Published online May 26, 2023. doi: 10.4252/wjsc.v15.i5.323
Peer-review started: December 28, 2022
First decision: January 31, 2023
Revised: February 14, 2023
Accepted: April 12, 2023
Article in press: April 12, 2023
Published online: May 26, 2023
Processing time: 149 Days and 1.2 Hours
Surgical resection, chemotherapy, and radiation are the standard therapeutic modalities for treating cancer. These approaches are intended to target the more mature and rapidly dividing cancer cells. However, they spare the relatively quiescent and intrinsically resistant cancer stem cells (CSCs) subpopulation residing within the tumor tissue. Thus, a temporary eradication is achieved and the tumor bulk tends to revert supported by CSCs' resistant features. Based on their unique expression profile, the identification, isolation, and selective targeting of CSCs hold great promise for challenging treatment failure and reducing the risk of cancer recurrence. Yet, targeting CSCs is limited mainly by the irrelevance of the utilized cancer models. A new era of targeted and personalized anti-cancer therapies has been developed with cancer patient-derived organoids (PDOs) as a tool for establishing pre-clinical tumor models. Herein, we discuss the updated and presently available tissue-specific CSC markers in five highly occurring solid tumors. Additionally, we highlight the advantage and relevance of the three-dimensional PDOs culture model as a platform for modeling cancer, evaluating the efficacy of CSC-based therapeutics, and predicting drug response in cancer patients.
Core Tip: Therapeutic approaches targeting cancer stem cell (CSC) markers hold great promise toward developing effective anti-cancer treatment. Tissue-specific CSCs (TSCSCs) possess unique expression profile that allows for their identification, isolation, and targeting. TSCSCs, isolated from patient tumor tissues, were shown to form organ analogs or patient-derived organoids (PDOs) under specific culturing conditions in vitro. These models simulate the original tumor characteristics in a three-dimensional culture dish. As such, PDOs have the potential to be used in patient-specific in vitro drug clinical trials and proof-of-concept studies on CSC-targeted therapies.
- Citation: Yehya A, Youssef J, Hachem S, Ismael J, Abou-Kheir W. Tissue-specific cancer stem/progenitor cells: Therapeutic implications. World J Stem Cells 2023; 15(5): 323-341
- URL: https://www.wjgnet.com/1948-0210/full/v15/i5/323.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i5.323
Cancer disease remains a leading cause of death worldwide. Despite significant progress directed toward developing anti-cancer therapies, the successful management of cancer remains impeded by multiple challenges, including metastatic dissemination, conventional-therapy resistance, and disease relapse[1,2]. Accumulating evidence suggests that the cancer stem cells (CSCs) subpopulation plays a vigorous role in sustaining the tumorigenic properties, thus contributing to tumor re-growth and progression[3] (Figure 1). This subpopulation of multipotent cells possesses unique properties of self-renewal and differentiation and is capable of extensively proliferating and generating different lineages of cancerous cells, which constitute the tumor bulk and contribute to the heterogeneous phenotype found in tumors[2,4].
CSCs may arise from the transformation of normal stem cells (SCs) found within tissues or from the de-differentiation of differentiated cells[5]. They were first identified in acute myeloid leukemia[6], and compelling evidence later showed that they exist in a variety of solid tumors where they act as key drivers of tumor progression and metastasis[7,8]
CSCs harbor multiple resistance mechanisms that enrich cancer hallmarks and result in the failure of conventional anti-cancer therapies. One underlying mechanism is the disrupted intracellular pathways that profoundly control CSCs behavior. For instance, overexpression of the Notch pathway plays a dual role that is context and cell-type-dependent, acting either as an oncogene or tumor suppressor[9-11]. In the context of CSCs, the Notch pathway has been implicated in proliferation, angiogenesis, metastasis, stemness maintenance, tumor immune evasion, and resistance to radiation[9,11-13]. Moreover, the Wnt pathway has been linked to the activation of dormant CSCs, their proliferation, maintenance, and inhibition of apoptosis. This pathway also plays a role in the metastasis and de-differentiation of CSCs[14,15]. Besides, the Hedgehog pathway is associated with increased proliferation, maintenance, and self-renewal of CSCs, as well as their migration, invasiveness, and resistance to chemotherapy[14,16,17]. Additionally, the NF-κB pathway is implicated in self-renewal, maintenance, and inhibition of apoptosis of CSCs, as well as regulation of epithelial to mesenchymal transition (EMT), angiogenesis, and metastasis[18]. Finally, the aberrant expression of the JAK/STAT3 pathway promotes cell survival and stemness properties, as well as metastasis and resistance to chemotherapy[14,19]. The intrinsic regulation of CSCs also occurs at the level of stemness-related transcription factors (TFs) such as OCT-4, SOX2, KLF4, c-MYC, STAT3, and NANOG, as well as epigenetics and epi-transcriptomics, which contribute to stemness maintenance and plasticity of CSCs[11]. Additionally, CSCs are regulated at an extrinsic level by their microenvironment, specifically by cancer-associated fibroblasts and tumor-associated macrophages. The tumor microenvironment is a major player in modulating CSCs resistance, metastasis, and heterogeneity[11,20].
The resistance mechanisms of CSCs further include their overexpression of DNA repair genes, resulting in resistance to radiotherapy and other DNA-damaging agents[21]. Also, they express upregulated multidrug efflux pumps such as ATP-binding cassette (ABC) transporters that mediate the active transport of chemotherapeutic drugs out of the cell[22]. CSCs were shown as well to overexpress aldehyde dehydrogenases (ALDHs) which are enzymes involved in the detoxification of aldehydes, chemotherapeutic agents, and reactive oxygen species[23]. Another mechanism that promotes the survival of CSCs is their ability to exist at a reversible quiescent state in the G0 phase, which contributes to their drug resistance since most chemotherapeutic agents target highly proliferative tumor cells[24]. Thus, standard therapies succeed at reducing tumor size but tend to spare the highly resistant CSCs subpopulation. The successful elimination of tumors, therefore, necessitates targeting the residual dormant CSCs to yield long-lasting eradication of cancer and prevent relapse.
In this review, we provide a recapitulation of the main tissue-specific CSC (TSCSC) biomarkers in five of the most diagnosed solid tumors. Importantly, we highlight the beneficial role of these CSCs in providing relevant preclinical cancer models and thus improving CSC-targeted therapies.
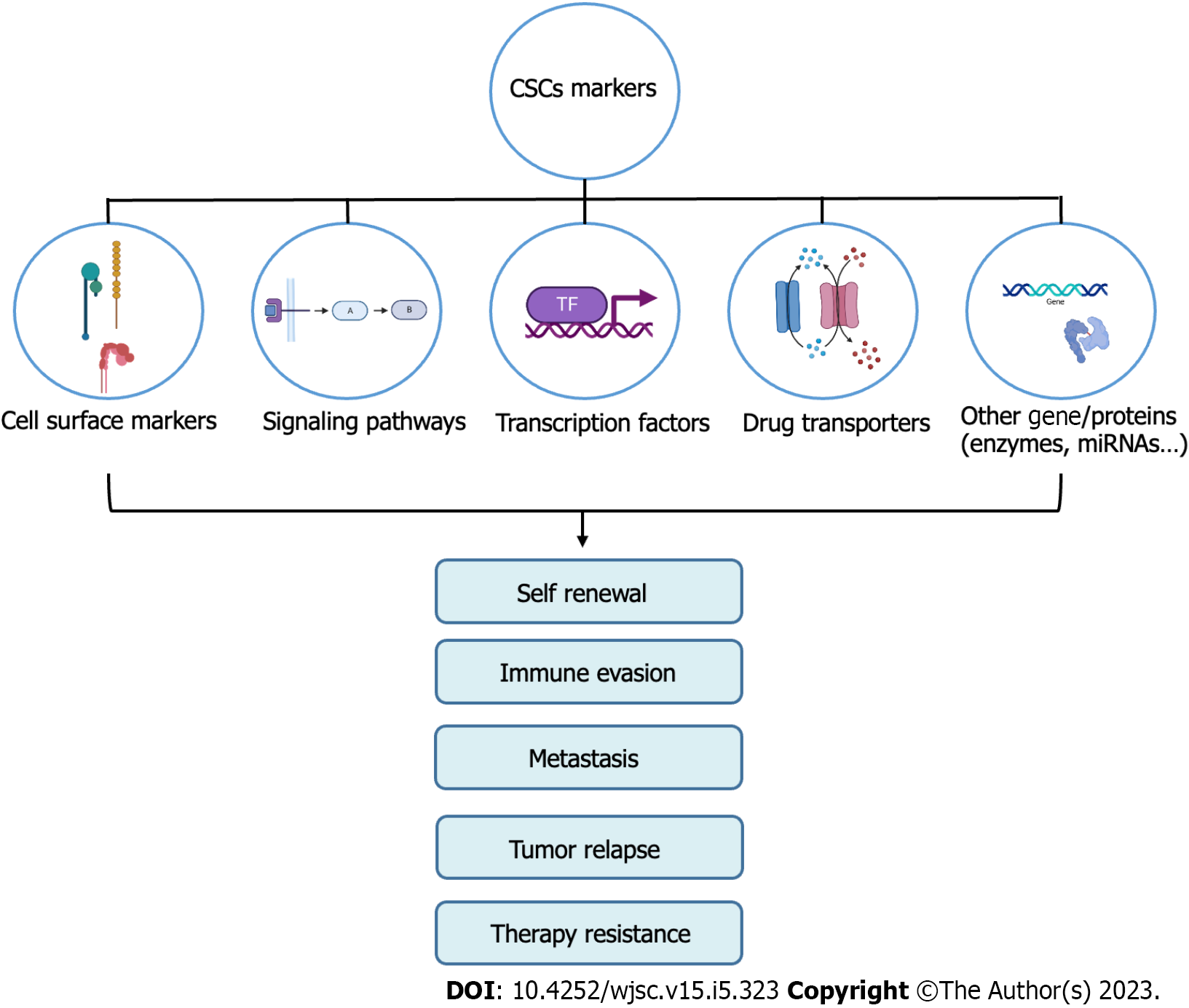
Given the importance of CSCs in tumor progression and prognosis, several attempts were made to identify and isolate CSCs from the tumor mass based on the markers they express. CSCs express a wide spectrum of markers, some of them being more universal than others. Several markers, mostly located on the cell surface, are often used in combination to ensure a more tissue-specific isolation of targeted CSCs. Here we provide an updated overview of the most prominent TSCSC surface markers, focusing on five solid cancers (prostate, colon, bladder, breast, and lung). Refer to Table 1 for the full list of markers.
| TSCSCs markers | PCSCs | CCSCs | BCSCs | BrCSCs | LCSCs | Ref. |
| CD24 | - | + | + | - | [64,90,159,160] | |
| CD26 | + | [161] | ||||
| CD29 | + | + | + | + | [99,162-164] | |
| CD44 | + | + | + | + | + | [31,63,74,108,165] |
| CD47 | + | + | + | [78,166,167] | ||
| CD49b (integrin α2 or ITGA2) | + | + | - | + | + | [168-171] |
| CD49f (integrin α6 or ITGA6) | + | + | + | + | + | [99,169,172,173] |
| CD51 | + | + | [69,174] | |||
| CD61 | + | [99] | ||||
| CD66c | + | - | [84,175] | |||
| CD67LR | + | [84] | ||||
| CD87 | + | [116] | ||||
| CD90 | + | + | + | + | [99,110,176,177] | |
| CD117 | + | + | + | [38,116,178] | ||
| CD126 | + | + | + | [179-181] | ||
| CD133 | + | + | + | + | + | [28,51,87,99,107] |
| CD151 | + | [35] | ||||
| CD166 | + | + | + | + | [46,104,182,183] | |
| CD326 (EpCAM or ESA) | + | + | + | + | + | [48,56,116,184,185] |
| Integrin α2β1 | + | + | [27,186] | |||
| TRA-1-60 | + | + | [35,187] | |||
| Trop2 | + | [45] | ||||
| CXCR4 | + | + | + | + | [102,162,188,189] | |
| ABCB5 | + | [73] | ||||
| ABCG2 | + | + | + | + | + | [49,102,177,190,191] |
| MAGE-A3 | + | [177] | ||||
| GLDC | + | [102] | ||||
| ALDH | + | + | + | + | + | [44,68,96,102,177] |
| BCMab1 | + | [79] | ||||
| Lgr5 | + | + | + | [53,99,192] | ||
| Prox1 | + | + | [70,193] | |||
| EMA (MUC1) | + | + | - | [77,194,195] | ||
| E-cadherin | + | + | [196,197] | |||
| ZEB-1 | + | + | + | + | [198-200] | |
| PSA | - | [201] | ||||
| CK5 | + | + | + | + | [117,202-204] | |
| CK17 | + | [89] | ||||
| CK18 | - | - | - | [89,205,206] | ||
| CK20 | - | [89] | ||||
| Ar-v7 | + | [207] |
The presence of prostate CSCs (PCSCs) was identified by Collins et al[25] using SCs markers (integrin α2β1 and CD133) that were previously identified in the normal prostate epithelium[25,26]. This subpopulation of PCSCs isolated from human prostate cancer (PC) biopsies showed a high expression of CD44, CD133, and integrin α2β1. The isolated cells exhibited high proliferative ability and were highly invasive on MatrigelTM. Moreover, they possessed a high self-renewal ability and could also differentiate into cells expressing the same phenotype as PC cells, thus re-establishing the original heterogeneous tumor from which they were isolated[27].
CD133 (Prominin-1), a cell surface glycoprotein, remains one of the most used biomarkers to identify and isolate PCSCs either alone or in combination with other markers. In fact, CD133+ PC cells that were isolated from human PC cell line exhibited self-renewal ability, which was correlated with their expression of stemness genes[28]. These cells could also generate a heterogeneous tumor mass when transplanted into immunocompromised mice. Moreover, they displayed high clonogenic abilities and led to the formation of tumor spheres (prostaspheres) that were more malignant than the ones formed by CD133- PC cells. Furthermore, the CD133+ cells were chemo-resistant and demonstrated high proliferation[28]. Interestingly, a well-established combination of CD133+ and CD44+ PC cells allowed the isolation of PCSCs and the formation of spheroids characterized by heterogeneous PC cells[29].
CD44 (also referred to as P-Glycoprotein 1) is a transmembrane glycoprotein that interacts with several extracellular matrix components, such as collagen, hyaluronic acid, osteopontin, and matrix metalloproteinases. It is one of the most conventional markers used to identify and isolate PCSCs. The expression of CD44 allowed the isolation of cells that were able to differentiate into all types of PC epithelium leading to complete reconstitution of the original tumor bulk when injected into immunocompromised mice[30]. Notably, CD44+ PC-derived cells expressed elevated levels of several mRNAs associated with stemness[31]. This marker was also associated with several aspects of PC tumorigenesis including proliferation, invasion, adhesion, EMT initiation, metastasis, and therapy resistance[32].
T cell receptor alpha locus (TRA-1-60) is a carbohydrate addition to podocalyxinis, which is a cell surface antigen that belongs to the CD36 family. TRA-1-60 is expressed on pluripotent SCs conferring them the ability to induce differentiation. TRA-1-60 was shown to be overexpressed in PC cells as compared to the adjacent normal prostate tissue, which qualifies it as a favorable marker to specifically target PCSCs while sparing normal cells[33]. Moreover, it was detected in the peripheral blood of patients with metastatic PC[34]. The isolation of TRA-1-60+ cells led to the generation of spheres and initiation of PC in a more efficient manner as compared to other known PCSCs markers. TRA-1-60 was then combined with two other markers of PCSCs (CD166 and CD151) leading to a more enhanced sphere-forming ability. Furthermore, the injection of the triple-marker-positive cells was able to form tumors with at least 5-fold more efficiency as compared to TRA-1-60+ cells alone[35].
CD117 (also termed c-Kit) is a member of the Type-III tyrosine kinase receptors known to be involved in several cancer mechanisms by binding to its stem cell factor (SCF) ligand[36]. CD117 overexpression was detected in PC[37]. A recent study suggested that CD117 may be considered a potential marker for PCSCs because it was shown to display a broad spectrum of tumorigenic abilities[38]. In fact, CD117 stimulated PC cell proliferation and migration. Moreover, CD117+ cells were able to form 1.35-fold larger prostaspheres as compared to CD117- cells. Most importantly, CD117+ cells expressed stemness genes and their implantation into immunocompromised mice led to PC initiation[38].
CD49f (integrin α6 or ITGA6) is a transmembrane glycoprotein that was demonstrated to be a putative marker of PCSCs. CD49fhigh cells were shown to be tumor-initiating cells in the Pten-null PC model[39]. Moreover, CD49f was shown to be the most selective marker for targeting colony-forming cells[40]. Additionally, it was expressed on the surface as well as in the middle of prostatospheres[41]. Importantly, the expression of CD49f allowed the isolation of sphere-forming SCs[42].
In addition to the ones discussed above, there are several markers that can be used to target PCSCs including ALDH1A1 (ALDH 1 family member A1)[43,44], trop-2 (Tumor-associated calcium signal transducer 2)[45], CD166 (activated leukocyte cell adhesion molecule)[46,47], EpCAM (Epithelial cell adhesion molecule)[48], and ABCG2 (ATP binding cassette super-family G member 2)[49].
Colon CSCs (CCSCs) were first identified and isolated by Ricci-Vitiani et al[50] after the injection of colon cancer (CC) CD133+ cells into immunocompromised mice, which led to the generation of the original tumor mass contrary to their CD133- counterparts. The CD133+ cells were able to exponentially grow in vitro as undifferentiated spheres while preserving the same phenotypic properties of the initial colon tumor[50]. O'Brien et al[51] in 2007 also showed that all CC-initiating cells were CD133+ cells that were able to either maintain themselves as undifferentiated CCSCs or to differentiate and therefore sustain the tumor heterogeneity[51].
Leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5) (also recognized as FEX; HG38; GPR49; GPR67) is a seven-transmembrane G-protein coupled receptor. LGR5 is an “orphan” receptor abundantly expressed in active SCs of the intestinal crypts[52]. LGR5 was shown to be overexpressed in CC[53]. A growing body of evidence supports the idea that LGR5 is a main marker of CCSCs. For instance, human LGR5+ CC cells were visualized as the CSC pool in proliferating CC tissue[54]. Furthermore, LGR5 was demonstrated to be a marker of tumor-initiating cells, where implantation of LGR5+ cells was able to form colon tumors, indicating that LGR5 provides a dynamic stemness characteristic in CC[55]. Additionally, LGR5 was correlated with tumor proliferation due to the ability of LGR5+ cells to form more multipotent spheres as compared to LGR5- cells[56]. Notably, LGR5 was shown to be involved in the colony formation capacity of CCSCs[56,57]. Importantly, LGR5 was found to have an essential role in CC metastasis where organoids derived from LGR5+ cells led to liver cancer formation in the absence of a primary tumor[55]. In addition, LRG5 was selected to be the most suitable CSC marker that identifies immature cancer cells in regional lymph nodes of CC patients[58].
EpCAM (also known as CD326) is a Type-I transmembrane glycoprotein that serves as an epithelial cell adhesion molecule. Interestingly, EpCAM along with its reprogramming TFs were shown to be overexpressed in CC-initiating cells leading to a high self-renewal ability and increased invasiveness[59]. In fact, EpCAM was considered to be a robust CCSCs marker[60]. Indeed, it was used along with CD133 and CD44 to initiate CC in mice[61]. Furthermore, EpCAM provided more enhanced CSC-like properties when combined with LRG5 and CD44[56]. Moreover, EpCAM was proven to promote CC invasion and metastasis, as EpCAMhigh/CD44+ cells were visible in corresponding liver metastasis regions of CC patients[62].
CD44 was also shown to be a robust marker for CCSCs. In fact, a single CD44+ cell was able not only to generate a sphere, but also to form a tumor with similar characteristics as the primary one from which it was isolated[63]. Moreover, the expression of CD44 was correlated with CC proliferation[4]. Furthermore, CD44 was reported as a stemness marker in spherical clusters[64]. In addition, CD44 was considered a reliable marker for the prediction of hepatic cancer metastasis in CC patients[65].
ALDH1 is also selected as a potential marker for CCSCs. ALDH1 expression increased during CC tumorigenesis and the implantation of only 25 ALDH1+ cells into immunocompromised mice led to the generation of xenograft tumors even in the absence of other CCSCs markers such as CD133 and CD44[66]. Furthermore, ALDH1 expression conferred high tumorigenic abilities and chemo-resistance to CC cell lines[67]. Interestingly, ALDH1 was linked to lymph node and vascular invasion in CC patients[68].
Among the most specific CSCs related to CC are LGR5, CD44 and EpCAM. However, the combination of multiple markers allows more accurate detection of CSCs which was proven when LGR5, CD44 and EpCAM resulted in more potent CSCs properties as compared to each marker alone[56]. Other markers are also attributed to CCSCs such as CD59[69], Prox1 a regulator of Notch-independent LGR5+ SCs[70,71], CD24[4,64], CD166[72], and ABCB5 (ATP binding cassette super-family B member 5)[73].
Bladder CSCs (BCSCs) were first isolated in 2009 by using markers for normal basal bladder SCs (CD44+). It was found that the CD44+ subpopulation of bladder cancer (BC) cells was 10 to 200 more likely to form tumors in immunocompromised mice in comparison with their CD44 counterparts[74]. Additionally, CD44+ BCSCs efficiently maintained the heterogeneity of the initial tumor mass after serial transplantation[74].
Epithelial membrane antigen (EMA, also known as MUC1) is a membrane-bound glycoprotein that belongs to the family of mucins[75]. EMA+ bladder cells are usually located in the mature differentiated layer of the urothelium, whereas EMA- cells are found in the basal layers, where SCs reside. It was demonstrated that EMA- BC cells had a greater colony-forming ability when compared with the unsorted BC population[75,76]. BCSCs can thus be identified through the combination of EMA- and CD44+ BC cells[77].
CD47 (also known as integrin associated protein) is a transmembrane protein overexpressed on the surface of CD44+ BCSCs compared to the CD44- subpopulation and was thus hypothesized to be a BCSCs marker[78,79]. CD47 acts like a “don’t eat me” signal by interacting with the signal regulatory protein-1 receptor on the surface of macrophages and neutrophils. Thus, CD47 has an immunosuppressive role, protecting the BSCSC from phagocytosis[78,79], that makes it a promising target for cancer therapy[80,81].
ALDH1A1 has also been used to isolate BCSCs. In fact, ALDH1A1+ cells retained the stem-cell ability to divide asymmetrically, yielding both ALDH1A1+ and ALDH1A1- cells[82]. Additionally, ALDH1A1+ BCSCs exhibited a greater tumorigenic potential both in vitro (sphere formation ability) and in vivo (xenografts in immunocompromised mice) compared to ALDH1A1- BC cells[82]. Knocking down the ALDH1A1 gene in BCSCs reduced their proliferation, confirming the key role played by the ALDH enzyme in BCSCs division and renewal[83]. Furthermore, ALDH1A1 BCSCs maintained the original tumor heterogeneity after sequential transplantations into immunocompromised mice[83]. Finally, ALDH+ BCSCs demonstrated an enhanced ability to migrate and invade tissues contrary to ALDH- BC cells[82].
67LR+ (67KDa Laminin Receptor)/ CD66c- (also known as CEACAM6) BC cells were demonstrated to have stemness properties. These markers, similar to CD44, are also present in normal bladder SCs[84]. He et al[85] showed that 67LR+ BCSCs were 5 to 10 times more potent in initiating tumors in vivo compared to 67LR- ones[85]. In addition, 67LR+ BCSCs expressed a panel of genes involved in stemness and resistance to chemotherapy and radiation[85,86]. Similarly, CD66c- cells were demonstrated to be more tumorigenic than the CD66c+ counterparts[85].
CD133+ BC cells were shown to upregulate the expression of genes involved in pluripotency. This subpopulation of BC cells was also more resistant to the chemotherapeutic agent cisplatin and to radiation. Additionally, CD133+ BCSCs exhibited a greater tumorigenicity both in vitro and in vivo, as well as a more aggressive proliferation in immunocompromised mice in comparison to CD133- BC cells[87].
Additional markers are also used for the identification of BSCSC namely MAGE-A3 (Melanoma antigen family A, 3)[88], BCMab1[79], and several members of the cytokeratin family of proteins (CK5+, CK17+, CK18-, CK20-)[89].
The importance of breast CSCs (BrCSCs) markers was first demonstrated by Al-Hajj et al[90] only a subpopulation of human breast cancer (BrC) cells appeared to lead to the formation of tumors in immunocompromised mice. Al-Hajj et al[90] isolated ESA+CD44+CD24-/Low cells from human BrC tissue, and showed that as low as 200 of these cells were enough to initiate cancer in immunocompromised mice, whereas more than 50000 BrC cells with a different phenotype were unable to form tumors[90].
CD44 and CD24 are often used in combination to detect and isolate BrCSCs[91]. In addition to its key role in adhesion, cell survival, metastasis and angiogenesis, CD44 act as a TF to regulate metastasis and stemness of BrCSCs[92,93]. On the other hand, CD24 is a cell surface adhesion glycoprotein which plays a key role in cell-cell and cell-extracellular matrix (ECM) interactions[94,95]. Even though CD24 is overexpressed in a number of cancers (including BrC), only CD44+CD24-/Low BrCSCs were able to form tumors in immunocompromised mice[90]. CD44+CD24-/Low BrCSCs were also shown to be more resistant to chemotherapy[91].
ALDH1 has also been used to target BrCSCs, as it was shown that ALDH1+ BrC cells were more resistant to chemotherapy and were able to form tumors in immunocompromised mice in comparison to ALDH1- cells[96]. ALDH1 is essential for the early development of the stemness properties of BrCSCs[97]. Interestingly, the subpopulation of BrC cells expressing ALDH1 is distinct from the CD44+CD24-/Low BrCSCs, with minimal overlap between the two (approximately 1%)[91]. Moreover, ALDH1+/CD44+ BrCSCs were highly tumorigenic, with a higher metastatic potential, and greater resistance to cancer therapies[91].
To date, CD44, CD24 and ALDH1 remain the most used biomarkers to isolate BrCSCs. Although there is little overlap between CD44+CD24-/Low and ALDH1+ BrCSCs, cells that share all three markers were more tumorigenic[98]. Moreover, the CD44/CD24 markers were more associated with cell proliferation and tumorigenesis while the ALDH1 marker was positively correlated with tumor metastasis[98]. Nonetheless, other markers have been studied and found suitable for the identification of BrCSCs, such as CD133 (in triple negative BrC; TNBC), GD2 (ganglioside in TNBC), CD49f, CD61+ (β3 integrin in Her2 BrC), CD29 (β1 integrin), CD90, and EpCAM[99-101].
Lung cancer is histologically divided into non-small cell lung carcinoma cells (NSCLC) and small cell lung carcinoma (SCLC)[102]. Due to a higher incidence and the greater ease to obtain NSCLC tissue, NSCLC CSCs (referred to afterward as lung CSCs; LCSCs) markers have been better characterized.
CD166 (also known as ALCAM) has also been associated with stemness properties of NSCLC. CD166 is a member of the immunoglobulin superfamily of cell adhesion molecules and participates in both homophilic and heterophilic interactions. Additionally, CD166 plays an important role in migration and invasion of LCSCs[103]. CD166 was characterized by Zhang et al[104] as the most robust cell marker for isolating LCSCs among other candidates (CD44, EpCAM and CD133)[104]. In contrast to CD166- NSCLC cells which failed to form tumors in vivo, CD166+ LCSCs were able to initiate tumors in immunocompromised mice. Furthermore, CD166+ NSCLC cells had enhanced self-renewal properties and were able to consistently form spheres in vitro.
The CD133+ subpopulation of NSCLC cells were able to indefinitely divide and form spheres in an in vitro setting, whereas CD133- NSCLC cells were characterized by a slow growth and an inability to form spheres[105]. These results also parallel the in vivo ability of CD133+ LCSCs to form tumors in immunocompromised mice compared to CD133- cells; the CD133+ xenografts were histologically similar to the initial cancer mass[105,106]. Moreover, the expression of CD133 in LCSCs was associated with increased resistance to chemotherapy and radiation[105,107]. Finally, CD133+ LCSCs are more prone to metastasize than their CD133- counterparts, especially to lymphoid organs. In fact, detection of CD133+ metastatic NSCLC in lymph nodes is indicative of a poor prognosis[107].
CD44 has also been studied as a marker to isolate LCSCs. Accordingly, CD44+ NSCLC cells demonstrated a greater ability to form spheres in vitro and to initiate tumors in immunocompromised mice in comparison to CD44- cells. Additionally, CD44+ LCSCs upregulated several stemness TFs to maintain their pluripotent properties. CD44+ LCSCs were also more resistant to the chemotherapeutic agent cisplatin compared to CD44- cells[108]. Moreover, the expression of CD44 in LCSCs was associated with an enhanced ability to metastasize and invade tissues[20].
CD90 (also known as Thy-1) is a glycosylphosphatidylinositol-anchored surface protein that is involved in cell-cell as well as cell-ECM interactions[109]. Initial studies have shown that CD90+ NSCLC cells demonstrated greater self-renewal and proliferative properties and expressed a higher level of stemness genes. Additionally, when compared to a control, as few as 5000 CD90+ LCSCs were able to initiate tumors in immunocompromised mice, indicating the stronger tumorigenicity associated with CD90[110].
ALDH1 was also suggested to be a LCSCs marker. Indeed, ALDH1+ LCSCs exhibited enhanced proliferative abilities and self-renewal properties[111,112]. Accordingly, knocking down the ALDH1A3 gene greatly reduced the tumorigenicity and clonogenicity of LCSCs[113]. In addition, ALDH1high LCSCs also showed greater resistance to chemotherapeutic drugs in comparison to ALDH1low cells[112]. Interestingly, the overexpression of the TAZ oncogene induces the formation of LCSCs by activating the ALDH1 gene[114] ALDH1 also appears to play a key role in chemoresistance as its inhibition leads to the re-sensitization of LCSCs to cisplatin[115].
Of note, additional markers have been used to isolate LCSCs. These include but are not limited to CD47, CD87, CD117, EpCAM, and CK5[116,117].
The conception of CSCs-targeted therapies relies on employing the above-mentioned CSCs' resistant characteristics and markers, which allows for CSCs' isolation, enrichment, characterization, and targeting[118]. CSCs-based therapeutic strategies include selectively targeting the stemness markers, such as the TSCSCs' surface markers, TFs, ABC transporters, and ALDHs[14,119]. As well as, the disrupted signaling pathways that enrich CSCs'-resistant features and contribute to their survival, proliferation, self-renewal, and differentiation. Also, targeting the tumor microenvironment components which acts as a foster niche in protecting CSCs[14,119].
In spite of the significant advances in CSCs' research and the great interest in drug discovery, there are currently few therapeutic approaches that have reached the late clinical stages. Many CSCs-targeting therapeutics performing remarkably in vitro and in vivo cultures have faced multiple hurdles in clinical trials[14,120]. One major reason behind this is the irrelevance of the preclinical cancer models being used[121-123]. Thus, more relevant CSCs models, that reflect the original tumor behavior of the individual patients, might strengthen the rationale for developing effective CSCs-targeted therapeutic modalities and complement more conventional cancer therapies.
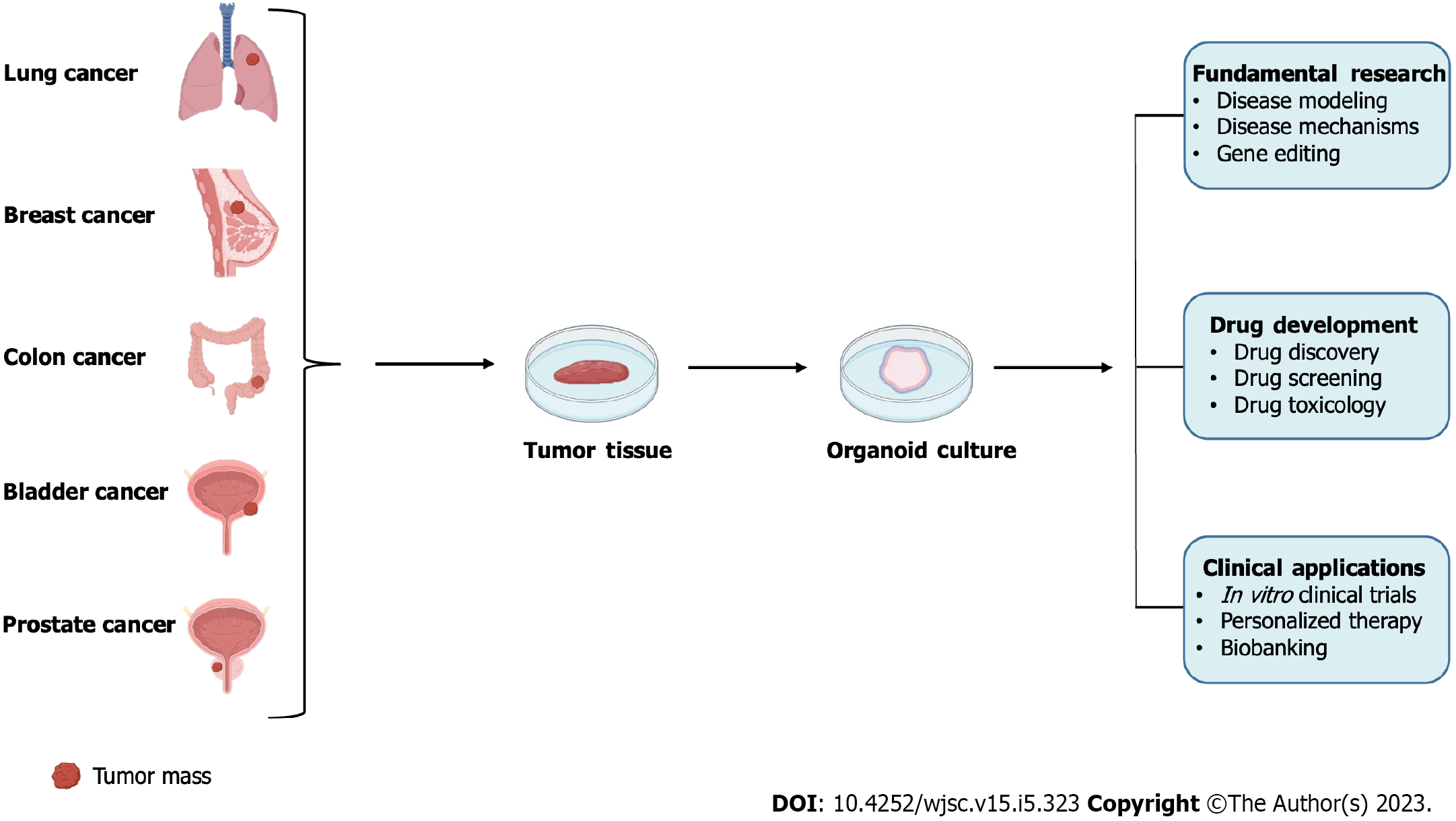
A new era of targeted and personalized anti-cancer therapies has evolved with the three-dimensional (3D) patient-derived organoids (PDOs)[124]. This versatile technique relies on the exclusive ability of SCs to give rise to organ-like structures known as organoids[125]. Sato et al[126] established the first organoid model with small intestinal crypt LGR5+ SCs[126]. Subsequently, models of normal and cancer PDOs from multiple tissues were derived successfully[127-134].
The formation of the 3D microscopic organoids from patient tumor tissues is accomplished using specific culturing conditions that are designed to preserve the CSCs component of the patient's tumor[135]. The formed PDOs, hence, recapitulate the structural and functional complexity constituting the originating tumor, mediated by the CSCs’ ability for self-renewal and differentiation into multiple cell types[136,137]. PDOs tool allows the modeling of human carcinogenesis in an in vitro culture dish[138,139]. Precisely, the process followed to generate cancer PDOs includes utilizing a tumor tissue sample, surgically isolated from a cancer patient, and dissociating it into single-cell suspension using mechanical dissociation and enzymatic digestion methods. The heterogeneous population of cells obtained, containing TSCSCs, is then cultured in proper culturing conditions to allow the self-organization of cells into functional units or tissue-specific architectures; organ analogs. The suspended culturing system includes the usage of biological or synthetic hydrogel scaffolds that mimic the natural ECM components. In addition to using a specific culturing medium that contains a cocktail of growth factors and inhibitors to imitate the organ stem cell niche, allow the generation of distinct component lineages, and stimulate the long-term expansion of organoids[140,141].
As PDOs are CSCs-based structures and replicate faithfully the heterogeneity and histological characteristics of the original cancers, they gain superiority over other models in terms of mimicking tumor microenvironments, facilitating the formation of ECM, exhibiting adequate proliferation rates with representative cellular morphology, maintaining the expression of ‘stemness-related’ markers and genes, and demonstrating a realistic individualized drug response[142-144]. This nominates PDOs to be ideal preclinical drug-response models for providing perspectives for testing novel CSCs-targeted therapies and evaluating the potential drug effectiveness in cancer patients (Figure 2).
PDOs technique generally shares several main steps but differs in varying degrees depending on the type of tissue being processed. Scaffold-based techniques are mostly adopted in culturing PDOs, where MatrigelTM is commonly used. The latter is a mixture of heterogeneous and gelatinous proteins secreted by mouse sarcoma cells. It comprises mainly adhesive proteins such as laminin, collagen IV, entactin, and heparin sulphate glycoprotein, which resemble the ECM and provide interactive and structural support to the cells[145-148]. Moreover, the universal organoid medium used in the culturing system adopts the first protocol developed by Sato et al[126] which includes advanced DMEM/F12 medium supplemented with epidermal growth factor, Noggin (NOG), and Wnt agonist R-spondin-1[126,127]. Other factors were then added including anaplastic lymphoma kinase 3/4/5 inhibitor A83-01, dihydrotestosterone, fibroblast growth factor-10, fibroblast growth factor-2, prostaglandin E2, nicotinamide (NAM), and p38 inhibitor SB202190, N-acetylcysteine (NAC), B27 supplement and Rho kinase inhibitor Y-27632 to culture PDOs successfully[149].
To date, organoids derivation from multiple human tumors including prostate, colon, bladder, breast, and lung cancers has been described, with varying success rates[133,150-155]. The established PDOs are subjected to tissue-specific genes and lineage markers expression studies to confirm that they represent the original tumor of the patient. Importantly, the cancerous origin of these organoids is confirmed by checking for the CSCs markers specific to each tumor tissue. The patient drug response to the therapy of interest can then be evaluated primarily by assessing the organoids' formation efficiency and size.
For example, a study done by Cheaito et al[150] established a minimum of 5-factor medium including NAC, NOG, A83-01, B27, and NAM to grow and maintain PC PDOs. Histopathological, transcriptomic, immunofluorescent, and immunohistochemical studies showed that the formed PDOs mimicked the histological architecture and prostate lineage profiles of their corresponding tissue specimens. This was confirmed by the presence of both prostate epithelial lineages, as the organoids stained positive for the luminal- (CK8, AR, and PSA) and basal- (CK5, CK14, and p63) specific markers. In addition, an intermediate cell population, co-expressing luminal CK8 and basal CK5 markers was also detected. Interestingly, CSCs markers, CD44 and CD49f, positive staining demonstrated the existence of putative stemlike cells within the bulk of the PDOs. Furthermore, differential drug response, between different patient samples, was recognized upon treatment with chemo-, radio-, and androgen-deprivation therapies[150]. In another study, Monzer et al[151] succeeded in establishing and propagating PDOs that model CC disease. The formed organoids recapitulated the architecture and the characteristics of CC tissues as revealed by the co-expression of the epithelial marker lineage CK19 and the CSC surface marker CD44. The organoids derived from different patients showed to exhibit different responses to Diiminoquinone treatment tested alone or in combination with Fluorouracil (5FU) chemotherapeutic drug. Similarly, Al Bitar et al’s study showed different responses to individual and combination treatments of radiation and Thymoquinone in CC PDOs[152].
Moreover, Yu et al[153] utilized BC PDOs to evaluate chimeric antigen receptor (CAR)-T cell-mediated cytotoxicity against BC. Analysis was done to confirm that the established organoids recapitulate the heterogeneity and the key features of the parental BCs. Based on a set of luminal (CK20, uroplakin II, and GATA3) and basal markers (CK5, P63, and CD44), the formed organoids were classified into luminal or basal subtypes, respectively. All the BC PDOs and their corresponding tumors expressed Ki67 and E-cadherin, confirming their epithelial origin and high proliferative ability. Additionally, the specific surface antigen profiling of each tumor sample was analyzed, and the MUC1 antigen was shown to be highly expressed among all tested antigens, in both the cancer tissues and their derived organoids. MUC1 was then used as a putative target to test the efficacy of second-generation CAR-T cells in BC PDOs[153]. Furthermore, a promising study done by Chen et al[154], showed the significance and applicability of using BrC PDOs as pre-clinical models for broader cancer studies, and more specifically as a tool to provide personalized therapy recommendations for patients with advanced refractory disease. This study focused mainly on deriving PDOs from specimens isolated from patients with advanced clinical features, including drug-resistant and metastatic BrC. The histopathological, immunohistochemical, and genomic characteristics were shown to be well inherited by the formed PDOs from the drug-treated as well as treatment-naïve tumors. Distinctive drug responses were also observed[154]. Furthermore, Kim et al[133] demonstrated the distinctive therapeutic responses of LC and normal bronchial PDOs, derived from patient tissues comprising five histological subtypes of LC and non-neoplastic bronchial mucosa. The differential responses to the tested drugs were shown to be affected by the individual genomic alterations profile. The PDOs were also proved to duplicate the tissue architecture and maintain the genomic alterations of the parental lung tumors during long-term expansion in vitro[133].
In this review, we have discussed briefly some of the CSC features that are known to account for cancer resistance and relapse and make CSCs promising anti-cancer targets. Additionally, we have summarized the updated list of the TSCSC molecular markers in prostate, colon, bladder, and lung tumors that are significant to selectively isolate and therapeutically target the CSCs subpopulation. Besides, we highlighted the advantage of utilizing the CSC-based PDO models to simulate carcinogenesis and predict patient-specific drug responses in vitro.
Despite the present challenges[156,157], PDOs are highly credible models that possess more physiological and pathological relevance than traditional ones. This robust method proved to faithfully maintain the histological, genetic, and stemness characteristics of their respective native tissues. Interestingly, the CSCs profile mimicked by the PDOs can serve as a platform for testing CSCs-targeted therapeutics. To our knowledge, there are no clinical trials discussing cancer PDOs in a preclinical context for testing CSC-targeted therapeutics[158].
Indeed, PDOs have prospective applications in patient-specific in vitro drug clinical trials and proof-of-concept studies on CSC-targeted therapies and -resistance mechanisms. If remarkable advancements are made, cancer patients will ultimately benefit from this radical technology.
We want to thank the members of the Abou-Kheir’s Laboratory (The WAKers).
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: Lebanon
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Crocé LS, Italy; Elbadawy M, Egypt S-Editor: Li L L-Editor: A P-Editor: Zhang XD
| 1. | Sun Y. Translational horizons in the tumor microenvironment: harnessing breakthroughs and targeting cures. Med Res Rev. 2015;35:408-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 2. | Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1234] [Cited by in RCA: 1880] [Article Influence: 235.0] [Reference Citation Analysis (0)] |
| 3. | Chang JC. Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine (Baltimore). 2016;95:S20-S25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 312] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 4. | Huang JL, Oshi M, Endo I, Takabe K. Clinical relevance of stem cell surface markers CD133, CD24, and CD44 in colorectal cancer. Am J Cancer Res. 2021;11:5141-5154. [PubMed] |
| 5. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47023] [Article Influence: 3358.8] [Reference Citation Analysis (5)] |
| 6. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3316] [Cited by in RCA: 3382] [Article Influence: 109.1] [Reference Citation Analysis (0)] |
| 7. | Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1987] [Cited by in RCA: 2134] [Article Influence: 118.6] [Reference Citation Analysis (0)] |
| 8. | Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 562] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 9. | Wang J, Sullenger BA, Rich JN. Notch signaling in cancer stem cells. Adv Exp Med Biol. 2012;727:174-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 10. | Rampias T, Vgenopoulou P, Avgeris M, Polyzos A, Stravodimos K, Valavanis C, Scorilas A, Klinakis A. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014;20:1199-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 152] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 11. | Naz F, Shi M, Sajid S, Yang Z, Yu C. Cancer stem cells: a major culprit of intra-tumor heterogeneity. Am J Cancer Res. 2021;11:5782-5811. [PubMed] |
| 12. | Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, Kuppanna G, Kallappa CG, Basalingappa KM. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 2018;5:5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 209] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 13. | Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat Rev Clin Oncol. 2020;17:204-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 500] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 14. | Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 896] [Cited by in RCA: 1199] [Article Influence: 239.8] [Reference Citation Analysis (0)] |
| 15. | Su YJ, Chang YW, Lin WH, Liang CL, Lee JL. An aberrant nuclear localization of E-cadherin is a potent inhibitor of Wnt/β-catenin-elicited promotion of the cancer stem cell phenotype. Oncogenesis. 2015;4:e157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 16. | Sari IN, Phi LTH, Jun N, Wijaya YT, Lee S, Kwon HY. Hedgehog Signaling in Cancer: A Prospective Therapeutic Target for Eradicating Cancer Stem Cells. Cells. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 139] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 17. | Usui T, Sakurai M, Umata K, Elbadawy M, Ohama T, Yamawaki H, Hazama S, Takenouchi H, Nakajima M, Tsunedomi R, Suzuki N, Nagano H, Sato K, Kaneda M, Sasaki K. Hedgehog Signals Mediate Anti-Cancer Drug Resistance in Three-Dimensional Primary Colorectal Cancer Organoid Culture. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 18. | Kaltschmidt C, Banz-Jansen C, Benhidjeb T, Beshay M, Förster C, Greiner J, Hamelmann E, Jorch N, Mertzlufft F, Pfitzenmaier J, Simon M, Schulte Am Esch J, Vordemvenne T, Wähnert D, Weissinger F, Wilkens L, Kaltschmidt B. A Role for NF-κB in Organ Specific Cancer and Cancer Stem Cells. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 19. | Wang T, Fahrmann JF, Lee H, Li YJ, Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, Somlo G, Jandial R, Ann D, Hanash S, Jove R, Yu H. JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 2018;27:136-150.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 547] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 20. | Heng WS, Gosens R, Kruyt FAE. Lung cancer stem cells: origin, features, maintenance mechanisms and therapeutic targeting. Biochem Pharmacol. 2019;160:121-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 21. | Desai A, Webb B, Gerson SL. CD133+ cells contribute to radioresistance via altered regulation of DNA repair genes in human lung cancer cells. Radiother Oncol. 2014;110:538-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 22. | McIntosh K, Balch C, Tiwari AK. Tackling multidrug resistance mediated by efflux transporters in tumor-initiating cells. Expert Opin Drug Metab Toxicol. 2016;12:633-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 23. | Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2:138-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 294] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 24. | Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522-526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1518] [Cited by in RCA: 1753] [Article Influence: 134.8] [Reference Citation Analysis (0)] |
| 25. | Collins AT, Habib FK, Maitland NJ, Neal DE. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114:3865-3872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 249] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 26. | Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539-3545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 562] [Cited by in RCA: 551] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 27. | Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946-10951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1998] [Cited by in RCA: 2033] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 28. | Kanwal R, Shukla S, Walker E, Gupta S. Acquisition of tumorigenic potential and therapeutic resistance in CD133+ subpopulation of prostate cancer cells exhibiting stem-cell like characteristics. Cancer Lett. 2018;430:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 29. | Acikgoz E, Soner BC, Ozdil B, Guven M. CD133+/CD44+ prostate cancer stem cells exhibit embryo-like behavior patterns. Acta Histochem. 2021;123:151743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 30. | Gu G, Yuan J, Wills M, Kasper S. Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo. Cancer Res. 2007;67:4807-4815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 265] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 31. | Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696-1708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 724] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 32. | Ni J, Cozzi PJ, Hao JL, Beretov J, Chang L, Duan W, Shigdar S, Delprado WJ, Graham PH, Bucci J, Kearsley JH, Li Y. CD44 variant 6 is associated with prostate cancer metastasis and chemo-/radioresistance. Prostate. 2014;74:602-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 33. | Heath EI, Heilbrun LK, Smith D, Schopperle WM, Ju Y, Bolton S, Ahmed Q, Sakr WA. Overexpression of the Pluripotent Stem Cell Marker Podocalyxin in Prostate Cancer. Anticancer Res. 2018;38:6361-6366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Schäfer C, Ju Y, Tak Y, Vazquez C, Han SJ, Tan E, Shay JW, Holmqvist M, Danuser G, Schopperle WM, Bubley G. TRA-1-60-positive/CD45(low) cells found in the peripheral blood of prostate cancer patients with metastatic disease - A proof-of-concept study. Heliyon. 2020;6:e03263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 35. | Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-κB signalling. Nat Commun. 2011;2:162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 209] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 36. | Wiesner C, Nabha SM, Dos Santos EB, Yamamoto H, Meng H, Melchior SW, Bittinger F, Thüroff JW, Vessella RL, Cher ML, Bonfil RD. C-kit and its ligand stem cell factor: potential contribution to prostate cancer bone metastasis. Neoplasia. 2008;10:996-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 37. | Kerr BA, Miocinovic R, Smith AK, West XZ, Watts KE, Alzayed AW, Klink JC, Mir MC, Sturey T, Hansel DE, Heston WD, Stephenson AJ, Klein EA, Byzova TV. CD117⁺ cells in the circulation are predictive of advanced prostate cancer. Oncotarget. 2015;6:1889-1897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 38. | Harris KS, Shi L, Foster BM, Mobley ME, Elliott PL, Song CJ, Watabe K, Langefeld CD, Kerr BA. CD117/c-kit defines a prostate CSC-like subpopulation driving progression and TKI resistance. Sci Rep. 2021;11:1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 39. | Mulholland DJ, Xin L, Morim A, Lawson D, Witte O, Wu H. Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res. 2009;69:8555-8562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 149] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 40. | Yamamoto H, Masters JR, Dasgupta P, Chandra A, Popert R, Freeman A, Ahmed A. CD49f is an efficient marker of monolayer- and spheroid colony-forming cells of the benign and malignant human prostate. PLoS One. 2012;7:e46979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 41. | Bahmad HF, Cheaito K, Chalhoub RM, Hadadeh O, Monzer A, Ballout F, El-Hajj A, Mukherji D, Liu YN, Daoud G, Abou-Kheir W. Sphere-Formation Assay: Three-Dimensional in vitro Culturing of Prostate Cancer Stem/Progenitor Sphere-Forming Cells. Front Oncol. 2018;8:347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 146] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 42. | Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci U S A. 2008;105:20882-20887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 277] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 43. | Matsika A, Srinivasan B, Day C, Mader SA, Kiernan DM, Broomfield A, Fu J, Hooper JD, Kench JG, Samaratunga H. Cancer stem cell markers in prostate cancer: an immunohistochemical study of ALDH1, SOX2 and EZH2. Pathology. 2015;47:622-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 44. | Gorodetska I, Offermann A, Püschel J, Lukiyanchuk1 V, Gaete G, Kurzyukova A, Schwarz1 F, Lange T, Knopf F, Wielockx B, Krause1 M, Perner S, Dubrovska1 A. The distinct role of ALDH1A1 and ALDH1A3 in the regulation of prostate cancer metastases. BioRxiv. 2021;. [DOI] [Full Text] |
| 45. | Trerotola M, Rathore S, Goel HL, Li J, Alberti S, Piantelli M, Adams D, Jiang Z, Languino LR. CD133, Trop-2 and alpha2beta1 integrin surface receptors as markers of putative human prostate cancer stem cells. Am J Transl Res. 2010;2:135-144. [PubMed] |
| 46. | Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, Lawson D, Chen D, Li Y, Guo C, Zhang B, Fazli L, Gleave M, Witte ON, Garraway IP, Wu H. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One. 2012;7:e42564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 47. | Hansen AG, Arnold SA, Jiang M, Palmer TD, Ketova T, Merkel A, Pickup M, Samaras S, Shyr Y, Moses HL, Hayward SW, Sterling JA, Zijlstra A. ALCAM/CD166 is a TGF-β-responsive marker and functional regulator of prostate cancer metastasis to bone. Cancer Res. 2014;74:1404-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 48. | Ni J, Cozzi P, Hao J, Beretov J, Chang L, Duan W, Shigdar S, Delprado W, Graham P, Bucci J, Kearsley J, Li Y. Epithelial cell adhesion molecule (EpCAM) is associated with prostate cancer metastasis and chemo/radioresistance via the PI3K/Akt/mTOR signaling pathway. Int J Biochem Cell Biol. 2013;45:2736-2748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 49. | Wang L, Stadlbauer B, Lyu C, Buchner A, Pohla H. Shikonin enhances the antitumor effect of cabazitaxel in prostate cancer stem cells and reverses cabazitaxel resistance by inhibiting ABCG2 and ALDH3A1. Am J Cancer Res. 2020;10:3784-3800. [PubMed] |
| 50. | Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2952] [Cited by in RCA: 3033] [Article Influence: 159.6] [Reference Citation Analysis (0)] |
| 51. | O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2977] [Cited by in RCA: 3048] [Article Influence: 160.4] [Reference Citation Analysis (0)] |
| 52. | Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3854] [Cited by in RCA: 4327] [Article Influence: 240.4] [Reference Citation Analysis (0)] |
| 53. | Takahashi H, Ishii H, Nishida N, Takemasa I, Mizushima T, Ikeda M, Yokobori T, Mimori K, Yamamoto H, Sekimoto M, Doki Y, Mori M. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann Surg Oncol. 2011;18:1166-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 54. | Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T, Sato T. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature. 2017;545:187-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 544] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 55. | de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, Dijkgraaf GJ, Piskol R, de Sauvage FJ. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 579] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 56. | Leng Z, Xia Q, Chen J, Li Y, Xu J, Zhao E, Zheng H, Ai W, Dong J. Lgr5+CD44+EpCAM+ Strictly Defines Cancer Stem Cells in Human Colorectal Cancer. Cell Physiol Biochem. 2018;46:860-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 57. | Kemper K, Prasetyanti PR, De Lau W, Rodermond H, Clevers H, Medema JP. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378-2386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 212] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 58. | AbdelMageed M, Ismail HTH, Olsson L, Lindmark G, Hammarström ML, Hammarström S, Sitohy B. Clinical Significance of Stem Cell Biomarkers EpCAM, LGR5 and LGR4 mRNA Levels in Lymph Nodes of Colon Cancer Patients. Int J Mol Sci. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 59. | Lin CW, Liao MY, Lin WW, Wang YP, Lu TY, Wu HC. Epithelial cell adhesion molecule regulates tumor initiation and tumorigenesis via activating reprogramming factors and epithelial-mesenchymal transition gene expression in colon cancer. J Biol Chem. 2012;287:39449-39459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 60. | Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158-10163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1541] [Cited by in RCA: 1661] [Article Influence: 92.3] [Reference Citation Analysis (0)] |
| 61. | Roy K, Kanwar RK, Kanwar JR. LNA aptamer based multi-modal, Fe3O4-saturated lactoferrin (Fe3O4-bLf) nanocarriers for triple positive (EpCAM, CD133, CD44) colon tumor targeting and NIR, MRI and CT imaging. Biomaterials. 2015;71:84-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 62. | Liu D, Sun J, Zhu J, Zhou H, Zhang X, Zhang Y. Expression and clinical significance of colorectal cancer stem cell marker EpCAM(high)/CD44(+) in colorectal cancer. Oncol Lett. 2014;7:1544-1548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 63. | Du L, Wang H, He L, Zhang J, Ni B, Wang X, Jin H, Cahuzac N, Mehrpour M, Lu Y, Chen Q. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751-6760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 470] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 64. | Kapeleris J, Zou H, Qi Y, Gu Y, Li J, Schoning J, Monteiro MJ, Gu W. Cancer stemness contributes to cluster formation of colon cancer cells and high metastatic potentials. Clin Exp Pharmacol Physiol. 2020;47:838-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 65. | Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH, Kim HR. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int J Oncol. 2015;46:1582-1588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 66. | Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382-3389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 831] [Cited by in RCA: 828] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 67. | Kozovska Z, Patsalias A, Bajzik V, Durinikova E, Demkova L, Jargasova S, Smolkova B, Plava J, Kucerova L, Matuskova M. ALDH1A inhibition sensitizes colon cancer cells to chemotherapy. BMC Cancer. 2018;18:656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 68. | Holah NS, Aiad HA, Asaad NY, Elkhouly EA, Lasheen AG. Evaluation of the Role of ALDH1 as Cancer Stem Cell Marker in Colorectal Carcinoma: An Immunohistochemical Study. J Clin Diagn Res. 2017;11:EC17-EC23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 69. | Wang J, Zhang B, Wu H, Cai J, Sui X, Wang Y, Li H, Qiu Y, Wang T, Chen Z, Zhu Q, Xia H, Song W, Xiang AP. CD51 correlates with the TGF-beta pathway and is a functional marker for colorectal cancer stem cells. Oncogene. 2017;36:1351-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 70. | Wiener Z, Högström J, Hyvönen V, Band AM, Kallio P, Holopainen T, Dufva O, Haglund C, Kruuna O, Oliver G, Ben-Neriah Y, Alitalo K. Prox1 promotes expansion of the colorectal cancer stem cell population to fuel tumor growth and ischemia resistance. Cell Rep. 2014;8:1943-1956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 71. | Abdelrahman AE, El-Azony A, Elsebai E, Ibrahim HM. Prognostic Impact of LGR5, Prox1, and Notch1 Biomarkers in Stage II to III Colon Cancer. Appl Immunohistochem Mol Morphol. 2022;30:126-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 72. | Shafaei S, Sharbatdaran M, Kamrani G, Khafri S. The association between CD166 detection rate and clinicopathologic parameters of patients with colorectal cancer. Caspian J Intern Med. 2013;4:768-772. [PubMed] |
| 73. | Guo Q, Grimmig T, Gonzalez G, Giobbie-Hurder A, Berg G, Carr N, Wilson BJ, Banerjee P, Ma J, Gold JS, Nandi B, Huang Q, Waaga-Gasser AM, Lian CG, Murphy GF, Frank MH, Gasser M, Frank NY. ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer. J Biol Chem. 2018;293:11166-11178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 74. | Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, Gill H, Presti J Jr, Chang HY, van de Rijn M, Shortliffe L, Weissman IL. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci U S A. 2009;106:14016-14021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 516] [Cited by in RCA: 494] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 75. | Yang YM, Chang JW. Bladder cancer initiating cells (BCICs) are among EMA-CD44v6+ subset: novel methods for isolating undetermined cancer stem (initiating) cells. Cancer Invest. 2008;26:725-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 76. | van der Horst G, Bos L, van der Pluijm G. Epithelial plasticity, cancer stem cells, and the tumor-supportive stroma in bladder carcinoma. Mol Cancer Res. 2012;10:995-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 77. | Aghaalikhani N, Rashtchizadeh N, Shadpour P, Allameh A, Mahmoodi M. Cancer stem cells as a therapeutic target in bladder cancer. J Cell Physiol. 2019;234:3197-3206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 78. | Ferreira-Teixeira M, Parada B, Rodrigues-Santos P, Alves V, Ramalho JS, Caramelo F, Sousa V, Reis F, Gomes CM. Functional and molecular characterization of cancer stem-like cells in bladder cancer: a potential signature for muscle-invasive tumors. Oncotarget. 2015;6:36185-36201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 79. | Li Y, Lin K, Yang Z, Han N, Quan X, Guo X, Li C. Bladder cancer stem cells: clonal origin and therapeutic perspectives. Oncotarget. 2017;8:66668-66679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 80. | Tan J, Wang Y, Sun L, Xu S, Li C, Jin X. The Origin and Evolution of Bladder Cancer Stem Cells. Front Cell Dev Biol. 2022;10:950241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 81. | Kong F, Gao F, Li H, Liu H, Zhang Y, Zheng R, Chen J, Li X, Liu G, Jia Y. CD47: a potential immunotherapy target for eliminating cancer cells. Clin Transl Oncol. 2016;18:1051-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 82. | Falso MJ, Buchholz BA, White RW. Stem-like cells in bladder cancer cell lines with differential sensitivity to cisplatin. Anticancer Res. 2012;32:733-738. [PubMed] |
| 83. | Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z, Stass SA, Jiang F. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:327-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 84. | Brandt WD, Matsui W, Rosenberg JE, He X, Ling S, Schaeffer EM, Berman DM. Urothelial carcinoma: stem cells on the edge. Cancer Metastasis Rev. 2009;28:291-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 85. | He X, Marchionni L, Hansel DE, Yu W, Sood A, Yang J, Parmigiani G, Matsui W, Berman DM. Differentiation of a highly tumorigenic basal cell compartment in urothelial carcinoma. Stem Cells. 2009;27:1487-1495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 86. | Hatina J, Schulz WA. Stem cells in the biology of normal urothelium and urothelial carcinoma. Neoplasma. 2012;59:728-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 87. | Huang P, Watanabe M, Kaku H, Ueki H, Noguchi H, Sugimoto M, Hirata T, Yamada H, Takei K, Zheng S, Xu K, Nasu Y, Fujii Y, Liu C, Kumon H. Cancer stem cell-like characteristics of a CD133(+) subpopulation in the J82 human bladder cancer cell line. Mol Clin Oncol. 2013;1:180-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 88. | Yin B, Zeng Y, Liu G, Wang X, Wang P, Song Y. MAGE-A3 is highly expressed in a cancer stem cell-like side population of bladder cancer cells. Int J Clin Exp Pathol. 2014;7:2934-2941. [PubMed] |
| 89. | Xia P, Liu DH, Xu ZJ, Ren F. Cancer Stem Cell Markers for Urinary Carcinoma. Stem Cells Int. 2022;2022:3611677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 90. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7721] [Article Influence: 351.0] [Reference Citation Analysis (0)] |
| 91. | DA Cruz Paula A, Lopes C. Implications of Different Cancer Stem Cell Phenotypes in Breast Cancer. Anticancer Res. 2017;37:2173-2183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 92. | Miletti-González KE, Murphy K, Kumaran MN, Ravindranath AK, Wernyj RP, Kaur S, Miles GD, Lim E, Chan R, Chekmareva M, Heller DS, Foran D, Chen W, Reiss M, Bandera EV, Scotto K, Rodríguez-Rodríguez L. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J Biol Chem. 2012;287:18995-19007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 101] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 93. | Cho Y, Lee HW, Kang HG, Kim HY, Kim SJ, Chun KH. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget. 2015;6:8709-8721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 94. | Fang X, Zheng P, Tang J, Liu Y. CD24: from A to Z. Cell Mol Immunol. 2010;7:100-103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 229] [Cited by in RCA: 218] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 95. | Jaggupilli A, Elkord E. Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin Dev Immunol. 2012;2012:708036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 358] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 96. | Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3173] [Cited by in RCA: 3057] [Article Influence: 169.8] [Reference Citation Analysis (0)] |
| 97. | Ginestier C, Wicinski J, Cervera N, Monville F, Finetti P, Bertucci F, Wicha MS, Birnbaum D, Charafe-Jauffret E. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle. 2009;8:3297-3302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 162] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 98. | Li W, Ma H, Zhang J, Zhu L, Wang C, Yang Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci Rep. 2017;7:13856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 304] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 99. | Zhang X, Powell K, Li L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 100. | Butti R, Gunasekaran VP, Kumar TVS, Banerjee P, Kundu GC. Breast cancer stem cells: Biology and therapeutic implications. Int J Biochem Cell Biol. 2019;107:38-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 101. | Ali HR, Dawson SJ, Blows FM, Provenzano E, Pharoah PD, Caldas C. Cancer stem cell markers in breast cancer: pathological, clinical and prognostic significance. Breast Cancer Res. 2011;13:R118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 102. | Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 256] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 103. | Ferragut F, Vachetta VS, Troncoso MF, Rabinovich GA, Elola MT. ALCAM/CD166: A pleiotropic mediator of cell adhesion, stemness and cancer progression. Cytokine Growth Factor Rev. 2021;61:27-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 104. | Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, Bhakoo KK, Jayapal SR, Nichane M, Yu Q, Ahmed DA, Tan C, Sing WP, Tam J, Thirugananam A, Noghabi MS, Pang YH, Ang HS, Mitchell W, Robson P, Kaldis P, Soo RA, Swarup S, Lim EH, Lim B. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 460] [Cited by in RCA: 525] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 105. | Zhang DG, Jiang AG, Lu HY, Zhang LX, Gao XY. Isolation, cultivation and identification of human lung adenocarcinoma stem cells. Oncol Lett. 2015;9:47-54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 106. | Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1158] [Cited by in RCA: 1255] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 107. | Wang S, Xu ZY, Wang LF, Su W. CD133+ cancer stem cells in lung cancer. Front Biosci (Landmark Ed). 2013;18:447-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 108. | Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, Sihoe AD, Fink LM, Ma Y, Wong MP. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 2010;5:e14062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 309] [Cited by in RCA: 368] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 109. | Sauzay C, Voutetakis K, Chatziioannou A, Chevet E, Avril T. CD90/Thy-1, a Cancer-Associated Cell Surface Signaling Molecule. Front Cell Dev Biol. 2019;7:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 71] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 110. | Yan X, Luo H, Zhou X, Zhu B, Wang Y, Bian X. Identification of CD90 as a marker for lung cancer stem cells in A549 and H446 cell lines. Oncol Rep. 2013;30:2733-2740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 111. | Ucar D, Cogle CR, Zucali JR, Ostmark B, Scott EW, Zori R, Gray BA, Moreb JS. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact. 2009;178:48-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 125] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 112. | Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, Katz RL. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 622] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 113. | Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, Liu H, Behrens C, Shay JW, Wistuba II, Minna JD. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res. 2014;20:4154-4166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 114. | Yu J, Alharbi A, Shan H, Hao Y, Snetsinger B, Rauh MJ, Yang X. TAZ induces lung cancer stem cell properties and tumorigenesis by up-regulating ALDH1A1. Oncotarget. 2017;8:38426-38443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 115. | MacDonagh L, Gallagher MF, Ffrench B, Gasch C, Breen E, Gray SG, Nicholson S, Leonard N, Ryan R, Young V, O'Leary JJ, Cuffe S, Finn SP, O'Byrne KJ, Barr MP. Targeting the cancer stem cell marker, aldehyde dehydrogenase 1, to circumvent cisplatin resistance in NSCLC. Oncotarget. 2017;8:72544-72563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 116. | Nimmakayala RK, Batra SK, Ponnusamy MP. Unraveling the journey of cancer stem cells from origin to metastasis. Biochim Biophys Acta Rev Cancer. 2019;1871:50-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 117. | Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973-978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1548] [Cited by in RCA: 1611] [Article Influence: 89.5] [Reference Citation Analysis (0)] |
| 118. | Herreros-Pomares A. Identification, Culture and Targeting of Cancer Stem Cells. Life (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 119. | Dragu DL, Necula LG, Bleotu C, Diaconu CC, Chivu-Economescu M. Therapies targeting cancer stem cells: Current trends and future challenges. World J Stem Cells. 2015;7:1185-1201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 130] [Article Influence: 13.0] [Reference Citation Analysis (2)] |
| 120. | Hait WN. Anticancer drug development: the grand challenges. Nat Rev Drug Discov. 2010;9:253-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 121. | Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12:786-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 672] [Cited by in RCA: 730] [Article Influence: 56.2] [Reference Citation Analysis (0)] |
| 122. | Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 1177] [Article Influence: 130.8] [Reference Citation Analysis (0)] |
| 123. | Miserocchi G, Mercatali L, Liverani C, De Vita A, Spadazzi C, Pieri F, Bongiovanni A, Recine F, Amadori D, Ibrahim T. Management and potentialities of primary cancer cultures in preclinical and translational studies. J Transl Med. 2017;15:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 124. | Dzobo K, Rowe A, Senthebane DA, AlMazyadi MAM, Patten V, Parker MI. Three-Dimensional Organoids in Cancer Research: The Search for the Holy Grail of Preclinical Cancer Modeling. OMICS. 2018;22:733-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 125. | Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1502] [Cited by in RCA: 1906] [Article Influence: 173.3] [Reference Citation Analysis (0)] |
| 126. | Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4394] [Cited by in RCA: 5153] [Article Influence: 322.1] [Reference Citation Analysis (0)] |
| 127. | Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762-1772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2253] [Cited by in RCA: 2715] [Article Influence: 193.9] [Reference Citation Analysis (0)] |
| 128. | Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, Little MH. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 904] [Cited by in RCA: 1070] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 129. | Broutier L, Andersson-Rolf A, Hindley CJ, Boj SF, Clevers H, Koo BK, Huch M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat Protoc. 2016;11:1724-1743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 536] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 130. | Hoang P, Wang J, Conklin BR, Healy KE, Ma Z. Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat Protoc. 2018;13:723-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 117] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 131. | Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, Ratjen F, Ellis J, Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol. 2012;30:876-882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 311] [Cited by in RCA: 309] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 132. | Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, Wongvipat J, Kossai M, Ramazanoglu S, Barboza LP, Di W, Cao Z, Zhang QF, Sirota I, Ran L, MacDonald TY, Beltran H, Mosquera JM, Touijer KA, Scardino PT, Laudone VP, Curtis KR, Rathkopf DE, Morris MJ, Danila DC, Slovin SF, Solomon SB, Eastham JA, Chi P, Carver B, Rubin MA, Scher HI, Clevers H, Sawyers CL, Chen Y. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 1148] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 133. | Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, Jung DJ, Shin TH, Jeong GS, Kim DK, Choi EK, Jeong SY, Taylor AM, Jain S, Meyerson M, Jang SJ. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun. 2019;10:3991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 431] [Cited by in RCA: 507] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 134. | Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, Korving J, van Boxtel R, Duarte AA, Lelieveld D, van Hoeck A, Ernst RF, Blokzijl F, Nijman IJ, Hoogstraat M, van de Ven M, Egan DA, Zinzalla V, Moll J, Boj SF, Voest EE, Wessels L, van Diest PJ, Rottenberg S, Vries RGJ, Cuppen E, Clevers H. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 2018;172:373-386.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 784] [Cited by in RCA: 1226] [Article Influence: 153.3] [Reference Citation Analysis (0)] |
| 135. | Abugomaa A, Elbadawy M. Patient-derived organoid analysis of drug resistance in precision medicine: is there a value? In: Expert Review of Precision Medicine and Drug Developmen. London: Taylor & Francis; 2020, pp. 1-5. [RCA] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 136. | Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15:647-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 535] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 137. | Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017;7:462-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 710] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 138. | Porter RJ, Murray GI, McLean MH. Current concepts in tumour-derived organoids. Br J Cancer. 2020;123:1209-1218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 139. | Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 838] [Cited by in RCA: 998] [Article Influence: 110.9] [Reference Citation Analysis (0)] |
| 140. | Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc. 2020;15:3380-3409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 395] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 141. | Elbadawy M, Sato Y, Mori T, Goto Y, Hayashi K, Yamanaka M, Azakami D, Uchide T, Fukushima R, Yoshida T, Shibutani M, Kobayashi M, Shinohara Y, Abugomaa A, Kaneda M, Yamawaki H, Usui T, Sasaki K. Anti-tumor effect of trametinib in bladder cancer organoid and the underlying mechanism. Cancer Biol Ther. 2021;22:357-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 142. | Xu H, Lyu X, Yi M, Zhao W, Song Y, Wu K. Organoid technology and applications in cancer research. J Hematol Oncol. 2018;11:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 214] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 143. | Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. 2018;19:671-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 697] [Article Influence: 99.6] [Reference Citation Analysis (0)] |
| 144. | Liu L, Yu L, Li Z, Li W, Huang W. Patient-derived organoid (PDO) platforms to facilitate clinical decision making. J Transl Med. 2021;19:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 145. | Orkin RW, Gehron P, McGoodwin EB, Martin GR, Valentine T, Swarm R. A murine tumor producing a matrix of basement membrane. J Exp Med. 1977;145:204-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 440] [Cited by in RCA: 478] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 146. | Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15:378-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1020] [Cited by in RCA: 1130] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 147. | Lee HJ, Mun S, Pham DM, Kim P. Extracellular Matrix-Based Hydrogels to Tailoring Tumor Organoids. ACS Biomater Sci Eng. 2021;7:4128-4135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 148. | Benton G, Arnaoutova I, George J, Kleinman HK, Koblinski J. Matrigel: from discovery and ECM mimicry to assays and models for cancer research. Adv Drug Deliv Rev. 2014;79-80:3-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 320] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 149. | Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, Clevers H. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11:347-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 483] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 150. | Cheaito K, Bahmad HF, Hadadeh O, Msheik H, Monzer A, Ballout F, Dagher C, Telvizian T, Saheb N, Tawil A, El-Sabban M, El-Hajj A, Mukherji D, Al-Sayegh M, Abou-Kheir W. Establishment and characterization of prostate organoids from treatment-naïve patients with prostate cancer. Oncol Lett. 2022;23:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 151. | Monzer A, Wakimian K, Ballout F, Al Bitar S, Yehya A, Kanso M, Saheb N, Tawil A, Doughan S, Hussein M, Mukherji D, Faraj W, Gali-Muhtasib H, Abou-Kheir W. Novel therapeutic diiminoquinone exhibits anticancer effects on human colorectal cancer cells in two-dimensional and three-dimensional in vitro models. World J Gastroenterol. 2022;28:4787-4811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 152. | Al Bitar S, Ballout F, Monzer A, Kanso M, Saheb N, Mukherji D, Faraj W, Tawil A, Doughan S, Hussein M, Abou-Kheir W, Gali-Muhtasib H. Thymoquinone Radiosensitizes Human Colorectal Cancer Cells in 2D and 3D Culture Models. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 153. | Yu L, Li Z, Mei H, Li W, Chen D, Liu L, Zhang Z, Sun Y, Song F, Chen W, Huang W. Patient-derived organoids of bladder cancer recapitulate antigen expression profiles and serve as a personal evaluation model for CAR-T cells in vitro. Clin Transl Immunology. 2021;10:e1248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 154. | Chen P, Zhang X, Ding R, Yang L, Lyu X, Zeng J, Lei JH, Wang L, Bi J, Shao N, Shu D, Wu B, Wu J, Yang Z, Wang H, Wang B, Xiong K, Lu Y, Fu S, Choi TK, Lon NW, Zhang A, Tang D, Quan Y, Meng Y, Miao K, Sun H, Zhao M, Bao J, Zhang L, Xu X, Shi Y, Lin Y, Deng C. Patient-Derived Organoids Can Guide Personalized-Therapies for Patients with Advanced Breast Cancer. Adv Sci (Weinh). 2021;8:e2101176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 155. | Cheaito K, Bahmad HF, Jalloul H, Hadadeh O, Msheik H, El-Hajj A, Mukherji D, Al-Sayegh M, Abou-Kheir W. Epidermal Growth Factor Is Essential for the Maintenance of Novel Prostate Epithelial Cells Isolated From Patient-Derived Organoids. Front Cell Dev Biol. 2020;8:571677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 156. | Huang Y, Huang Z, Tang Z, Chen Y, Huang M, Liu H, Huang W, Ye Q, Jia B. Research Progress, Challenges, and Breakthroughs of Organoids as Disease Models. Front Cell Dev Biol. 2021;9:740574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 157. | Bose S, Clevers H, Shen X. Promises and Challenges of Organoid-Guided Precision Medicine. Med. 2021;2:1011-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 158. | Foo MA, You M, Chan SL, Sethi G, Bonney GK, Yong WP, Chow EK, Fong ELS, Wang L, Goh BC. Clinical translation of patient-derived tumour organoids- bottlenecks and strategies. Biomark Res. 2022;10:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 159. | Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44+ CD24(-) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98:756-765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 333] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 160. | Ooki A, VandenBussche CJ, Kates M, Hahn NM, Matoso A, McConkey DJ, Bivalacqua TJ, Hoque MO. CD24 regulates cancer stem cell (CSC)-like traits and a panel of CSC-related molecules serves as a non-invasive urinary biomarker for the detection of bladder cancer. Br J Cancer. 2018;119:961-970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 161. | Vázquez-Iglesias L, Barcia-Castro L, Rodríguez-Quiroga M, Páez de la Cadena M, Rodríguez-Berrocal J, Cordero OJ. Surface expression marker profile in colon cancer cell lines and sphere-derived cells suggests complexity in CD26(+) cancer stem cells subsets. Biol Open. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 162. | El-Benhawy SA, Morsi MI, Fahmy EI, Soula MA, Khalil FAZF, Arab AR. Role of Resveratrol as Radiosensitizer by Targeting Cancer Stem Cells in Radioresistant Prostate Cancer Cells (PC-3). Asian Pac J Cancer Prev. 2021;22:3823-3837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 163. | Zou J, Yu XF, Bao ZJ, Dong J. Proteome of human colon cancer stem cells: a comparative analysis. World J Gastroenterol. 2011;17:1276-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 164. | Gardelli C, Russo L, Cipolla L, Moro M, Andriani F, Rondinone O, Nicotra F, Sozzi G, Bertolini G, Roz L. Differential glycosylation of collagen modulates lung cancer stem cell subsets through β1 integrin-mediated interactions. Cancer Sci. 2021;112:217-230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 165. | Senbanjo LT, Chellaiah MA. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front Cell Dev Biol. 2017;5:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 577] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 166. | Tan W, Tang H, Jiang X, Ye F, Huang L, Shi D, Li L, Huang X, Xie X. Metformin mediates induction of miR-708 to inhibit self-renewal and chemoresistance of breast cancer stem cells through targeting CD47. J Cell Mol Med. 2019;23:5994-6004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 167. | Liu L, Zhang L, Yang L, Li H, Li R, Yu J, Wei F, Yan C, Sun Q, Zhao H, Yang F, Jin H, Wang J, Wang SE, Ren X. Anti-CD47 Antibody As a Targeted Therapeutic Agent for Human Lung Cancer and Cancer Stem Cells. Front Immunol. 2017;8:404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 168. | Hoogland AM, Verhoef EI, Roobol MJ, Schröder FH, Wildhagen MF, van der Kwast TH, Jenster G, van Leenders GJ. Validation of stem cell markers in clinical prostate cancer: α6-integrin is predictive for non-aggressive disease. Prostate. 2014;74:488-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 169. | Botchkina IL, Rowehl RA, Rivadeneira DE, Karpeh MS Jr, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y, Botchkina GI. Phenotypic subpopulations of metastatic colon cancer stem cells: genomic analysis. Cancer Genomics Proteomics. 2009;6:19-29. [PubMed] |
| 170. | Martin TA, Jiang WG. Evaluation of the expression of stem cell markers in human breast cancer reveals a correlation with clinical progression and metastatic disease in ductal carcinoma. Oncol Rep. 2014;31:262-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 171. | Wang P, Gao Q, Suo Z, Munthe E, Solberg S, Ma L, Wang M, Westerdaal NA, Kvalheim G, Gaudernack G. Identification and characterization of cells with cancer stem cell properties in human primary lung cancer cell lines. PLoS One. 2013;8:e57020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 172. | Marcinkiewicz K, Scotland KB, Boorjian SA, Nilsson EM, Persson JL, Abrahamsson PA, Allegrucci C, Hughes IA, Gudas LJ, Mongan NP. The androgen receptor and stem cell pathways in prostate and bladder cancers (review). Int J Oncol. 2012;40:5-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 173. | Zhang Y, Xu W, Guo H, Zhang Y, He Y, Lee SH, Song X, Li X, Guo Y, Zhao Y, Ding C, Ning F, Ma Y, Lei QY, Hu X, Li S, Guo W. NOTCH1 Signaling Regulates Self-Renewal and Platinum Chemoresistance of Cancer Stem-like Cells in Human Non-Small Cell Lung Cancer. Cancer Res. 2017;77:3082-3091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 174. | Sui X, Cai J, Li H, He C, Zhou C, Dong Y, Chen L, Zhang B, Wang Y, Zhang Y, Qiu Y, Zhao Y, Huang Y, Shen Y, Wu H, Xiao J, Mason C, Zhu Q, Han S. p53-dependent CD51 expression contributes to characteristics of cancer stem cells in prostate cancer. Cell Death Dis. 2018;9:523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 175. | Gemei M, Mirabelli P, Di Noto R, Corbo C, Iaccarino A, Zamboli A, Troncone G, Galizia G, Lieto E, Del Vecchio L, Salvatore F. CD66c is a novel marker for colorectal cancer stem cell isolation, and its silencing halts tumor growth in vivo. Cancer. 2013;119:729-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 176. | Dzobo K, Ganz C, Thomford NE, Senthebane DA. Cancer Stem Cell Markers in Relation to Patient Survival Outcomes: Lessons for Integrative Diagnostics and Next-Generation Anticancer Drug Development. OMICS. 2021;25:81-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 177. | Abugomaa A, Elbadawy M, Yamawaki H, Usui T, Sasaki K. Emerging Roles of Cancer Stem Cells in Bladder Cancer Progression, Tumorigenesis, and Resistance to Chemotherapy: A Potential Therapeutic Target for Bladder Cancer. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 178. | Foster BM, Zaidi D, Young TR, Mobley ME, Kerr BA. CD117/c-kit in Cancer Stem Cell-Mediated Progression and Therapeutic Resistance. Biomedicines. 2018;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 179. | Ying J, Tsujii M, Kondo J, Hayashi Y, Kato M, Akasaka T, Inoue T, Shiraishi E, Hiyama S, Tsujii Y, Maekawa A, Kawai S, Fujinaga T, Araki M, Shinzaki S, Watabe K, Nishida T, Iijima H, Takehara T. The effectiveness of an anti-human IL-6 receptor monoclonal antibody combined with chemotherapy to target colon cancer stem-like cells. Int J Oncol. 2015;46:1551-1559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 180. | Sorrentino C, Ciummo SL, D'Antonio L, Fieni C, Lanuti P, Turdo A, Todaro M, Di Carlo E. Interleukin-30 feeds breast cancer stem cells via CXCL10 and IL23 autocrine loops and shapes immune contexture and host outcome. J Immunother Cancer. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 181. | Zhang S, Yang X, Wang L, Zhang C. Interplay between inflammatory tumor microenvironment and cancer stem cells. Oncol Lett. 2018;16:679-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 182. | Mărgaritescu C, Pirici D, Cherciu I, Bărbălan A, Cârtână T, Săftoiu A. CD133/CD166/Ki-67 triple immunofluorescence assessment for putative cancer stem cells in colon carcinoma. J Gastrointestin Liver Dis. 2014;23:161-170. [PubMed] |
| 183. | Gao X, Dong QZ. Advance in metabolism and target therapy in breast cancer stem cells. World J Stem Cells. 2020;12:1295-1306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 184. | Bryan RT. Cell adhesion and urothelial bladder cancer: the role of cadherin switching and related phenomena. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 185. | Wang R, Yang L, Li S, Ye D, Liu Q, Zhao Z, Cai Q, Tan J, Li X. Quercetin Inhibits Breast Cancer Stem Cells via Downregulation of Aldehyde Dehydrogenase 1A1 (ALDH1A1), Chemokine Receptor Type 4 (CXCR4), Mucin 1 (MUC1), and Epithelial Cell Adhesion Molecule (EpCAM). Med Sci Monit. 2018;24:412-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 186. | Kirkland SC. Type I collagen inhibits differentiation and promotes a stem cell-like phenotype in human colorectal carcinoma cells. Br J Cancer. 2009;101:320-326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 187. | Hirashima K, Yue F, Kobayashi M, Uchida Y, Nakamura S, Tomotsune D, Matsumoto K, Takizawa-Shirasawa S, Yokoyama T, Kanno H, Sasaki K. Cell biological profiling of reprogrammed cancer stem cell-like colon cancer cells maintained in culture. Cell Tissue Res. 2019;375:697-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 188. | Serna N, Álamo P, Ramesh P, Vinokurova D, Sánchez-García L, Unzueta U, Gallardo A, Céspedes MV, Vázquez E, Villaverde A, Mangues R, Medema JP. Nanostructured toxins for the selective destruction of drug-resistant human CXCR4(+) colorectal cancer stem cells. J Control Release. 2020;320:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 189. | Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle T, Etzkorn M, Seo HC, Nagiec M, Luna RE, Reinherz EL, Blenis J, Gygi SP, Wagner G. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc Natl Acad Sci U S A. 2014;111:E2182-E2190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 99] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 190. | Xie ZY, Lv K, Xiong Y, Guo WH. ABCG2-meditated multidrug resistance and tumor-initiating capacity of side population cells from colon cancer. Oncol Res Treat. 2014;37:666-668, 670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 191. | Das S, Mukherjee P, Chatterjee R, Jamal Z, Chatterji U. Enhancing Chemosensitivity of Breast Cancer Stem Cells by Downregulating SOX2 and ABCG2 Using Wedelolactone-encapsulated Nanoparticles. Mol Cancer Ther. 2019;18:680-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 192. | Green R, Howell M, Khalil R, Nair R, Yan J, Foran E, Katiri S, Banerjee J, Singh M, Bharadwaj S, Mohapatra SS, Mohapatra S. Actinomycin D and Telmisartan Combination Targets Lung Cancer Stem Cells Through the Wnt/Beta Catenin Pathway. Sci Rep. 2019;9:18177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 193. | Lin CJ, Yun EJ, Lo UG, Tai YL, Deng S, Hernandez E, Dang A, Chen YA, Saha D, Mu P, Lin H, Li TK, Shen TL, Lai CH, Hsieh JT. The paracrine induction of prostate cancer progression by caveolin-1. Cell Death Dis. 2019;10:834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 194. | Wong N, Major P, Kapoor A, Wei F, Yan J, Aziz T, Zheng M, Jayasekera D, Cutz JC, Chow MJ, Tang D. Amplification of MUC1 in prostate cancer metastasis and CRPC development. Oncotarget. 2016;7:83115-83133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 195. | Guo M, Luo B, Pan M, Li M, Zhao F, Dou J. MUC1 plays an essential role in tumor immunity of colorectal cancer stem cell vaccine. Int Immunopharmacol. 2020;85:106631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 196. | Bae KM, Parker NN, Dai Y, Vieweg J, Siemann DW. E-cadherin plasticity in prostate cancer stem cell invasion. Am J Cancer Res. 2011;1:71-84. [PubMed] |
| 197. | Tamura S, Isobe T, Ariyama H, Nakano M, Kikushige Y, Takaishi S, Kusaba H, Takenaka K, Ueki T, Nakamura M, Akashi K, Baba E. Ecadherin regulates proliferation of colorectal cancer stem cells through NANOG. Oncol Rep. 2018;40:693-703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 198. | Yun EJ, Baek ST, Xie D, Tseng SF, Dobin T, Hernandez E, Zhou J, Zhang L, Yang J, Sun H, Xiao G, He D, Kittler R, Hsieh JT. DAB2IP regulates cancer stem cell phenotypes through modulating stem cell factor receptor and ZEB1. Oncogene. 2015;34:2741-2752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 199. | Yuan W, Ji J, Shu Y, Chen J, Liu S, Wu L, Zhou Z, Liu Z, Tang Q, Zhang X, Shu X. Downregulation of DAPK1 promotes the stemness of cancer stem cells and EMT process by activating ZEB1 in colorectal cancer. J Mol Med (Berl). 2019;97:89-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 200. | Preca BT, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, Wellner U, Hopt UT, Brummer T, Brabletz S, Brabletz T, Stemmler MP. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int J Cancer. 2015;137:2566-2577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 201. | Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, Calhoun-Davis T, Li H, Palapattu GS, Pang S, Lin K, Huang J, Ivanov I, Li W, Suraneni MV, Tang DG. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 256] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 202. | Garraway IP, Sun W, Tran CP, Perner S, Zhang B, Goldstein AS, Hahm SA, Haider M, Head CS, Reiter RE, Rubin MA, Witte ON. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate. 2010;70:491-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 117] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 203. | Chan KS, Volkmer JP, Weissman I. Cancer stem cells in bladder cancer: a revisited and evolving concept. Curr Opin Urol. 2010;20:393-397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 204. | Ricardo S, Vieira AF, Gerhard R, Leitão D, Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F, Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011;64:937-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 448] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 205. | Chen X, Rycaj K, Liu X, Tang DG. New insights into prostate cancer stem cells. Cell Cycle. 2013;12:579-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 206. | Shi R, Liu L, Wang F, He Y, Niu Y, Wang C, Zhang X, Zhang H, Chen M, Wang Y. Downregulation of cytokeratin 18 induces cellular partial EMT and stemness through increasing EpCAM expression in breast cancer. Cell Signal. 2020;76:109810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 207. | Chen Y, Lan T. Molecular Origin, Expression Regulation, and Biological Function of Androgen Receptor Splicing Variant 7 in Prostate Cancer. Urol Int. 2021;105:337-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |