Published online May 26, 2023. doi: 10.4252/wjsc.v15.i5.302
Peer-review started: December 10, 2022
First decision: January 23, 2023
Revised: February 3, 2023
Accepted: March 8, 2023
Article in press: March 8, 2023
Published online: May 26, 2023
Processing time: 166 Days and 22.8 Hours
Literature data on glioblastoma ongoingly underline the link between metabolism and cancer stemness, the latter is one responsible for potentiating the resistance to treatment, inter alia due to increased invasiveness. In recent years, glioblastoma stemness research has bashfully introduced a key aspect of cytoskeletal rearrangements, whereas the impact of the cytoskeleton on invasiveness is well known. Although non-stem glioblastoma cells are less invasive than glioblastoma stem cells (GSCs), these cells also acquire stemness with greater ease if characterized as invasive cells and not tumor core cells. This suggests that glioblastoma stemness should be further investigated for any phenomena related to the cytoskeleton and metabolism, as they may provide new invasion-related insights. Previously, we proved that interplay between metabolism and cytoskeleton existed in glioblastoma. Despite searching for cytoskeleton-related processes in which the investigated genes might have been involved, not only did we stumble across the relation to metabolism but also reported genes that were found to be implicated in stemness. Thus, dedicated research on these genes in GSCs seems justifiable and might reveal novel directions and/or biomarkers that could be utilized in the future. Herein, we review the previously identified cyto
Core Tip: Glioblastoma stemness intensifies the resistance to treatment via increased invasiveness. Among the processes crucial for glioblastoma stem cells, metabolism is known to influence invasion. However, the cytoskeleton is currently negligent in glioblastoma stemness research, while it also regulates invasion. Herein, we review the link between stemness and cytoskeleton/metabolism-related genes that we previously identified in glioblastoma. These genes influence stemness via numerous biological processes; for some genes, clinical trials are currently ongoing. Others were connected to glioblastoma stemness for the first time. Future glioblastoma-related research should delve into the cytoskeleton since the concept is already encouraging.
- Citation: Kałuzińska-Kołat Ż, Kołat D, Kośla K, Płuciennik E, Bednarek AK. Delineating the glioblastoma stemness by genes involved in cytoskeletal rearrangements and metabolic alterations. World J Stem Cells 2023; 15(5): 302-322
- URL: https://www.wjgnet.com/1948-0210/full/v15/i5/302.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i5.302
Glioblastoma (GBM) has remained an incurable condition with increasing incidence in many countries[1,2]. Although GBM is less prevalent than breast, colon, or lung cancer, it outperforms other tumors by affecting patients in the prime of their lives and causing them to lose many years of life[3]. The initial intervention in newly diagnosed GBM includes a surgical approach, with post-surgery temozolomide (TMZ) and radiation therapy[4]. Adding tumor-treating electric fields (TTFields) to maintenance TMZ chemotherapy was found to prolong progression-free and overall survival, but is currently limited due to the lack of methods to predict or quantify the efficacy of TTFields (the imaging features associated with treatment response are unclear and there are no predictive neuroimaging markers). Moreover, the treatment device is required to be worn for a predetermined period (typically approximately 75% of the time) or until there is a clinical progression of the disease, which introduces a delay in getting used to the device and makes patients anxious with regard to the intended therapy effect[5]. Strong motivation to predict TTField efficacy in a patient-specific manner was provided[6]. Nevertheless, glioblastoma recurrence is practically inevitable which, combined with a grim prognosis and ineffective treatment, underlines the importance of further research into this deadliest tumor[3,7].
One of the GBM traits that implicate the lack of effective treatment is the heterogeneity that can be explained by both clonal evolution and the presence of stem cells[8]. Stemness refers to the molecular events that underlie the essential characteristics of self-renewal and differentiation into daughter cells[9]. On the cellular level, some processes were indicated as crucial for GBM stemness, namely epigenomic regulation, posttranscriptional regulation, and metabolism[10]. Glioblastoma stemness research in recent years has also bashfully introduced a key aspect of cytoskeletal rearrangements [11,12] while it has been long time since this machinery is well-known for controlling two processes that influence cancer malignant behavior, i.e., cellular division and invasion[13]. The stemness itself is also responsible for potentiating the resistance to treatment[14,15], inter alia due to increased invasiveness[16]. In addition, more recent studies have identified the role of metabolism in GBM invasion[17]. Although non-stem glioblastoma cells are less invasive than GBM stem cells (confirmed by sevenfold reduced cell migration through the Matrigel, or 3.8-times and 6.8-times lower expression of matrix metalloproteinase-14 and -16)[18], the same cells also acquire stemness with greater ease if they are characterized as invasive cells and not tumor core cells[19,20].
The above-mentioned data imply that GBM stemness should be further explored for any phenomena related to the cytoskeleton and metabolism, as they may provide the missing puzzle from the point-of-view of invasion. Moreover, the cytoskeleton and metabolism are related; for instance, the cytoskeleton is involved in carbohydrate metabolism[21] and at the same time the actin and tubulin require energy from nucleotide hydrolysis to maintain structural dynamics[22]. Cytoskeletal rearrangements and metabolic alterations are important not only for GBM cells but also for neuronal and glial progenitors. For example, cytoskeleton dynamics underlie the cellular asymmetry while metabolic reprogramming ensures a transition in energy production from glycolytic to oxidative[23,24]. Nevertheless, it is possible to discriminate normal glial cells from glioblastoma; the cancerous cells present decreased cortical but increased intracellular stiffness, and preferentially metabolized glucose into lactate despite the abundance of oxygen[17,25]. Stiffness and metabolic adaptations can also influence stem cell differentiation[26,27]. Moreover, the cellular cross-talk that utilizes cytoskeleton or metabolites affects physical dynamics and signaling of various cell types including astrocytes, neurons, and oligodendrocytes[28,29]. In cancers, such cross-talk renders abnormal protrusions or extensions termed as tumor microtubes that contribute to glioma resistance[30]. These structures are rich in cytoskeletal proteins, such as actin and tubulin, and are known to modify energetic metabolism of the receiving cells via transport of mitochondria[31].
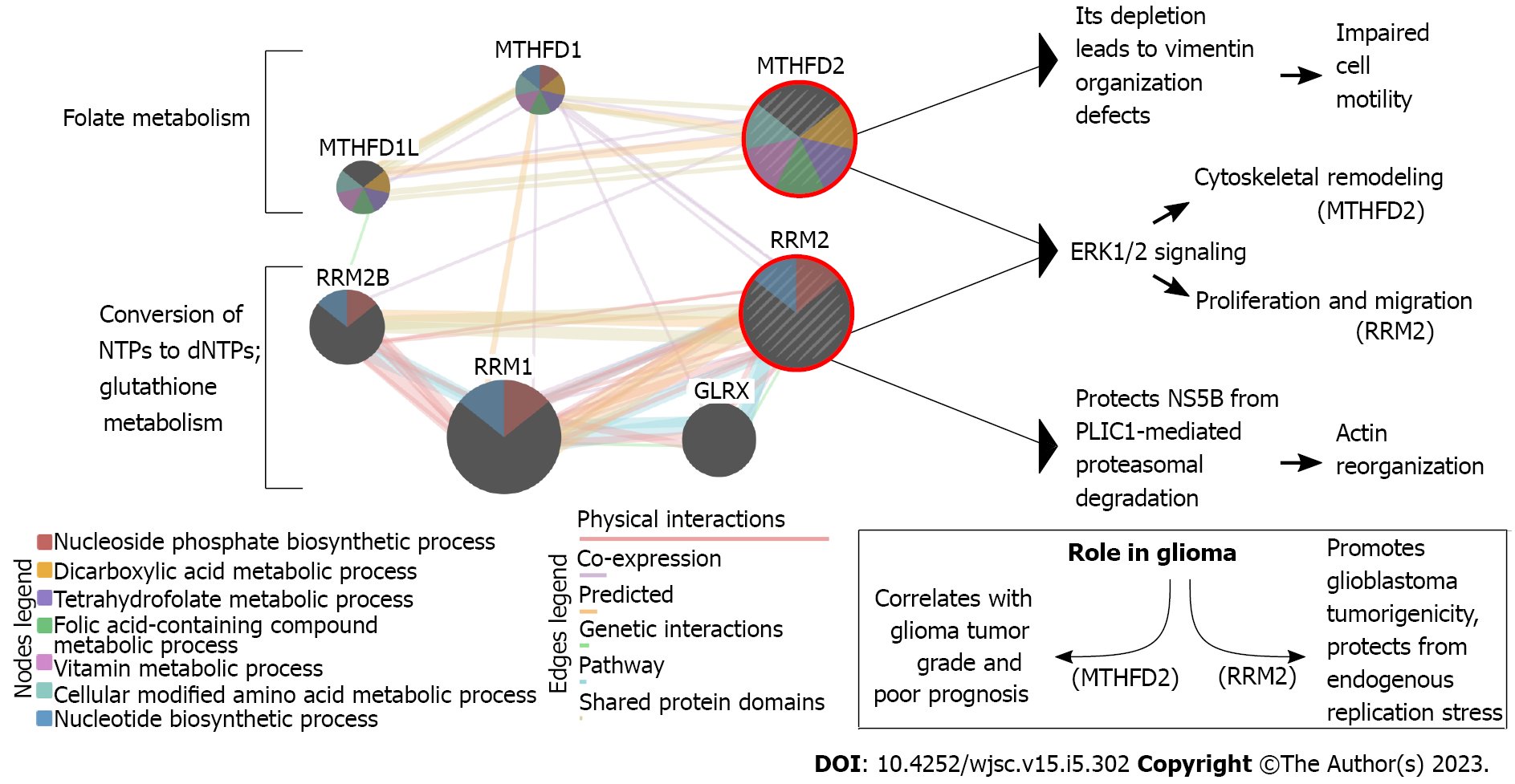
Our previous research has proved that interplay between metabolic alterations and cytoskeletal rearrangements exists in GBM[32]. Of genes described below in the present review (some previously identified genes were not included if their implication in stemness was not found in the literature) (Supplementary Table 1)[33-37], the example of a relationship between metabolism and cytoskeleton can be visualized (Figure 1) based on the literature on methylenetetrahydrofolate dehydrogenase 2 (MTHFD2)[38-41] and ribonucleotide reductase subunit M2 (RRM2)[42-45]. In our previous research, despite searching for cytoskeleton-related processes in which the investigated genes might have been involved, not only did we stumble across the relation to metabolism, but we also reported some genes which were found to be implicated in glioblastoma stemness. Thus, the dedicated work on these genes in the GBM stem cells (GSCs) seems justifiable and might reveal novel therapeutic directions and/or biomarkers that could be utilized in the future. Herein, we review the previously identified cytoskeleton/metabolism-related genes through the prism of GBM stemness. Literature screening allowed the decision to split these genes based on whether their role in stemness is known from GBM or another tumor, the latter suggesting an urgent need to experimentally verify the observations in the glioblastoma context.
Based on the literature abundance, the best-known from its implication in glioblastoma stemness is bone morphogenetic protein 4 (BMP4). The bone morphogenetic proteins are growth factors from the TGF-β superfamily that undergo expression during embryogenesis and control development. Initially denoted as crucial for osteogenesis, they are now described as regulators of gastrulation, neurulation, mesoderm patterning, proliferation, and differentiation in many tissues[46]. About 15 years ago, it was found that the signaling via BMPs and their cognate receptors (BMPRs) influenced the activity of normal brain stem cells but could also inhibit the cancer-initiating GBM stem-like cells[47]. Later the same year, these authors confirmed that in vivo delivery of BMP4 blocked the tumor growth and associated mortality, which occurred in all mice following intracerebral grafting of human glioblastoma[48]. This protein was suggested as a non-cytotoxic therapeutic agent that can be utilized in combination with stem cell-based therapy[49]; this complements its usage as an agent used to differentiate GSCs into normal glial cells[50]. BMP4 has been found promising to the extent that it entailed the development of novel therapies. For example, one that utilizes the oncolytic vaccinia virus was developed to alleviate glioblastoma and prevent its recurrence[51]. Later on, the cell-based treatment option of BMP4-secreting human adipose-derived mesenchymal stem cells was found to reduce proliferation and migration both in vitro and in vivo, as well as prolong survival in a murine model[52]. Still, Videla Richardson et al[53] admitted that little is known about this morphogen regarding triggered cellular events, which prompted the authors to establish several GSC-enriched cell lines growing as adherent monolayers and not floating neurospheres. Distinct lineage preferences were noticed depending on the expression pattern of BMP signaling-astrocyte fate or neuronal commitment was noticed and, under certain conditions, even a smooth muscle-like phenotype[53]. Providing new findings to the available data, BMP4-overexpressing neural stem cells were found to promote GSCs apoptosis via Smad1/5/8 signaling[54]. Moreover, recent studies indicate a formerly underestimated link between BMP4 and metabolism or mechanotransduction which affects oxygen consumption or matrix stiffness[55]. The latter is known to be associated with cytoskeletal remodeling[56,57]. With regard to the cytoskeleton, BMP4 was found to re-organize actin dynamics via activation of Rac1, Rho, and Cdc42[58]. The impact of BMP4 in inducing asymmetric cell division was also noted, limiting the GSCs expansion[59]. The newest literature data on BMP4 consider it on a broader scale, either evaluating other GBM aspects and referring to BMP4, or investigating upstream/downstream molecules. Ciechomska et al[60] explored EGFR alterations in glioblastoma since GSCs with various EGFR levels respond differently to therapy; the authors found that EGFR/FOXO3a/BIM signaling pathway determined chemosensitivity of BMP4-differentiated GSCs to TMZ. On the other hand, Wu et al[61] identified BIRC3 as an inducer of glioblastoma stemness via downstream BMP4 inactivation. At last, the most recent paper by Verploegh et al[62] summarized the cellular viability variance in response to BMP4 and proposed early-response markers for sensitivity to BMP4. Three cultures with the highest sensitivity for BMP4 revealed a new cell subpopulation that presented a reduced cell proliferation but an elevation of apoptosis. These changes in composition correlated with treatment efficacy; the latter was predicted using OLIG1/2 expression. Furthermore, upregulated RPL27A and RPS27 were considered early-response markers. Interestingly, RPS27 is one of the genes identified in our previous study that prompted us to investigate the aspects presented in this review. This gene will be described below in a separate subsection.
Glutamate ionotropic receptor NMDA type subunit 2B (GRIN2B) encodes one subtype of glutamate-binding GluN2 subunit, which is a part of the N-methyl-D-aspartate receptor (NMDAR). Ionotropic glutamate receptors from this family mediate Ca2+, i.e., the permeable component of excitatory synaptic transmission in the central nervous system (CNS)[63]. NMDARs assemble from four subunits: two GluN1 and two GluN2. The former subunits are widely expressed in the nervous system, while four subtypes of GluN2 subunits (from “A” to “D”) are characterized by various expression patterns[64]. GRIN2B encodes the GluN2B subunit, which is abundantly expressed in the prenatal period, then declines in most brain parts[65]. The presence of GluN2B in such an early stage implies that it contributes to brain development, circuit formation, synaptic plasticity, as well as migration and differentiation[66]. Glutamate-dependent synaptic transmission is frequently dysfunctional in gliomas[67], and regarding this specific subunit, an enrichment of expression was noticed in GSCs[68]. In our previous research, with the use of literature data, we related this gene with the cytoskeleton since GluN2B interacts with cytoskeletal protein α-actinin-2 via the carboxyl-terminal domain[63]. It might be of importance as α-actinin-2 is closely associated with multimerins which are possible markers and therapeutic targets in low-grade glioma[69]. Moreover, one of the multimerins encoded by the MMRN1 gene was found to be correlated to stemness and chemoresistance, although these observations were based on the leukemia model[70]. Nevertheless, GRIN2B is confirmed to influence stemness not only in glioblastoma but also in lung cancer. She et al[71] identified GRIN2B expression to be higher in primary tumors than in normal tissues, and at the same time higher in metastatic lesions than in primary tumors which contributed to poorer prognosis. Moreover, the same authors observed inhibition of tumorsphere formation during GRIN2B silencing.
The homeotic genes, in vertebrates denoted as homeobox, are highly conserved and regulate the proper development of various body segments during ontogeny[72]. Homeobox protein A10 (HOXA10) is implicated in the embryogenesis of the uterine epithelium, stroma, and muscle[73]. In response to hormones, it undergoes periodical expression in the mature endometrium, controlling receptivity during the implantation window[74]. Concerning GBM stemness, the functionality of HOXA10 was presented as a direct result of the activation of protein from the Trithorax family, which serves as a histone methyltransferase, i.e., MLL. Afterward, HOXA10 activated other HOXA genes, such as HOXA7 and HOXC10[75]. In another study, HOXA10 was marked as one of the strongest candidates (alongside the HOX -A9, -C4, and -D9 genes), having value as a therapeutic target and biomarker for both GBM and GSCs[76]. Our previous research echoed the data that HOXA10 facilitated cytoskeleton remodeling (via CK15)[77], promoted tumorigenesis in glioma[78], and regulated homologous recombinant DNA repair and subsequently TMZ resistance in GBM[79]. Since stemness also contributes to treatment resistance[14], the last two events complement each other mutually. Another homeotic gene that we identified in our previous study was HOXA1, a homeobox that is abundantly expressed in the mesoderm and neuroectoderm at the level of the brainstem precursor[80]. Upregulation of HOXA1 was noted in GBM, which inversely correlated with the survival of patients[81]. This homeotic member was also implicated in regulating the cytoskeleton via E-cadherin. Namely, CDH1-dependent signaling was found to increase HOXA1 expression through Rac1, i.e., the same pathway that regulates actin cytoskeleton at cadherin adhesive contacts[79]. With regard to GBM stemness, Schmid et al[82] observed upregulated HoxA locus (encompassing, e.g., HOXA1) after they dedifferentiated murine astrocytes into GSCs via Rb knockout, Kras activation, and Pten deletion. These cells were sufficient to form GBMs in their transplant mouse model. Although the insights did not provide further mechanistic details, the regulation loop of HOXA1 and HOXA transcript antisense RNA (HOTAIRM1) was found to be involved in stemness maintenance[81,83]. This was presented in colorectal carcinoma and uveal melanoma. Still, taking into account the study by Schmid et al[82], the profound investigation of HOXA1 in GSCs in this aspect should be considered.
Matrix metalloproteinases are constituents of extracellular matrix (ECM) belonging to the zinc-containing endopeptidases family that encompasses 23 members[84]. Functionally, these calcium-dependent molecules are responsible for the degradation and remodeling of other proteins that constitute ECM. Moreover, their roles in various biological and physiological processes dependent on hormones, growth factors, and cytokines were described[85]. It is known that different ECM components modulate cancer stem cells’ properties; regarding glioblastoma, the confirmed ones were type I collagen, laminin α2, fibronectin, periostin, decorin, and lumican[86]. Matrix metalloproteinase 13 (MMP13) is a collagenase almost universally upregulated in the pan-cancer view[87]; in GBM, its overexpression increases migration and invasion[88], as well as confers poor prognosis[89]. The relationships between MMP13 and the cytoskeleton[33] or metabolism[90] are known. In terms of stemness, Inoue et al[91] suggested that highly invasive potential GSCs depended on MMP13 enzymatic activity; the authors also proposed MMP13 as a potential therapeutic target.
The folate cycle is responsible for appropriate cellular metabolism by regulating ATP production, methylation reactions for DNA/protein synthesis, or developing immunomodulatory molecules that orchestrate signaling and cytotoxicity[92]. The differences between MTHFD1 and MTHFD2, two enzymes implicated in the folate pathway, include the use of different co-enzyme (NADP vs NAD), functionality (MTHFD1 has three distinct enzymatic activities while MTHFD2 is bifunctional), and location (cytoplasm vs mitochondria). Compared to MTHFD1, which generates NADPH and formate for purine biosynthesis, MTHFD2 is overexpressed in rapidly proliferating malignant tumors. It is considered the “main switch” that enables mitochondria to produce additional growth-facilitating one-carbon units and generates NADH necessary for protection from reactive oxygen species[93]. MTHFD2 is also an excellent example to present the link between metabolism and cytoskeleton. Lehtinen et al[39] have found that MTHFD2 depletion leads to vimentin organization defects, and identified this gene as a regulator of cell migration and invasion. Regarding glioma, MTHFD2 was found to be associated with tumor grade and prognosis[38]. Inhibition of this enzyme in GSCs induced apoptosis and affected not only central carbon metabolic pathways (e.g., glycolysis, pentose phosphate pathway, and tricarboxylic acid cycle) but also unfolded protein response, highlighting a novel connection between one-carbon metabolism and reaction to cellular stress[94]. Nishimura et al[95] suggested that the purine synthesis pathway, as well as folate-mediated one-carbon metabolism, seem to be crucial for the maintenance of tumor-initiating cells. The same authors also concluded that EGF-induced expression of MTHFD2 may be mediated by Myc, with the latter regulating the expression of metabolic enzymes for the maintenance of brain tumor-initiating cells.
Alternative splicing maintains post-transcriptional gene regulation, which enables a single gene to be transcribed into various RNAs, diversifying the proteome. Abnormal splicing function can lead to tumor-related processes, e.g., proliferation, angiogenesis, and metastasis[96]. Spliceosome, a dynamic machinery responsible for splicing, is made of small nuclear ribonucleoproteins (snRNPs; five molecules are known: U1, U2, U4, U5, and U6) and numerous non-snRNP proteins[97,98]. U2 snRNP comprises U2 snRNA, SF3a complex, and SF3b complex, which are responsible for recognizing branchpoint sequences during initial spliceosome assembly stages[99]. Splicing factors comprising the SF3b complex include plant homeodomain (PHD) finger-like domain-containing protein 5A (PHF5A), which facilitates interactions between the U2 snRNP and RNA helicases[100] but can also bind chromatin via its PHD that is composed of a small zinc finger structural fold[101,102]. The knockdown of PHF5A results in reduced GBM viability and cell cycle arrest[103]. Trappe et al[104] revealed that systematic deletion of its yeast homolog is lethal, showing that PHF5A is crucial for cell viability. The flagship paper on PHF5A in brain tumor[105] indicates that the gene is required to expand GSCs and that in these tumor-initiating cells, but not untransformed neural stem cells, PHF5A contribute to the identification of exons having unusual C-rich 3’ splice sites in thousands of essential genes. The same authors inhibited PHF5A, which reduced GSCs-driven tumor formation in vivo and inhibited the growth of established GBM patient-derived xenograft tumors.
One of the most dynamic and largest molecular motors (driven by a complex thermal ratchet translocation mechanism) are ribosomes[106]. Metallopanstimulin-1, also known as ribosomal protein S27 (RPS27), is a constituent of the human 40S ribosome that is mainly found in the cytoplasm while it can also relocate to the nucleus[107] or even extracellular space[108]. Regarding the nuclear location, it is able to interact with DNA via its C4-type zinc finger[109]. In glioblastoma, RPS27 was found to be correlated with age in IDH-mutated glioma patients and with Ki67 in GBM patients. Interestingly, it is detected in astrocytic tumors but not in normal astrocytes unless the tissue was inflamed[109]. This allowed the same authors to emphasize that in comparison to inflammatory tissue (in which only a small number of macrophages were positive for RPS27), almost all macrophages in tumor tissue were distinctly enriched in RPS27 expression. As for GSCs, the ribosomes and related proteins were generally found to reprogram glioma cells to induce plasticity and stemness[110]. Among these molecules, RPS27 was considered oncogenic with higher expression at the GSC-dominant area[111]. Inquisitive findings revealed that RPS27 is also detected in the microvascular proliferation area and pseudopalisading cells around necrosis[110]. It is worth underlining that aberrant vessels are crucial for the formation of pseudopalisading necrotic regions that provide shelter for residing cancer stem cells from anti-tumor agents, which enable these cells to expand and promote proliferation and growth[112]. As mentioned above, upregulated RPL27A and RPS27 were considered to be early-response markers related to the presence of BMP4. This suggests a link that should be further investigated since the signaling of ribosome translation was found to be overexpressed during the response to stress in glioblastoma.
A balanced supply of deoxyribonucleotide triphosphates (dNTPs) is a prerequisite of DNA synthesis. Still, de novo synthesis of dNTP is also possible via the reaction catalyzed by the ribonucleotide reductase (RR) that reduces the C2’-OH bond of the four ribonucleotides triphosphates to form corresponding dNTPs[113]. RRM2 encodes the β subunit of RR; each RRM2 monomer contains the tyrosyl radical and non-heme iron[114]. Since a sufficient supply of dNTPs drives an uncontrolled DNA replication in cancer[115], it is not surprising that RRM2 was frequently subjected to molecular therapy[116,117]. Currently, several RRM2 inhibitors have been developed, e.g., radical scavengers, iron chelators, subunit polymerization inhibitors, or expression silencers[118-120]; this is to inhibit proliferation, division, but also invasion[32]. In glioblastoma, RRM2 is responsible for the advancement of GBM tumorigenicity and protection from endogenous replication stress via the BRCA1-RRM2 axis[45]. For glioma in general, regulation of proliferation and migration via ERK1/2 and AKT signaling was noted[44]. Available literature also links the RRM2 to the cytoskeleton via hPLIC1; the latter decreases during RRM2 downregulation, which entails actin cytoskeleton re-organization[42]. Perrault et al[121] have suggested that RRM2 can be a chemoresistance driver that dictates how GBM cells respond to TMZ. The same authors further verified that RRM2-overexpressing cells had enhanced DNA repair efficiency. Moreover, the use of a selective FDA-approved RRM2 inhibitor, 3-AP Triapine, enabled Perrault et al[121] to observe that in comparison to both TMZ and control, glioblastoma treated with the 3AP + TMZ formed fewer neurospheres that were also significantly smaller. Another group found that RRM2 expression dramatically declined after 12 d of dasatinib treatment compared to naïve GSCs of the GSC8 cell line[122].
In order to re-establish homeostasis, both adaptable and primordial mechanisms exist; the latter comprises the acute-phase response (APR) that is a set of changes that occur after inflammation, infection, or trauma[123]. During APR, the changes include the altered levels of serum proteins, with the most notable being C-reactive protein and serum amyloid A (SAA)[124]. Being an apolipoprotein, SAA is related to plasma high-density lipoprotein and is implicated in the cholesterol transport to the liver for excretion as bile[125]. Its other functions include regulation of amyloidogenesis, tumor pathogenesis, anti-bacterial events, and inflammatory response[126]. The role of SAA in tumor progression was suggested owing to its cytokine-like properties that influence the course of inflammation[127]. SAA2 is one of the paralogs of the family and was investigated as a lung cancer biomarker a few years ago[128]. The description of its role in glioblastoma is limited, yet it is already known that SAA2 increases GBM proliferation and invasion[129]. Knebel et al[130] have confirmed that SAA production occurs not only in the liver but also in tumor cells; the authors emphasized that exploring the SAA influence on the cytoskeleton and invasiveness using more complex assays is needed. In terms of GBM stemness, Adamski et al[131] recently have compiled the literature data and stated that SAA2 is implicated in a drug-promoted cellular dormancy, with the latter being closely connected to stem cell characteristics. The group also indicated the ability of SAA2 to sustain inflammatory conditions in the brain, which consequently supports TMZ resistance and induces the expression of stemness markers in glioblastoma.
The 5-methylcytosine (5mC) and its derivatives have altered patterns in a range of tumors. 5mC can be recognized and oxidized to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine by Ten-Eleven translocation (TET) enzymes[132,133]. One of the transcription factors that directly interacts with TET proteins is Wilms’ tumor protein 1 (WT1): A master regulator essential for urogenital, epicardium, and kidney development that can act as a tumor suppressor or oncoprotein in multiple tumors[134,135]. Initially cloned as a suppressor of Wilms’ tumor, WT1 is now considered to be an oncoprotein in hematologic malignancies and a variety of solid tumors, as well as the protein with the highest potential for cancer immunotherapy[136-138]. According to the phase I/II clinical trial, WT1 peptide-based vaccine for glioblastoma patients was considered safe and induced cellular and humoral immune response[139]. This is important due to the fact that WT1 is involved in GBM tumorigenicity via increasing proliferation and decreasing apoptosis[140]. As for the impact on the cytoskeleton, this protein was found to interact with actin both in the cytoplasm and nucleus, as well as supposedly binds to RNA in a cytoskeleton-dependent regulation manner[141]. Focusing on GBM stemness, Mao et al[142] found that WT1 was expressed predominantly in mesenchymal GSCs which, compared to proneural stem cells subtype, are characterized by higher proliferation, greater radioresistance, and implication in worse patients’ prognosis. Uribe et al[143] reviewed that mesenchymal GSCs develop tumors having more blood vessels, hemorrhagic lesions, and necrotic areas; the expression pattern in these stem cells generally facilitates inflammation, angiogenesis, migration, invasion, and glycolysis-mediated metabolism. Undoubtedly, more insights are needed concerning GBM molecular pathways in which WT1 is implicated.
Cytokines are soluble proteins that are secreted by immune and non-immune cells in response to stimulants such as immunogens or mitogens; this allows them to maintain the immune response and homeostasis[144]. Chemokines constitute a specific type of small (8-13 kDa) cytokines that promote the directed chemotaxis of nearby cells[145]. Consisting of nine members, the chemokine-like factor superfamily (CMTM) is expressed throughout the human tissues and regulates immune, circulatory and muscular systems, as well as the hematopoiesis[146-149]. The aberrant CMTM expression is implicated in various diseases, e.g., rheumatoid arthritis, atopic dermatitis, focal cerebral ischemia, male infertility, as well as tumorigenesis and metastasis[150-153]. The influence of CMTM6 on glioblastoma is known, but the research in this entity seems to be in the initial state. Guan et al[154] revealed that the highest CMTM6 expression was noted in the glioblastoma (WHO grade IV) compared with WHO grade II and III gliomas. Enrichment was also observed in both microvascular proliferation and hyperplastic blood vessels, which are both essential for tumor progression. In GBM, CMTM6 was also associated with one of the genes of immune checkpoints, i.e., TIM-3. From a broader glioma scale, the same authors summarized it as a molecule diminishing T-lymphocyte-dependent anti-tumor immunity, reducing patient survival and indicating poor prognosis. However, it is still yet to be elucidated what role CMTM6 may play in the GBM stemness. Currently, its contribution to such characteristics is confirmed on the basis of data from head-and-neck squamous cell carcinoma. Chen et al[155] observed poorer patient prognosis during CMTM6 overexpression that correlated with overactive Wnt/β-catenin signaling, i.e., the pathway crucial for tumorigenesis, epithelial-to-mesenchymal transition (EMT) and cancer stem cells maintenance. Silencing of CMTM6 led to PD-L1 downregulation, decreased tumor growth, and increased CD8+ and CD4+ T-cell infiltration. Eventually, the authors not only suggested the therapeutic suitability of CMTM6 but also concluded that this protein is implicated in EMT, stemness, and T-cell dysfunction. Similar research in the glioblastoma context is advisable, especially since CMTM6 can stabilize PD-L1 protein to impair T-cell function[156,157], as well as their combined expression had prognostic significance in pancreatic ductal adenocarcinoma and triple-negative breast cancer[158]. Nowadays, the role of PD-L1 in cancer and immunotherapy is unquestionable[159]; focusing on another protein related to this well-established molecule might bring novel strategies.
Signal transduction is based on phosphorylation and dephosphorylation events performed by kinases and phosphatases, leading to a cellular program relevant to the encountered stimulus[160]. Dual specificity phosphatases (DUSP) are responsible for the dephosphorylation of threonine and tyrosine residues on mitogen-activated protein kinases, rendering them inactive[161]. Even if DUSP7 was only noted as downregulated in glioblastoma, whereas DUSP1, DUSP5, and DUSP6 were induced within pseudopalisading and perinecrotic GBM regions[162], the role of DUSP7 in preserving the pluripotency of non-cancerous stem cells was certified in a murine model[163]. However, its contribution could be distinct from DUSP1, DUSP5, and DUSP6 but similar to DUSP2, DUSP8, and DUSP9 which were clustered together with DUSP7 in the study of Mills et al[162]. At last, it is worth noting that DUSP7 guides chromosome dynamics which is known for being regulated by cytoskeletal proteins[164,165]. The study linking this phosphatase to metabolism revealed that DUSP7 knockout accelerates metabolic disorder and insulin resistance in mice with a high-fat diet[166].
Cytoskeletal elements that act as scaffolds for intracellular cargo transport are microtubules. Motor proteins known as kinesins and dyneins orchestrate microtubule-related transport that is essential for cell differentiation or survival[167]. Kinesins constitute a large superfamily responsible for cargo trafficking, as well as controlling microtubule growth and stability[168]. Increased expression of kinesin superfamily representatives KIF4A, -9, -18A, and -23 was associated with poor prognosis in low-grade glioma and glioblastoma[169]. The pro-cancerous characteristics of Kinesin family member 20A (KIF20A) were noted more than 15 years ago in pancreatic cancer, which presented a reduction of proliferation once KIF20A was downregulated[170]. Currently, accumulating evidence shows that this kinesin is overexpressed in multiple tumors[171]. In glioblastoma, KIF20A downregulation induces cell cycle arrest and apoptosis via suppressing PI3K/AKT pathway[172]. Regarding cytoskeleton-related events, it is not only essential for cytokinesis but also interacts with Rab6 to regulate Golgi-related vesicle trafficking[173]. Although the role of KIF20A in GBM stemness has not yet been confirmed, it was suggested outside of the glioblastoma context in a study by Qiu et al[174]. The authors conceived the importance of KIF20A in controlling proliferation vs differentiation of tumor-initiating cells, based on both the fact that cancer stem cells share many mechanisms with neural progenitors, as well as their observations where KIF20A was implicated in balancing symmetric and asymmetric divisions during cerebral cortical development[175]. The KIF20A inactivation affected cortical neural progenitor cells that switched from proliferative to differentiative mode. During divisions, daughter cell-fate specification was controlled by KIF20A in coordination with RGS39 and SEPT710[174,176].
Neurofibromatoses (type 1, type 2, schwannomatosis) are distinct, dominantly inherited disorders that have in common the occurrence of nerve sheath tumors[177]. Type 1 neurofibromatosis presents with neurofibromas, cafe-au-lait spots/macules, freckling, and optic gliomas, whereas type 2 neuro
The signal transduction molecules being vitamin A derivatives are retinoids, they regulate cellular differentiation and proliferation via members of the nuclear receptors superfamily, including retinoic acid receptors (RARs) and retinoid X receptors (RXRs)[197]. The RXR family members (RXRA, RXRB, and RXRG) form heterodimers within the superfamily, e.g., with vitamin D, retinoic acid, or peroxisome proliferator-activated types of receptors[198,199]. RXRs have tumor suppressor properties and, as partners of RARA and RARB, they are implicated in the anti-proliferative effects of retinoic acid[197]. RXRG was found to modulate differentiation and apoptosis in various tumors, indicating its function in cancer pathogenesis[200]. Glioblastoma-related research certifies the general view that RXRG contributes to anti-neoplastic effect via its ligands; in study by Papi et al[201], the treatment of GBM with 6-OH-11-O-hydroxyfenantrene had anti-proliferative and anti-invasive effects. However, the literature data on glioblastoma stemness seem to focus on RARs rather than RXRs. Ying et al[202] evaluated the cellular and molecular responses of GSCs to all-trans retinoic acid; this treatment changed cells morphology (e.g., decreased neurosphere-forming capacity), caused growth arrest at G1/G0 to S transition, reduced cyclin D1 expression, and elevated p27 expression. Moreover, differentiation markers such as Tuj1 and GFAP were induced, while stem cell markers, such as CD133, Msi-1, Nestin, and Sox-2, had decreased expression. Friedman et al[203] provided similar observations with regard to Nestin level or neurosphere formation but also indicated that GBM differentiation induced by all-trans retinoic acid is executed via the ERK1/2 pathway. Evidently, retinoid-related research in the GBM context frequently focuses on all-trans retinoic acid while this isomer is bound only by RARs and not by both RARs and RXRs, as is the case with another retinoic lipid: 9-cis[204]. Even if two of the best-known retinoid receptors (RARA and RXRA) are described in detail by Rodriguez et al[205] in the GBM stemness context, the data on RXRG is still lacking and should begin with evaluation of whether 9-cis retinoid acid is able to manifest the anti-glioblastoma effects via RXRG and subsequently ERK1/2 pathway.
ECM is a component containing elastin, collagen, laminins, glycoproteins, fibronectin, and proteoglycans. Together, these elements bind via cell adhesion receptors and form a complex macromolecular network[206]. Matricellular proteins are made of matrix-binding proteins and cytokines that can be located within the cell or secreted outside[207]. SPARC/Osteonectin, CWCV, Kazal-like domains 1 (SPOCK1), also referred to as testican-1, is an ECM proteoglycan from a matricellular family of proteins that regulate matrix remodeling and affects tumor progression[208-210]. As the interplay between ECM and cytoskeleton is known[211], it is not surprising that changes in SPOCK1 lead to alterations in cytoskeletal components. For example, Schulz et al[212] noticed that SPOCK1 upregulation paralleled that of EPB41L4B, the latter being a cortical cytoskeleton protein underlying cellular membrane. With regard to brain tumors, testican-1 contributes to GBM metastasis and resistance to TMZ, as well as promotes glioma invasion, migration, and proliferation via Wnt/β-catenin and PI3K/AKT pathways[213,214]. Mediating TMZ chemoresistance via SPOCK1 in GBM was independently confirmed by Sun et al[215]. Although not yet directly concluded by any scientific group, it is conceivable that the impact of SPOCK1 on TMZ resistance renders a similar GSCs-related effect as SAA2 which was described in one of the previous sections.
The proteins’ turnover and degradation depend on ubiquitination that is orchestrated by the ubiquitin-proteasome system (UPS)[216], of which alterations can lead to several tumor types[217,218]. One of the ubiquitin-protein ligases responsible for the UPS specificity is ubiquitin-like with PHD and ring finger domains 1 (UHRF1)[219], a molecule also interacting with DNA methyltransferase 1, which together constitute the main regulatory axis of cellular senescence[220]. UHRF1 was already identified as a novel oncogene and/or druggable epigenetic target for various tumors[221-223], and Jung et al[220] suggested its role as a switch molecule between senescence and cancer. In GBM, UHRF1 is overexpressed by upstream CD47 and regulates downstream silencing of tumor suppressor gene p16INK4A, leading to increased proliferation[224]. Regarding cytoskeleton, UHRF1 contributes to microtubule organization through its downstream targets: BRCA2, HOOK1, KIF11, and KIF18A[225]. The role of UHRF1 in different types of stem cells is documented but overlooks GSCs. Namely, it was found to be required for the proliferative potential of basal stem cells in response to airway injury[226], as well as regulate the transcriptional marks at bivalent domains in pluripotent stem cells[227]. On the other hand, UHRF1 decrease was found to be a major cause of DNA demethylation in embryonic stem cells[228] and led to the activation of retroviral elements and delayed neurodegeneration[229]. It is evident that research in the glioblastoma context should be pursued in the future, especially since some epigenetic features, next to transcriptional ones, are unique in GSCs compared to neural stem cells and may include druggable targets for new therapeutic approaches[230].
Despite molecular advancements, there is still a considerable need for glioblastoma biomarkers[231], especially since the relatively ineffective treatment leaves the patients with a very dismal chance of survival[232]. One of the glioblastoma traits involved in the absence of effective treatment is tumor heterogeneity which can be explained by clonal evolution and the presence of stem cells[8].
Many independent studies on various tumor types have reported common genes as potential therapeutic or diagnostic biomarkers[233]. Al-Fatlawi et al[234] contemplated that biomarker signatures for different cancer types should be similar, due to the fundamental mechanisms shared between tumors, e.g., survival, tumor growth, or invasion. Thus, we presume that our description of stemness-related genes, especially those still unconfirmed in GBM, adds significant value to the current knowledge and provide insights into novel therapeutic or diagnostic directions.
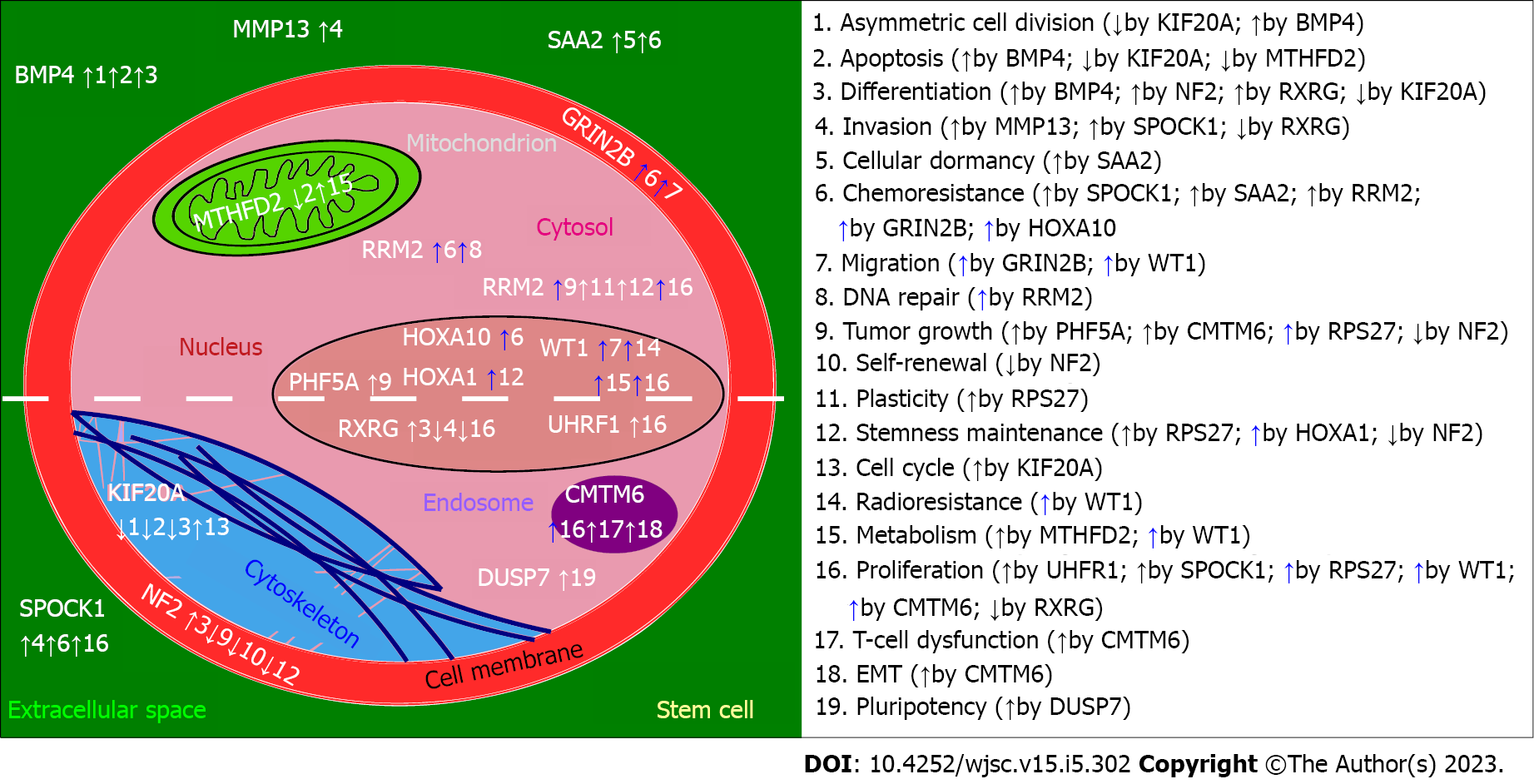
For clarity, a graphical presentation was prepared to emphasize the role of described genes specifically in stem cells, setting aside the rest of the information provided for each gene (Figure 2). At first glance, the most frequently regulated processes are proliferation and chemoresistance, followed by differentiation, tumor growth, invasion, and apoptosis. Except for BMP4 (increase in asymmetric cell division and apoptosis), NF2 (reduced self-renewal, tumor growth, stemness maintenance), RXRG (decrease in invasion and proliferation), and DUSP7 (insufficient data for a definite conclusion), the remaining genes exhibit pro-cancerous properties. This corresponds to what was described in subsections, separately for each gene. Interestingly, two genes that promote invasiveness of stem cells (SPOCK1, MMP13) are known to affect the cytoskeleton[33,212] and, in terms of MMP13, also the metabolism[90]. Two genes that were also found to regulate both the cytoskeleton and metabolism were MTHFD2 and RRM2. On the one hand, they control the organization of vimentin and actin; these proteins are known for influencing glioblastoma migratory potential[235,236]. On the other hand, the contribution of MTHFD2 and RRM2 to metabolism is related to folate and glutathione cycles that are implicated in the resistance of GBM to therapy[237,238].
In order to gravitate towards the link between metabolism, cytoskeleton, and GBM stemness, the appropriate representatives of each process (including the most frequently regulated processes that were mentioned above), were compiled into a cross-talk network. This allowed us to integrate the aim of our review with the main processes that are regulated by genes described in this work, additionally with the inclusion of GBM biomarkers (acquired from review by Sasmita et al[231]). Prevalent interaction types include co-expression and physical interaction between these representatives, there is also a high interconnectivity of the entire network, confirming that these molecular events are related. The cross-talk is visualized in Supplementary Figure 1, whereas the datasets used in the workflow are summarized in Supplementary Table 2.
The narrative of this review was intended to elaborate on the background of the biological machinery in which each successive gene is involved, then proceed with details regarding the regulation of glioblastoma, cytoskeleton/metabolism, and stemness (GBM-related or, if not present in the literature, any available). It is worth emphasizing that the herein described genes constitute more than half of the “top genes” that we established in our previous in silico study via a multi-stage methodology that included, e.g., enrichment analysis, machine learning algorithm, and differential expression analysis[32]. The remainder was not presented due to a lack of stemness-related literature data (Supp
| Gene | Compound | Condition | Trial number and phase | Intervention details |
| BMP4 | hrBMP4 | Glioblastoma | NCT02869243 (phase I) | Delivery of human recombinant BMP4 |
| GRIN2B | EVT 101 | Healthy volunteers (brain function assessment) | NCT00526968 (phase I) | Delivery of selective GRIN2B antagonist |
| RRM2 | COH29 | Solid tumors | NCT02112565 (phase I) | Delivery of ribonucleotide reductase inhibitor |
| WT1 | DSP-7888 | Gliomas (incl. GBM) | NCT02750891 (phase I/II) | Delivery of WT1 peptide-based cancer vaccine |
| KIF20A | KIF20A peptide | Small cell lung cancer | NCT01069653 (phase I) | Delivery of KIF20A peptide-based vaccination |
| NF2 | IAG933 | Solid tumors | NCT04857372 (phase I) | Not yet disclosed (the drug presumably counteracts the YAP/TAZ hyperactivity that occur following NF2 loss) |
| RXRG | 9-cis retinoic acid | Breast cancer | NCT00001504 (phase I) | Delivery of RXRG ligand |
Taken together, a promising set of genes involved in cytoskeletal rearrangements and metabolic alterations were found to influence glioblastoma stemness via a plethora of biological processes. Most of the described genes exhibit pro-cancerous properties; among them, clinical trials on GRIN2B, RRM2, WT1, and KIF20A are ongoing and focus on selective inhibitors or peptide-based vaccines. Concerning tumor suppressors, the anti-cancer effect can also be achieved via delivery of recombinant proteins (BMP4), ligands for tumor suppressors (RXRG), or counteracting the pathways that become hyperactive following an anti-oncogene loss (NF2). The cytoskeletal phenomena currently linked to the described genes require experimental verification of their contribution to GSCs expansion. Future GBM stemness-related research should generally delve into cytoskeleton and related molecular events, since the concept is already encouraging.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Poland
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Li C, United States; Shao AW, China; Ventura C, Italy S-Editor: Chen YL L-Editor: Ma JY P-Editor: Chen YL
| 1. | Gesundheit B, Ben-David E, Posen Y, Ellis R, Wollmann G, Schneider EM, Aigner K, Brauns L, Nesselhut T, Ackva I, Weisslein C, Thaller A. Effective Treatment of Glioblastoma Multiforme With Oncolytic Virotherapy: A Case-Series. Front Oncol. 2020;10:702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 2. | Grech N, Dalli T, Mizzi S, Meilak L, Calleja N, Zrinzo A. Rising Incidence of Glioblastoma Multiforme in a Well-Defined Population. Cureus. 2020;12:e8195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 3. | Oronsky B, Reid TR, Oronsky A, Sandhu N, Knox SJ. A Review of Newly Diagnosed Glioblastoma. Front Oncol. 2020;10:574012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 4. | Fernandes C, Costa A, Osório L, Lago RC, Linhares P, Carvalho B, Caeiro C. Current Standards of Care in Glioblastoma Therapy. In: Glioblastoma [Internet]. Brisbane (AU): Codon Publications; 2017-Sep-27. [PubMed] |
| 5. | Soni VS, Yanagihara TK. Tumor treating fields in the management of Glioblastoma: opportunities for advanced imaging. Cancer Imaging. 2019;19:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Vymazal J, Wong ET. Response patterns of recurrent glioblastomas treated with tumor-treating fields. Semin Oncol. 2014;41 Suppl 6:S14-S24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 7. | van Linde ME, Brahm CG, de Witt Hamer PC, Reijneveld JC, Bruynzeel AME, Vandertop WP, van de Ven PM, Wagemakers M, van der Weide HL, Enting RH, Walenkamp AME, Verheul HMW. Treatment outcome of patients with recurrent glioblastoma multiforme: a retrospective multicenter analysis. J Neurooncol. 2017;135:183-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 135] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 8. | Dymova MA, Kuligina EV, Richter VA. Molecular Mechanisms of Drug Resistance in Glioblastoma. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 9. | Mushtaq M, Kovalevska L, Darekar S, Abramsson A, Zetterberg H, Kashuba V, Klein G, Arsenian-Henriksson M, Kashuba E. Cell stemness is maintained upon concurrent expression of RB and the mitochondrial ribosomal protein S18-2. Proc Natl Acad Sci U S A. 2020;117:15673-15683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019;33:591-609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 336] [Cited by in RCA: 345] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 11. | Zhang C, Hai L, Zhu M, Yu S, Li T, Lin Y, Liu B, Zhou X, Chen L, Zhao P, Zhou H, Huang Y, Zhang K, Ren B, Yang X. Actin cytoskeleton regulator Arp2/3 complex is required for DLL1 activating Notch1 signaling to maintain the stem cell phenotype of glioma initiating cells. Oncotarget. 2017;8:33353-33364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Keller M, Blom M, Conze LL, Guo M, Hägerstrand D, Aspenström P. Altered cytoskeletal status in the transition from proneural to mesenchymal glioblastoma subtypes. Sci Rep. 2022;12:9838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 13. | Cardelli J, Skalli O. Divide and Invade: The Dynamic Cytoskeleton of Glioblastoma Cells. Glioblastoma. 2010;167-183. [DOI] [Full Text] |
| 14. | Harland A, Liu X, Ghirardello M, Galan MC, Perks CM, Kurian KM. Glioma Stem-Like Cells and Metabolism: Potential for Novel Therapeutic Strategies. Front Oncol. 2021;11:743814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 15. | Singh SX, Yang R, Roso K, Hansen LJ, Du C, Chen LH, Greer PK, Pirozzi CJ, He Y. Purine Synthesis Inhibitor L-Alanosine Impairs Mitochondrial Function and Stemness of Brain Tumor Initiating Cells. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Velásquez C, Mansouri S, Mora C, Nassiri F, Suppiah S, Martino J, Zadeh G, Fernández-Luna JL. Molecular and Clinical Insights into the Invasive Capacity of Glioblastoma Cells. J Oncol. 2019;2019:1740763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 17. | Garcia JH, Jain S, Aghi MK. Metabolic Drivers of Invasion in Glioblastoma. Front Cell Dev Biol. 2021;9:683276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 18. | Cheng L, Wu Q, Guryanova OA, Huang Z, Huang Q, Rich JN, Bao S. Elevated invasive potential of glioblastoma stem cells. Biochem Biophys Res Commun. 2011;406:643-648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 158] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 19. | Molina JR, Hayashi Y, Stephens C, Georgescu MM. Invasive glioblastoma cells acquire stemness and increased Akt activation. Neoplasia. 2010;12:453-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 20. | Hoelzinger DB, Demuth T, Berens ME. Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J Natl Cancer Inst. 2007;99:1583-1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 286] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 21. | Masters C. On the role of the cytoskeleton in metabolic compartmentation. Role in Cell Physiology. The Cytoskeleton. 1995;2:1-30. [DOI] [Full Text] |
| 22. | Marelli-Berg FM, Jangani M. Metabolic regulation of leukocyte motility and migration. J Leukoc Biol. 2018;104:285-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, Hamm M, Gage FH, Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 462] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 24. | Compagnucci C, Piemonte F, Sferra A, Piermarini E, Bertini E. The cytoskeletal arrangements necessary to neurogenesis. Oncotarget. 2016;7:19414-19429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 25. | Alibert C, Pereira D, Lardier N, Etienne-Manneville S, Goud B, Asnacios A, Manneville JB. Multiscale rheology of glioma cells. Biomaterials. 2021;275:120903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Wang C, Sinha S, Jiang X, Murphy L, Fitch S, Wilson C, Grant G, Yang F. Matrix Stiffness Modulates Patient-Derived Glioblastoma Cell Fates in Three-Dimensional Hydrogels. Tissue Eng Part A. 2021;27:390-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 27. | Angelopoulos I, Gakis G, Birmpas K, Kyrousi C, Habeos EE, Kaplani K, Lygerou Z, Habeos I, Taraviras S. Metabolic regulation of the neural stem cell fate: Unraveling new connections, establishing new concepts. Front Neurosci. 2022;16:1009125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 28. | Li J, Zou Y, Li Z, Jiu Y. Joining actions: crosstalk between intermediate filaments and actin orchestrates cellular physical dynamics and signaling. Sci China Life Sci. 2019;62:1368-1374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | Weigel M, Wang L, Fu MM. Microtubule organization and dynamics in oligodendrocytes, astrocytes, and microglia. Dev Neurobiol. 2021;81:310-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 30. | Weil S, Osswald M, Solecki G, Grosch J, Jung E, Lemke D, Ratliff M, Hänggi D, Wick W, Winkler F. Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol. 2017;19:1316-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 203] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 31. | Roehlecke C, Schmidt MHH. Tunneling Nanotubes and Tumor Microtubes in Cancer. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 32. | Kałuzińska Ż, Kołat D, Bednarek AK, Płuciennik E. PLEK2, RRM2, GCSH: A Novel WWOX-Dependent Biomarker Triad of Glioblastoma at the Crossroads of Cytoskeleton Reorganization and Metabolism Alterations. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Toriseva MJ, Ala-aho R, Karvinen J, Baker AH, Marjomäki VS, Heino J, Kähäri VM. Collagenase-3 (MMP-13) enhances remodeling of three-dimensional collagen and promotes survival of human skin fibroblasts. J Invest Dermatol. 2007;127:49-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | Wang Z, Yang X, Liu C, Li X, Zhang B, Wang B, Zhang Y, Song C, Zhang T, Liu M, Liu B, Ren M, Jiang H, Zou J, Liu X, Zhang H, Zhu WG, Yin Y, Zhang Z, Gu W, Luo J. Acetylation of PHF5A Modulates Stress Responses and Colorectal Carcinogenesis through Alternative Splicing-Mediated Upregulation of KDM3A. Mol Cell. 2019;74:1250-1263.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 35. | Dai Y, Pierson SE, Dudney WC, Stack BC Jr. Extraribosomal function of metallopanstimulin-1: reducing paxillin in head and neck squamous cell carcinoma and inhibiting tumor growth. Int J Cancer. 2010;126:611-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 36. | Connolly M, Veale DJ, Fearon U. Acute serum amyloid A regulates cytoskeletal rearrangement, cell matrix interactions and promotes cell migration in rheumatoid arthritis. Ann Rheum Dis. 2011;70:1296-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 37. | Paul S, Gangwar A, Arya A, Bhargava K, Ahmad Y. Modulation of lung cytoskeletal remodeling, RXR based metabolic cascades and inflammation to achieve redox homeostasis during extended exposures to lowered pO(2). Apoptosis. 2021;26:431-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 38. | Zhu Z, Leung GKK. More Than a Metabolic Enzyme: MTHFD2 as a Novel Target for Anticancer Therapy? Front Oncol. 2020;10:658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 39. | Lehtinen L, Ketola K, Mäkelä R, Mpindi JP, Viitala M, Kallioniemi O, Iljin K. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 2013;4:48-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 40. | Huang M, Xue J, Chen Z, Zhou X, Chen M, Sun J, Xu Z, Wang S, Xu H, Du Z, Liu M. MTHFD2 suppresses glioblastoma progression via the inhibition of ERK1/2 phosphorylation. Biochem Cell Biol. 2023;101:112-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 41. | Wang J, Luo J, Sun Z, Sun F, Kong Z, Yu J. Identification of MTHFD2 as a novel prognosis biomarker in esophageal carcinoma patients based on transcriptomic data and methylation profiling. Medicine (Baltimore). 2020;99:e22194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 42. | Kitab B, Satoh M, Ohmori Y, Munakata T, Sudoh M, Kohara M, Tsukiyama-Kohara K. Ribonucleotide reductase M2 promotes RNA replication of hepatitis C virus by protecting NS5B protein from hPLIC1-dependent proteasomal degradation. J Biol Chem. 2019;294:5759-5773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | Tarangelo A, Rodencal J, Kim JT, Magtanong L, Long JZ, Dixon SJ. Nucleotide biosynthesis links glutathione metabolism to ferroptosis sensitivity. Life Sci Alliance. 2022;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 44. | Sun H, Yang B, Zhang H, Song J, Zhang Y, Xing J, Yang Z, Wei C, Xu T, Yu Z, Xu Z, Hou M, Ji M. RRM2 is a potential prognostic biomarker with functional significance in glioma. Int J Biol Sci. 2019;15:533-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 45. | Rasmussen RD, Gajjar MK, Tuckova L, Jensen KE, Maya-Mendoza A, Holst CB, Møllgaard K, Rasmussen JS, Brennum J, Bartek J Jr, Syrucek M, Sedlakova E, Andersen KK, Frederiksen MH, Bartek J, Hamerlik P. BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat Commun. 2016;7:13398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 99] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 46. | Nixon TRW, Richards A, Towns LK, Fuller G, Abbs S, Alexander P, McNinch A, Sandford RN, Snead MP. Bone morphogenetic protein 4 (BMP4) loss-of-function variant associated with autosomal dominant Stickler syndrome and renal dysplasia. Eur J Hum Genet. 2019;27:369-377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 47. | Piccirillo SG, Vescovi AL. Bone morphogenetic proteins regulate tumorigenicity in human glioblastoma stem cells. Ernst Schering Found Symp Proc. 2006;59-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 48. | Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 898] [Cited by in RCA: 894] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 50. | Cho DY, Lin SZ, Yang WK, Lee HC, Hsu DM, Lin HL, Chen CC, Liu CL, Lee WY, Ho LH. Targeting cancer stem cells for treatment of glioblastoma multiforme. Cell Transplant. 2013;22:731-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 51. | Duggal R, Geissinger U, Zhang Q, Aguilar J, Chen NG, Binda E, Vescovi AL, Szalay AA. Vaccinia virus expressing bone morphogenetic protein-4 in novel glioblastoma orthotopic models facilitates enhanced tumor regression and long-term survival. J Transl Med. 2013;11:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Li Q, Wijesekera O, Salas SJ, Wang JY, Zhu M, Aprhys C, Chaichana KL, Chesler DA, Zhang H, Smith CL, Guerrero-Cazares H, Levchenko A, Quinones-Hinojosa A. Mesenchymal stem cells from human fat engineered to secrete BMP4 are nononcogenic, suppress brain cancer, and prolong survival. Clin Cancer Res. 2014;20:2375-2387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 53. | Videla Richardson GA, Garcia CP, Roisman A, Slavutsky I, Fernandez Espinosa DD, Romorini L, Miriuka SG, Arakaki N, Martinetto H, Scassa ME, Sevlever GE. Specific Preferences in Lineage Choice and Phenotypic Plasticity of Glioma Stem Cells Under BMP4 and Noggin Influence. Brain Pathol. 2016;26:43-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 54. | Liu S, Yin F, Zhao M, Zhou C, Ren J, Huang Q, Zhao Z, Mitra R, Fan W, Fan M. The homing and inhibiting effects of hNSCs-BMP4 on human glioma stem cells. Oncotarget. 2016;7:17920-17931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 55. | Hughes JH, Ewy JM, Chen J, Wong SY, Tharp KM, Stahl A, Kumar S. Transcriptomic analysis reveals that BMP4 sensitizes glioblastoma tumor-initiating cells to mechanical cues. Matrix Biol. 2020;85-86:112-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 56. | Zhou C, Duan M, Guo D, Du X, Zhang D, Xie J. Microenvironmental stiffness mediates cytoskeleton re-organization in chondrocytes through laminin-FAK mechanotransduction. Int J Oral Sci. 2022;14:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 57. | Shen K, Kenche H, Zhao H, Li J, Stone J. The role of extracellular matrix stiffness in regulating cytoskeletal remodeling via vinculin in synthetic smooth muscle cells. Biochem Biophys Res Commun. 2019;508:302-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 58. | Thériault BL, Shepherd TG, Mujoomdar ML, Nachtigal MW. BMP4 induces EMT and Rho GTPase activation in human ovarian cancer cells. Carcinogenesis. 2007;28:1153-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 59. | Koguchi M, Nakahara Y, Ito H, Wakamiya T, Yoshioka F, Ogata A, Inoue K, Masuoka J, Izumi H, Abe T. BMP4 induces asymmetric cell division in human glioma stem-like cells. Oncol Lett. 2020;19:1247-1254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 60. | Ciechomska IA, Gielniewski B, Wojtas B, Kaminska B, Mieczkowski J. EGFR/FOXO3a/BIM signaling pathway determines chemosensitivity of BMP4-differentiated glioma stem cells to temozolomide. Exp Mol Med. 2020;52:1326-1340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 61. | Wu Q, Berglund AE, MacAulay RJ, Etame AB. A Novel Role of BIRC3 in Stemness Reprogramming of Glioblastoma. Int J Mol Sci. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 62. | Verploegh ISC, Conidi A, Brouwer RWW, Balcioglu HE, Karras P, Makhzami S, Korporaal A, Marine JC, Lamfers M, Van IJcken WFJ, Leenstra S, Huylebroeck D. Comparative single-cell RNA-sequencing profiling of BMP4-treated primary glioma cultures reveals therapeutic markers. Neuro Oncol. 2022;24:2133-2145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 63. | Hu C, Chen W, Myers SJ, Yuan H, Traynelis SF. Human GRIN2B variants in neurodevelopmental disorders. J Pharmacol Sci. 2016;132:115-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 169] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 64. | Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2476] [Cited by in RCA: 2677] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 65. | Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol. 1994;347:150-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 369] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 66. | Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 2008;24:183-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 345] [Cited by in RCA: 327] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 67. | Tuncbag N, Milani P, Pokorny JL, Johnson H, Sio TT, Dalin S, Iyekegbe DO, White FM, Sarkaria JN, Fraenkel E. Network Modeling Identifies Patient-specific Pathways in Glioblastoma. Sci Rep. 2016;6:28668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 68. | Pollak J, Rai KG, Funk CC, Arora S, Lee E, Zhu J, Price ND, Paddison PJ, Ramirez JM, Rostomily RC. Ion channel expression patterns in glioblastoma stem cells with functional and therapeutic implications for malignancy. PLoS One. 2017;12:e0172884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 69. | Zhao Y, Zhang X, Yao J, Jin Z, Liu C. Expression patterns and the prognostic value of the EMILIN/Multimerin family members in low-grade glioma. PeerJ. 2020;8:e8696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 70. | Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, Schuh AC, Yee KW, Bullinger L, Herold T, Görlich D, Büchner T, Hiddemann W, Berdel WE, Wörmann B, Cheok M, Preudhomme C, Dombret H, Metzeler K, Buske C, Löwenberg B, Valk PJ, Zandstra PW, Minden MD, Dick JE, Wang JC. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 606] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 71. | She X, Gao Y, Zhao Y, Yin Y, Dong Z. A high-throughput screen identifies inhibitors of lung cancer stem cells. Biomed Pharmacother. 2021;140:111748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 72. | Zanatta A, Rocha AM, Carvalho FM, Pereira RM, Taylor HS, Motta EL, Baracat EC, Serafini PC. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: a review. J Assist Reprod Genet. 2010;27:701-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 73. | Taylor HS, Vanden Heuvel GB, Igarashi P. A conserved Hox axis in the mouse and human female reproductive system: late establishment and persistent adult expression of the Hoxa cluster genes. Biol Reprod. 1997;57:1338-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 309] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 74. | Taylor HS, Arici A, Olive D, Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. J Clin Invest. 1998;101:1379-1384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 136] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 75. | Gallo M, Ho J, Coutinho FJ, Vanner R, Lee L, Head R, Ling EK, Clarke ID, Dirks PB. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 2013;73:417-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 76. | Arunachalam E, Rogers W, Simpson GR, Möller-Levet C, Bolton G, Ismael M, Smith C, Keegen K, Bagwan I, Brend T, Short SC, Hong B, Otani Y, Kaur B, Annels N, Morgan R, Pandha H. HOX and PBX gene dysregulation as a therapeutic target in glioblastoma multiforme. BMC Cancer. 2022;22:400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 77. | Dong CY, Cui J, Li DH, Li Q, Hong XY. HOXA10AS: A novel oncogenic long noncoding RNA in glioma. Oncol Rep. 2018;40:2573-2583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 78. | Kim JW, Kim JY, Kim JE, Kim SK, Chung HT, Park CK. HOXA10 is associated with temozolomide resistance through regulation of the homologous recombinant DNA repair pathway in glioblastoma cell lines. Genes Cancer. 2014;5:165-174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 79. | Zhang X, Emerald BS, Mukhina S, Mohankumar KM, Kraemer A, Yap AS, Gluckman PD, Lee KO, Lobie PE. HOXA1 is required for E-cadherin-dependent anchorage-independent survival of human mammary carcinoma cells. J Biol Chem. 2006;281:6471-6481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 80. | Makki N, Capecchi MR. Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev Biol. 2011;357:295-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 81. | Shi T, Guo D, Xu H, Su G, Chen J, Zhao Z, Shi J, Wedemeyer M, Attenello F, Zhang L, Lu W. HOTAIRM1, an enhancer lncRNA, promotes glioma proliferation by regulating long-range chromatin interactions within HOXA cluster genes. Mol Biol Rep. 2020;47:2723-2733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 82. | Schmid RS, Simon JM, Vitucci M, McNeill RS, Bash RE, Werneke AM, Huey L, White KK, Ewend MG, Wu J, Miller CR. Core pathway mutations induce de-differentiation of murine astrocytes into glioblastoma stem cells that are sensitive to radiation but resistant to temozolomide. Neuro Oncol. 2016;18:962-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 83. | Li F, Xu Y, Xu X, Ge S, Zhang F, Zhang H, Fan X. lncRNA HotairM1 Depletion Promotes Self-Renewal of Cancer Stem Cells through HOXA1-Nanog Regulation Loop. Mol Ther Nucleic Acids. 2020;22:456-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 84. | Cui N, Hu M, Khalil RA. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog Mol Biol Transl Sci. 2017;147:1-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 844] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 85. | Kapoor C, Vaidya S, Wadhwan V; Hitesh, Kaur G, Pathak A. Seesaw of matrix metalloproteinases (MMPs). J Cancer Res Ther. 2016;12:28-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 154] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 86. | Nallanthighal S, Heiserman JP, Cheon DJ. The Role of the Extracellular Matrix in Cancer Stemness. Front Cell Dev Biol. 2019;7:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 243] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 87. | Gobin E, Bagwell K, Wagner J, Mysona D, Sandirasegarane S, Smith N, Bai S, Sharma A, Schleifer R, She JX. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer. 2019;19:581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 212] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 88. | Kobayashi K, Takahashi H, Inoue A, Harada H, Toshimori S, Kobayashi Y, Goto K, Sugimoto K, Yano H, Ohnishi T, Tanaka J. Oct-3/4 promotes migration and invasion of glioblastoma cells. J Cell Biochem. 2012;113:508-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 89. | Wang J, Li Y, Wang J, Li C, Yu K, Wang Q. Increased expression of matrix metalloproteinase-13 in glioma is associated with poor overall survival of patients. Med Oncol. 2012;29:2432-2437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 90. | Li Y, Tang L, Duan Y, Ding Y. Upregulation of MMP-13 and TIMP-1 expression in response to mechanical strain in MC3T3-E1 osteoblastic cells. BMC Res Notes. 2010;3:309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 91. | Inoue A, Takahashi H, Harada H, Kohno S, Ohue S, Kobayashi K, Yano H, Tanaka J, Ohnishi T. Cancer stem-like cells of glioblastoma characteristically express MMP-13 and display highly invasive activity. Int J Oncol. 2010;37:1121-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 92. | Bayer AL, Fraker CA. The Folate Cycle As a Cause of Natural Killer Cell Dysfunction and Viral Etiology in Type 1 Diabetes. Front Endocrinol (Lausanne). 2017;8:315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 93. | Tedeschi PM, Vazquez A, Kerrigan JE, Bertino JR. Mitochondrial Methylenetetrahydrofolate Dehydrogenase (MTHFD2) Overexpression Is Associated with Tumor Cell Proliferation and Is a Novel Target for Drug Development. Mol Cancer Res. 2015;13:1361-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 94. | Zhu Z, Kiang KM, Li N, Liu J, Zhang P, Jin L, He X, Zhang S, Leung GK. Folate enzyme MTHFD2 links one-carbon metabolism to unfolded protein response in glioblastoma. Cancer Lett. 2022;549:215903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 95. | Nishimura T, Nakata A, Chen X, Nishi K, Meguro-Horike M, Sasaki S, Kita K, Horike SI, Saitoh K, Kato K, Igarashi K, Murayama T, Kohno S, Takahashi C, Mukaida N, Yano S, Soga T, Tojo A, Gotoh N. Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene. 2019;38:2464-2481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 96. | Chang Y, Zhao Y, Wang L, Wu M, He C, Huang M, Lei Z, Yang J, Han S, Wang B, Chen Y, Liu C, Yu H, Xue L, Geng J, Dai T, Ren L, Wang Q, Liu X, Chu X, Chen C. PHF5A promotes colorectal cancerprogression by alternative splicing of TEAD2. Mol Ther Nucleic Acids. 2021;26:1215-1227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 97. | Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 884] [Cited by in RCA: 1166] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 98. | Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1406] [Cited by in RCA: 1568] [Article Influence: 104.5] [Reference Citation Analysis (0)] |
| 99. | Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015;84:291-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 906] [Article Influence: 90.6] [Reference Citation Analysis (0)] |
| 100. | Rzymski T, Grzmil P, Meinhardt A, Wolf S, Burfeind P. PHF5A represents a bridge protein between splicing proteins and ATP-dependent helicases and is differentially expressed during mouse spermatogenesis. Cytogenet Genome Res. 2008;121:232-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 101. | Zheng YZ, Xue MZ, Shen HJ, Li XG, Ma D, Gong Y, Liu YR, Qiao F, Xie HY, Lian B, Sun WL, Zhao HY, Yao L, Zuo WJ, Li DQ, Wang P, Hu X, Shao ZM. PHF5A Epigenetically Inhibits Apoptosis to Promote Breast Cancer Progression. Cancer Res. 2018;78:3190-3206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 102. | Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36:364-372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 244] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 103. | Mhyre AJ, Turnbaugh S, Morris SM, Xin H, Paddison PJ, Ferrer M, Olson JM. Abstract 3200: Targeting PHF5A for the treatment of glioblastoma and other Myc-driven cancers. Cancer Res. 2017;77:3200. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 104. | Trappe R, Ahmed M, Gläser B, Vogel C, Tascou S, Burfeind P, Engel W. Identification and characterization of a novel murine multigene family containing a PHD-finger-like motif. Biochem Biophys Res Commun. 2002;293:816-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 105. | Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Girard EJ, Davison J, Berndt J, Corrin P, Hardcastle J, Basom R, Delrow JJ, Webb T, Pollard SM, Lee J, Olson JM, Paddison PJ. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 106. | Opron K, Burton ZF. Ribosome Structure, Function, and Early Evolution. Int J Mol Sci. 2018;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 107. | Yang ZY, Qu Y, Zhang Q, Wei M, Liu CX, Chen XH, Yan M, Zhu ZG, Liu BY, Chen GQ, Wu YL, Gu QL. Knockdown of metallopanstimulin-1 inhibits NF-κB signaling at different levels: the role of apoptosis induction of gastric cancer cells. Int J Cancer. 2012;130:2761-2770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 108. | Dai Y, Pierson S, Dudney C, Zeng Y, Macleod V, Shaughnessy JD, Stack BC Jr. Ribosomal protein metallopanstimulin-1 impairs multiple myeloma CAG cells growth and inhibits fibroblast growth factor receptor 3. Clin Lymphoma Myeloma Leuk. 2011;11:490-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 109. | Feldheim J, Kessler AF, Schmitt D, Salvador E, Monoranu CM, Feldheim JJ, Ernestus RI, Löhr M, Hagemann C. Ribosomal Protein S27/Metallopanstimulin-1 (RPS27) in Glioma-A New Disease Biomarker? Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 110. | Hide T, Shibahara I, Inukai M, Shigeeda R, Kumabe T. Ribosomes and Ribosomal Proteins Promote Plasticity and Stemness Induction in Glioma Cells via Reprogramming. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 111. | Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon JG, Smith KA, Lankerovich M, Bertagnolli D, Bickley K, Boe AF, Brouner K, Butler S, Caldejon S, Chapin M, Datta S, Dee N, Desta T, Dolbeare T, Dotson N, Ebbert A, Feng D, Feng X, Fisher M, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Hejazinia N, Hohmann J, Hothi P, Howard R, Joines K, Kriedberg A, Kuan L, Lau C, Lee F, Lee H, Lemon T, Long F, Mastan N, Mott E, Murthy C, Ngo K, Olson E, Reding M, Riley Z, Rosen D, Sandman D, Shapovalova N, Slaughterbeck CR, Sodt A, Stockdale G, Szafer A, Wakeman W, Wohnoutka PE, White SJ, Marsh D, Rostomily RC, Ng L, Dang C, Jones A, Keogh B, Gittleman HR, Barnholtz-Sloan JS, Cimino PJ, Uppin MS, Keene CD, Farrokhi FR, Lathia JD, Berens ME, Iavarone A, Bernard A, Lein E, Phillips JW, Rostad SW, Cobbs C, Hawrylycz MJ, Foltz GD. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360:660-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 401] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 112. | Huang WJ, Chen WW, Zhang X. Glioblastoma multiforme: Effect of hypoxia and hypoxia inducible factors on therapeutic approaches. Oncol Lett. 2016;12:2283-2288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 113. | Torrents E. Ribonucleotide reductases: essential enzymes for bacterial life. Front Cell Infect Microbiol. 2014;4:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 114. | Liu X, Peng J, Zhou Y, Xie B, Wang J. Silencing RRM2 inhibits multiple myeloma by targeting the Wnt/βcatenin signaling pathway. Mol Med Rep. 2019;20:2159-2166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 115. | Zou Y, Zhou J, Xu B, Li W, Wang Z. Ribonucleotide reductase subunit M2 as a novel target for clear-cell renal cell carcinoma. Onco Targets Ther. 2019;12:3267-3275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 116. | Shao J, Liu X, Zhu L, Yen Y. Targeting ribonucleotide reductase for cancer therapy. Expert Opin Ther Targets. 2013;17:1423-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 117. | Fatkhutdinov N, Sproesser K, Krepler C, Liu Q, Brafford PA, Herlyn M, Aird KM, Zhang R. Targeting RRM2 and Mutant BRAF Is a Novel Combinatorial Strategy for Melanoma. Mol Cancer Res. 2016;14:767-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 118. | Aye Y, Long MJC, Stubbe J. Mechanistic studies of semicarbazone triapine targeting human ribonucleotide reductase in vitro and in mammalian cells: tyrosyl radical quenching not involving reactive oxygen species. J Biol Chem. 2012;287:35768-35778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 119. | Chaston TB, Lovejoy DB, Watts RN, Richardson DR. Examination of the antiproliferative activity of iron chelators: multiple cellular targets and the different mechanism of action of triapine compared with desferrioxamine and the potent pyridoxal isonicotinoyl hydrazone analogue 311. Clin Cancer Res. 2003;9:402-414. [PubMed] |
| 120. | Cooperman BS, Gao Y, Tan C, Kashlan OB, Kaur J. Peptide inhibitors of mammalian ribonucleotide reductase. Adv Enzyme Regul. 2005;45:112-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 121. | Perrault EN, Shireman JM, Ali ES, Preddy I, Lin P, Park C, Tomes L, Zolp AJ, Budhiraja S, Baisiwala S, James CD, Ben-Sahra I, Pott S, Basu A, Ahmed AU. Ribonucleotide Reductase Regulatory Subunit M2 as a Driver of Glioblastoma TMZ-Resistance through Modulation of dNTP Production. November 24, 2021. [cited 14 December 2022]. Available from: https://www.biorxiv.org/content/10.1101/2021.11.23.469785v1#page. |
| 122. | Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, Venteicher AS, Hebert CH, Carey CD, Rodig SJ, Shareef SJ, Najm FJ, van Galen P, Wakimoto H, Cahill DP, Rich JN, Aster JC, Suvà ML, Patel AP, Bernstein BE. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell. 2017;20:233-246.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 379] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 123. | Gruys E, Toussaint MJ, Niewold TA, Koopmans SJ. Acute phase reaction and acute phase proteins. J Zhejiang Univ Sci B. 2005;6:1045-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 682] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 124. | Sack GH Jr. Serum amyloid A - a review. Mol Med. 2018;24:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 343] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 125. | Malle E, De Beer FC. Human serum amyloid A (SAA) protein: a prominent acute-phase reactant for clinical practice. Eur J Clin Invest. 1996;26:427-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 269] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 126. | Sun L, Ye RD. Serum amyloid A1: Structure, function and gene polymorphism. Gene. 2016;583:48-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 127. | Upragarin N, Landman WJ, Gaastra W, Gruys E. Extrahepatic production of acute phase serum amyloid A. Histol Histopathol. 2005;20:1295-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 47] [Reference Citation Analysis (0)] |
| 128. | Kim YJ, Gallien S, El-Khoury V, Goswami P, Sertamo K, Schlesser M, Berchem G, Domon B. Quantification of SAA1 and SAA2 in lung cancer plasma using the isotype-specific PRM assays. Proteomics. 2015;15:3116-3125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 129. | Ana C, Gilberto K, Raquel H, Luziane B, Franciele K. Effect of SAA1, SAA2 and SAA4 knockdown on proliferation and invasion of glioblastomas multiformes cells. [cited 14 December 2022]. Available from: https://www.frontiersin.org/10.3389/conf.fimmu.2013.02.00949/event_abstract. |
| 130. | Knebel FH, Albuquerque RC, Massaro RR, Maria-Engler SS, Campa A. Dual effect of serum amyloid A on the invasiveness of glioma cells. Mediators Inflamm. 2013;2013:509089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 131. | Adamski V, Hattermann K, Kubelt C, Cohrs G, Lucius R, Synowitz M, Sebens S, Held-Feindt J. Entry and exit of chemotherapeutically-promoted cellular dormancy in glioblastoma cells is differentially affected by the chemokines CXCL12, CXCL16, and CX3CL1. Oncogene. 2020;39:4421-4435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 132. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4860] [Cited by in RCA: 4373] [Article Influence: 273.3] [Reference Citation Analysis (0)] |
| 133. | Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2733] [Cited by in RCA: 2601] [Article Influence: 185.8] [Reference Citation Analysis (0)] |
| 134. | Ramsawhook A, Ruzov A, Coyle B. Wilms' Tumor Protein 1 and Enzymatic Oxidation of 5-Methylcytosine in Brain Tumors: Potential Perspectives. Front Cell Dev Biol. 2018;6:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 135. | Szemes M, Dallosso AR, Melegh Z, Curry T, Li Y, Rivers C, Uney J, Mägdefrau AS, Schwiderski K, Park JH, Brown KW, Shandilya J, Roberts SG, Malik K. Control of epigenetic states by WT1 via regulation of de novo DNA methyltransferase 3A. Hum Mol Genet. 2013;22:74-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 136. | Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms' tumor locus. Cell. 1990;60:509-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1353] [Cited by in RCA: 1352] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 137. | Qi XW, Zhang F, Wu H, Liu JL, Zong BG, Xu C, Jiang J. Wilms' tumor 1 (WT1) expression and prognosis in solid cancer patients: a systematic review and meta-analysis. Sci Rep. 2015;5:8924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 138. | Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL, Weiner LM, Matrisian LM. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323-5337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 1053] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 139. | Oji Y, Hashimoto N, Tsuboi A, Murakami Y, Iwai M, Kagawa N, Chiba Y, Izumoto S, Elisseeva O, Ichinohasama R, Sakamoto J, Morita S, Nakajima H, Takashima S, Nakae Y, Nakata J, Kawakami M, Nishida S, Hosen N, Fujiki F, Morimoto S, Adachi M, Iwamoto M, Oka Y, Yoshimine T, Sugiyama H. Association of WT1 IgG antibody against WT1 peptide with prolonged survival in glioblastoma multiforme patients vaccinated with WT1 peptide. Int J Cancer. 2016;139:1391-1401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 140. | Kijima N, Hosen N, Kagawa N, Hashimoto N, Kinoshita M, Oji Y, Sugiyama H, Yoshimine T. Wilms' tumor 1 is involved in tumorigenicity of glioblastoma by regulating cell proliferation and apoptosis. Anticancer Res. 2014;34:61-67. [PubMed] |
| 141. | Dudnakova T, Spraggon L, Slight J, Hastie N. Actin: a novel interaction partner of WT1 influencing its cell dynamic properties. Oncogene. 2010;29:1085-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 142. | Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, Wang M, Hu B, Cheng SY, Sobol RW, Nakano I. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644-8649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 525] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 143. | Uribe D, Niechi I, Rackov G, Erices JI, San Martín R, Quezada C. Adapt to Persist: Glioblastoma Microenvironment and Epigenetic Regulation on Cell Plasticity. Biology (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 144. | Cai X, Deng J, Ming Q, Cai H, Chen Z. Chemokine-like factor 1: A promising therapeutic target in human diseases. Exp Biol Med (Maywood). 2020;245:1518-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 145. | Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1834] [Cited by in RCA: 1942] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 146. | Tian L, Li W, Wang J, Zhang Y, Zheng Y, Qi H, Guo X, Ma D, Shen H, Wang Y. The CKLF1-C19 peptide attenuates allergic lung inflammation by inhibiting CCR3- and CCR4-mediated chemotaxis in a mouse model of asthma. Allergy. 2011;66:287-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 147. | Morrison AC, Felix JF, Cupples LA, Glazer NL, Loehr LR, Dehghan A, Demissie S, Bis JC, Rosamond WD, Aulchenko YS, Wang YA, Haritunians T, Folsom AR, Rivadeneira F, Benjamin EJ, Lumley T, Couper D, Stricker BH, O'Donnell CJ, Rice KM, Chang PP, Hofman A, Levy D, Rotter JI, Fox ER, Uitterlinden AG, Wang TJ, Psaty BM, Willerson JT, van Duijn CM, Boerwinkle E, Witteman JC, Vasan RS, Smith NL. Genomic variation associated with mortality among adults of European and African ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2010;3:248-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 148. | Zhang T, Zhang X, Yu W, Chen J, Li Q, Jiao Y, He P, Shen C. Effects of chemokine-like factor 1 on vascular smooth muscle cell migration and proliferation in vascular inflammation. Atherosclerosis. 2013;226:49-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 149. | Chrifi I, Louzao-Martinez L, Brandt M, van Dijk CGM, Burgisser P, Zhu C, Kros JM, Duncker DJ, Cheng C. CMTM3 (CKLF-Like Marvel Transmembrane Domain 3) Mediates Angiogenesis by Regulating Cell Surface Availability of VE-Cadherin in Endothelial Adherens Junctions. Arterioscler Thromb Vasc Biol. 2017;37:1098-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 150. | Tao K, Tang X, Wang B, Li RJ, Zhang BQ, Lin JH, Li H. Distinct expression of chemokine-like factor 1 in synovium of osteoarthritis, rheumatoid arthritis and ankylosing spondylitis. J Huazhong Univ Sci Technolog Med Sci. 2016;36:70-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 151. | Yang GY, Chen X, Sun YC, Ma CL, Qian G. Chemokine-like factor 1 (CLFK1) is over-expressed in patients with atopic dermatitis. Int J Biol Sci. 2013;9:759-765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 152. | Kong LL, Wang ZY, Han N, Zhuang XM, Wang ZZ, Li H, Chen NH. Neutralization of chemokine-like factor 1, a novel C-C chemokine, protects against focal cerebral ischemia by inhibiting neutrophil infiltration via MAPK pathways in rats. J Neuroinflammation. 2014;11:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 153. | Li M, Luo F, Tian X, Yin S, Zhou L, Zheng S. Chemokine-Like Factor-Like MARVEL Transmembrane Domain-Containing Family in Hepatocellular Carcinoma: Latest Advances. Front Oncol. 2020;10:595973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 154. | Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine. 2018;35:233-243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 155. | Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, Xiao Y, Bu LL, Zhang WF, Sun ZJ. Targeting CMTM6 Suppresses Stem Cell-Like Properties and Enhances Antitumor Immunity in Head and Neck Squamous Cell Carcinoma. Cancer Immunol Res. 2020;8:179-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 156. | Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, Broeks A, Horlings HM, Wessels LFA, Blank CU, Xiao Y, Heck AJR, Borst J, Brummelkamp TR, Schumacher TNM. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 338] [Cited by in RCA: 536] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 157. | Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, Gilan O, Bloor S, Noori T, Morgens DW, Bassik MC, Neeson PJ, Behren A, Darcy PK, Dawson SJ, Voskoboinik I, Trapani JA, Cebon J, Lehner PJ, Dawson MA. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 686] [Article Influence: 85.8] [Reference Citation Analysis (0)] |
| 158. | Mamessier E, Birnbaum DJ, Finetti P, Birnbaum D, Bertucci F. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann Transl Med. 2018;6:54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 159. | Jiang Y, Chen M, Nie H, Yuan Y. PD-1 and PD-L1 in cancer immunotherapy: clinical implications and future considerations. Hum Vaccin Immunother. 2019;15:1111-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 413] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 160. | Nguyen LK, Matallanas D, Croucher DR, von Kriegsheim A, Kholodenko BN. Signalling by protein phosphatases and drug development: a systems-centred view. FEBS J. 2013;280:751-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 161. | Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov. 2007;6:391-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 404] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 162. | Mills BN, Albert GP, Halterman MW. Expression Profiling of the MAP Kinase Phosphatase Family Reveals a Role for DUSP1 in the Glioblastoma Stem Cell Niche. Cancer Microenviron. 2017;10:57-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 163. | Chappell J, Sun Y, Singh A, Dalton S. MYC/MAX control ERK signaling and pluripotency by regulation of dual-specificity phosphatases 2 and 7. Genes Dev. 2013;27:725-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 164. | Tischer T, Schuh M. The Phosphatase Dusp7 Drives Meiotic Resumption and Chromosome Alignment in Mouse Oocytes. Cell Rep. 2016;17:1426-1437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 165. | Spichal M, Fabre E. The Emerging Role of the Cytoskeleton in Chromosome Dynamics. Front Genet. 2017;8:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 166. | Wu L, Liu Y, Zhao Y, Li M, Guo L. Targeting DUSP7 signaling alleviates hepatic steatosis, inflammation and oxidative stress in high fat diet (HFD)-fed mice via suppression of TAK1. Free Radic Biol Med. 2020;153:140-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 167. | Konjikusic MJ, Gray RS, Wallingford JB. The developmental biology of kinesins. Dev Biol. 2021;469:26-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 168. | Kevenaar JT, Bianchi S, van Spronsen M, Olieric N, Lipka J, Frias CP, Mikhaylova M, Harterink M, Keijzer N, Wulf PS, Hilbert M, Kapitein LC, de Graaff E, Ahkmanova A, Steinmetz MO, Hoogenraad CC. Kinesin-Binding Protein Controls Microtubule Dynamics and Cargo Trafficking by Regulating Kinesin Motor Activity. Curr Biol. 2016;26:849-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 169. | Cho SY, Kim S, Kim G, Singh P, Kim DW. Integrative analysis of KIF4A, 9, 18A, and 23 and their clinical significance in low-grade glioma and glioblastoma. Sci Rep. 2019;9:4599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 170. | Taniuchi K, Nakagawa H, Nakamura T, Eguchi H, Ohigashi H, Ishikawa O, Katagiri T, Nakamura Y. Down-regulation of RAB6KIFL/KIF20A, a kinesin involved with membrane trafficking of discs large homologue 5, can attenuate growth of pancreatic cancer cell. Cancer Res. 2005;65:105-112. [PubMed] |
| 171. | Zhao X, Zhou LL, Li X, Ni J, Chen P, Ma R, Wu J, Feng J. Overexpression of KIF20A confers malignant phenotype of lung adenocarcinoma by promoting cell proliferation and inhibiting apoptosis. Cancer Med. 2018;7:4678-4689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 172. | Wang M LK, Zhou XL, Mei SY, Zhang CJ, Zhang TG. Downregulation of KIF20A induces cell cycle arrest and apoptosis by suppressing PI3K/AKT in human glioblastoma. Int J Clin Exp Med. 2017;10:16133-16143. |
| 173. | Groth-Pedersen L, Aits S, Corcelle-Termeau E, Petersen NH, Nylandsted J, Jäättelä M. Identification of cytoskeleton-associated proteins essential for lysosomal stability and survival of human cancer cells. PLoS One. 2012;7:e45381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 174. | Qiu R, Runxiang Q, Geng A, Liu J, Xu CW, Menon MB, Gaestel M, Lu Q. SEPT7 Interacts with KIF20A and Regulates the Proliferative State of Neural Progenitor Cells During Cortical Development. Cereb Cortex. 2020;30:3030-3043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 175. | Qiu R, Wu J, Gudenas B, Northcott PA, Wechsler-Reya RJ, Lu Q. Depletion of kinesin motor KIF20A to target cell fate control suppresses medulloblastoma tumour growth. Commun Biol. 2021;4:552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 176. | Geng A, Qiu R, Murai K, Liu J, Wu X, Zhang H, Farhoodi H, Duong N, Jiang M, Yee JK, Tsark W, Lu Q. KIF20A/MKLP2 regulates the division modes of neural progenitor cells during cortical development. Nat Commun. 2018;9:2707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 177. | Korf BR. Neurofibromatosis. Handb Clin Neurol. 2013;111:333-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 178. | Le C, Bedocs PM. Neurofibromatosis. 2022 Apr 9. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022 Jan-. [PubMed] |
| 179. | Guerrero PA, Yin W, Camacho L, Marchetti D. Oncogenic role of Merlin/NF2 in glioblastoma. Oncogene. 2015;34:2621-2630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 180. | Lau YK, Murray LB, Houshmandi SS, Xu Y, Gutmann DH, Yu Q. Merlin is a potent inhibitor of glioma growth. Cancer Res. 2008;68:5733-5742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 84] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 181. | Reed N, Gutmann DH. Tumorigenesis in neurofibromatosis: new insights and potential therapies. Trends Mol Med. 2001;7:157-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 182. | Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1018] [Cited by in RCA: 1065] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 183. | Cole BK, Curto M, Chan AW, McClatchey AI. Localization to the cortical cytoskeleton is necessary for Nf2/merlin-dependent epidermal growth factor receptor silencing. Mol Cell Biol. 2008;28:1274-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 184. | Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol. 1998;140:885-895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 478] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 185. | Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391-401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 593] [Cited by in RCA: 621] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 186. | Bai Y, Liu YJ, Wang H, Xu Y, Stamenkovic I, Yu Q. Inhibition of the hyaluronan-CD44 interaction by merlin contributes to the tumor-suppressor activity of merlin. Oncogene. 2007;26:836-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 187. | Stamenkovic I, Yu Q. Shedding light on proteolytic cleavage of CD44: the responsible sheddase and functional significance of shedding. J Invest Dermatol. 2009;129:1321-1324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 188. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6844] [Cited by in RCA: 6909] [Article Influence: 287.9] [Reference Citation Analysis (0)] |
| 189. | Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2752] [Cited by in RCA: 2777] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 190. | Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 891] [Cited by in RCA: 904] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 191. | Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol. 2008;26:2806-2812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 284] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 192. | Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70:2455-2464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 193. | Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074-1078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 744] [Cited by in RCA: 811] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 194. | Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1301] [Cited by in RCA: 1218] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 195. | Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 547] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 196. | Larsson J, Ohishi M, Garrison B, Aspling M, Janzen V, Adams GB, Curto M, McClatchey AI, Schipani E, Scadden DT. Nf2/merlin regulates hematopoietic stem cell behavior by altering microenvironmental architecture. Cell Stem Cell. 2008;3:221-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 197. | Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 460] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 198. | Long MD, Campbell MJ. Pan-cancer analyses of the nuclear receptor superfamily. Nucl Receptor Res. 2015;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 199. | Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 831] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 200. | Joseph C, Al-Izzi S, Alsaleem M, Kurozumi S, Toss MS, Arshad M, Goh FQ, Alshankyty IM, Aleskandarany MA, Ali S, Ellis IO, Mongan NP, Green AR, Rakha EA. Retinoid X receptor gamma (RXRG) is an independent prognostic biomarker in ER-positive invasive breast cancer. Br J Cancer. 2019;121:776-785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 201. | Papi A, Tatenhorst L, Terwel D, Hermes M, Kummer MP, Orlandi M, Heneka MT. PPARgamma and RXRgamma ligands act synergistically as potent antineoplastic agents in vitro and in vivo glioma models. J Neurochem. 2009;109:1779-1790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 202. | Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, Guerrero-Cazares H, Quinones-Hinojosa A, Laterra J, Xia S. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30:3454-3467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 203. | Friedman MD, Jeevan DS, Tobias M, Murali R, Jhanwar-Uniyal M. Targeting cancer stem cells in glioblastoma multiforme using mTOR inhibitors and the differentiating agent all-trans retinoic acid. Oncol Rep. 2013;30:1645-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 204. | Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 2000;19:2592-2601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 263] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 205. | Rodriguez V, Bailey R, Larion M, Gilbert MR. Retinoid receptor turnover mediated by sumoylation, ubiquitination and the valosin-containing protein is disrupted in glioblastoma. Sci Rep. 2019;9:16250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 206. | Ye Z, Chen J, Hu X, Yang S, Xuan Z, Lu X, Zhao Q. SPOCK1: a multi-domain proteoglycan at the crossroads of extracellular matrix remodeling and cancer development. Am J Cancer Res. 2020;10:3127-3137. [PubMed] |
| 207. | Murphy-Ullrich JE, Sage EH. Revisiting the matricellular concept. Matrix Biol. 2014;37:1-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 304] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 208. | Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest. 2001;107:1049-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 496] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 209. | Wu T, Ouyang G. Matricellular proteins: multifaceted extracellular regulators in tumor dormancy. Protein Cell. 2014;5:249-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 210. | Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol. 2012;44:480-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 203] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 211. | Rayego-Mateos S, Campillo S, Rodrigues-Diez RR, Tejera-Muñoz A, Marquez-Exposito L, Goldschmeding R, Rodríguez-Puyol D, Calleros L, Ruiz-Ortega M. Interplay between extracellular matrix components and cellular and molecular mechanisms in kidney fibrosis. Clin Sci (Lond). 2021;135:1999-2029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 212. | Schulz WA, Ingenwerth M, Djuidje CE, Hader C, Rahnenführer J, Engers R. Changes in cortical cytoskeletal and extracellular matrix gene expression in prostate cancer are related to oncogenic ERG deregulation. BMC Cancer. 2010;10:505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 213. | Yu F, Li G, Gao J, Sun Y, Liu P, Gao H, Li P, Lei T, Chen Y, Cheng Y, Zhai X, Sayari AJ, Huang H, Mu Q. SPOCK1 is upregulated in recurrent glioblastoma and contributes to metastasis and Temozolomide resistance. Cell Prolif. 2016;49:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 214. | Yang J, Yang Q, Yu J, Li X, Yu S, Zhang X. SPOCK1 promotes the proliferation, migration and invasion of glioma cells through PI3K/AKT and Wnt/β-catenin signaling pathways. Oncol Rep. 2016;35:3566-3576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 215. | Sun LR, Li SY, Guo QS, Zhou W, Zhang HM. SPOCK1 Involvement in Epithelial-to-Mesenchymal Transition: A New Target in Cancer Therapy? Cancer Manag Res. 2020;12:3561-3569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 216. | Micel LN, Tentler JJ, Smith PG, Eckhardt GS. Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J Clin Oncol. 2013;31:1231-1238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 217. | Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 454] [Cited by in RCA: 460] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 218. | Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 219. | Naujokat C, Sarić T. Concise review: role and function of the ubiquitin-proteasome system in mammalian stem and progenitor cells. Stem Cells. 2007;25:2408-2418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 220. | Jung HJ, Byun HO, Jee BA, Min S, Jeoun UW, Lee YK, Seo Y, Woo HG, Yoon G. The Ubiquitin-like with PHD and Ring Finger Domains 1 (UHRF1)/DNA Methyltransferase 1 (DNMT1) Axis Is a Primary Regulator of Cell Senescence. J Biol Chem. 2017;292:3729-3739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 221. | Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, Lachenmayer A, Revill K, Alsinet C, Sachidanandam R, Desai A, SenBanerjee S, Ukomadu C, Llovet JM, Sadler KC. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 259] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 222. | Sidhu H, Capalash N. UHRF1: The key regulator of epigenetics and molecular target for cancer therapeutics. Tumour Biol. 2017;39:1010428317692205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 223. | Reardon ES, Shukla V, Xi S, Gara SK, Liu Y, Straughan D, Zhang M, Hong JA, Payabyab EC, Kumari A, Richards WG, De Rienzo A, Hassan R, Miettinen M, Xi L, Raffeld M, Uechi LT, Li X, Wang R, Chen H, Hoang CD, Bueno R, Schrump DS. UHRF1 Is a Novel Druggable Epigenetic Target in Malignant Pleural Mesothelioma. J Thorac Oncol. 2021;16:89-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 224. | Boukhari A, Alhosin M, Bronner C, Sagini K, Truchot C, Sick E, Schini-Kerth VB, André P, Mély Y, Mousli M, Gies JP. CD47 activation-induced UHRF1 over-expression is associated with silencing of tumor suppressor gene p16INK4A in glioblastoma cells. Anticancer Res. 2015;35:149-157. [PubMed] |
| 225. | Matsushita R, Yoshino H, Enokida H, Goto Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M, Seki N. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget. 2016;7:28460-28487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 226. | Xiang H, Yuan L, Gao X, Alexander PB, Lopez O, Lau C, Ding Y, Chong M, Sun T, Chen R, Liu SQ, Wu H, Wan Y, Randell SH, Li QJ, Wang XF. UHRF1 is required for basal stem cell proliferation in response to airway injury. Cell Discov. 2017;3:17019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 227. | Kim KY, Tanaka Y, Su J, Cakir B, Xiang Y, Patterson B, Ding J, Jung YW, Kim JH, Hysolli E, Lee H, Dajani R, Kim J, Zhong M, Lee JH, Skalnik D, Lim JM, Sullivan GJ, Wang J, Park IH. Uhrf1 regulates active transcriptional marks at bivalent domains in pluripotent stem cells through Setd1a. Nat Commun. 2018;9:2583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 228. | Alhosin M, Omran Z, Zamzami MA, Al-Malki AL, Choudhry H, Mousli M, Bronner C. Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J Exp Clin Cancer Res. 2016;35:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 229. | Ramesh V, Bayam E, Cernilogar FM, Bonapace IM, Schulze M, Riemenschneider MJ, Schotta G, Götz M. Loss of Uhrf1 in neural stem cells leads to activation of retroviral elements and delayed neurodegeneration. Genes Dev. 2016;30:2199-2212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 230. | Valor LM, Hervás-Corpión I. The Epigenetics of Glioma Stem Cells: A Brief Overview. Front Oncol. 2020;10:602378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 231. | Sasmita AO, Wong YP, Ling APK. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol. 2018;14:40-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 232. | Bryukhovetskiy I. Cell-based immunotherapy of glioblastoma multiforme. Oncol Lett. 2022;23:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 233. | Essaghir A, Demoulin JB. A minimal connected network of transcription factors regulated in human tumors and its application to the quest for universal cancer biomarkers. PLoS One. 2012;7:e39666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 234. | Al-Fatlawi A, Afrin N, Ozen C, Malekian N, Schroeder M. NetRank Recovers Known Cancer Hallmark Genes as Universal Biomarker Signature for Cancer Outcome Prediction. Front Bioinform. 2022;2:780229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 235. | Nowicki MO, Hayes JL, Chiocca EA, Lawler SE. Proteomic Analysis Implicates Vimentin in Glioblastoma Cell Migration. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 236. | Zhao J, Zhang L, Dong X, Liu L, Huo L, Chen H. High Expression of Vimentin is Associated With Progression and a Poor Outcome in Glioblastoma. Appl Immunohistochem Mol Morphol. 2018;26:337-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 237. | Okada M, Suzuki S, Togashi K, Sugai A, Yamamoto M, Kitanaka C. Targeting Folate Metabolism Is Selectively Cytotoxic to Glioma Stem Cells and Effectively Cooperates with Differentiation Therapy to Eliminate Tumor-Initiating Cells in Glioma Xenografts. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 238. | Zhu Z, Du S, Du Y, Ren J, Ying G, Yan Z. Glutathione reductase mediates drug resistance in glioblastoma cells by regulating redox homeostasis. J Neurochem. 2018;144:93-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 239. | Barry ER, Simov V, Valtingojer I, Venier O. Recent Therapeutic Approaches to Modulate the Hippo Pathway in Oncology and Regenerative Medicine. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |