Published online Apr 26, 2023. doi: 10.4252/wjsc.v15.i4.235
Peer-review started: December 24, 2022
First decision: January 31, 2023
Revised: February 12, 2023
Accepted: March 20, 2023
Article in press: March 20, 2023
Published online: April 26, 2023
Processing time: 122 Days and 20.1 Hours
Different fates of neural stem/progenitor cells (NSPCs) and their progeny are determined by the gene regulatory network, where a chromatin-remodeling complex affects synergy with other regulators. Here, we review recent research progress indicating that the BRG1/BRM-associated factor (BAF) complex plays an important role in NSPCs during neural development and neural developmental disorders. Several studies based on animal models have shown that mutations in the BAF complex may cause abnormal neural differentiation, which can also lead to various diseases in humans. We discussed BAF complex subunits and their main characteristics in NSPCs. With advances in studies of human pluripotent stem cells and the feasibility of driving their differentiation into NSPCs, we can now investigate the role of the BAF complex in regulating the balance between self-renewal and differentiation of NSPCs. Considering recent progress in these research areas, we suggest that three approaches should be used in investigations in the near future. Sequencing of whole human exome and genome-wide asso
Core Tip: There are several reviews in the literature contributed to the role of BRG1/BRM-associated factor (BAF) complex in neural cell specification and neural development diseases. We review recent progress indicating that BAF complex plays an important role in neural stem/progenitor cells (NSPCs) during neural development and neural developmental disorders. More progresses in the role of BAF complex subunits in balancing self-renewal and differentiation of NSPCs and neurodevelopment could finally be involved in highlighting new methods for clinical application.
- Citation: Ke NY, Zhao TY, Wang WR, Qian YT, Liu C. Role of brahma-related gene 1/brahma-associated factor subunits in neural stem/progenitor cells and related neural developmental disorders. World J Stem Cells 2023; 15(4): 235-247
- URL: https://www.wjgnet.com/1948-0210/full/v15/i4/235.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i4.235
Neural stem/progenitor cells (NSPCs) are self-renewing neural cells capable of differentiating into neurons, astrocytes and/or oligodendrocytes[1-3]. During development, NSPCs are critical for the establishment of the central nervous system (CNS)[4-6]. In the adult central neural system, neurogenesis plays a key role in fundamental processes, for example, memory, learning, the maintenance of normal tissue homeostasis, and the autonomous repair of pathological brain tissues[7,8]. NSPCs can be isolated from three canonical neurogenic niches in the spinal cord and brain, namely, the central canal (CC) in the spinal cord, the subgranular zone (SGZ) of the dentate gyrus (DG) and the subventricular zone (SVZ) of the lateral ventricle (LV) in the brain[9-13]. In addition, NSPCs have been identified in the developing cerebral cortex[14], olfactory epithelium[15] and outside the CC[16]. The fact that NSPCs can be isolated and propagated in vitro opens up new opportunities for medical research, and we hope they can be used to compensate for cell loss that features in several serious neurological disorders. Moreover, NSPCs can be created in vitro through induced differentiation from induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs), expanding the pool of models for studying stem cells in health and illness[17,18]. For cell-based therapy approaches targeting the brain and spinal cord, NSPCs have emerged as focal points. However, the small amount of NSPCs present in this tissue has restricted the clinical uses of these cells. Although recent advancements in ESCs and iPSC research have meant that these cells can be novel sources of NSPCs, understanding NSC molecular regulation and NSPC applications, which are important for disease modeling and regenerative medicine, remain challenges[19-21].
The BRG1/BRM-associated factor (BAF) complex, which is a mammalian chromatin remodeler, regulates chromatin structure and transcription by providing subunit organization and nucleosome recognition in an ATP-dependent manner. Structurally, the BAF complex contains three modules, an ATPase, an actin-related protein (ARP) and a base module[22]. More than 30 major subunits of these modules can combine to form the complex[22-24].
By controlling chromatin structure, controlling the differentiation of NSPCs to produce different types of brain cells, and regulating transdifferentiation between cell types, BAF complexes perform crucial roles in the maintenance of a gene expression program. At various phases of brain formation, BAF complexes have specific roles and seem to be produced by the combinatorial assembly of their subunits. Moreover, intimate interactions between the BAF chromatin-remodeling complex and transcriptional machinery control the development of NSPCs[25]. With the combinatorial assembly of homologous component families, which enables their nonredundant functions, distinct BAF complex structures are made possible. ESCs, NSPCs, and postmitotic neurons all exhibit developmental stage-specific BAF assemblages during mammalian brain development. Neuronal BAF (nBAF) function is required for the development of mature postmitotic neuronal features along with long-term memory, and neural progenitor-special BAF (npBAF) structures are particularly important for modulating the rates and modalities of neural progenitor cell replication. Given the high prevalence of BAF subunit mutagenesis in neurological illnesses, it is obvious that BAF complexes perform a crucial role in controlling the rate of neuronal development, homeostasis, and plasticity. Understanding how BAF complex subunits influence BAF complex function and the roles played by different subunits will reveal disease pathogenesis and lead to novel treatments for related human disorders ultimately[26,27].
Here, we summarize recent progress in comprehending the role of the BAF complex in NSPCs. We focus on NSC-/NPC-specific BAF complex subunits and describe how they determine the balance between the self-renewal and differentiation of NSCs and are involved in several human neural developmental disorders. Supplementary Table 1 shows the abbreviations and their descriptions.
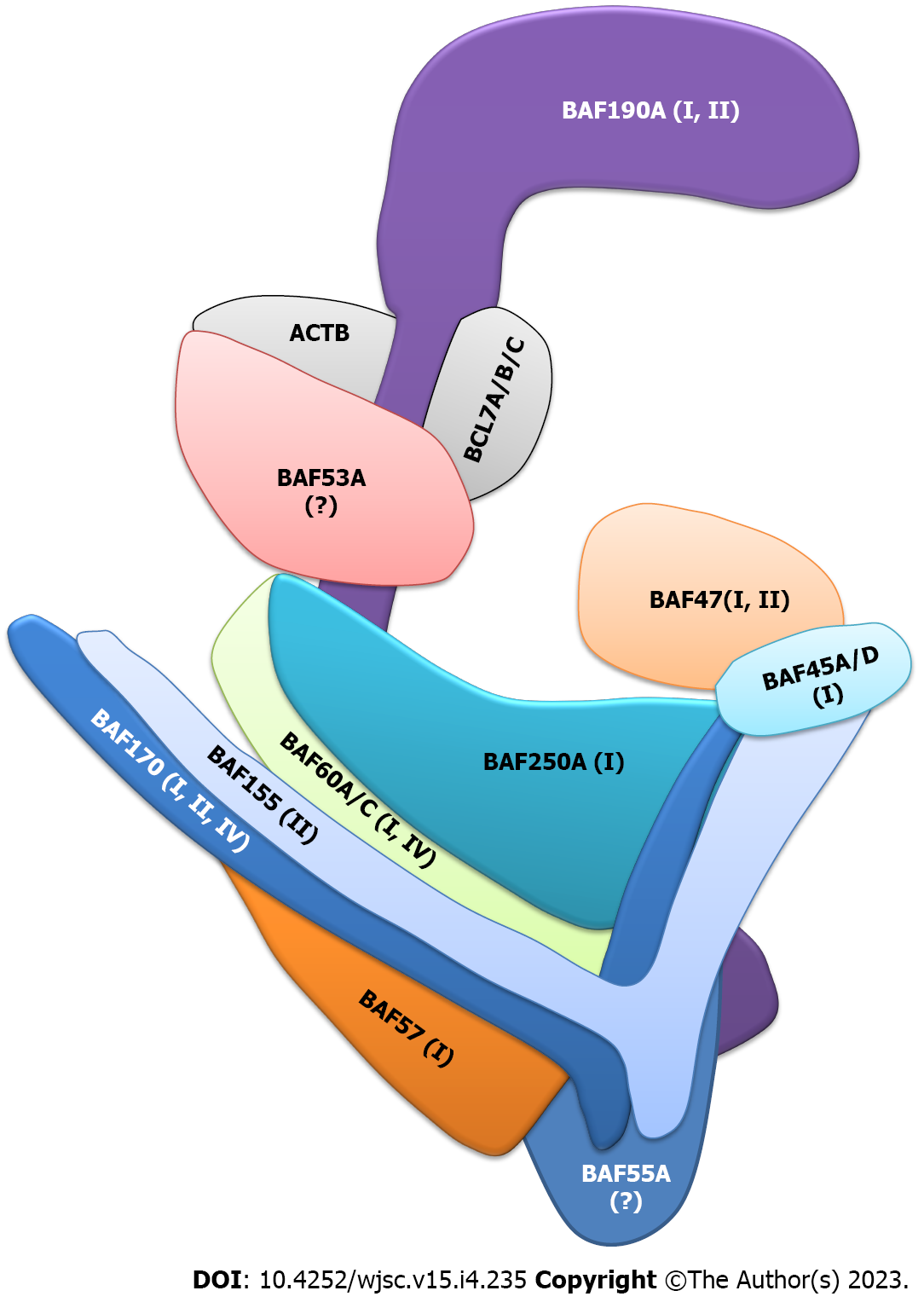
Among the major BAF complex subunits, there are 12 subunits expressed in NSPCs in the embryo cortex, olfactory bulb (OB) and spinal cord, adult brain and spinal cord, and human and mouse retina (detailed information and references are shown in Table 1).
| BAF complex subunits (aliases) | NSPC types | Function | Interaction signalings/factors | Related neural developmental diseases | Ref. |
| BAF45A (PHF10) | oNSC, E10.5-E11.5 cortical NPCs, E10.5-E16.5 spinal cord VZ | NSPC proliferation | BRG1 | - | Bachmann et al[15], 2016; MacDonald et al[89], 2022; Lessard et al[41], 2007 |
| BAF45D (DPF2) | SGZ, SVZ, spinal cord central canal GFAP + radial glial cells | PAX6 expression, contributes to NSPC induction | PAX6, BRG1 | CSS | Liu et al[42], 2017; Wang et al[44], 2019; Chen et al[43], 2022; Vasileiou et al[69], 2018; Knapp et al[70], 2019; Milone et al[71], 2020 |
| BAF47 (SMARCB1) | oNSC | - | BRG1 | CSS, kleefstra syndrome, Nicolaides-Baraitser syndrome | Bachmann et al[15], 2016; Tsurusaki et al[72], 2012; Santen et al[74], 2013; Wieczorek et al[75], 2013; Kleefstra et al[88], 2012; Gossai et al[73], 2015 |
| BAF53A (ACTL6A) | oNSC, NPCS in the neural tubes of E11.5 embryos | BAF53A is essential for NPC proliferation | miR-9 and miR-124 | - | Bachmann et al[15], 2016; Lessard et al[41], 2007; Yoo et al[45], 2009 |
| BAF55A (SS18) | NPCs in VZ | SS18 is required for NSC self-renewal | BRG1 | - | Staahl et al[48], 2013 |
| BAF57 (SMARCE1) | Adult human OBNSCs | NSC proliferation | Neuregulin-1 | CSS | Wieczorek et al[75], 2013; Kosho et al[67], 2014; Marei et al[52], 2012; Zarate et al[76], 2016; Pirotte et al[63], 2010 |
| BAF60A (SMARCD1) | Oligodendrocyte precursors | - | BRG1, BAF155 and BAF170 | CSS, Nicolaides-Baraitser syndrome | (Yu et al[54], 2013; Hsiao et al[53], 2003; Machol et al[78], 2019; Nixon et al[77], 2019 |
| BAF60C (SMARCD3) | NPCs in human retinas as well as mouse retina, cortex and spinal cord | Keeps the progenitors in a proliferative state - | Notch signaling | - | Lamba et al[55], 2008 |
| BAF155 (SMARCC1) | oNSC, adult human OBNSCs | Proliferation and maintenance of ONSCs forebrain development | BRG1, PAX6 | ASD | Lessard et al[41], 2007; Marei et al[52], 2012; Narayanan et al[33], 2015; Neale et al[84], 2012 |
| BAF170 (SMARCC2) | oNSC, in postnatal DG, in RGL progenitors | oNSC proliferation, affects NSC proliferation, differentiation, forebrain development | BRG1 | CSS, ASD, Nicolaides-Baraitser syndrome | Lessard et al[41], 2007; Machol et al[78], 2019; Narayanan et al[33], 2015; Neale et al[84], 2012; Tuoc et al[38], 2017 |
| BAF190A (BRG1/SMARCA4) | Mouse VZ, SGZ, SEZ, NSCs/NPCs, NCC | NSC maintenance and neuronal differentiation | PAX6, BAF45D | CSS, ASD | Tsurusaki et al[72], 2012; De Rubeis et al[85], 2014; Matsumoto et al[28], 2006; Petrik et al[29], 2015; Ninkovic et al[30], 2013 |
| BAF250A (ARID1A) | Developing cortex | Regulates NSPC proliferation and differentiation during cortical development | BRG1 | CSS | Tsurusaki et al[72], 2012; Liu et al[40], 2021 |
Brg1 (Brahma-related gene 1) is necessary for NSPCs to remain in a state where they can react to gliogenic signals and to suppress neuronal commitment. Lack of Brg1 in NSPCs led to precocious neurogenesis, such that cells in the ventricular zone developed into postmitotic neurons before the start of gliogenesis in conditional brg1- mutant mice. As a result, these animals significantly lost the ability to differentiate astrocytes and oligodendrocytes. In addition, in vitro Brg1 deletion reduced growth factor-induced astrocyte development in gliogenic progenitors. Furthermore, it has been discovered that the levels of proteins associated with stem cell maintenance, such as Pax6, Sox1, and Musashi-1, are significantly lower in the ventricular zones of the mice with brg1-mutations[28]. The hippocampus experienced abnormal adult neurogenesis as a result of Brg1 deletion in nestin-expressing NSPCs. This abnormal adult neurogenesis initially decreased the number of adult NSPCs in the hippocampus, inhibited progenitor maintenance, and later decreased NSPC responsiveness to physiological stimulation. Brg1 deletion appears to hinder cell cycle progression mechanistically, which is partly because of increased p53 pathway activation and p21 expression. Defects in neurosphere formation brought on by Brg1 deletion were repaired by knocking down p53. These findings suggest that aNSPC and progenitor cell maintenance and responsiveness during neurogenesis are determined by epigenetic chromatin remodeling via a Brg1- and p53/p21-dependent mechanism[29].
Many transcription factors manipulating neuronogenesis have been ascertained in the adult and developing brain, and notably, the neurogenic transcriptional regulation factor Pax6, which can directly interact with the Brg1-carrying BAF complex in adult neural progenitor cells. Abolition of either Pax6 or Brg1 in the subependymal zone (SEZ) caused the offspring of adult NSPCs to switch to the SEZ ependymal cell lineage. In the interim, shifting neuroblasts changed to distinct glial cell lineages during or after reaching the OB. Several studies have revealed that there is a network with tripartite effector programing neuronal fate, which can be activated by Pax6-BAF and promote neurogenesis and the transformation from glia to neurons. According to the whole-genome analysis, the downstream regulatory factors of Pax6-BAF include Nfib, Sox11 and Pou3f4. While Brg1 is absent in SEZ and OB, the binding sites of Pax6 carried by Brg1 and most down-regulated genes will be down-regulated at the same time[30]. In the study of Jayaprakash et al[31], they find that Brg1 appears as a slender spherical structure so that it can provide a lager surface and form the further complex by binding to BAF57 and BAF60A. Furthermore, a neural Brg1 isoform interacts with the central neurodevelopmental transcriptional repressor REST/NRSF, suggesting that Brg1 can interact with transcription regulators and factors to conduct the BAF complex to specific sites[31].
Recent study found that conditional deletion of BAF155 can lead to the decrease of basal intermediate progenitors (bIPs), while the delamination of apical radial glial progenitors (RGs) can cause the increase of basal RGs (bRGs). During the progress, BAF155 has been proved necessary for the normal activity of PAX6 to regulate the gene expression. BAF155 can control the expression of the CDC42 effecting protein CEP4 in the Pax6-dependent way in order to regulate the stratification of progenitor. In addition, BAF155-dependent chromatin remodeling is also involved in regulating several human RG-specific genes, thereby acting on the generation of basal progenitors (BPs)[32].
The study of Narayanan et al[33] in 2015 revealed a molecular mechanism of controlling the global chromatin state and transcriptional program during development, which is mediated by the BAF complex. They found that BAF155 and BAF170 knockout can cause elimination of the entire BAF complex with all kinds of BAF subunits in cortical development, with the overall increase of the abundance of the repressive marks (H3K27me2/3), decrease of active chromatin mark H3K9Ac, and down-regulation of gene expression. Both BAF155 and BAF170 subunits are expressed in early cortical progenitors (E10.5-E14.5), and BAF170 is replaced by BAF155 in late cortical progenitors[34]. Specifically, between E12.5 and E14.5, apical progenitors express BAF170 and BAF155. They exhibit comparable expression patterns at the height of upper layer neurogenesis (E15.5), with BAF170 being missing from VZ progenitors but expressed in the cortical plate but expressed in the cortical plate[34]. While BAF170 overexpression resulted in the acquisition of a feature similar to that acquired by the Pax6-loss brain, with depletion of overlying layer neurons, BAF170 loss in Emx1t cortical progenitors exerted an effect opposite to that of Pax6 elimination and caused an aberrant growth of IPs and a bias toward the acquisition of an overlying layer identity[34]. By the recruitment of the transcriptional suppressor REST to Pax6 targets, the BAF170-containing npBAF complex indirectly suppresses neurogenesis in its early stages[34,35]. Interestingly, the transcription of BAF170 and Brm subunits, which replace BAF155 and Brg1 subunits in the esBAF, respectively, begins at the same time when ES cells commit to developing into neural progenitors[33,36]. According to a recent functional investigation, Pax6 is directly associated to the BAF complex in aNSCs that contained the Brg1, and its neurogenic activity was abolished in the lack of the Brg1[30]. BAF170 and BAF45 also carry domains with putative DNA-binding capacities[37]. In the neonatal DG, the BAF170 member of the complex is found in radial glial-like (RGL) cells and cell types implicated in following stages of adult neurogenesis together with mature astrocytes. The pool of RGL cells in the DG is reduced by conditional knockout of BAF170 during late cortical neurogenesis and also adult brain neurogenesis, which encourages terminal astrocyte differentiation. A moderate loss in spatial learning was produced by the induced abolition of BAF170 in the DG throughout adult brain neurogenesis, with the reversal test showing the greatest impact on spatial learning. These findings indicate a specialized role for adult neurogenesis in the DG regulating adaptive behavior and show the engagement of BAF170-reliant chromatin modification in neurogenesis and cognition of hippocampus[38].
The core ATPase, Brg1 or Brm, BAF250A or BAF250B, a homologous dimer of BAF155, or a heterologous dimer of BAF170 and BAF155 are just a few examples of the different subunits that npBAF can contain[27]. Mammal BAF is significantly different from its yeast analogue, losing some members while obtaining new core members, such as BAF45a/b/c/d, BAF57, BAF250A/B, SS18/CREST, beta-actin, BRD7, and BRD9. Notably, BAF250A/B are the most commonly mutated subunits of BAF complex in related human neurological diseases[36]. Loss of BAF250A or BAF250B in mouse models leads to early embryonic lethality[39], indicating that, despite their homology, both genes are essential for early development.
Recent genome sequencing and many clinical studies have revealed that mutations of BAF250A is closely related to microcephaly and mental retardation. Liu et al[40] generated a baf250a conditional knockout mouse line and found that cortical thickness was reduced in the developing cortex. Radial glial cell proliferation is inhibited by loss of BAF250A function, which also increases the rate of cell damage during late cortical neurogenesis and causes abnormal expression of genes involved in proliferation and differentiation. Therefore, for the purpose of better understanding the pathogenic mechanisms and creating new treatments for neurodevelopmental disorders brought on by its mutations, BAF250A may be one of the gene possibilities that is worthwhile being researched[40].
BAF45 family proteins, including BAF45A, BAF45B, BAF45C and BAF45D, are subunits of the BAF complex[41]. BAF45A, an npBAF, is required for the self-renewal and proliferative features of neural cells[41]. BAF45A is identified in olfactory neural stem cells (oNSCs), E10.5-E11.5 cortical NPCs, and E10.5-E16.5 spinal cord ventricular zone NSCs[15,41]. BAF45D is expressed in the SGZ of the LV, SGZ of the DG, and the CC of the adult spinal cord. Co-expression of BAF45D and glial fibrillary acidic protein (GFAP), a marker protein of radial glial cell-like cell was found in the SGZ, SVZ, and CC of the adult spinal cord. According to the results of quantitative examination, BAF45D is selectively expressed in the adult neurogenic regions of neurons in CNS. What’s more, BAF45D is necessary for the induction of PAX6, a determinant for neuroectoderm that regulates adult neural stem/progenitor cell self-renewal and specific neural fate, during neuroectodermal differentiation of H9 cells[42].
Recently, BAF45D has been shown to regulate spinal cord neural stem/progenitor cell (SCNSC/SCNPC) fate mediated through the SMAD-PAX6 axis. SCNSCs exhibit increased DNA-BAF45D compared to that in human ESCs[43]. It has been reported that postmitotic neurons express BAF45B/C protein but lack the paralogous BAF45A/D, which confers neuronal properties[25-27,41]. The protein expression of BAF45D in adult rat NSPCs and neurons but not astrocytes may imply a role for BAF45D in the differentiation of NSPCs[44].
The BAF53 subunit is encoded by the BAF53A (ACTL6A) or BAF53B (ACTL6B) gene, with the latter exclusively expressed in differentiated neurons[41].
BAF53A is specific to the npBAF complex and can be replaced by BAF53B in the nBAF complex in neurons[26,45]. miR-9 and miR-124 control the transition of postmitotic neurons from the BAF complex’s BAF53A to BAF53B subunit[46]. These miRNAs’ expression is derepressed in neurons during cell development, which encourages the replacement of BAF53B in the nBAF complex by suppressing the expression of BAF53A. Increased progenitor proliferation can result from BAF53A’s continued expression[46]. These miRNAs have an interacting location in the 3′UTR of BAF53A, and their expression is constrained in progenitors by REST and its corepressors[47]. As a result of decreased chromatin accessibility at particular neural transcription factor-binding sites brought on by BAF53A loss, cell cycle-related genes are repressed, preventing the advancement of the cell cycle and cell differentiation[48].
BAF57, subunit in the BAF complex core, is highly conserved in the BAF complexes of vertebrates[49]. The DNA-binding capabilities of its primary structural characteristic, a high-mobility group domain, imply that BAF57 may perform topological functions when the BAF complex moves in or out a nucleosome. BAF57 specifically interacts with a variety of proteins which aren’t part of the BAF complex. BAF57 demonstrates particular functionalities as a result of these interactions. For example, in the embryo, it interacts with the transcriptional cosuppressor Co-REST to enable the neuronal genes inactivation in nonneuronal cells throughout development[50]. Interestingly, global gene expression profiling results indicated that BAF57A was specifically upregulated in human NSPCs of OB (OBNSPCs) but not in human embryonic NSPCs (hENSC)[51]. In cultured NSCs, BAF57 interacts with the overexpressed intracellular domain of neuregulin-1 and leads to a decrease in the NSC proliferation rate[52].
The BAF60A/B subunits define an ESC-specific assembly of BAF, named esBAF[27]. The BAF subunits BAF60A binds to BRG1, the primary factor that drives the BAF complex to particular genomic locations, and interacts with transcription regulators and factors[31]. Nuclear receptors and the BRG1 complex may have an additional important and direct relationship that is necessary for recruitment of promoter and subsequent chromatin modification[53]. Low levels of BAF60A and BAF45B/D expression are found in oligodendrocyte precursors, and these expression levels are upregulated upon differentiation[54]. However, their specific roles in NSPCs are still unclear.
During development, another crucial BAF complex component called BAF60C also has neuronal progenitor-specific actions. BAF60C is expressed in neural progenitors in the mouse and human retina, as well as the human cortex and spinal cord[55]. BAF60C expression decreases during neural differentiation, and through its interaction with the Notch pathway, its overexpression keeps progenitors in a proliferative condition. Finally, Muller glia that resume the cell cycle following neurotoxic damage express BAF60C once more[55].
BAF55A, also called SS18, is a member of the npBAF complex. In NSCs, SS18 knockdown results in cell cycle exit and self-renewal failure[48]. Moreover, SS18 is adaptively activated to respond to ethanol exposure, protecting fetal NSCs against complete loss of miR-9-2[49].
BAF47, a ubiquitous BAF complex subunit, is expressed in oNSCS[15]. However, the exact function of BAF47 remains largely unknown.
The number of neural stem cells expands through symmetrical division, and through asymmetrical divisions, they maintain the capability to self-renew and produce other differentiated cells[56]. Normal neural development requires the maintenance of the dynamic balance between the proliferative cells and differentiated cells of neural stem cells, which controls the number of stem cells and neurons and protects the body from diseases such as glioblastoma[57]. Furthermore, neural stem cells respond to several molecular signals during neurogenesis, especially those mediated by the Wnt and Notch signaling pathways[58]. For instance, Notch signaling is a significant mechanism regulating the balance between the quiescence and differentiation of neural stem cells[59]. By suppressing the Notch pathway, the homeostasis of neural stem cells can be altered, promoting neural differentiation[60].
Several BAF family members, which act as components or modulators of the Notch and Wnt/β-catenin signaling pathways, have been shown to exert either positive or negative effects on the differentiation of neural stem cells[55,61]. On the one hand, neural differentiation can be promoted by BAF155 and BAF170, which act on regulators of Wnt/β-catenin signaling-related genes, including H3K27me3 and H3K4me2[15,62]. On the other hand, BAF45a and BAF53a contribute to the proliferation and self-renewal of neural stem cells[41]. The repression of BAF53a during neural differentiation is realized through its 3’-UTR and the cooperative influence of miR-9 and miR-124[45]. Brm and BAF57 interact with the neuregulin-1 (Nrg-1) intracellular domain, reduce proliferation and promote differentiation of neural stem cells in vitro, possibly by reversing the direction of signaling in a pathway[63]. Furthermore, it has been shown that the overexpression of BAF60c maintains the proliferation of neural progenitors by interacting with the Notch signaling pathway[55]. The BAF complex may also make direct contact with the neurogenic transcription factors that are critical for neural stem cell specification or differentiation. Notably, the subunit composition of BAF complexes undergoes a profound switching during differentiation; for instance, BAF40c is replaced by BAF40a, the function of which is activated by Pax6, Tbr1, and Tbr2[64]. Moreover, BAF155 in the primate brain has been shown to be involved in the normal activity of Pax6, thereby regulating the specification of progenitors during cortex development[32].
Overall, BAF complexes function as both agonists and inhibitors of neural differentiation. Notably, certain similarities and differences have been revealed through the approaches used in the aforementioned studies. A study by Narayanan et al[32] was carried out by generating cortex-specific BAF155-knockout mice, while in another study, Nguyen et al[62] adopted a transgenic mouse model in which both BAF155 and BAF170 were knocked out, eliminating the entire BAF complex during late cortical neurogenesis. Their double-knockout model offered a novel and practical method for examining the function of complete BAF complexes during cerebral development[65]. Furthermore, a study by Bi-Lin et al[66] was conducted after the neural crest-specific knockout of BAF155 and BAF170 in Wnt1Cre/+ and Pax3Cre/+ mice, and the results demonstrated that the BAF complex modulated the gene expression network and pathways, including the Notch signaling pathway, critical for neural crest development.
Given the importance of BAF complexes in creating and sustaining a chromatin state that keeps proper transcriptional output during NSPC proliferation, differentiation and responses to exogenous or endogenous stimuli, it is clear that several neural developmental disorders have been linked to NSPC-related BAF complex subunits (Table 1 and Figure 1).
Coffin-Siris syndrome (CSS) is a unusual congenital neural developmental disorder characterized clinically by intellectual disability (ID), progressive facial thickening, hirsutism, recurrent infections, difficult feeding, and restricted growth of the distal fifth phalanx and nails[67,68]. Genes encoding NSPC-related BAF complex subunits have been found to have mutations recently thanks to extensive human exome sequencing and genome-wide association research, including BAF45D[69-71], BAF47[72-74], BAF57[67,75,76], BAF60A[77], BAF170[78], BAF190A/BRG1[72], and BAF250A[72]. Different brain midline abnormalities, similar to those seen in people with CSS, were shown to be caused specifically by BAF47, BAF57, and BAF250B mutations in mice with a heterozygous neural system-specific SMARCB1 defect and a partial dysfunctional mutation in a BAF core member gene, indicating significant clinical suggestions for BAF complex-related ID/neurodevelopmental diseases[79].
Within a cohort of 15 unrelated individuals on the CSS pathway disorders registry, who were carriers of a SMARCA4 variant, they showed differences in the severity and number of learning disabilities and health problems. Two of them with novel nonsense variants appeared to acquire a non-organ/system affected phenotype with minor learning/behavioral differences[80]. A recent report described a Chinese woman presenting with on the CSS pathway disorders registry, who were carriers of a SMARCA4 variant, they showed differences in the severity and number of learning disabilities and health problems. Two of them with novel nonsense variants appeared to acquire a non-organ/system affected phenotype with minor learning/behavioral differences and endocrine dysfunction. Whole-exome sequencing was used and led to the identification of a heterozygous missense variant in the SMARCC2 gene of the proband[81].
The clinical features of patients with nicolaides-Baraitser syndrome (NCBRS) caused by mutations in the SMARCA2 gene, remarkably resemble those of CSS patients[82]. The characteristics of hands and feet are where NCBRS and CSS most clearly diverge. While typical CSS patients have hypoplasia or agenesis of the fifth fingernail with or without engagement of the nail phalanges, typical NCBRS patients have pronounced distal phalanges and interphalangeal joints[75]. Five individuals with mutations in BAF60A exhibited developmental delay, small hands and feet, hypotonia, ID, and difficult feeding. According to TRIO exome sequencing, these mutations developed in four of the five patients[77]. A missense mutation (p. Arg366Cys) of BAF47 also has been reported to be associated with NCBRS[75]. Furthermore, variants in BAF170 have also been found in patients presenting with overlapping ID syndromes related to other BAF subunits, such as those identified in CSS and NCBRS[78].
Autism is a neurodevelopmental disease which is caused by both environmental and genetic factors, presented as speech problems, limited and repetitive behaviors and social skills barrier[83]. Recent exome-sequencing studies on individuals with autism have found mutations in the BAF155, BAF170[84] and BAF190A[85] genes. But at least seventy percent of the cases, the fundamental genetic cause is still unknown.
Kleefstra syndrome spectrum (KSS) is a recognizable syndrome characterized by ID and induced by a hybrid mutation in the euchromatin histone methyltransferase 1 (EHMT1) gene[86,87]. According to the research of Kleefstra et al[88], KSS could result from mutations in any complex protein including MLL3, MBD5, NR1I3, BAF47, or EHMT1. The EHMT1 mutation is not present in every patient with the primary symptoms of Kleefstra syndrome. In fact, de novo mutations in four genes that code for epigenetic regulators, including BAF47 (SMARCB1), were found in four of the nine individuals who did not have EHMT1 mutations[88]. And their research using a Drosophila model showed that MLL3, MBD5, NR1I3, BAF47, and EHMT1 directly interact with one another[88].
Considering recent research progress, we suggest that three aspects of BAF complexes should be investigated in the near future. First, BAF complexes are crucial for development of neural system and differentiation of neural cells, including the specification of neural fate and functionality[4]. BAF complex subunits play key roles in the manipulation of gene expression, and the distinct ontogenetic stage-specific BAF complex functions in neural stem/progenitors and postmitotic neuronal cells that are derived from the combinatorial organization of these subunits[26]. Sequencing of human whole-exome and genome-wide association studies have recently demonstrated the link between mutants of BAF complex subunits and neurodevelopmental illnesses like CSS, NCBS, KSS and ASD[89-92]. Focus must be placed on the roles played by BAF complex organization during neural development as well as the ways in which mutations in well-known BAF complex subunits contribute to certain neurodevelopmental illnesses[93]. Secondly, both loss- and gain-of-function techniques to evaluating neural network development at the molecular, single-cell, and network activity levels should be used in order to comprehend how BAF complex gene mutations might cause the phenotypic convergence of CSS, NCBS, KSS, and ASD. For example, ASD and KSS genes focus primarily on the level of neural network communication and provide insight into the pathophysiology of phenotypic consistency disorders[79,94]. Thirdly, recent investigations have shown that BAF complexes are frequently involved in human cerebral development abnormalities, providing fresh mechanisms and corresponding paths for therapeutic intervention, all of which should be looked into. In recent years, small compounds that target the BAF complex subunits have become viable therapeutic agents. Researchers in the field have been motivated to recognize and take use of the whole spectrum of therapeutic options as a result of the creation and evaluation of novel drugs that target a variety of BAF complex subunits[95].
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Neurosciences
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cardile V, Italy; Xuei X, United States S-Editor: Chen YL L-Editor: A P-Editor: Chen YL
| 1. | Radecki DZ, Samanta J. Endogenous Neural Stem Cell Mediated Oligodendrogenesis in the Adult Mammalian Brain. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 2. | Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 415] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 3. | Ladran I, Tran N, Topol A, Brennand KJ. Neural stem and progenitor cells in health and disease. Wiley Interdiscip Rev Syst Biol Med. 2013;5:701-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | D’Souza L, Channakkar AS, Muralidharan B. Chromatin remodelling complexes in cerebral cortex development and neurodevelopmental disorders. Neurochem Int. 2021;147:105055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 5. | Bergström T, Forsberg-Nilsson K. Neural stem cells: brain building blocks and beyond. Ups J Med Sci. 2012;117:132-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Andreotti JP, Silva WN, Costa AC, Picoli CC, Bitencourt FCO, Coimbra-Campos LMC, Resende RR, Magno LAV, Romano-Silva MA, Mintz A, Birbrair A. Neural stem cell niche heterogeneity. Semin Cell Dev Biol. 2019;95:42-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 7. | Obernier K, Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 368] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 8. | Finkel Z, Esteban F, Rodriguez B, Fu T, Ai X, Cai L. Diversity of Adult Neural Stem and Progenitor Cells in Physiology and Disease. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Schiro LE, Bauer US, Sandvig A, Sandvig I. Isolation and comparison of neural stem cells from the adult rat brain and spinal cord canonical neurogenic niches. STAR Protoc. 2022;3:101426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 10. | Hugnot JP. Isolate and Culture Neural Stem Cells from the Mouse Adult Spinal Cord. Methods Mol Biol. 2022;2389:45-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 11. | Suk Y, Kieliszek A, Mobilio D, Venugopal C, Singh SK. Derivation and culturing of neural stem cells from human embryonic brain tissue. STAR Protoc. 2022;3:101628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 12. | Chicheportiche A, Ruat M, Boussin FD, Daynac M. Isolation of Neural Stem and Progenitor Cells from the Adult Brain and Live Imaging of Their Cell Cycle with the FUCCI System. Methods Mol Biol. 2018;1686:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Stenudd M, Sabelström H, Llorens-Bobadilla E, Zamboni M, Blom H, Brismar H, Zhang S, Basak O, Clevers H, Göritz C, Barnabé-Heider F, Frisén J. Identification of a discrete subpopulation of spinal cord ependymal cells with neural stem cell properties. Cell Rep. 2022;38:110440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Katada S, Takouda J, Nakagawa T, Honda M, Igarashi K, Imamura T, Ohkawa Y, Sato S, Kurumizaka H, Nakashima K. Neural stem/precursor cells dynamically change their epigenetic landscape to differentially respond to BMP signaling for fate switching during brain development. Genes Dev. 2021;35:1431-1444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 15. | Bachmann C, Nguyen H, Rosenbusch J, Pham L, Rabe T, Patwa M, Sokpor G, Seong RH, Ashery-Padan R, Mansouri A, Stoykova A, Staiger JF, Tuoc T. mSWI/SNF (BAF) Complexes Are Indispensable for the Neurogenesis and Development of Embryonic Olfactory Epithelium. PLoS Genet. 2016;12:e1006274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 16. | Shu M, Xue X, Nie H, Wu X, Sun M, Qiao L, Li X, Xu B, Xiao Z, Zhao Y, Fan Y, Chen B, Zhang J, Shi Y, Yang Y, Lu F, Dai J. Single-cell RNA sequencing reveals Nestin(+) active neural stem cells outside the central canal after spinal cord injury. Sci China Life Sci. 2022;65:295-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Yao J, Mu Y, Gage FH. Neural stem cells: mechanisms and modeling. Protein Cell. 2012;3:251-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Kumamaru H, Kadoya K, Adler AF, Takashima Y, Graham L, Coppola G, Tuszynski MH. Generation and post-injury integration of human spinal cord neural stem cells. Nat Methods. 2018;15:723-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 19. | Zhao X, Moore DL. Neural stem cells: developmental mechanisms and disease modeling. Cell Tissue Res. 2018;371:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 20. | Baggiani M, Dell’Anno MT, Pistello M, Conti L, Onorati M. Human Neural Stem Cell Systems to Explore Pathogen-Related Neurodevelopmental and Neurodegenerative Disorders. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Fernandez-Muñoz B, Garcia-Delgado AB, Arribas-Arribas B, Sanchez-Pernaute R. Human Neural Stem Cells for Cell-Based Medicinal Products. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y. Structure of nucleosome-bound human BAF complex. Science. 2020;367:875-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 228] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 23. | Varga J, Kube M, Luck K, Schick S. The BAF chromatin remodeling complexes: structure, function, and synthetic lethalities. Biochem Soc Trans. 2021;49:1489-1503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 24. | El Hadidy N, Uversky VN. Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 25. | Narayanan R, Tuoc TC. Roles of chromatin remodeling BAF complex in neural differentiation and reprogramming. Cell Tissue Res. 2014;356:575-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Alfert A, Moreno N, Kerl K. The BAF complex in development and disease. Epigenetics Chromatin. 2019;12:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 176] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 27. | Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 2014;166C:333-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 28. | Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 116] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 29. | Petrik D, Latchney SE, Masiulis I, Yun S, Zhang Z, Wu JI, Eisch AJ. Chromatin Remodeling Factor Brg1 Supports the Early Maintenance and Late Responsiveness of Nestin-Lineage Adult Neural Stem and Progenitor Cells. Stem Cells. 2015;33:3655-3665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Ninkovic J, Steiner-Mezzadri A, Jawerka M, Akinci U, Masserdotti G, Petricca S, Fischer J, von Holst A, Beckers J, Lie CD, Petrik D, Miller E, Tang J, Wu J, Lefebvre V, Demmers J, Eisch A, Metzger D, Crabtree G, Irmler M, Poot R, Götz M. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 31. | Jayaprakash S, Drakulic S, Zhao Z, Sander B, Golas MM. The ATPase BRG1/SMARCA4 is a protein interaction platform that recruits BAF subunits and the transcriptional repressor REST/NRSF in neural progenitor cells. Mol Cell Biochem. 2019;461:171-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | Narayanan R, Pham L, Kerimoglu C, Watanabe T, Castro Hernandez R, Sokpor G, Ulmke PA, Kiszka KA, Tonchev AB, Rosenbusch J, Seong RH, Teichmann U, Frahm J, Fischer A, Bonn S, Stoykova A, Staiger JF, Tuoc T. Chromatin Remodeling BAF155 Subunit Regulates the Genesis of Basal Progenitors in Developing Cortex. iScience. 2018;4:109-126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 33. | Narayanan R, Pirouz M, Kerimoglu C, Pham L, Wagener RJ, Kiszka KA, Rosenbusch J, Seong RH, Kessel M, Fischer A, Stoykova A, Staiger JF, Tuoc T. Loss of BAF (mSWI/SNF) Complexes Causes Global Transcriptional and Chromatin State Changes in Forebrain Development. Cell Rep. 2015;13:1842-1854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 34. | Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 137] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 35. | Battaglioli E, Andrés ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, Mandel G. REST repression of neuronal genes requires components of the hSWI.SNF complex. J Biol Chem. 2002;277:41038-41045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 160] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181-5186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 443] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 37. | Ho PJ, Lloyd SM, Bao X. Unwinding chromatin at the right places: how BAF is targeted to specific genomic locations during development. Development. 2019;146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 38. | Tuoc T, Dere E, Radyushkin K, Pham L, Nguyen H, Tonchev AB, Sun G, Ronnenberg A, Shi Y, Staiger JF, Ehrenreich H, Stoykova A. Ablation of BAF170 in Developing and Postnatal Dentate Gyrus Affects Neural Stem Cell Proliferation, Differentiation, and Learning. Mol Neurobiol. 2017;54:4618-4635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Celen C, Chuang JC, Luo X, Nijem N, Walker AK, Chen F, Zhang S, Chung AS, Nguyen LH, Nassour I, Budhipramono A, Sun X, Bok LA, McEntagart M, Gevers EF, Birnbaum SG, Eisch AJ, Powell CM, Ge WP, Santen GW, Chahrour M, Zhu H. Arid1b haploinsufficient mice reveal neuropsychiatric phenotypes and reversible causes of growth impairment. Elife. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 40. | Liu X, Dai SK, Liu PP, Liu CM. Arid1a regulates neural stem/progenitor cell proliferation and differentiation during cortical development. Cell Prolif. 2021;54:e13124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 41. | Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201-215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 638] [Cited by in RCA: 587] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 42. | Liu C, Sun R, Huang J, Zhang D, Huang D, Qi W, Wang S, Xie F, Shen Y, Shen C. The BAF45D Protein Is Preferentially Expressed in Adult Neurogenic Zones and in Neurons and May Be Required for Retinoid Acid Induced PAX6 Expression. Front Neuroanat. 2017;11:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (1)] |
| 43. | Chen XY, Hu XJ, Jiang J, Tao J, Liu LH, Fang SY, Shen YX, Hu QS, Liu C. Baf45d regulates spinal cord neural stem/progenitor cell fate through the smad-pax6 axis. May 20, 2022. [cited 12 February 2023]. Available from: https://www.researchgate.net/publication/360834915_BAF45D_regulates_spinal_cord_neural_stemprogenitor_cell_fate_through_the_SMAD-PAX6_axis. |
| 44. | Wang Z, Huang J, Liu C, Liu L, Shen Y, Shen C. BAF45D Downregulation in Spinal Cord Ependymal Cells Following Spinal Cord Injury in Adult Rats and Its Potential Role in the Development of Neuronal Lesions. Front Neurosci. 2019;13:1151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 45. | Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642-646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 497] [Cited by in RCA: 482] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 46. | Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422-2427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 561] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 47. | Alfert A, Walter C, Moreno N, Melcher V, Graf M, Hotfilder M, Dugas M, Albert T, Kerl K. Smarcb1 Loss Results in a Deregulation of esBAF Binding and Impacts the Expression of Neurodevelopmental Genes. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 48. | Staahl BT, Tang J, Wu W, Sun A, Gitler AD, Yoo AS, Crabtree GR. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J Neurosci. 2013;33:10348-10361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 49. | Burrowes SG, Salem NA, Tseng AM, Balaraman S, Pinson MR, Garcia C, Miranda RC. The BAF (BRG1/BRM-Associated Factor) chromatin-remodeling complex exhibits ethanol sensitivity in fetal neural progenitor cells and regulates transcription at the miR-9-2 encoding gene locus. Alcohol. 2017;60:149-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 50. | Heo Y, Park JH, Kim J, Han J, Yun JH, Lee W. Crystal structure of the HMG domain of human BAF57 and its interaction with four-way junction DNA. Biochem Biophys Res Commun. 2020;533:919-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 51. | Lomelí H, Castillo-Robles J. The developmental and pathogenic roles of BAF57, a special subunit of the BAF chromatin-remodeling complex. FEBS Lett. 2016;590:1555-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 52. | Marei HE, Ahmed AE, Michetti F, Pescatori M, Pallini R, Casalbore P, Cenciarelli C, Elhadidy M. Gene expression profile of adult human olfactory bulb and embryonic neural stem cell suggests distinct signaling pathways and epigenetic control. PLoS One. 2012;7:e33542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 53. | Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210-6220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 144] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 54. | Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, Chan JR, Wu J, Lu QR. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 293] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 55. | Lamba DA, Hayes S, Karl MO, Reh T. Baf60c is a component of the neural progenitor-specific BAF complex in developing retina. Dev Dyn. 2008;237:3016-3023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 56. | Wang J, Zhu HH, Chu M, Liu Y, Zhang C, Liu G, Yang X, Yang R, Gao WQ. Symmetrical and asymmetrical division analysis provides evidence for a hierarchy of prostate epithelial cell lineages. Nat Commun. 2014;5:4758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 57. | Shen F, Song C, Liu Y, Zhang J, Wei Song S. IGFBP2 promotes neural stem cell maintenance and proliferation differentially associated with glioblastoma subtypes. Brain Res. 2019;1704:174-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 58. | Traiffort E, Ferent J. [Neural stem cells and Notch signalling]. Med Sci (Paris). 2015;31:1115-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 59. | Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17:329-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 596] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 60. | Kim TJ, Kwon HS, Kang M, Leem HH, Lee KH, Kim DY. The Antitumor Natural Compound Falcarindiol Disrupts Neural Stem Cell Homeostasis by Suppressing Notch Pathway. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 61. | Vasileiou G, Ekici AB, Uebe S, Zweier C, Hoyer J, Engels H, Behrens J, Reis A, Hadjihannas MV. Chromatin-Remodeling-Factor ARID1B Represses Wnt/β-Catenin Signaling. Am J Hum Genet. 2015;97:445-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 62. | Nguyen H, Kerimoglu C, Pirouz M, Pham L, Kiszka KA, Sokpor G, Sakib MS, Rosenbusch J, Teichmann U, Seong RH, Stoykova A, Fischer A, Staiger JF, Tuoc T. Epigenetic Regulation by BAF Complexes Limits Neural Stem Cell Proliferation by Suppressing Wnt Signaling in Late Embryonic Development. Stem Cell Reports. 2018;10:1734-1750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 63. | Pirotte D, Wislet-Gendebien S, Cloes JM, Rogister B. Neuregulin-1 modulates the differentiation of neural stem cells in vitro through an interaction with the Swi/Snf complex. Mol Cell Neurosci. 2010;43:72-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 64. | Elsen GE, Bedogni F, Hodge RD, Bammler TK, MacDonald JW, Lindtner S, Rubenstein JLR, Hevner RF. The Epigenetic Factor Landscape of Developing Neocortex Is Regulated by Transcription Factors Pax6→ Tbr2→ Tbr1. Front Neurosci. 2018;12:571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 65. | Nguyen H, Sokpor G, Pham L, Rosenbusch J, Stoykova A, Staiger JF, Tuoc T. Epigenetic regulation by BAF (mSWI/SNF) chromatin remodeling complexes is indispensable for embryonic development. Cell Cycle. 2016;15:1317-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 66. | Bi-Lin KW, Seshachalam PV, Tuoc T, Stoykova A, Ghosh S, Singh MK. Critical role of the BAF chromatin remodeling complex during murine neural crest development. PLoS Genet. 2021;17:e1009446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 67. | Kosho T, Miyake N, Carey JC. Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: historical review and recent advances using next generation sequencing. Am J Med Genet C Semin Med Genet. 2014;166C:241-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 68. | Schrier SA, Bodurtha JN, Burton B, Chudley AE, Chiong MA, D’avanzo MG, Lynch SA, Musio A, Nyazov DM, Sanchez-Lara PA, Shalev SA, Deardorff MA. The Coffin-Siris syndrome: a proposed diagnostic approach and assessment of 15 overlapping cases. Am J Med Genet A. 2012;158A:1865-1876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 69. | Vasileiou G, Vergarajauregui S, Endele S, Popp B, Büttner C, Ekici AB, Gerard M, Bramswig NC, Albrecht B, Clayton-Smith J, Morton J, Tomkins S, Low K, Weber A, Wenzel M, Altmüller J, Li Y, Wollnik B, Hoganson G, Plona MR, Cho MT; Deciphering Developmental Disorders Study, Thiel CT, Lüdecke HJ, Strom TM, Calpena E, Wilkie AOM, Wieczorek D, Engel FB, Reis A. Mutations in the BAF-Complex Subunit DPF2 Are Associated with Coffin-Siris Syndrome. Am J Hum Genet. 2018;102:468-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 70. | Knapp KM, Poke G, Jenkins D, Truter W, Bicknell LS. Expanding the phenotypic spectrum associated with DPF2: A new case report. Am J Med Genet A. 2019;179:1637-1641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 71. | Milone R, Gnazzo M, Stefanutti E, Serafin D, Novelli A. A new missense mutation in DPF2 gene related to Coffin Siris syndrome 7: Description of a mild phenotype expanding DPF2-related clinical spectrum and differential diagnosis among similar syndromes epigenetically determined. Brain Dev. 2020;42:192-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 72. | Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, Kaname T, Naritomi K, Kawame H, Wakui K, Fukushima Y, Homma T, Kato M, Hiraki Y, Yamagata T, Yano S, Mizuno S, Sakazume S, Ishii T, Nagai T, Shiina M, Ogata K, Ohta T, Niikawa N, Miyatake S, Okada I, Mizuguchi T, Doi H, Saitsu H, Miyake N, Matsumoto N. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 404] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 73. | Gossai N, Biegel JA, Messiaen L, Berry SA, Moertel CL. Report of a patient with a constitutional missense mutation in SMARCB1, Coffin-Siris phenotype, and schwannomatosis. Am J Med Genet A. 2015;167A:3186-3191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 74. | Santen GW, Aten E, Vulto-van Silfhout AT, Pottinger C, van Bon BW, van Minderhout IJ, Snowdowne R, van der Lans CA, Boogaard M, Linssen MM, Vijfhuizen L, van der Wielen MJ, Vollebregt MJ; Coffin-Siris consortium, Breuning MH, Kriek M, van Haeringen A, den Dunnen JT, Hoischen A, Clayton-Smith J, de Vries BB, Hennekam RC, van Belzen MJ. Coffin-Siris syndrome and the BAF complex: genotype-phenotype study in 63 patients. Hum Mutat. 2013;34:1519-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 75. | Wieczorek D, Bögershausen N, Beleggia F, Steiner-Haldenstätt S, Pohl E, Li Y, Milz E, Martin M, Thiele H, Altmüller J, Alanay Y, Kayserili H, Klein-Hitpass L, Böhringer S, Wollstein A, Albrecht B, Boduroglu K, Caliebe A, Chrzanowska K, Cogulu O, Cristofoli F, Czeschik JC, Devriendt K, Dotti MT, Elcioglu N, Gener B, Goecke TO, Krajewska-Walasek M, Guillén-Navarro E, Hayek J, Houge G, Kilic E, Simsek-Kiper PÖ, López-González V, Kuechler A, Lyonnet S, Mari F, Marozza A, Mathieu Dramard M, Mikat B, Morin G, Morice-Picard F, Ozkinay F, Rauch A, Renieri A, Tinschert S, Utine GE, Vilain C, Vivarelli R, Zweier C, Nürnberg P, Rahmann S, Vermeesch J, Lüdecke HJ, Zeschnigk M, Wollnik B. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet. 2013;22:5121-5135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 76. | Zarate YA, Bhoj E, Kaylor J, Li D, Tsurusaki Y, Miyake N, Matsumoto N, Phadke S, Escobar L, Irani A, Hakonarson H, Schrier Vergano SA. SMARCE1, a rare cause of Coffin-Siris Syndrome: Clinical description of three additional cases. Am J Med Genet A. 2016;170:1967-1973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 77. | Nixon KCJ, Rousseau J, Stone MH, Sarikahya M, Ehresmann S, Mizuno S, Matsumoto N, Miyake N; DDD Study, Baralle D, McKee S, Izumi K, Ritter AL, Heide S, Héron D, Depienne C, Titheradge H, Kramer JM, Campeau PM. A Syndromic Neurodevelopmental Disorder Caused by Mutations in SMARCD1, a Core SWI/SNF Subunit Needed for Context-Dependent Neuronal Gene Regulation in Flies. Am J Hum Genet. 2019;104:596-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 78. | Machol K, Rousseau J, Ehresmann S, Garcia T, Nguyen TTM, Spillmann RC, Sullivan JA, Shashi V, Jiang YH, Stong N, Fiala E, Willing M, Pfundt R, Kleefstra T, Cho MT, McLaughlin H, Rosello Piera M, Orellana C, Martínez F, Caro-Llopis A, Monfort S, Roscioli T, Nixon CY, Buckley MF, Turner A, Jones WD, van Hasselt PM, Hofstede FC, van Gassen KLI, Brooks AS, van Slegtenhorst MA, Lachlan K, Sebastian J, Madan-Khetarpal S, Sonal D, Sakkubai N, Thevenon J, Faivre L, Maurel A, Petrovski S, Krantz ID, Tarpinian JM, Rosenfeld JA, Lee BH; Undiagnosed Diseases Network, Campeau PM. Expanding the Spectrum of BAF-Related Disorders: De Novo Variants in SMARCC2 Cause a Syndrome with Intellectual Disability and Developmental Delay. Am J Hum Genet. 2019;104:164-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 79. | Filatova A, Rey LK, Lechler MB, Schaper J, Hempel M, Posmyk R, Szczaluba K, Santen GWE, Wieczorek D, Nuber UA. Mutations in SMARCB1 and in other Coffin-Siris syndrome genes lead to various brain midline defects. Nat Commun. 2019;10:2966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 80. | Li D, Ahrens-Nicklas RC, Baker J, Bhambhani V, Calhoun A, Cohen JS, Deardorff MA, Fernández-Jaén A, Kamien B, Jain M, Mckenzie F, Mintz M, Motter C, Niles K, Ritter A, Rogers C, Roifman M, Townshend S, Ward-Melver C, Schrier Vergano SA. The variability of SMARCA4-related Coffin-Siris syndrome: Do nonsense candidate variants add to milder phenotypes? Am J Med Genet A. 2020;182:2058-2067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 81. | Zou Y, Yin Y, Xiao Z, Zhao Y, Han J, Chen B, Xu B, Cui Y, Ma X, Dai J. Transplantation of collagen sponge-based three-dimensional neural stem cells cultured in a RCCS facilitates locomotor functional recovery in spinal cord injury animals. Biomater Sci. 2022;10:915-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 82. | Aref-Eshghi E, Bend EG, Hood RL, Schenkel LC, Carere DA, Chakrabarti R, Nagamani SCS, Cheung SW, Campeau PM, Prasad C, Siu VM, Brady L, Tarnopolsky MA, Callen DJ, Innes AM, White SM, Meschino WS, Shuen AY, Paré G, Bulman DE, Ainsworth PJ, Lin H, Rodenhiser DI, Hennekam RC, Boycott KM, Schwartz CE, Sadikovic B. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat Commun. 2018;9:4885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 83. | Chaste P, Leboyer M. Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin Neurosci. 2012;14:281-292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 355] [Cited by in RCA: 442] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 84. | Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH Jr, Devlin B, Gibbs RA, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242-245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1412] [Cited by in RCA: 1309] [Article Influence: 100.7] [Reference Citation Analysis (0)] |
| 85. | De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimäki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnström K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Rüther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209-215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1776] [Cited by in RCA: 1978] [Article Influence: 179.8] [Reference Citation Analysis (0)] |
| 86. | Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Geneviève D, Cormier-Daire V, van Esch H, Fryns JP, Hamel BC, Sistermans EA, de Vries BB, van Bokhoven H. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79:370-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 285] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 87. | Kleefstra T, van Zelst-Stams WA, Nillesen WM, Cormier-Daire V, Houge G, Foulds N, van Dooren M, Willemsen MH, Pfundt R, Turner A, Wilson M, McGaughran J, Rauch A, Zenker M, Adam MP, Innes M, Davies C, López AG, Casalone R, Weber A, Brueton LA, Navarro AD, Bralo MP, Venselaar H, Stegmann SP, Yntema HG, van Bokhoven H, Brunner HG. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J Med Genet. 2009;46:598-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 155] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 88. | Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, Vissers LE, Wissink-Lindhout W, Fenckova M, van den Akker WM, Kasri NN, Nillesen WM, Prescott T, Clark RD, Devriendt K, van Reeuwijk J, de Brouwer AP, Gilissen C, Zhou H, Brunner HG, Veltman JA, Schenck A, van Bokhoven H. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet. 2012;91:73-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 199] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 89. | MacDonald SK, Marshall AE, Lemire G, Hartley T; Care4Rare Canada Consortium, Kernohan KD, Boycott KM. A novel intragenic DPF2 deletion identified by genome sequencing in an adult with clinical features of Coffin-Siris syndrome. Am J Med Genet A. 2022;188:2493-2496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 90. | Foley R, Duignan S, McArdle L, Betts DR, Green A, McMahon CJ. Nicolaides-Baraitser syndrome in a patient with hypertrophic cardiomyopathy and SMARCA2 gene deletion. Cardiol Young. 2022;32:821-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 91. | Lo T, Kushima I, Aleksic B, Kato H, Nawa Y, Hayashi Y, Otgonbayar G, Kimura H, Arioka Y, Mori D, Ozaki N. Sequencing of selected chromatin remodelling genes reveals increased burden of rare missense variants in ASD patients from the Japanese population. Int Rev Psychiatry. 2022;34:154-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 92. | Aydin H, Bucak IH, Bagis H. Kleefstra Syndrome. J Coll Physicians Surg Pak. 2022;32:S76-S78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 93. | Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front Mol Neurosci. 2017;10:243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 94. | Frega M, Selten M, Mossink B, Keller JM, Linda K, Moerschen R, Qu J, Koerner P, Jansen S, Oudakker A, Kleefstra T, van Bokhoven H, Zhou H, Schubert D, Nadif Kasri N. Distinct Pathogenic Genes Causing Intellectual Disability and Autism Exhibit a Common Neuronal Network Hyperactivity Phenotype. Cell Rep. 2020;30:173-186.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 95. | Centore RC, Sandoval GJ, Soares LMM, Kadoch C, Chan HM. Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet. 2020;36:936-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 239] [Article Influence: 47.8] [Reference Citation Analysis (0)] |