Published online Jun 26, 2023. doi: 10.4252/wjsc.v15.i6.589
Peer-review started: February 9, 2023
First decision: April 10, 2023
Revised: April 18, 2023
Accepted: May 5, 2023
Article in press: May 5, 2023
Published online: June 26, 2023
Processing time: 137 Days and 8.9 Hours
Accumulating evidence suggests that the maxillary process, to which cranial crest cells migrate, is essential to tooth development. Emerging studies indicate that Cd271 plays an essential role in odontogenesis. However, the underlying mecha
To establish the functionally heterogeneous population in the maxillary process, elucidate the effects of Cd271 deficiency on gene expression differences.
p75NTR knockout (Cd271-/-) mice (from American Jackson laboratory) were used to collect the maxillofacial process tissue of p75NTR knockout mice, and the wild-type maxillofacial process of the same pregnant mouse wild was used as control. After single cell suspension, the cDNA was prepared by loading the single cell suspension into the 10x Genomics Chromium system to be sequenced by NovaSeq6000 sequencing system. Finally, the sequencing data in Fastq format were obtained. The FastQC software is used to evaluate the quality of data and CellRanger analyzed the data. The gene expression matrix is read by R software, and Seurat is used to control and standardize the data, reduce the dimension and cluster. We search for marker genes for subgroup annotation by consulting literature and database; explore the effect of p75NTR knockout on mesenchymal stem cells (MSCs) gene expression and cell proportion by cell subgrouping, differential gene analysis, enrichment analysis and protein-protein interaction network analysis; understand the interaction between MSCs cells and the differentiation trajectory and gene change characteristics of p75NTR knockout MSCs by cell communication analysis and pseudo-time analysis. Last we verified the findings single cell sequencing in vitro.
We identified 21 cell clusters, and we re-clustered these into three subclusters. Importantly, we revealed the cell–cell communication networks between clusters. We clarified that Cd271 was significantly associated with the regulation of mineralization.
This study provides comprehensive mechanistic insights into the maxillary- process-derived MSCs and demonstrates that Cd271 is significantly associated with the odontogenesis in mesenchymal populations.
Core Tip: Our study reveals the following findings: (1) High cellular heterogeneity and molecular details; (2) Significant functional and signaling differences between cell types; (3) Novel subclusters of mesenchymal stem cells; and (4) Crucial cell-cell interactions of mesenchymal subpopulations. We provided new insights into the biological features of mesenchymal stem cells at the single cell level. Our findings contribute to thorough exploration of the mechanism of Cd271 in regulating odontogenesis and osteogenesis which add to the theory of tooth development.
- Citation: Zhang YY, Li F, Zeng XK, Zou YH, Zhu BB, Ye JJ, Zhang YX, Jin Q, Nie X. Single cell RNA sequencing reveals mesenchymal heterogeneity and critical functions of Cd271 in tooth development. World J Stem Cells 2023; 15(6): 589-606
- URL: https://www.wjgnet.com/1948-0210/full/v15/i6/589.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i6.589
During the early stage of embryogenesis, cranial neural crest cells migrate throughout the maxillary and mandibular processes, which are defined as ecto-mesenchymal stem cells (MSCs)[1,2]. Ecto-MSCs were regarded as the primitive dental cells in the classical theory of tooth development. To date, in-depth studies of the odontogenesis and osteogenesis of MSCs are still lacking.
Cd271 (low-affinity nerve growth factor receptor, p75 neurotrophic receptor) is a member of the tumor necrosis factor receptor superfamily, and is implicated in various biological functions[3]. It is used as a specific cell surface marker for purifying and identifying MSCs[4]. Previous studies focused on cellular physiological functions in processes, such as migration, proliferation, differentiation, survival and apoptosis[5-8]. Cd271 is involved in the regulation of morphogenesis and the development of various tissues, including nerves, fat, liver and teeth[9-13]. In addition to these features, emerging studies indicate that Cd271 plays a critical role in initiating tooth development, differentiation and mineralization of odontogenic stem cells. Mitsiadis et al[14] indicated that Cd271 surrounded the developing tooth germ within nerve fibers and was highly expressed in the epithelium and mesenchyme in the early stages of odontogenesis within the cells of origin[14]. The spatial–temporal expression of Cd271 was similar to the mineralization factor Runx2 during the early development of tooth germ[15], which was supported by other studies[16,17]. These studies indicate that Cd271 is involved in the physiological processes of tooth development and biomineralization. However, the molecular mechanisms of Cd271, as a transmembrane signaling molecule, in the morphogenesis and development of teeth remains largely unknown.
Conventionally, MSCs were considered a unified group of fusiform cells. Nevertheless, accumulating evidence suggests that MSCs are functionally and morphologically heterogeneous in essence[18,19]. High-throughput single cell RNA sequencing (scRNA-Seq) is powerful for disclosing the complexity and diversity of cells and relationships among genes involved in tissues, and offers an opportunity to explore unbiased gene expression profiling of cells[20]. scRNA-Seq also provides insights into specific changes in cell lineages, trajectory inference, and the identification of biomarkers[21,22]. scRNA-seq has been used to study MSCs derived from adipose tissue, bone marrow, endometrium, placenta and dental pulp[23-27]. However, application of scRNA-seq in maxillary-process-derived MSCs is still absent.
Herein, we used the maxillary process from mouse embryos as a model to understand the development of maxillary-process-derived MSCs. We applied scRNA-Seq analysis to elucidate the cellular heterogeneity and explore molecular details better compared with conventional methods. Our study provides novel insights into the biological features of MSCs at the single cell level and the mechanism of Cd271 in regulating odontogenesis and osteogenesis.
Cd271 knockout (Cd271-/-) and wild-type (Cd271+/+) mice were used in this study. The Cd271-/- mice were gifts from The Jackson Laboratory (Bar Harbor, ME, United States). These mutant mice that exhibit the targeted deletion of exon III of the Cd271 locus do not express functional full-length Cd271. All animal experiments were performed according to the protocols approved by the Medical Ethics Committee of Wenzhou Medical University (No. wydw2019-0224). We completed experimental steps under ethical guidelines. The animal protocol was designed to minimize pain or discomfort to the animals. The animals were housed to laboratory conditions (23 °C, 12 h/12 h light/dark, 50% humidity, ad libitum access to food and water) prior to experimentation. The homozygotes of Cd271+/+ and Cd271-/- mice were mated to produce heterozygous offspring. The heterozygous mice were mated to generate three types of genotype embryos, Cd271+/+ and Cd271-/-and Cd271+/-. We obtained embryos through abdominal surgery, and chose homozygous embryos from embryonic day 16.5. The embryos were placed in a 6-cm Petri dish and washed with Phosphate buffer saline (PBS). We cut the amniotic membrane with ophthalmic scissors to separate the fetal mice and washed them with PBS again. After cutting the head of the fetus, we cut the maxillary process tissue under a stereomicroscope. After the removal of nonpurpose tissues like blood stains and fatty layers, we rinsed the tissue twice in PBS. The genotype of wild-type and Cd271 knockout mice was confirmed with a one-step genotyping kit (Vazyme, Nanjing, China).
Maxillary processes were dissociated into single cells in dissociation solution (0.35% collagenase IV5, 2 mg/mL papain, and 120 U/mL DNase I) in 37 °C water bath with shaking for 20 min at 100 rpm. Single cell suspensions were washed and resuspended to load to the 10X Chromium platform. More than 25000 single cells were captured and subjected to 10X Chromium Controller machine to generate gel beads-in-emulsion (GEMs). mRNA was prepared using the 10X Genomics Chromium Single Cell 3′ Reagent Kit V2 (10X Genomics, Pleasanton, CA, United States). Cells were divided by partitions into the GEMs along with GelBeads coated with oligos in this step. These oligos utilized poly-dT sequences to capture mRNAs and cell-specific and transcript-specific barcodes. The following cDNA amplification generated adequate quantities for library construction according to the standard protocol. Libraries were sequenced on an Illumina NovaSeq 6000 sequencing system (paired-end multiplexing run, 150 bp) by the LC-Bio Technology (Hangzhou, China) at a minimum depth of 20000 reads per cell.
We processed sequencing reads using CellRanger (version 6.0.1). Then we created objects by Seurat (version 4.0.5) R package, and merged data using the function of Merge[28]. The ratio of mitochondrial genes to all genetic material was used to judge whether a cell was in a steady state. Generally, it was thought that a cell might be in a state of stress when it had a higher proportion of mitochondrial genes. Therefore, we filtered cells with > 10% mitochondrial gene content. Considering potential cellular diploidy, we filtered cells with < 1000 or > 6000 genes. After the above steps, we finally obtained 17426 cells.
Data were normalized using the log-normalization method. After controlling for the relationship between mean expression and dispersion, we identified highly variable genes in individual cells. We input variable genes to perform principal component analysis (PCA) and identified significant principal components based on the function of JackStraw[29]. A total of 20 principal components were selected as statistically significant inputs to the Uniform Manifold Approximation and Projection (UMAP). We examined the distribution of UMAP and PCA between these samples. The data showed correlations. We compared the mean expression of genes between samples and found an excellent Pearson correlation between them. We divided the cells clustered by the FindCluster into 21 clusters.
We identified cell types based on specific maker genes. The MSCs lineage was identified by Col1a1 and Col3a1. Lgals7 and Krtdap marked the epithelial cell lineage; muscle cell marker genes included Actc1 and Tnnt1; macrophage marker genes included Pf4 and C1qb; glial marker genes included Dct and Ptgds; T cell marker genes included Cma1 and Cpa3; endothelial cell marker genes included Egfl7 and Cdh5; and perivascular cell marker genes included Rgs5 and Ndufa4 L2. The FindAllMarkers function was used to find differentially expressed genes between each cell type.
Gene set enrichment analysis (GSEA) can identify whether predefined gene sets show significant differences between biological processes by a computational method. Typically, GSEA is used to estimate expression in dataset samples. To study differences in the biological processes between the two groups, we downloaded the reference gene set c2.cp.kegg.v7.4.entrez.gmt from the MSigDB database on the basis of the gene expression profiling dataset[30]. We used GSEA included in the R package ClusterProfiler to perform enrichment analysis and visualization of the dataset. The cut-off standards were set as nominal P < 0.05, FDR q < 0.25 and normalized enrichment score (NES) > 0.6.
On account of our main focus on MSCs, we extracted a subpopulation of MSCs. Based on hierarchical clustering and defined marker genes, we reclustered the MSCs using the FindClusters function and assigned the MSCs to 13 cell subpopulations. The progenitor subpopulation was identified by Cdk1 and Dkk1, the osteoblasts subpopulation was identified by Runx2 and Sp7, and the fibroblasts subpopulation was determined by maker genes Dlk1 and Shox2. We next counted the proportions and number of these cell subpopulations in wild-type or Cd271 knockout mice.
Gene Ontology (GO) is an approach for functional enrichment analysis of genes in diverse levels and dimensions. GO analysis comprises three levels: Biological process, molecular function and cellular components. The Wilcoxon rank-sum test was used to identify genes differentially expressed in MSCs between wild-type and Cd271 knockout samples (logFC > 0.25, P < 0.05). Differentially expressed genes that were identified between wild-type and Cd271 knockout samples were subjected to GO functional annotation by the ClusterProfiler (version 4.2.0) R package to identify the significantly enriched biological processes[31].The enrichment results were visualized in the form of a lollipop plot, and the significance threshold for the enrichment analysis was set at a corrected P < 0.05.
The STRING online database (https://string-db.org/) analyzed the interaction between the marker genes. We constructed a protein–protein interaction (PPI) network for the results obtained by Cytoscape (3.9.0). The functional interactions between the proteins expressed by the genes were mapped, including direct physical interactions and indirect functional correlations.
Cell differentiation of MSCs (progenitors, osteoblasts and fibroblasts) was inferred by the Monocle (2.22.0) R package and default parameters recommended by the developers[32]. The integrated gene expression profiling of each cell type was exported from Seurat into Monocle to establish the cellular gene dataset. The variable genes were defined by the process of DispersionTable, and the cells were sorted with the function of setOrderingFilter. The DDRTree method was used to reduce the dimension, and the orderCells function was used to estimate the cell arrangement along the trajectory. Based on clustering characteristics and marker gene analysis, we obtained the trajectory map of the differentiation time of MSCs. The study of each trajectory used a standard protocol with default parameters.
We identified the underlying interactions between MSCs and other cell populations by CellChat (version 1.1.3) (http://www.cellchat.org/) R package, which is commonly used to analyze cell–cell communication networks from single cell transcriptome sequencing profiling[33]. Taking advantage of CellChat, we inferred the scRNA-Seq data quantitatively and researched cell–cell communication networks. We predicted the main ingoing and outgoing signal patterns by network analysis and pattern recognition methods, as well as the coordination function between cells and signals. We counted all the important receptor–ligand pairs in the intercellular signal transmission by bubble chart, and we selected the signaling with a higher contribution to the cell for network centrality analysis.
MSCs were isolated from embryonic maxillary processes. After washing with high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco, United States) three times, the cells were digested with trypsin–EDTA (Gibco) at 37 °C for 10 min, and centrifuged at 1000 rpm for 5 min. The cells were resuspended in a growth medium with DMEM, 10% fetal bovine serum (FBS) (Gibco), and 1% antibiotics (100 mg/mL penicillin and 100 µg/mL streptomycin) to generate primary MSCs. We maintained the cells at 37 °C in a 5% CO2 humidified incubator and replaced the medium every 3 d.
For mineralization induction, cells were seeded in DMEM supplemented with 10% FBS at 4 × 104 cells per well in 24-well plates. At 75%–80% confluence, we changed the medium to mineralization induction medium, which consisted of DMEM with 10% FBS, 100 IU/mL penicillin, 100 IU/mL streptomycin, 0.1 μM dexamethasone, 10 mmol/L β-glycerol phosphate (Sigma–Aldrich, St. Louis, MO, United States) and 50 μM ascorbic acid (Sigma–Aldrich). We cultured the cells for up to 7 d, and replaced the medium every 3 d.
The alkaline phosphatase (ALP) activity kit (Nanjing Jiancheng Biotech, China) was used to detect intracellular ALP activity. Cells were lysed in RIPA lysis buffer (Beyotime, China) without protease and phosphatase inhibitors to induce mineralization for 7 d. We centrifuged the lysate at 12000 rpm and 4 °C for 30 min, and incubated the supernatant with reaction buffer at 37 °C for 15 min. We stopped the color development and measured the absorbance at 520 nm. We measured the protein concentration of the lysate with a Bicinchoninic Acid Assay (BCA) protein assay kit (Beyotime). The ALP staining assay was performed with a Beyotime kit. On day 7 of induction, the cells were fixed for 30 min and stained with Alkaline Phosphatase Assay Kit (Beyotime, China) in the dark for 30 min. The cells were observed and imaged for histochemical detection of ALP with a Nikon microscope.
After osteogenic inducting for 14 d, the cells were fixed with 4% paraformaldehyde for 30 min. They were stained with 1% Alizarin Red (pH 4.3) (Beyotime) for 20 min at room temperature and washed three times with deionized water. We observed and imaged the calcium deposits under a Nikon microscope. To quantify calcium deposits, we destained the stained cells with 10% cetylpyridinium chloride monohydrate (Sigma–Aldrich) in 10 mmol/L sodium phosphate (pH 7.0) for 30 min. We transferred a 200-μL aliquot to a 96-well plate to measure the absorbance at 550 nm by a Varioskan Flash Spectral Scanning Multimode Reader (ThermoFisher Scientific, United States).
Total RNA was isolated from the cells by an RNA prep pure Cell Kit (TIANGEN, Beijing, China). cDNA was synthesized following the instructions for the HiScript® III RT Super Mix for quantitative real time polymerase chain reaction (qPCR) kit (Vazyme, Nanjing, China). Quantitative real-time polymerase chain reaction (PCR) was performed with ChamQ Universal SYBR qPCR Master Mix kit (Vazyme) and Real-Time PCR Detection System (Quantstudio5, United States). Gene-specific primer pairs are show in Supplementary Table 1.
Cell Counting Kit-8 (CCK-8; Dojindo Kagaku, Japan) was used to investigate the proliferation rate of E16.5d Cd271 knockout and wild-type MSCs. The cells were seeded at 2 × 103 cells/well in a 96-well plate. After mixing the CCK-8 solution, the cells were cultured at 37 °C for 2 h in the dark. Absorbance was detected for 7 d continuously by a microplate reader at 450 nm.
Except the scRNA Seq, all the experiments were repeated more than three times. A one-way analysis of variance (ANOVA) or t test (GraphPad Prism 9.0 software, La Jolla, CA, United States) was used to identify significant differences. P < 0.05 was considered statistically significant.
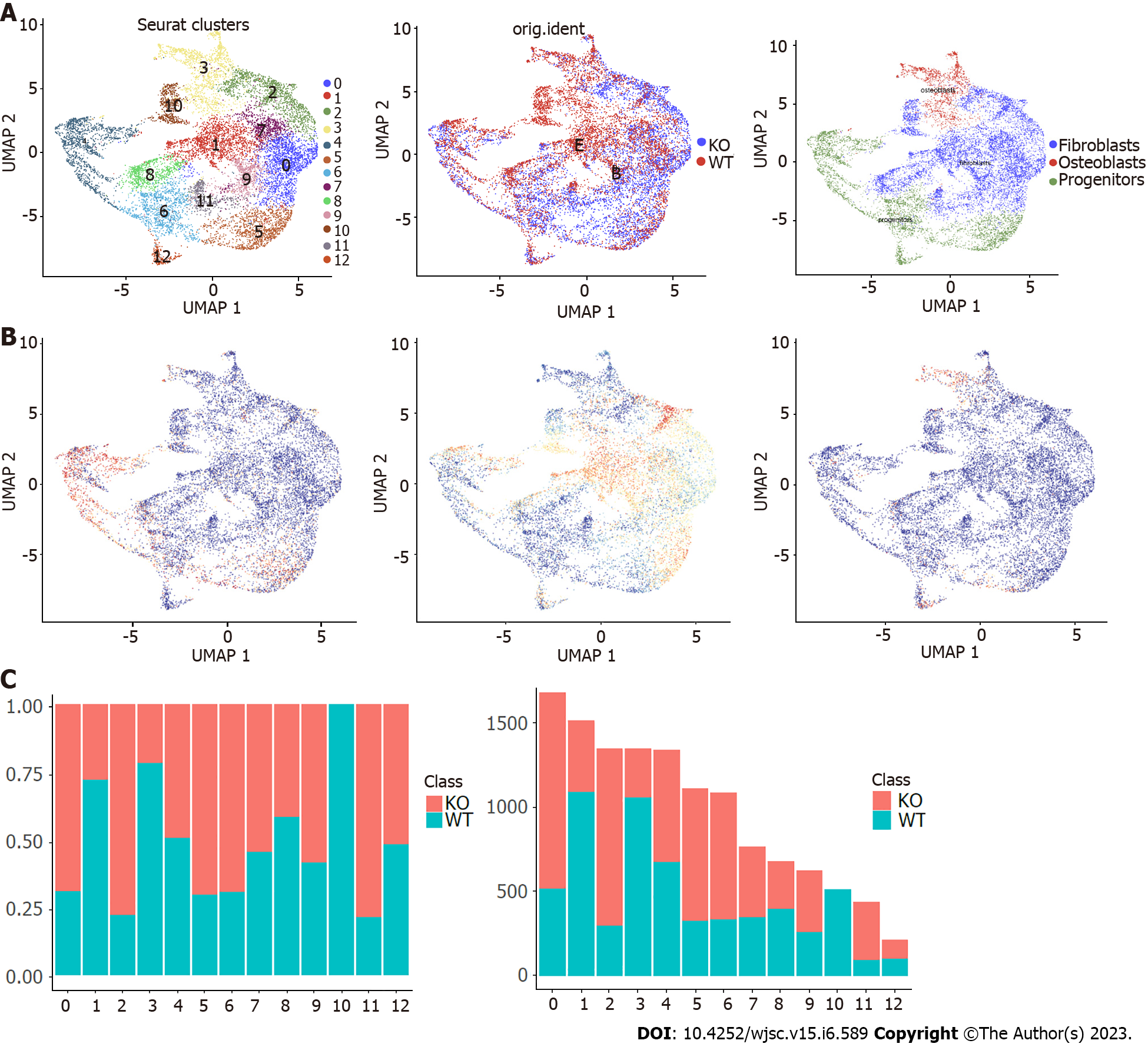
To reveal cellular heterogeneity, we performed scRNA-Seq with murine maxillary processes from wild-type and Cd271 knockout embryo tissues (Figure 1A and Supplementary Figure 1). After quality controlling and normalizing scRNA-Seq data, we obtained transcriptomes of 17426 cells (Supple
We also analyzed the single cell sequence profiling from wild-type and Cd271 knockout mice to establish the distribution of different types of cells and the source of samples. We counted the number and proportion of each cell type using histograms (Figure 1F). MSCs occupied most of the cell transcriptomes from the maxillary process, and the proportion of these cells between Cd271 knockout and wild-type was similar.
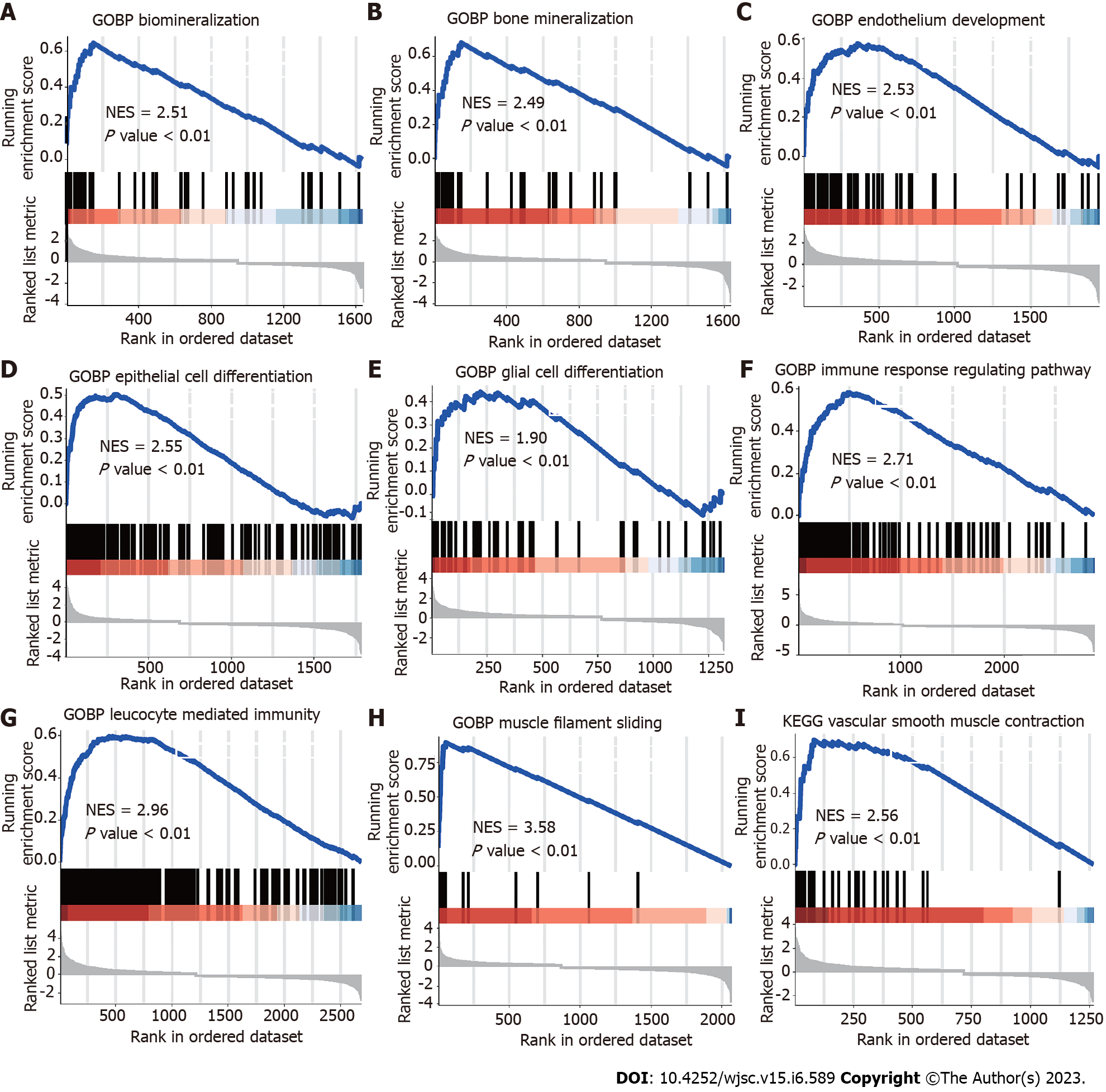
To identify the relevant molecular mechanisms and possible pathways, we subjected all expressions to GSEA. The results showed a high correlation with MSCs and a close relationship with biomineralization. The results suggested that biomineralization (NES = 2.51, P < 0.01) and bone mineralization (NES = 2.49, P < 0.001) associated pathways enriched in MSCs (Figure 2A and B). GSEA also revealed that the differentially expressed genes enriched endothelial development, epithelial and glial cell differentiation, immune response, myofilament sliding and vascular smooth muscle contraction (Figure 2C–I).
As MSCs contribute much to embryogenesis and are essential in odontogenesis and osteogenesis, we performed unsupervised reclustering of MSCs. We performed the Subcluster analysis to investigate the heterogeneity within the mesenchymal populations. We observed further heterogeneity in subclusters 0–12 (Figure 3A). We used the published markers to identify the subclusters in MSCs. For instance, Cdk1 and Dkk1 marked progenitors, Runx2 and sp7 marked osteoblasts, and Dlk1 and Shox2 marked fibroblasts. We scored these marker genes in MSC clusters (Figure 3B). In this way, MSC clusters could also be identified into three subpopulations: Progenitors, fibroblasts and osteoblasts. We counted the cell proportions of the MSCs subclusters from wild-type and Cd271 knockout maxillary processes. The results showed that the numbers of subclusters 1, 3 and 10 were lower, while the other subclusters were higher in Cd271 knockout compared with wild-type mice (Figure 3C).
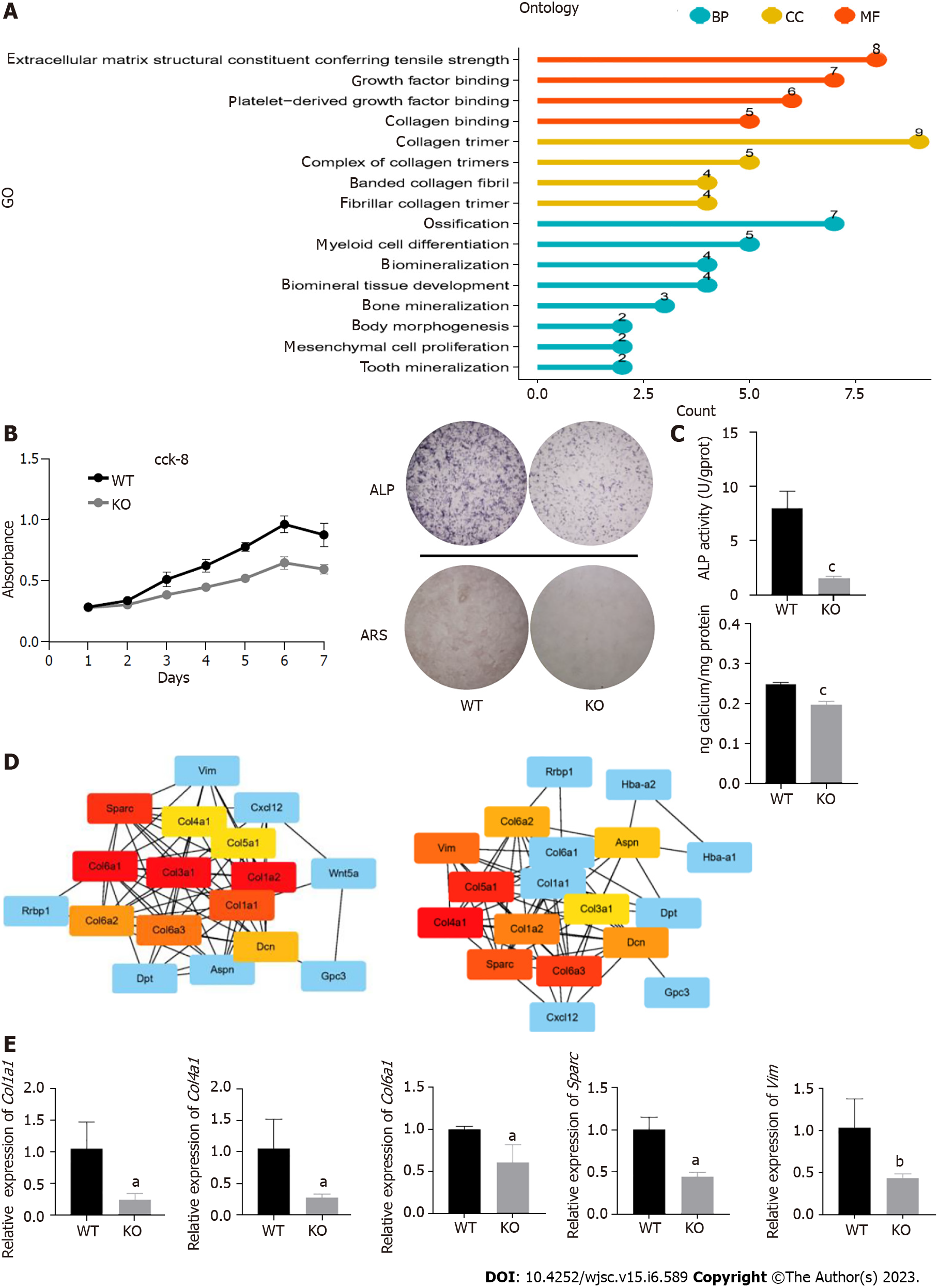
We compared MSCs between wild-type and Cd271 knockout maxillary processes. GO functional enrichment analysis revealed that differentially expressed genes were enriched in ossification, myeloid cell differentiation and biomineralization cell proliferation-related signaling (Figure 4A). To compare the differences in proliferation and osteogenic differentiation between wild-type and Cd271 knockout MSCs, we investigated the potential for MSC proliferation. As the CCK-8 assay showed, the Cd271 knockout MSCs exhibited weaker cell proliferation (Figure 4B). We induced the cells with an osteogenic induction medium and performed the mineralization assay. ALP staining and a quantitative assay revealed a lower and lighter mineralization level of Cd271 knockout than wild-type MSCs. Lighter and fewer mineralized nodules were observed by Alizarin Red staining in Cd271 knock out MSCs (Figure 4C). These results indicated more inadequate osteogenic differentiation potential in Cd271 knockout MSCs. We selected several differentially expressed genes with considerable interaction by Cytoscape, and measured the gene expression by real-time quantitative PCR (Figure 4D). Proliferation, ossification and osteoblast differentiation-related genes, such as Col4a1, Col6a1, Sparc, Vim, Col1a1 and Col3a1, were significantly decreased in Cd271 knockout MSCs (Figure 4E). Cd271 was involved in regulating osteogenic differentiation in MSCs, and our results implied a weaker potential of proliferation and osteogenic differentiation in Cd271 knockout MSCs.
We selected MSCs to institute a pseudotemporal trajectory map that contained terminals consistent with distinct cell fates. The progenitor cells aggregated at the root and branches, indicating a good cell cycle of the progenitor cells. Osteoblasts and fibroblasts were scattered in various branches, showing high invasive potential (Figure 5A–C). We extracted differentially expressed genes over pseudotemporal changes and plotted heatmaps. Different sets of genes were also found to be shifted during the progression of MSCs, stressing the actional change of the progenitor cells, which managed the cell fate transition in the mesenchyme. We clustered the differentially expressed genes into three and performed GO analysis. Clusters 1 and 2 were significantly enriched in biological processes of osteoblast differentiation, biomineralization and odontogenesis, while cluster 3 was mainly enriched in cell metabolism (Figure 5D).
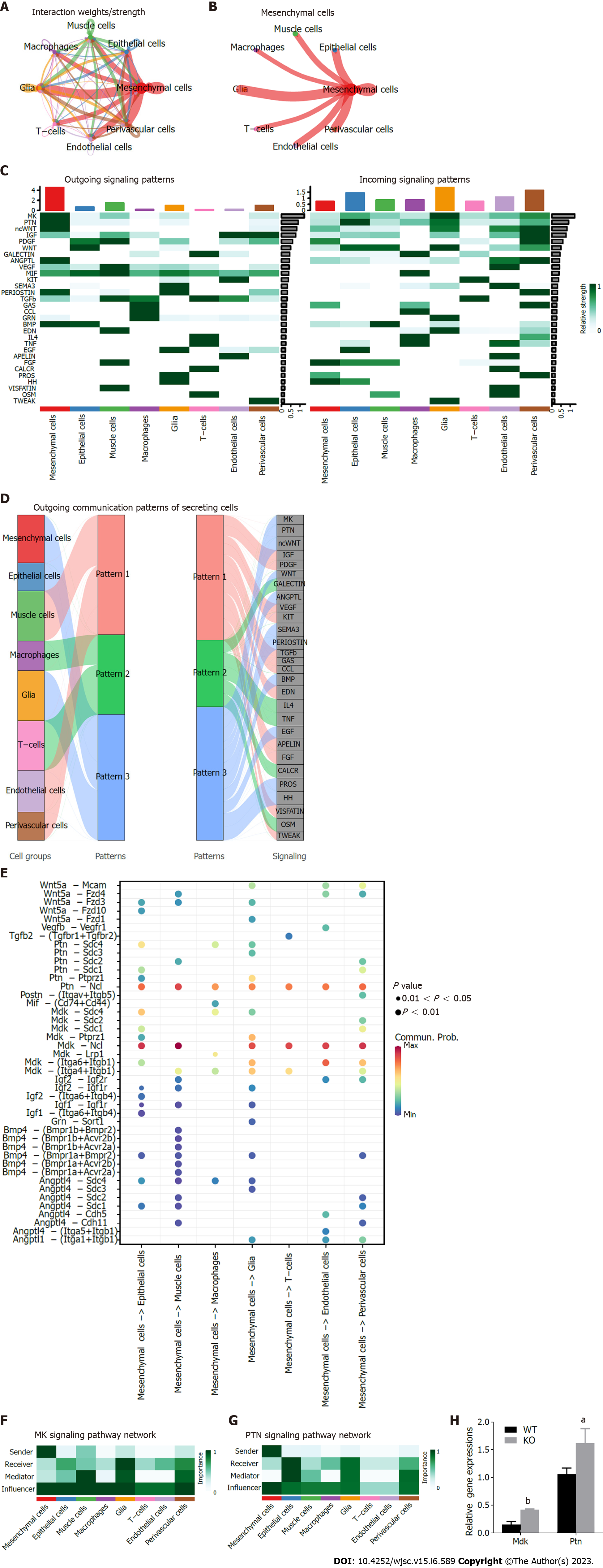
The dataset of scRNA-Seq provided an opportunity to analyze intercellular communication mediated by ligand–receptor interactions. To gain insights into potential signaling interactions between different cellular subpopulations, we interrogated our data with CellChat (version 1.1.3) (http://www.cellchat.org/) R package, which could predict the ligand and receptor interactions at single cell resolution[33]. We found dense communication between MSCs and other cells, and the most concentrated interactions occurred in the mesenchymal subpopulation (Figure 6A and B). It revealed the importance of mesenchymal interaction signaling. We detected 30 signaling pathways in eight cell groups, including MK, PTN, ncWNT, IGF, PDGF, WNT and VEGF signaling pathways. We compared the contribution of efferent (or afferent) signaling between cells, confirming that MSCs had higher interaction than other cell types while acting as outgoing signaling pathways (Figure 6C). We analyzed the potential signaling of MSCs, revealing a series of signaling pathways, such as odontogenesis-related signaling such as WNT, BMP and HH signaling (Figure 6D).
We explored ligand–receptor signals broadcast by MSCs, which revealed that MSCs might affect other cells in the ligand–receptor interaction of MK and PTN signaling (Figure 6E). We performed network centrality analysis on MK and PTN signaling, which confirmed that MSCs were important signal senders in intercellular communication (Figure 6F and G). The expression of Mdk and Ptn in wild-type and Cd271-/- MSCs exhibited significant differences by quantitative real-time PCR assay. The expression of Mdk and Ptn was higher in the Cd271 knockout MSCs (Figure 6H). These results disclosed underlying ligand–receptor interactions and suggested that MK and PTN signaling may be influenced by Cd271 in mesenchymal subpopulations.
Although MSCs have been extensively studied, there is still a lack of knowledge about the key mechanisms in tooth development. Researchers have found that the maxillary process included various cell subpopulations with diverse phenotypic and functional characteristics[34]. How this heterogeneity appears in osteogenesis and odontogenesis remains unclear. Cd271 used to be a marker of the MSCs, and it had been reported that Cd271 promotes differentiation in MSCs[35-37]. Hence, we used Cd271 knockout mouse embryos to investigate what happens to MSCs in their initial developmental stage. We constructed a single cell profiling of representative mouse embryo maxillary processes and selected MSCs to explore their characteristics and critical regulatory mechanism Cd271 related.
Maxillary-process-derived MSCs are recognized as primitive odontogenic stem cells. For instance, researchers obtained the MSCs from the first branchial arch of mice and the maxillary process tissue of rats in succession, and revealed the pluripotency of MSCs[38,39]. Wen et al[40] found that p75+/+ MSCs have good multidirectional differentiation potential[40]. We identified nine MSC clusters among 21 clusters identified using scRNA-Seq in mouse embryo maxillary processes. MSC clusters accounted for the majority, suggesting that MSCs contribute much to embryogenesis. Emerging studies have revealed that the marker genes of MSCs, Col1a1 and Col3a1, promote collagen production and influence odontogenesis or osteogenesis[41-43]. The decreased expression of these two genes in Cd271 knockout MSCs compared to wild-type, suggested that Cd271 was related to MSC regulation of odontogenesis or osteogenesis.
In the past, several studies concentrated on the whole population of MSCs rather than the relationships among the subclusters. Our subcluster analysis disclosed that these MSC clusters could be separated into three subpopulations: Fibroblast, progenitors and osteoblast, which had not been separated before. One of the characteristics of MSC clusters is the expression of osteoblast differentiation-associated genes such as Runx2, Sox9, Msx1 and Omd[44-47]. Previous studies showed that proteins such as Runx2 and Sox9 were positively regulated by Cd271 in MSCs[48-50]. However, these studies did not show the heterogeneity of MSCs, or which subpopulations of MSCs these proteins derived from. Our results revealed that these proteins were mainly expressed in the osteoblast subpopulation. The number of osteoblast subpopulations decreased while progenitor subpopulations increased. This suggested that some progenitors were hindered from differentiating into osteoblasts in the Cd271 knockout MSC clusters. Consistently, GO enrichment analysis and GSEA verified significant enrichment of the ossification, biomineralization and bone mineralization pathways in the Cd271 knockout and wild-type cells. Based on our scRNA-Seq data, we gained differentially expressed genes between two groups for the first time. We picked out several differentially expressed genes with extensive interactions; for instance, Sparc, Vim, Col4a1, Col6a1, Col1a1 and Col3a1. These genes are considered to relate to cell proliferation, bone mineralization and multipotent differentiation[41-43,51-54]. In Cd271 knockout MSCs, we verified by quantitative real-time PCR that these differentially expressed gene clusters had lower expression. Functional experiments also demonstrated a weaker odontogenic and osteogenic differentiation and proliferation in the Cd271 knockout MSCs. Consistent with previous studies, our results confirmed that the MSC clusters had an essential role in odontogenesis and osteogenesis and Cd271 possibly regulated it.
Pseudotemporal analysis uncovered a developmental trajectory among these three subpopulations. Significantly, the progenitor subpopulation was found to develop into the osteoblast and fibroblast subpopulations. We concluded that the progenitors were the base subpopulation and increasingly transformed into the osteoblast and fibroblast subpopulations in MSCs. Consistent with our GO and GSEA findings, the differentially expressed genes that emerged in pseudotemporal analysis showed a significant relation with ossification and osteoblast differentiation in MSCs. Collectively, our results revealed that MSCs were progressively diversified and determined to the odontogenic fate. Thus, the mesenchyme in the maxillary process provides a remarkable model to study the development of maxillary-process-derived MSCs.
It is known that the interactions between dental mesenchyme and epithelium play a crucial part in the integral tooth developmental process. Nevertheless, the cell–cell interactions within the maxillary process have not been extensively studied[55]. In our study, scRNA-Seq offered the opportunity to identify communicating pairs on the basis of the expression of their cell-surface receptors and ligands. CellChat showed that the most concentrated interactions occurred between mesenchymal and other cells. It indicated that MSCs play a vital role in cell communication. It is worth noting that MSCs mainly participated in MK, PTN, ncWNT, WNT and BMP signaling. Previous studies reported that Cd271 might regulate the odontogenic differentiation of MSCs through the BMP, WNT and PI3K pathways[17,49,56]. However, there are no reports that MDK signaling participates in regulating MSCs. In the CellChat, the receptor interactions of MDK and PTN appeared to be obvious. It reminded us that the Mdk and Ptn might play a primary role in the developmental process of MSCs. Mdk and Ptn expression notably increased in Cd271 knockout cells during induction of mineralization for 7 d. Our subsequent study will explore the molecular mechanism of how Cd271 regulates Mdk to influence osteogenesis.
Although our studies revealed several significant discoveries, there were some limitations. First, different clustering can generate different results; thus, further groupings are needed to perform future analysis. Second, the current study was based on scRNA-Seq. Therefore, we did some biological observation but did not illustrate the direct mechanisms of Cd271 involved in tooth development. Therefore, further studies about direct mechanisms and in vivo studies are needed.
Our study reveals high cellular heterogeneity, molecular details and cell-cell interactions in MSCs. It provides a valuable resource for understanding the development of maxillary-process-derived MSCs. And it enables the maxillary process to serve as an excellent model to explore tooth development and cell fate determinations. Furthermore, we have found significant functional and signaling differences between Cd271 knockout and wildtype MSCs. We have clarified that Cd271 is significantly associated with the regulation of mineralization. We tentatively propose that Mdk signaling is involved in the regulatory mechanism of mineralization. These findings contribute to thorough exploration of the mechanism of Cd271 in regulating odontogenesis and osteogenesis which add to the theory of tooth development.
Tooth loss has become a common problem in human life. Compared with traditional denture restoration, dental tissue engineering has become the most ideal means to solve this problem, and it is also one of the most active research fields of stomatology in recent years. The tooth development involves complex signal pathways. Ecto-mesenchymal stem cells (MSCs) were regarded as the primitive dental cells in the classical theory of tooth development. To date, in-depth studies of the odontogenesis and osteogenesis of MSCs are still lacking.
We contribute to thorough exploration of the mechanism of odontogenesis and osteogenesis to add to the theory of tooth development.
Our study provides novel insights into the biological features of MSCs at the single cell level and the mechanism of Cd271 in regulating odontogenesis and osteogenesis.
We used the maxillary process from mouse embryos as a model to understand the development of maxillary-process-derived MSCs. We applied single cell RNA sequence analysis to elucidate the cellular heterogeneity and explore molecular details. And we verified the findings from single cell sequencing in vitro by lab experience such as cell staining, cell counting and quantitative real time polymerase chain reaction.
Our study reveals: (1) High cellular heterogeneity and molecular details; (2) Significant functional and signaling differences between cell types; (3) Novel subclusters of mesenchymal stem cells; and (4) Crucial cell-cell interactions of mesenchymal subpopulations. Besides, we contribute to thorough exploration of the mechanism of Cd271 in regulating odontogenesis and osteogenesis.
Our study reveals high cellular heterogeneity, molecular details and cell-cell interactions in MSCs. We found significant functional and signaling differences between Cd271 knockout and wildtype MSCs. We clarified that Cd271 is significantly associated with the regulation of mineralization.
We need illustrate that Mdk signaling is involved in the regulatory mechanism of mineralization in future research. And direct mechanisms of Cd271 involved in tooth development are needed in further studies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Elchaninov AV, Russia; Stogov MV, Russia S-Editor: Li L L-Editor: A P-Editor: Ma YJ
| 1. | Egbuniwe O, Idowu BD, Funes JM, Grant AD, Renton T, Di Silvio L. P16/p53 expression and telomerase activity in immortalized human dental pulp cells. Cell Cycle. 2011;10:3912-3919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 2. | Strobl-Mazzulla PH, Bronner ME. Epithelial to mesenchymal transition: new and old insights from the classical neural crest model. Semin Cancer Biol. 2012;22:411-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 3. | Abbasian M, Langlois A, Gibon J. Sexual Dimorphism in Balance and Coordination in p75NTR(exonIII) Knock-Out Mice. Front Behav Neurosci. 2022;16:842552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 4. | Alvarez R, Lee HL, Hong C, Wang CY. Single CD271 marker isolates mesenchymal stem cells from human dental pulp. Int J Oral Sci. 2015;7:205-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 5. | Akiyama Y, Mikami Y, Watanabe E, Watanabe N, Toriumi T, Takahashi T, Komiyama K, Isokawa K, Shimizu N, Honda MJ. The P75 neurotrophin receptor regulates proliferation of the human MG63 osteoblast cell line. Differentiation. 2014;87:111-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Castellini C, Mattioli S, Cotozzolo E, Pistilli A, Rende M, Bartolini D, Di Sante G, Menchetti L, Dal Bosco A, Stabile AM. The Effect of Interaction NGF/p75(NTR) in Sperm Cells: A Rabbit Model. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Zanin JP, Friedman WJ. p75NTR prevents the onset of cerebellar granule cell migration via RhoA activation. Elife. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Zhao H, Fan S, Sun J. Delayed wound healing in the elderly and a new therapeutic target: CD271. Curr Stem Cell Res Ther. 2023;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 9. | Passino MA, Adams RA, Sikorski SL, Akassoglou K. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science. 2007;315:1853-1856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 144] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 10. | Wen X, Liu L, Deng M, Liu R, Zhang L, Nie X. In vitro cementoblast-like differentiation of postmigratory neural crest-derived p75(+) stem cells with dental follicle cell conditioned medium. Exp Cell Res. 2015;337:76-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Baeza-Raja B, Sachs BD, Li P, Christian F, Vagena E, Davalos D, Le Moan N, Ryu JK, Sikorski SL, Chan JP, Scadeng M, Taylor SS, Houslay MD, Baillie GS, Saltiel AR, Olefsky JM, Akassoglou K. p75 Neurotrophin Receptor Regulates Energy Balance in Obesity. Cell Rep. 2016;14:255-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Gonçalves NP, Mohseni S, El Soury M, Ulrichsen M, Richner M, Xiao J, Wood RJ, Andersen OM, Coulson EJ, Raimondo S, Murray SS, Vægter CB. Peripheral Nerve Regeneration Is Independent From Schwann Cell p75(NTR) Expression. Front Cell Neurosci. 2019;13:235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Smith RJP, Faroni A, Barrow JR, Soul J, Reid AJ. The angiogenic potential of CD271+ human adipose tissue-derived mesenchymal stem cells. Stem Cell Res Ther. 2021;12:160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Mitsiadis TA, Pagella P. Expression of Nerve Growth Factor (NGF), TrkA, and p75(NTR) in Developing Human Fetal Teeth. Front Physiol. 2016;7:338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Zhao X, Ning L, Xie Z, Jie Z, Li X, Wan X, Sun X, Huang B, Tang P, Shen S, Qin A, Ma Y, Song L, Fan S, Wan S. The Novel p38 Inhibitor, Pamapimod, Inhibits Osteoclastogenesis and Counteracts Estrogen-Dependent Bone Loss in Mice. J Bone Miner Res. 2019;34:911-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Colosimo A, Rofani C, Ciraci E, Salerno A, Oliviero M, Maio ED, Iannace S, Netti PA, Velardi F, Berardi AC. Osteogenic differentiation of CD271(+) cells from rabbit bone marrow cultured on three phase PCL/TZ-HA bioactive scaffolds: comparative study with mesenchymal stem cells (MSCs). Int J Clin Exp Med. 2015;8:13154-13162. [PubMed] |
| 17. | Xing Y, Nie X, Chen G, Wen X, Li G, Zhou X, Tian W, Liu L. Comparison of P75 NTR-positive and -negative etcomesenchymal stem cell odontogenic differentiation through epithelial-mesenchymal interaction. Cell Prolif. 2016;49:185-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Li Z, Zhang C, Weiner LP, Zhang Y, Zhong JF. Molecular characterization of heterogeneous mesenchymal stem cells with single-cell transcriptomes. Biotechnol Adv. 2013;31:312-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Chen Y, Zhang Z, Yang X, Liu A, Liu S, Feng J, Xuan K. Odontogenic MSC Heterogeneity: Challenges and Opportunities for Regenerative Medicine. Front Physiol. 2022;13:827470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 528] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 21. | Yan H, Ye Y, Zhao H, Zuo H, Li Y. Single-Cell RNA Sequencing for Analyzing the Intestinal Tract in Healthy and Diseased Individuals. Front Cell Dev Biol. 2022;10:915654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 22. | Li Y, Ju S, Li X, Li W, Zhou S, Wang G, Cai Y, Dong Z. Characterization of the microenvironment of diabetic foot ulcers and potential drug identification based on scRNA-seq. Front Endocrinol (Lausanne). 2022;13:997880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 23. | Liu X, Xiang Q, Xu F, Huang J, Yu N, Zhang Q, Long X, Zhou Z. Single-cell RNA-seq of cultured human adipose-derived mesenchymal stem cells. Sci Data. 2019;6:190031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 24. | Kirkwood PM, Gibson DA, Smith JR, Wilson-Kanamori JR, Kelepouri O, Esnal-Zufiaurre A, Dobie R, Henderson NC, Saunders PTK. Single-cell RNA sequencing redefines the mesenchymal cell landscape of mouse endometrium. FASEB J. 2021;35:e21285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 25. | Wang Z, Li X, Yang J, Gong Y, Zhang H, Qiu X, Liu Y, Zhou C, Chen Y, Greenbaum J, Cheng L, Hu Y, Xie J, Yang X, Li Y, Schiller MR, Tan L, Tang SY, Shen H, Xiao HM, Deng HW. Single-cell RNA sequencing deconvolutes the in vivo heterogeneity of human bone marrow-derived mesenchymal stem cells. Int J Biol Sci. 2021;17:4192-4206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 26. | Bayarsaihan D, Enkhmandakh B, Vijaykumar A, Robson P, Mina M. Single-cell transcriptome analysis defines mesenchymal stromal cells in the mouse incisor dental pulp. Gene Expr Patterns. 2022;43:119228. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 27. | Li J, Wang Q, An Y, Chen X, Xing Y, Deng Q, Li Z, Wang S, Dai X, Liang N, Hou Y, Yang H, Shang Z. Integrative Single-Cell RNA-Seq and ATAC-Seq Analysis of Mesenchymal Stem/Stromal Cells Derived from Human Placenta. Front Cell Dev Biol. 2022;10:836887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2637] [Cited by in RCA: 4125] [Article Influence: 412.5] [Reference Citation Analysis (0)] |
| 29. | Kim S, Kang D, Huo Z, Park Y, Tseng GC. Meta-analytic principal component analysis in integrative omics application. Bioinformatics. 2018;34:1321-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 30. | Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4238] [Cited by in RCA: 8353] [Article Influence: 835.3] [Reference Citation Analysis (0)] |
| 31. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22106] [Article Influence: 1700.5] [Reference Citation Analysis (0)] |
| 32. | Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309-315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 762] [Cited by in RCA: 1143] [Article Influence: 142.9] [Reference Citation Analysis (0)] |
| 33. | Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3052] [Cited by in RCA: 4019] [Article Influence: 1004.8] [Reference Citation Analysis (0)] |
| 34. | Fabian P, Crump JG. Reassessing the embryonic origin and potential of craniofacial ectomesenchyme. Semin Cell Dev Biol. 2023;138:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 35. | Colombo E, Romaggi S, Medico E, Menon R, Mora M, Falcone C, Lochmüller H, Confalonieri P, Mantegazza R, Morandi L, Farina C. Human neurotrophin receptor p75NTR defines differentiation-oriented skeletal muscle precursor cells: implications for muscle regeneration. J Neuropathol Exp Neurol. 2011;70:133-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 36. | Mikami Y, Suzuki S, Ishii Y, Watanabe N, Takahashi T, Isokawa K, Honda MJ. The p75 neurotrophin receptor regulates MC3T3-E1 osteoblastic differentiation. Differentiation. 2012;84:392-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Yang K, Wang Y, Ju Y, Li G, Liu C, Liu J, Liu Q, Wen X, Liu LC. p75 neurotrophin receptor regulates differential mineralization of rat ectomesenchymal stem cells. Cell Prolif. 2017;50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 38. | Deng MJ, Jin Y, Shi JN, Lu HB, Liu Y, He DW, Nie X, Smith AJ. Multilineage differentiation of ectomesenchymal cells isolated from the first branchial arch. Tissue Eng. 2004;10:1597-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Zhang J, Duan X, Zhang H, Deng Z, Zhou Z, Wen N, Smith AJ, Zhao W, Jin Y. Isolation of neural crest-derived stem cells from rat embryonic mandibular processes. Biol Cell. 2006;98:567-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Wen X, Liu L, Deng M, Zhang L, Liu R, Xing Y, Zhou X, Nie X. Characterization of p75(+) ectomesenchymal stem cells from rat embryonic facial process tissue. Biochem Biophys Res Commun. 2012;427:5-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 41. | Liu Y, Wang Z, Ju M, Zhao Y, Jing Y, Li J, Shao C, Fu T, Lv Z, Li G. Modification of COL1A1 in Autologous Adipose Tissue-Derived Progenitor Cells Rescues the Bone Phenotype in a Mouse Model of Osteogenesis Imperfecta. J Bone Miner Res. 2021;36:1521-1534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Malmgren B, Andersson K, Lindahl K, Kindmark A, Grigelioniene G, Zachariadis V, Dahllöf G, Åström E. Tooth agenesis in osteogenesis imperfecta related to mutations in the collagen type I genes. Oral Dis. 2017;23:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 43. | Boban I, Jacquin C, Prior K, Barisic-Dujmovic T, Maye P, Clark SH, Aguila HL. The 3.6 kb DNA fragment from the rat Col1a1 gene promoter drives the expression of genes in both osteoblast and osteoclast lineage cells. Bone. 2006;39:1302-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13:27-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 746] [Cited by in RCA: 830] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 45. | Liao J, Hu N, Zhou N, Lin L, Zhao C, Yi S, Fan T, Bao W, Liang X, Chen H, Xu W, Chen C, Cheng Q, Zeng Y, Si W, Yang Z, Huang W. Sox9 potentiates BMP2-induced chondrogenic differentiation and inhibits BMP2-induced osteogenic differentiation. PLoS One. 2014;9:e89025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 46. | Goto N, Fujimoto K, Fujii S, Ida-Yonemochi H, Ohshima H, Kawamoto T, Noshiro M, Shukunami C, Kozai K, Kato Y. Role of MSX1 in Osteogenic Differentiation of Human Dental Pulp Stem Cells. Stem Cells Int. 2016;2016:8035759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 47. | Lin W, Zhu X, Gao L, Mao M, Gao D, Huang Z. Osteomodulin positively regulates osteogenesis through interaction with BMP2. Cell Death Dis. 2021;12:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 48. | Chen J, Wang N, Zhang H, Zhang X, Zhao L, Zhu L, Li Z, Bei C. [Lentivirus-mediated silencing of P75 neurotrophin receptor combined with nerve growth factor overexpression and transfection of bone marrow mesenchymal stem cells combined with demineralized bone matrix for heterotopic osteogenesis]. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2020;34:1438-1445. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 49. | Wang Y, Yang K, Li G, Liu R, Liu J, Li J, Tang M, Zhao M, Song J, Wen X. p75NTR(-/-) mice exhibit an alveolar bone loss phenotype and inhibited PI3K/Akt/β-catenin pathway. Cell Prolif. 2020;53:e12800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 50. | Shan P, Wang X, Zhang Y, Teng Z, Jin Q, Liu J, Ma J, Nie X. P75 neurotrophin receptor positively regulates the odontogenic/osteogenic differentiation of ectomesenchymal stem cells via nuclear factor kappa-B signaling pathway. Bioengineered. 2022;13:11201-11213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 51. | Rosset EM, Bradshaw AD. SPARC/osteonectin in mineralized tissue. Matrix Biol. 2016;52-54:78-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 210] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 52. | Castro-Muñozledo F, Meza-Aguilar DG, Domínguez-Castillo R, Hernández-Zequinely V, Sánchez-Guzmán E. Vimentin as a Marker of Early Differentiating, Highly Motile Corneal Epithelial Cells. J Cell Physiol. 2017;232:818-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 53. | Wang H, Ning T, Song C, Luo X, Xu S, Zhang X, Deng Z, Ma D, Wu B. Priming integrin α5 promotes human dental pulp stem cells odontogenic differentiation due to extracellular matrix deposition and amplified extracellular matrix-receptor activity. J Cell Physiol. 2019;234:12897-12909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 54. | Du C, Li Y, Xia X, Du E, Lin Y, Lian J, Ren C, Li S, Wei W, Qin Y. Identification of a novel collagen-like peptide by high-throughput screening for effective wound-healing therapy. Int J Biol Macromol. 2021;173:541-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 55. | Mantesso A, Sharpe P. Dental stem cells for tooth regeneration and repair. Expert Opin Biol Ther. 2009;9:1143-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 56. | Li G, Liu J, Wang Y, Yang K, Zhao M, Xiao Y, Wen X, Liu L. LNGFR targets the Wnt/β-catenin pathway and promotes the osteogenic differentiation in rat ectomesenchymal stem cells. Sci Rep. 2017;7:11021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |