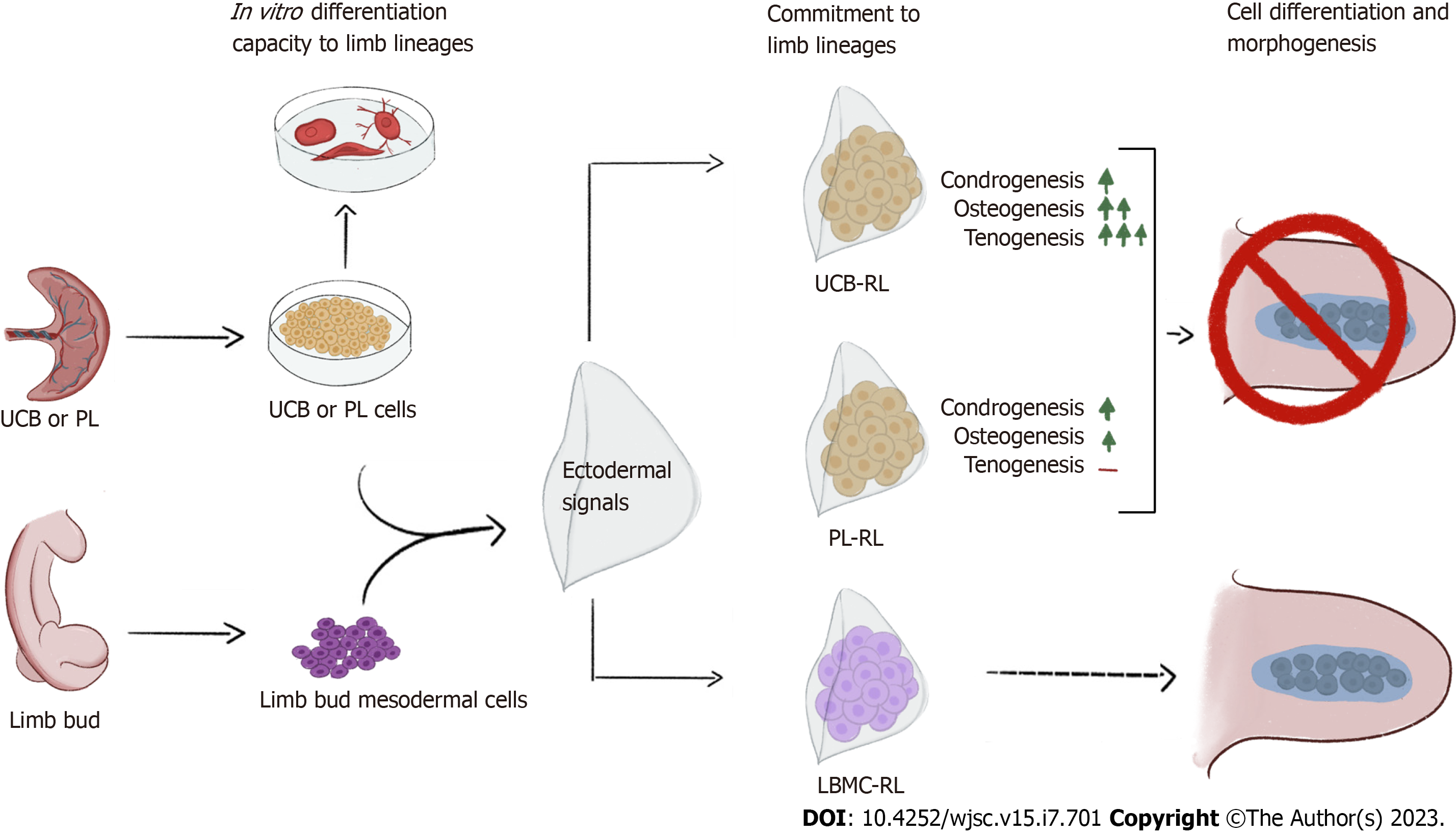
We found that human PL-MSCs and UCB-MSCs constituting the RLs expressed chondrogenic, osteogenic, and tenogenic molecular markers while differentially committing into limb lineages but could not generate complex structures in vivo. MSCs-RL from PL or UCB were committed early to chondrogenic lineage. Nevertheless, the UCB-RL osteogenic commitment was favored, although preferentially to a tenogenic cell fate. These findings suggest that the commitment of MSCs to differentiate into skeletal lineages differs according to the source and is independent of their capacity to generate skeletal elements or connective tissue in vivo. Our results suggest that the failure to form skeletal structures may be due to the intrinsic characteristics of MSCs. Thus, it is necessary to thoroughly evaluate the biological aspects of MSCs and how they respond to morphogenetic signals in an in vivo context.
PL-MSCs and UCB-MSCs express molecular markers of differentiation into skeletal lineages, but they are not sufficient to generate complex skeletal structures in vivo.
Core Tip: Human mesenchymal stromal cells (MSCs) from umbilical cord blood or placenta can differentiate into osteogenic and chondrogenic lineages in culture systems and have been used in regenerative medicine. Here, we used the recombinant limb (RL) model to provide evidence that MSCs do not have the ability to generate skeletal structures in vivo. MSCs received and responded to the ectoderm’s embryonic spatiotemporal signals in this RL system. However, the expression of differentiation markers of skeletal lineages was not sufficient to generate skeletal structures in vivo.
- Citation: Marín-Llera JC, García-García D, Garay-Pacheco E, Adrian Cortes-Morales V, Montesinos-Montesinos JJ, Chimal-Monroy J. Commitment of human mesenchymal stromal cells to skeletal lineages is independent of their morphogenetic capacity. World J Stem Cells 2023; 15(7): 701-712
- URL: https://www.wjgnet.com/1948-0210/full/v15/i7/701.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i7.701
The limbs have an intricate anatomy that results from highly coordinated morphogenesis and cellular differentiation processes, which establish the correct position and shape of muscles, bones, cartilage, tendons, skin, and nerves[1]. During embryonic development, limb bud mesodermal cells (LBMCs) form a bud covered by a layer of ectodermal cells. The morphogenesis and differentiation of limb tissues are controlled by the response to signals that emanate from three signaling centers: The apical ecto
SRY-box transcription factor 9 (SOX9) is the earliest molecular marker of chondrogenic commitment[3]. Besides, SOX9 induces the expression of extracellular matrix proteins such as collagen type 2 (Col2α) and aggrecan (ACAN)[4]. On the other hand, growth differentiation factor 5 (GDF5) promotes cell condensa
Regenerative medicine has emerged as an alternative treatment for degenerative diseases or an adjuvant therapy[15,16]. To this end, the potential use of mesenchymal stromal cells (MSCs) has garnered great interest. Because MSCs share characteristics with LBMCs and have the potential to differentiate into osteogenic or chondrogenic lineages[17-21], in this work, we used the recombinant limb (RL) model to evaluate the capacity of human MSCs to form complex skeletal structures[22]. The RL model recapitulates the differentiation, morphogenesis, and patterning programs that occur during normal limb development[23-25]. RLs consist of an ectodermal cover obtained from an early chicken LB (CK-LB) filled with dissociated-reaggregated cells grafted into the back of a donor chick embryo. Because of the high capacity of human MSCs from umbilical cord blood (UCB-MSCs) or placenta (PL-MSCs) to differentiate into osteogenic and chondrogenic lineages, we evaluated their ability to generate skeletal structures in vivo. Our results demonstrate that although MSCs receive and respond to the ectoderm’s embryonic spatiotemporal signals, they do not generate complex skeletal structures, possibly due to the intrinsic characteristics of MSCs.
MSCs were collected from Villa Coapa Hospital, Mexican Social Security Institute (IMSS; Mexico City, Mexico) according to their ethical guidelines, including informed consent. MSCs derived from bone marrow (BM; n = 3) were used to validate the specificity of all primers used in this study. BM cells were obtained from hematologically normal BM transplantation child donors at the Bernardo Sepulveda Hospital (National Medical Center, IMSS). A cell population enriched for MSCs was isolated according to Montesinos et al[25] using a negative selection procedure [RosetteSepTM system; STEMCELL Technologies Inc. (STI), Vancouver, Canada] and a Ficoll gradient. The cells were resuspended in low-glucose Dulbecco’s modified Eagle medium (Lg-DMEM; Gibco BRL, Rockville, MD, United States) supplemented with 10% fetal bovine serum (FBS) and seeded at a density of 0.2 × 106 cells/cm2 into T25 cell culture flasks (Corning Inc., Costar, New York, NY, United States). After 4 d, the nonadherent cells were removed by pipetting, and fresh medium was added. Every 5 d, a medium change was performed. When the cultures reached 80% confluence, they were trypsinized (0.05% trypsin, 0.53 mmol/L EDTA; Gibco BRL, New York, NY, United States) and subcultured at a density of 0.01 × 106 cells/cm2 into T75 flasks (Corning). In the second passage, cells were harvested and analyzed.
UCB-MSC and PL-MSC samples were obtained from two volunteer donors from normal full-term deliveries according to the institutional guidelines of Troncoso Hospital, IMSS, as previously described by Montesinos et al[25]. UCB-derived MSCs were obtained using a negative selection procedure (RosetteSepTM system; STI), as described for BM. Cells were resuspended in Lg-DMEM (Gibco) supplemented with 10% FBS (STI) and seeded at a density of 0.2 × 106 cells/cm2 in T25 culture flasks (corning), and subsequent cultures were manipulated as described for BM. PL-derived MSCs were obtained using an enzymatic digestion procedure. The internal area (about 2 cm3) of the central PL lobules was washed with phosphate-buffered saline (PBS) and dissected into small pieces with sterilized scissors and forceps. The chopped tissues were digested with trypsin-EDTA (Gibco) at 37 °C for 10 min. A single-cell suspension was collected by flushing the tissue parts through a 100-μm nylon filter (Falcon; Becton, Dickinson and Company, San Jose, CA, United States) with Lg-DMEM containing 10% FBS. The cell suspension was centrifuged for 10 min at 400 g to collect the cell pellet. Then, the cells were resu
RLs were established according to the protocol by Marín-Llera et al[24]. PL-MSCs, UCB-MSCs, or 22 Hamburger-Hamilton (HH) CK-LBMCs were collected to stuff 22 HH CK limb ectoderms, according to Hamburger and Hamilton[26]. After obtaining PL-MSCs, UCB-MSCs, or CK-LBMCs, they were centrifuged at 3000 rpm and incubated at 37 °C between 1.5 h and 2 h to form a compact pellet. Separately, to obtain ectoderms, LBs from about 10 of the 22 HH embryos were dissected in PBS, transferred to a tube, and digested in 0.5% trypsin in PBS for 30 min at 37 °C. Then, the LBs were transferred to PBS supplemented with 10% FBS, and the ectoderm was peeled off. Next, the pellet of PL-MSCs, UCB-MSCs, or CK-LBMCs was detached from the bottom of the tube and transferred to a small petri dish containing the ectoderms. Later, a pellet fragment was stuffed into each ectoderm. Ectoderms were individually transferred into a previously windowed 22 HH chick embryo, positioned between somites 15-20, and fixed with palladium wires over a previously scratched wound. Manipulated embryos were incubated for 24 h, 48 h, or 72 h at 37.5 °C until their collection and processing. The leftover pellet from three sources was independently frozen to evaluate its basal expression compared to RLs by quantitative polymerase chain reaction (qPCR). All RL experiments were performed in triplicate.
RNA from RLs was extracted with NucleoSpin RNA (Cat. No. 740955; Macherey-Nagel, Düren, Ger
For Alcian blue staining of RLs, samples were fixed in 5% trichloroacetic acid (Cat. No. A-5268; Sigma-Aldrich, St. Louis, MO, United States) for 24 h and stained with 1% Alcian blue in ethanol-hydrogen chloride for 24 h. Then, the RLs were transferred to 100% ethanol for 24 h and cleared with methyl salicylate (Cat. No. M-2047; Sigma-Aldrich) until the skeleton was observed. RL images were acquired with the AxioZoom v16 microscope (Carl Zeiss, Oberkochen, Germany) using ZEN lite software (Carl Zeiss). After image acquisition, Alcian blue-stained RLs were dehydrated in ethanol and xylol before embedding in paraffin. Ten-micrometer sections were obtained with a microtome (RM2125 RTS; Leica, Wetzlar, Germany). For hematoxylin and eosin (H&E) staining, samples were rehydrated with an ascending gradient of xylol-ethanol, followed by incubation with H&E dyes. Next, slides were dehydrated with ethanol-xylol and mounted with DPX medium (Cat. 44581; Sigma-Aldrich). Images were acquired with the Olympus BX51-WI microscope equipped with a fluorescence and gyratory disc unit (Tokyo, Japan) using Stereo Investigator 9 software (MicroBrightField Inc., Colchester, VT, United States).
RLs were incubated in 1 μM LysoTracker Red DND-99 (Cat. L7528; Molecular Probes, Eugene, OR, United States) at 37 °C for 15 min. Then, the samples were rinsed with PBS and fixed in 4% paraformaldehyde overnight at 4 °C. Next, the RLs were dehydrated in an increasing methanol-PBS-Tween series and cleared with 2:1 benzylic alcohol-benzyl benzoate solution for 1 h (following the 1999 recommendations by Parish). Images were acquired with an Olympus BX51-WI microscope equipped with a fluorescence and gyratory disc unit using Stereo Investigator 9 software (MicroBrightField).
RLs were washed twice with PBS and fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH = 7.2) for 2 h. The fixation solution was washed three times with 0.1 M sodium cacodylate at 4 °C. For postfixation, samples were immersed in 1% osmium tetroxide in cacodylate solution for 2 h before dehydration in graded concentrations of ethanol. The samples were placed in microporous capsules submerged in pure ethanol and placed in the Critical Point drying equipment (SPI) for desiccation. Ethanol was replaced with liquid carbon dioxide until its critical constants (31.3 °C and 1072 PSI) were met. After drying, the sample was mounted on an aluminum specimen holder with a silver-based adhesive. The coating was performed by ionizing the sample with a layer of gold in a low vacuum ionizer. Images were acquired using the DSM-950 Carl Zeiss microscope.
RNA antisense probes were labeled with UTP-digoxigenin (Cat. No. 11209256910; Roche Applied Science, Indianapolis, IN, United States) and used for whole-mount in situ hybridization as previously described[6]. Samples were treated with 15 μg/mL proteinase K for 20 min at 21 °C, and the hybridization temperature was 68 °C. The Fgf8 signal was visualized with BM Purple substrate for alkaline phosphatase (Roche). Images were acquired with the AxioZoom v16 microscope (Carl Zeiss).
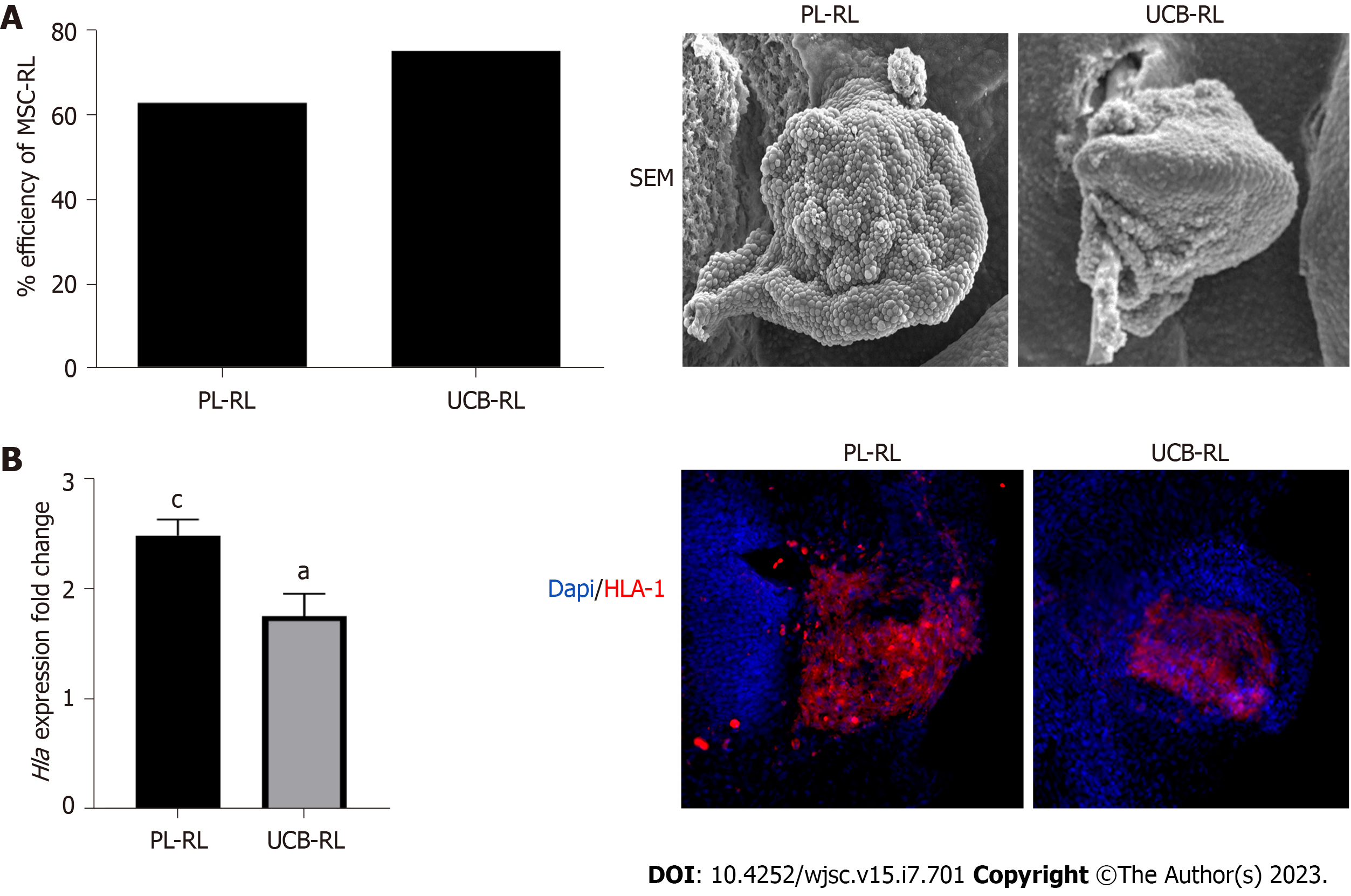
We used RLs as an experimental model to evaluate the capacity of PL-MSCs and UCB-MSCs to differentiate into limb tissues and to determine their capacity to generate well-organized tissues in an in vivo context. In the first instance, we evaluated the competence of human MSCs to respond to embryonic signals present in the limb ectoderm (Figure 1). The results showed that 24 h postimplantation (hpi), the PL-MSCs and UCB-MSCs were successfully integrated and formed RLs, with an efficiency of 62.95% and 76.87%, respectively (Figure 1A). Next, we measured the expression and distribution of major histocompatibility complex 1 (HLA1) to determine whether the RLs were exclusively formed by MSCs. We observed that both MSCs-RLs expressed significantly higher levels of HLA1 compared with their own pellet before transplantation. In addition, the cells in the center of the RLs were HLA1+ (Figure 1B). These results demonstrated the successful establishment of RLs composed of human MSCs for the evaluation of MSC behavior in vivo.
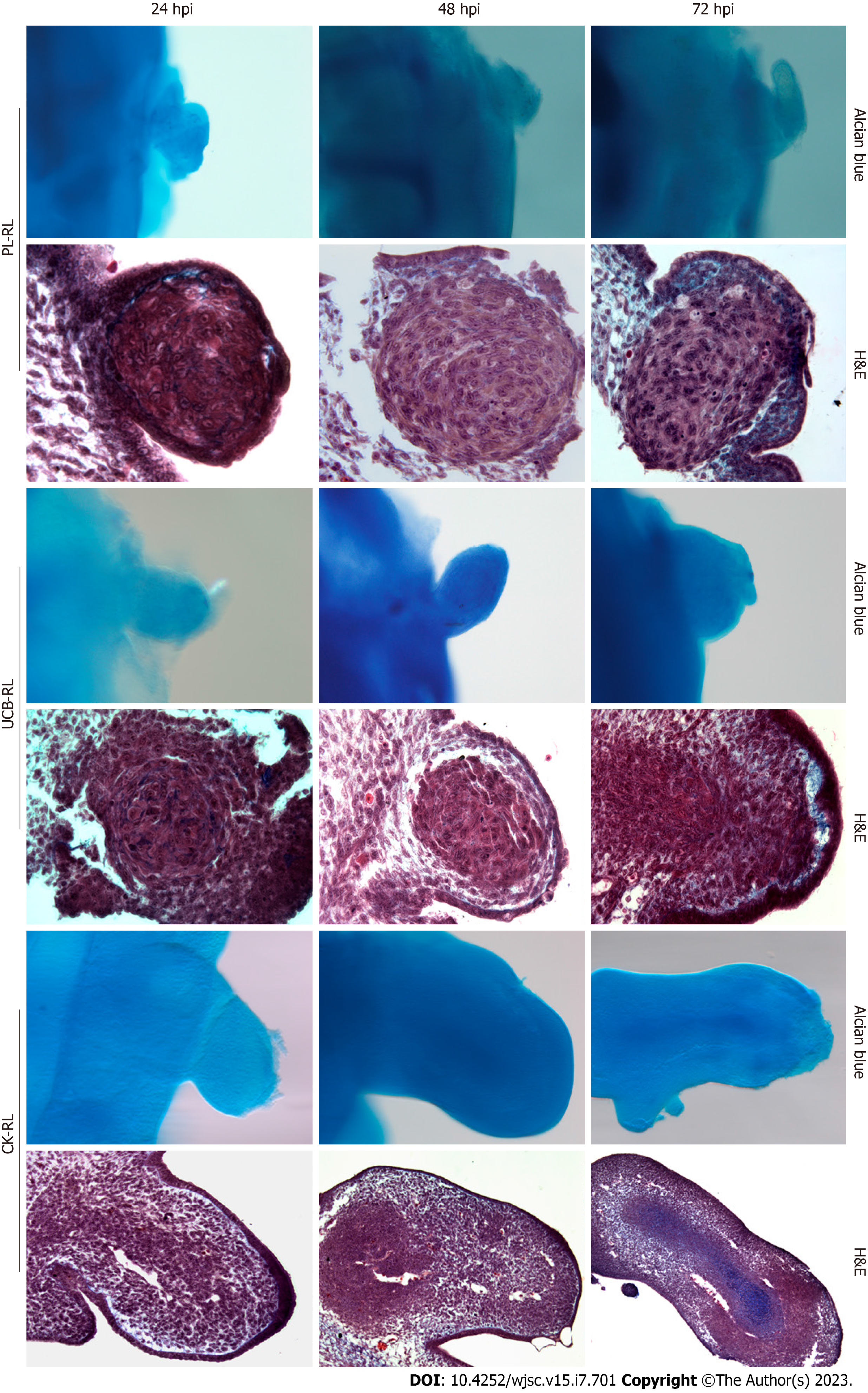
To assess the capacity of human MSCs to form well-organized tissues, the phenotype of MSC-RLs and their cell organization were evaluated at 24 hpi, 48 hpi, and 72 hpi and compared with RLs formed with CK-LBMCs as a reference of the morphogenetic behavior under RL conditions. In contrast to CK-RL, no evidence of central skeletal elements was observed in RLs from either MSC source (Figure 2). Histolo
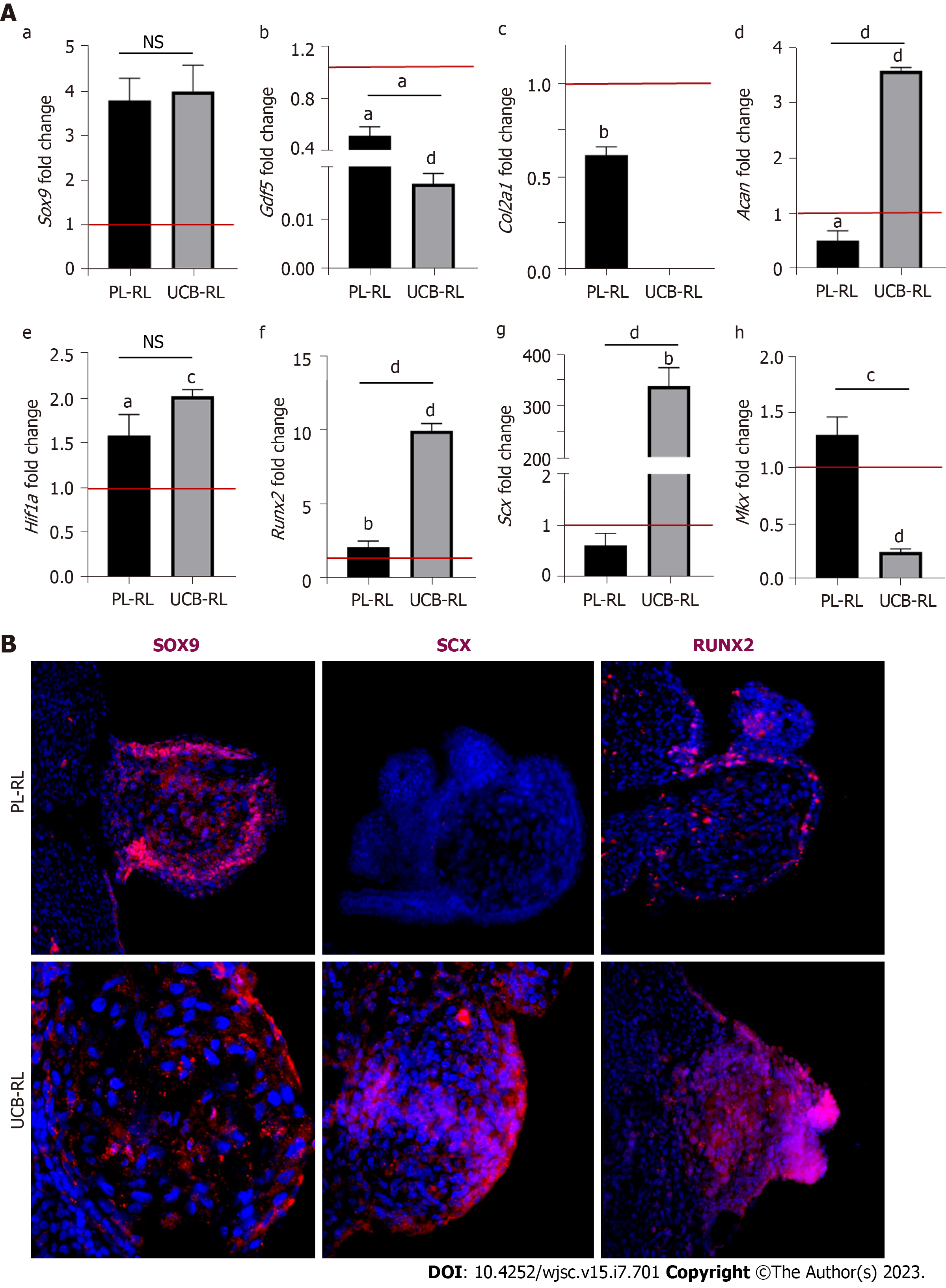
In contrast to CK-RL, histological analyses of MSCs-RLs did not indicate specific tissue formation at any evaluated time. Thus, to determine if the PL-MSCs and UCB-MSCs committed to limb cellular lineages during MSCs-RL formation, we evaluated the expression profile of chondrogenic (Sox9, Gdf5, Col2a1, Acan, and Hif1a), osteogenic (Runx2), and tenogenic (Scx and Mkx) differentiation genes at 24 hpi in MSCs-RLs from both sources (Figure 3A). The results confirmed that MSCs commit to limb lineages but respond differentially to embryonic ectodermal signals as the expression of molecular markers of chondrogenic, osteogenic, and tenogenic lineages was induced at different levels in MSCs-RLs from both sources compared with its initial pellet (Table 1).
| Source | Sox9 | Col2a1 | Acan | Gdf5 | Hif1α | Runx2 | Scx | Mkx |
| UCB-RL vs UCB-pellet | 3.99 ± 0.4, P < 0.0004 | ND | 3.58 ± 0.10, P < 0.0001 | 0.01 ± 00.003, P < 0.0001 | 1.58 ± 0.79, P < 0.0002 | 9.96 ± 0.65 P < 0.0001 | 377.61 ± 78.24, P < 0.002 | 0.24 ± 0.03, P < 0.0001 |
| PL-RL vs PL-pellet | 3.79 ± 0.4, P < 0.0007 | 0.61 ± 0.12, P < 0.002 | 0.55 ± 0.21, P < 0.052 | 0.56 ± 0.12, P < 0.01 | 1.61 ± 0.28, P < 0.03 | 2.18 ± 0.59, P < 0.03 | 0.61 ± 0.33, P < 0.14 | 1.29 ± 0.22, P < 0.21 |
Comparative analyses of Sox9 expression levels between MSCs-RLs showed no significant differences (P < 0.8421), but remarkably, it was highly induced in both MSCs after receiving ectodermal signals (Figure 3A, Table 1). By contrast, low Gdf5 expression was observed in both sources compared with its initial levels. In addition, it was significatively reduced in UCB-RLs compared with PL-RLs (P < 0.0205; Figure 3A, Table 1). Regarding later chondrogenic-associated genes, expression of Col2a1 was not detected in UCB-RLs, whereas its expression was significatively reduced in PL-RLs (Figure 3A, Table 1). On the other hand, Acan was overexpressed only in UCB-RLs and was significatively reduced in PL-RLs (P < 0.0001; Figure 3A, Table 1). Interestingly, the levels of Hif1a observed in both MSCs-RLs suggest that a hypoxic environment is favored under these conditions (Figure 3A, Table 1). Regarding osteo
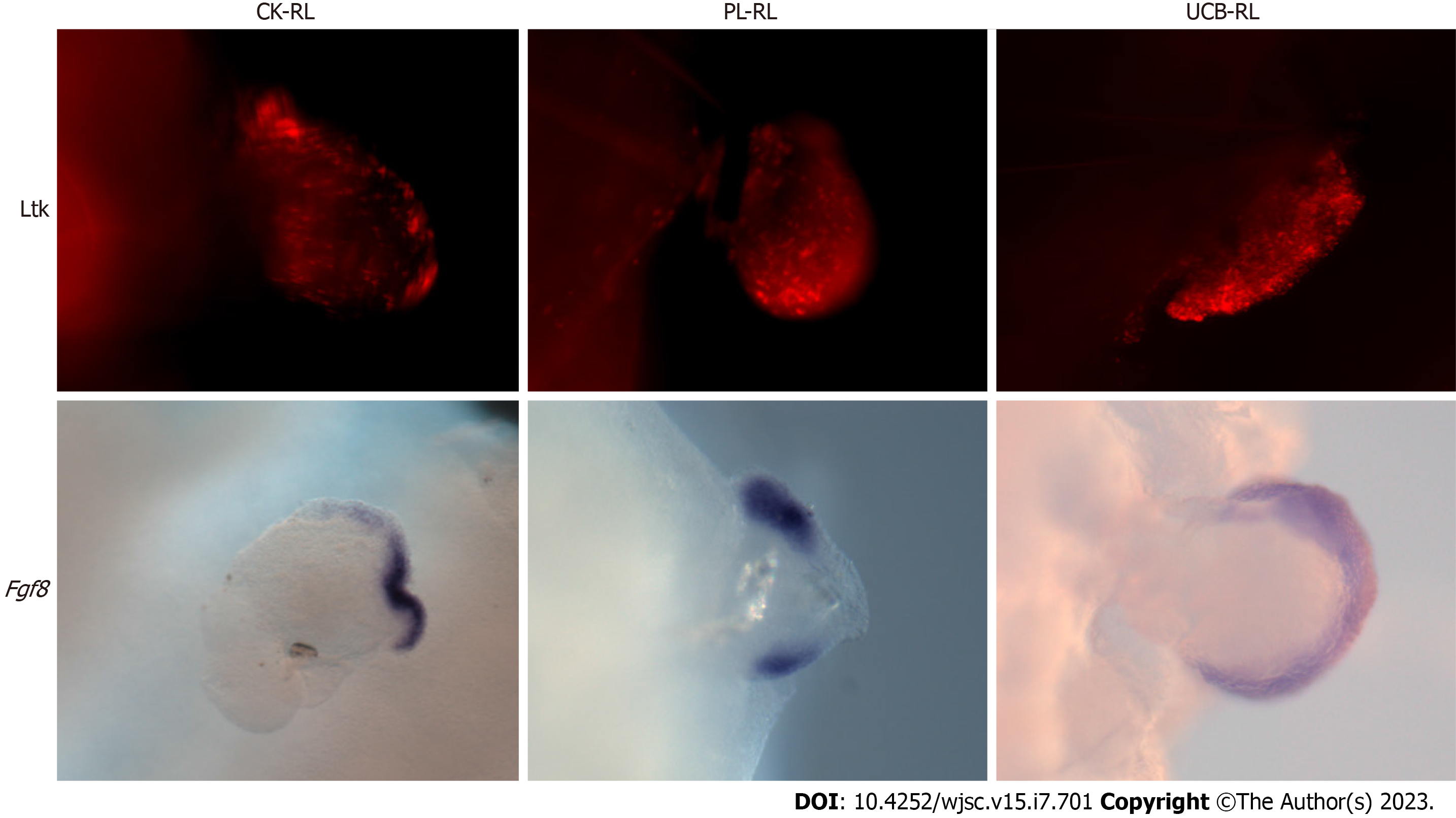
Signals from the ectoderm and AER are indispensable for maintaining cell survival and proliferation during limb development and RL formation[23]. During limb development, the undifferentiated cellular state and cell commitment balance depend on signals from the AER[2]. To determine whether the lack of ectodermal signals or massive cell death was the mechanism underlying the inability of MSC to form skeletal structures, ectoderm integrity by Fgf8 expression and lysotracker staining were evaluated in RLs (Figure 4). The results showed that the cell death process was present in the ectoderm of all evaluated RLs; UCB-RLs showed a higher number of dead cells than PL-RLs and CK-RLs. Interestingly, in PL-RLs and CK-RLs, dead cells were concentrated in the apical area of the ectoderm, whereas in UCB-RLs, dead cells were distributed homogeneously on the ectoderm of all samples evaluated (Figure 4). Despite the death of ectodermal cells, Fgf8 expression was maintained in some MSCs-RLs (see details in the figure legend) or its expression in PL-RLs was disrupted in the center of the ectoderm (Figure 4). These results suggest that at 24 hpi, signals continue to emanate from the AER, which are received by the MSCs or CK-LBMCs beneath the AER.
Cells commit to specific lineages once the expression of master genes occurs in response to inducing factors. However, the ability to interpret signals and acquire a particular fate and behavior depend on the developmental history of the cells. The capacity to organize complex structures depends on cell differentiation, cell recruitment, cell movement, differential cell proliferation, and cell death, which occur during the morphogenetic process of tissues and organs.
In this study, we used the RL model to evaluate the morphogenetic abilities of MSCs other than those observed in cell culture. Because RLs provide limb spatial-temporal signals, we evaluated the ability of MSCs to generate complex skeletal structures[23-25]. We found that human MSCs can integrate into an RL implanted in the CK embryo. PL- or UCB-MSCs from RLs respond differently to ectodermal signals, suggesting that the source of MSCs might be important to delineate a particular lineage in vivo (Figure 5). Although MSCs from both sources start the chondrogenic program with high levels of SOX9 expression, they do not necessarily equally follow and complete the cell differentiation process (Figure 5). In vitro studies have shown that UCB-MSCs preferentially differentiate into osteogenic lineages[17] and present a higher in vivo osteogenic and chondrogenic differentiation potential than BM-MSCs when grafted subcutaneously with a scaffold of hydroxyapatite/tricalcium phosphate[19]. By contrast, PL-MSCs preferentially differentiate into an osteogenic lineage when seeded into microcarriers or nanofiber scaffolds[20,21]. These findings suggest that depending on the tissue source, MSCs can represent a cell population with a different number of osteogenic or chondrogenic progenitors or both. Similarly, the molecular markers from chondrogenic and osteogenic lineages were induced at higher levels in UCB-RLs than in PL-RLs. However, the high expression levels of Runx2 and Scx genes did not lead to the formation of well-defined bone tissue or tendons. In addition, the gene expression pattern of Sox9, Scx, and Rux2 did not associate with a specific tissue pattern or arrangement inside the RLs. The expression of molecular markers was not sufficient to promote the formation of well-defined structures, likely because the morphogenetic program was not triggered. Interestingly, PL-RLs expressed high levels of Mkx while UCB-RL expressed high levels of Scx. It is unknown if in this system both genes are needed to form well-structured tendons. Nevertheless, the expression of Mkx or Scx can drive the in vitro differentiation of BM-MSCs to a tenocyte fate[13,27].
The inability of MSCs to generate complex structures suggests that MSC populations, in addition to being composed of a wide variety of cell types, are unable to initiate or follow a morphogenetic program in vivo. Although MSCs can differentiate into skeletal lineages, it does not guarantee that they will organize into skeletal elements. It is possible that the RL model lacks signals to support the formation of skeletal elements. However, CK-LBMCs respond to differentiation signals, resulting in the generation of skeletal elements. Thus, the RL system provides signals that instruct cells to form skeletal elements.
We did not observe exacerbated cell death in the MSCs-RLs, but in some MSCs-RLs the loss of Fgf8 expression in the AER was evident. Additional studies are needed to determine if this cell death results from the loss of FGF8 loop signaling between the AER and MSCs, or if MSCs cannot recover and maintain the loop to maintain FGF8 expression. Our data suggest that the differentiation process in MSCs is detached from the process of morphogenesis and patterning, although it is possible that addi
Although MSCs are considered relevant in regenerative medicine, their intrinsic cellular properties may explain why the reported therapeutic effects of MSCs are mostly indirect through immunomodulation or paracrine mechanisms rather than reliable integration into adult tissues or de novo tissue forma
This study demonstrates that the expression of differentiation markers of skeletal lineages in MSCs is not sufficient to generate skeletal structures in vivo, possibly due to the intrinsic characteristics of MSCs. In regenerative medicine, cells must be incorporated into well-defined tissues or generate new complex structures with a well-defined patterning. Thus, further application of MSCs in regenerative medicine needs to focus on understanding their biological characteristics to gain insights into how MSCs can integrate into adult tissues or properly rebuild tissues and organs.
Mesenchymal stem cells (MSCs) differentiate in vitro to different skeletal lineages; however, it is un
Although MSCs are considered relevant in regenerative medicine, reliable integration into adult tissues or de novo tissue formation has not been demonstrated. The application of MSCs in regenerative medicine needs to focus on understanding their biological characteristics to gain insights into how MSCs can integrate into adult tissues or properly rebuild tissues and organs.
To evaluate the ability of MSCs to organize and form complex skeletal structures in vivo under the influ
The recombinant limb (RL) is an experimental system that recapitulates the embryonic environment and its influence on cells to generate skeletal structures. Here, umbilical cord blood (UCB)-MSCs or placenta (PL)-MSCs were placed in an RL to assess their ability to form skeletal structures. The evaluation was conducted by Alcian blue staining, immunofluorescence, and quantitative polymerase chain reaction of molecular markers of skeletal lineages.
MSCs expressed molecular markers of skeletal lineages but were unable to generate complex skeletal structures. PL-MSCs or UCB-MSCs integrated into an RL implanted in a chicken embryo. They res
PL-MSCs or UCB-MSCs express molecular markers of skeletal lineages but do not organize into com
The application of MSCs to regenerative medicine needs to focus on understanding their biological characteristics to gain insights into how MSCs can integrate into adult tissues or properly rebuild tissues and organs.
The authors thank Maria Valeria Chimal-Montes de Oca for her artwork.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shamseldeen AM, Egypt; Yang YZ, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 341] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 2. | Marín-Llera JC, Garciadiego-Cázares D, Chimal-Monroy J. Understanding the Cellular and Molecular Mechanisms That Control Early Cell Fate Decisions During Appendicular Skeletogenesis. Front Genet. 2019;10:977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Chimal-Monroy J, Rodriguez-Leon J, Montero JA, Gañan Y, Macias D, Merino R, Hurle JM. Analysis of the molecular cascade responsible for mesodermal limb chondrogenesis: Sox genes and BMP signaling. Dev Biol. 2003;257:292-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 171] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 4. | Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9 Suppl A:S69-S75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 273] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 5. | Buxton P, Edwards C, Archer CW, Francis-West P. Growth/differentiation factor-5 (GDF-5) and skeletal development. J Bone Joint Surg Am. 2001;83-A Suppl 1:S23-S30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Merino R, Macias D, Gañan Y, Economides AN, Wang X, Wu Q, Stahl N, Sampath KT, Varona P, Hurle JM. Expression and function of Gdf-5 during digit skeletogenesis in the embryonic chick leg bud. Dev Biol. 1999;206:33-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 155] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134:3917-3928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 244] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 8. | Chen H, Ghori-Javed FY, Rashid H, Adhami MD, Serra R, Gutierrez SE, Javed A. Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation. J Bone Miner Res. 2014;29:2653-2665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 9. | Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2205] [Cited by in RCA: 2184] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 10. | Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3364] [Cited by in RCA: 3327] [Article Influence: 118.8] [Reference Citation Analysis (0)] |
| 11. | Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855-3866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 609] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 12. | Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697-2708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 433] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 13. | Ito Y, Toriuchi N, Yoshitaka T, Ueno-Kudoh H, Sato T, Yokoyama S, Nishida K, Akimoto T, Takahashi M, Miyaki S, Asahara H. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci U S A. 2010;107:10538-10542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 14. | Lejard V, Blais F, Guerquin MJ, Bonnet A, Bonnin MA, Havis E, Malbouyres M, Bidaud CB, Maro G, Gilardi-Hebenstreit P, Rossert J, Ruggiero F, Duprez D. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J Biol Chem. 2011;286:5855-5867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 15. | Huang Y, Wu Q, Tam PKH. Immunomodulatory Mechanisms of Mesenchymal Stem Cells and Their Potential Clinical Applications. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 109] [Reference Citation Analysis (0)] |
| 16. | Aguiar Koga BA, Fernandes LA, Fratini P, Sogayar MC, Carreira ACO. Role of MSC-derived small extracellular vesicles in tissue repair and regeneration. Front Cell Dev Biol. 2022;10:1047094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 17. | Lee RH, Kim B, Choi I, Kim H, Choi HS, Suh K, Bae YC, Jung JS. Characterization and expression analysis of mesenchymal stem cells from human bone marrow and adipose tissue. Cell Physiol Biochem. 2004;14:311-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 676] [Cited by in RCA: 684] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 18. | Sacchetti B, Funari A, Remoli C, Giannicola G, Kogler G, Liedtke S, Cossu G, Serafini M, Sampaolesi M, Tagliafico E, Tenedini E, Saggio I, Robey PG, Riminucci M, Bianco P. No Identical "Mesenchymal Stem Cells" at Different Times and Sites: Human Committed Progenitors of Distinct Origin and Differentiation Potential Are Incorporated as Adventitial Cells in Microvessels. Stem Cell Reports. 2016;6:897-913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 286] [Cited by in RCA: 321] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 19. | Igura K, Zhang X, Takahashi K, Mitsuru A, Yamaguchi S, Takashi TA. Isolation and characterization of mesenchymal progenitor cells from chorionic villi of human placenta. Cytotherapy. 2004;6:543-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 201] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 20. | Zhang D, Tong A, Zhou L, Fang F, Guo G. Osteogenic differentiation of human placenta-derived mesenchymal stem cells (PMSCs) on electrospun nanofiber meshes. Cytotechnology. 2012;64:701-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | García-García RD, Garay-Pacheco E, Marín-Llera JC, Chimal-Monroy J. Recombinant Limb Assay as in Vivo Organoid Model. Front Cell Dev Biol. 2022;10:863140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 22. | Elisa Piedra M, Borja Rivero1 F, Fernandez-Teran M, Ros MA. Pattern formation and regulation of gene expressions in chick recombinant limbs. Mech Dev. 2000;90:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 23. | Ros MA, Lyons GE, Mackem S, Fallon JF. Recombinant limbs as a model to study homeobox gene regulation during limb development. Dev Biol. 1994;166:59-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 56] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Marín-Llera JC, Fernández-Calderón M, Chimal-Monroy J. Chicken Recombinant Limbs Assay to Understand Morphogenesis, Patterning, and Early Steps in Cell Differentiation. J Vis Exp. 2022;2022:179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Montesinos JJ, Flores-Figueroa E, Castillo-Medina S, Flores-Guzmán P, Hernández-Estévez E, Fajardo-Orduña G, Orozco S, Mayani H. Human mesenchymal stromal cells from adult and neonatal sources: comparative analysis of their morphology, immunophenotype, differentiation patterns and neural protein expression. Cytotherapy. 2009;11:163-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1326] [Cited by in RCA: 1482] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 27. | Liu H, Zhang C, Zhu S, Lu P, Zhu T, Gong X, Zhang Z, Hu J, Yin Z, Heng BC, Chen X, Ouyang HW. Mohawk promotes the tenogenesis of mesenchymal stem cells through activation of the TGFβ signaling pathway. Stem Cells. 2015;33:443-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 131] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 28. | Almeida A, Lira R, Oliveira M, Martins M, Azevedo Y, Silva KR, Carvalho S, Cortez E, Stumbo AC, Carvalho L, Thole A. Bone marrow-derived mesenchymal stem cells transplantation ameliorates renal injury through anti-fibrotic and anti-inflammatory effects in chronic experimental renovascular disease. Biomed J. 2022;45:629-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Rackham CL, Chagastelles PC, Nardi NB, Hauge-Evans AC, Jones PM, King AJ. Co-transplantation of mesenchymal stem cells maintains islet organisation and morphology in mice. Diabetologia. 2011;54:1127-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 106] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 30. | Fu Z, Chu Y, Geng X, Ma Y, Chi K, Song C, Liao S, Hong Q, Wu D, Wang Y. Artificial Kidney Capsule Packed with Mesenchymal Stem Cell-Laden Hydrogel for the Treatment of Rhabdomyolysis-Induced Acute Kidney Injury. ACS Biomater Sci Eng. 2022;8:1726-1734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 31. | Kumar S, Wan C, Ramaswamy G, Clemens TL, Ponnazhagan S. Mesenchymal stem cells expressing osteogenic and angiogenic factors synergistically enhance bone formation in a mouse model of segmental bone defect. Mol Ther. 2010;18:1026-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 123] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 32. | Gao F, Chiu SM, Motan DA, Zhang Z, Chen L, Ji HL, Tse HF, Fu QL, Lian Q. Mesenchymal stem cells and immunomodulation: current status and future prospects. Cell Death Dis. 2016;7:e2062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 611] [Cited by in RCA: 826] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 33. | Sarsenova M, Kim Y, Raziyeva K, Kazybay B, Ogay V, Saparov A. Recent advances to enhance the immunomodulatory potential of mesenchymal stem cells. Front Immunol. 2022;13:1010399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 34. | Wang M, Yuan Q, Xie L. Mesenchymal Stem Cell-Based Immunomodulation: Properties and Clinical Application. Stem Cells Int. 2018;2018:3057624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 342] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 35. | Xiang XN, Zhu SY, He HC, Yu X, Xu Y, He CQ. Mesenchymal stromal cell-based therapy for cartilage regeneration in knee osteoarthritis. Stem Cell Res Ther. 2022;13:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 96] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 36. | Waterman RS, Tomchuck SL, Henkle SL, Betancourt AM. A new mesenchymal stem cell (MSC) paradigm: polarization into a pro-inflammatory MSC1 or an Immunosuppressive MSC2 phenotype. PLoS One. 2010;5:e10088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 798] [Cited by in RCA: 949] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 37. | Tomchuck SL, Zwezdaryk KJ, Coffelt SB, Waterman RS, Danka ES, Scandurro AB. Toll-like receptors on human mesenchymal stem cells drive their migration and immunomodulating responses. Stem Cells. 2008;26:99-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 346] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 38. | Fu Y, Karbaat L, Wu L, Leijten J, Both SK, Karperien M. Trophic Effects of Mesenchymal Stem Cells in Tissue Regeneration. Tissue Eng Part B Rev. 2017;23:515-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 193] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 39. | Hofer HR, Tuan RS. Secreted trophic factors of mesenchymal stem cells support neurovascular and musculoskeletal therapies. Stem Cell Res Ther. 2016;7:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 264] [Article Influence: 29.3] [Reference Citation Analysis (0)] |