Published online Dec 26, 2022. doi: 10.4252/wjsc.v14.i12.822
Peer-review started: August 4, 2022
First decision: September 29, 2022
Revised: October 7, 2022
Accepted: November 30, 2022
Article in press: November 30, 2022
Published online: December 26, 2022
Processing time: 138 Days and 20.1 Hours
Spermatogonial stem cells (SSCs) are the origin of male spermatogenesis, which can reconstruct germ cell lineage in mice. However, the application of SSCs for male fertility restoration is hindered due to the unclear mechanisms of proliferation and self-renewal in humans.
To investigate the role and mechanism of SPOC domain-containing protein 1 (SPOCD1) in human SSC proliferation.
We analyzed publicly available human testis single-cell RNA sequencing (RNA-seq) data and found that SPOCD1 is predominantly expressed in SSCs in the early developmental stages. Small interfering RNA was applied to suppress SPOCD1 expression to detect the impacts of SPOCD1 inhibition on SSC proliferation and apoptosis. Subsequently, we explored the target genes of SPOCD1 using RNA-seq and confirmed their role by restoring the expression of the target genes. In addition, we examined SPOCD1 expression in some non-obstructive azoospermia (NOA) patients to explore the correlation between SPOCD1 and NOA.
The uniform manifold approximation and projection clustering and pseudotime analysis showed that SPOCD1 was highly expressed in the early stages of SSC, and immunohistological results showed that SPOCD1 was mainly localized in glial cell line-derived neurotrophic factor family receptor alpha-1 positive SSCs. SPOCD1 knockdown significantly inhibited cell proliferation and promoted apoptosis. RNA-seq results showed that SPOCD1 knockdown significantly downregulated genes such as adenylate kinase 4 (AK4). Overexpression of AK4 in SPOCD1 knockdown cells partially reversed the phenotypic changes, indicating that AK4 is a functional target gene of SPOCD1. In addition, we found a significant downregulation of SPOCD1 expression in some NOA patients, suggesting that the downregulation of SPOCD1 may be relevant for NOA.
Our study broadens the understanding of human SSC fate determination and may offer new theories on the etiology of male infertility.
Core Tip: In this study, we reported the dominant expression of SPOC domain-containing protein 1 (SPOCD1) in human spermatogonial stem cells (SSCs). Knockdown of SPOCD1 in SSC caused a significant decrease in proliferation and self-renewal, and the induction of apoptosis. RNA sequencing showed that SPOCD1 knockdown caused significant downregulation of genes such as adenylate kinase 4 (AK4), and overexpression of AK4 in SPOCD1-knockdown cells reversed the phenotypic alterations induced by SPOCD knockdown. Additionally, we found significant downregulation of SPOCD1 in non-obstructive azoospermia patients. These results broaden our understanding of human SSC fate determination and provide new theories on the etiology of male infertility.
- Citation: Zhou D, Zhu F, Huang ZH, Zhang H, Fan LQ, Fan JY. SPOC domain-containing protein 1 regulates the proliferation and apoptosis of human spermatogonial stem cells through adenylate kinase 4. World J Stem Cells 2022; 14(12): 822-838
- URL: https://www.wjgnet.com/1948-0210/full/v14/i12/822.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i12.822
Infertility affects about 15% of couples worldwide, and about 50% of these cases are due to male factors[1]. Non-obstructive azoospermia (NOA) is the most severe cause of male infertility, for which there is a lack of effective treatment[2]. Thus, solving the fertility issues associated with NOA has been an important research direction in reproductive medicine.
Spermatogonial stem cells (SSCs) are responsible for initiating and maintaining adult spermatogenesis throughout life, which produces mature sperm through constant self-renewal and differentiation[3]. In rodents, long-term in vitro culture of mouse SSCs has been achieved, with reports showing the restoration of testicular transplantation in recipient mouse germline reconstitution[4,5]. However, the mouse SSC culture system is unsuitable for humans, and insufficient proliferation capacity in vitro is currently a significant problem encountered during human SSC culture[6]. Therefore, exploring the mechanism of human SSC proliferation and self-renewal is key to solving the long-term in vitro culture of human SSCs and the basis for using SSCs in treating male infertility.
Glial cell line-derived neurotrophic factor (GDNF) is a crucial growth factor for maintaining SSC proliferation and self-renewal[7]. GDNF binds to the GDNF family receptor alpha-1 (GFRA1)/c-Ret; activates downstream RAS, AKT, and mitogen-activated protein kinase (MAPK) pathways; and regulates the transcription of ETS variant transcription factor 5, B-cell CLL/lymphoma 6 member B protein, and LIM homeobox 1 to promote the self-renewal of SSCs[8]. Another essential growth factor is fibroblast growth factor 2, which regulates SSC self-renewal by activating the MAPK pathway[9]. Due to differences in species, sample sources, and ethical issues, only a few studies have been performed to investigate the regulations of human SSCs. Recently, microRNA-1908-3p (miR-1908-3p) was shown to enhance SSC proliferation by mediating the degradation of Krüppel-like factor 2 (KLF2) in humans[10]. miR-122-5p[11] and miR-663a[12] are also involved in the regulation of SSC proliferation. Calcium-responsive transcription factor (CARF) affects SSC functions in mice through the WNT pathway, and mutations in human CARF also cause male infertility[13]. Human SSCs have also been modulated by RNF144B through the Fc epsilon receptor II/neurogenic locus notch homolog protein 2/HES1 pathway[14]. In addition, we previously reported that transcription factor 3 is specifically localized in the nucleus of human SSCs, and promotes human SSC proliferation by regulating podocalyxin-like protein 1 expression[15]. However, the regulatory mechanisms of SSCs are poorly understood.
To further explore the developmental process of human SSCs, GSE149512[16] and GSE112013[17] databases containing adult testis single-cell data were analyzed. We found that SPOC domain-containing protein 1 (SPOCD1) was specifically expressed in a subpopulation of SSCs, and the result was validated by immunohistochemistry. SPOCD1 knockdown decreased the proliferation of immortalized human SSCs, with numerous genes downregulated, including adenylate kinase 4 (AK4), KLF8, and vesicular, overexpressed in cancer, prosurvival protein 1 (VOPP1). AK4 re-expression reversed the cell proliferation and apoptotic changes caused by SPOCD1 knockdown. Furthermore, the expression of SPOCD1 was significantly reduced in some NOA patients. Overall, these results describe a role for SPOCD1 in SSC proliferation and expand our understanding of SSC fate determination.
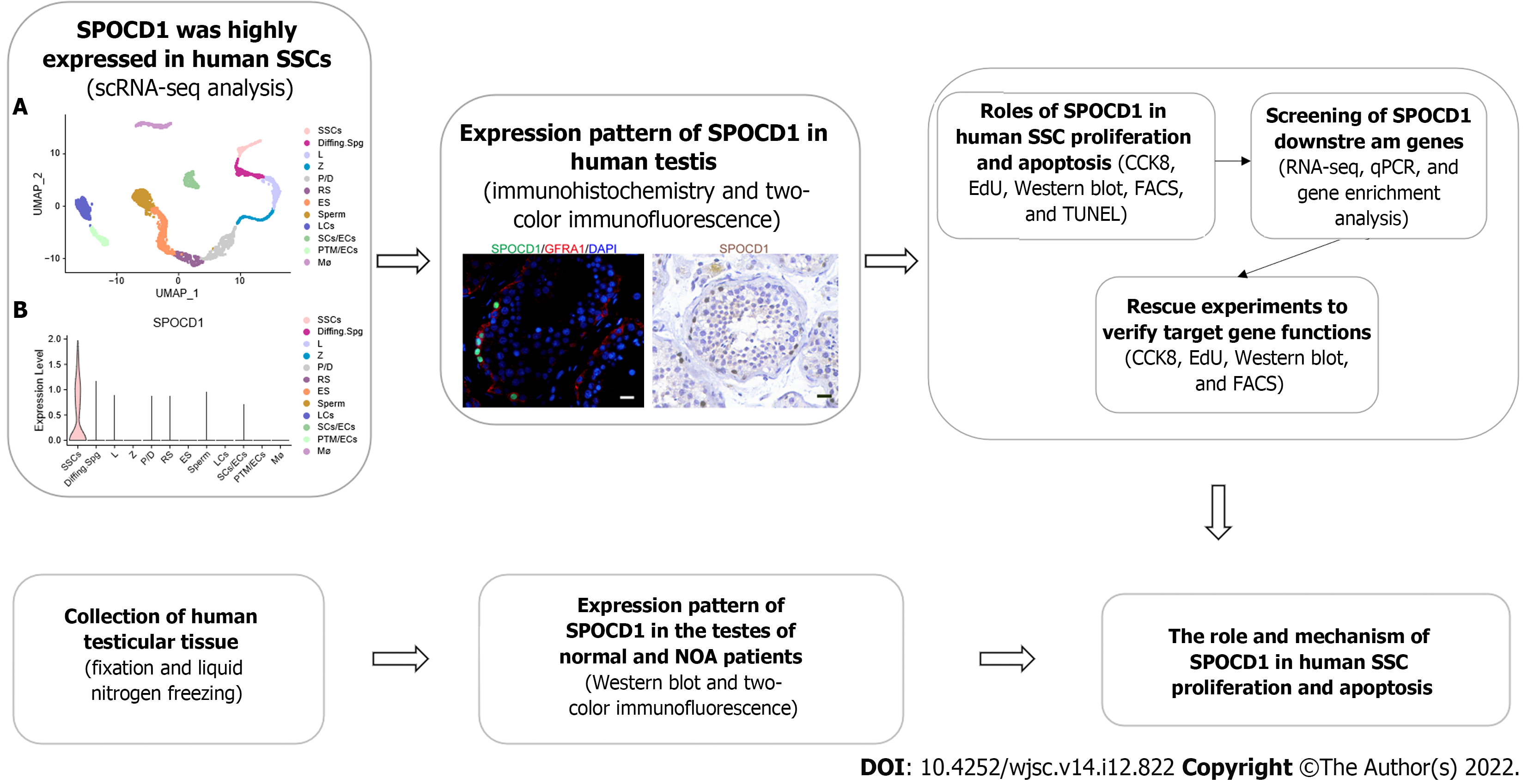
Our study was approved by the ethics committee of the Reproductive and Genetic Hospital of CITIC-Xiangya (LL-SC-2021-025). The overview of our research is illustrated in Figure 1. Each participant provided signed informed consent. Testicular tissues of 18 patients (6 OA and 12 NOA), aged between 28-years-old and 48-years-old who underwent testicular biopsy were collected, with approximately 25 mg each. Sterile phosphate-buffered saline (PBS) was used to wash the samples at least three times to remove blood cells. Subsequently, the tissues were frozen in liquid nitrogen or fixed in 40 g/L paraformaldehyde (PFA).
To analyze single-cell RNA sequencing (scRNA-seq) datasets (GSE149512 and GSE112013) of the normal adult testis, Seurat 4.2 (https://github.com/satijalab/seurat/) program in R was employed. In the first step, we used the Read.10X function to load the expression matrix data in R and created the Seurat object. Cells with gene expression numbers between 500 and 4000 and with less than 15% of mitochondrial genes were retained. Then, each Seurat object was processed using the NormalizeData and FindVariableFeatures functions. Next, all Seurat objects were merged using the FindIntegrationAnchors and IntegrateData functions. The combined data were clustered using the uniform manifold approximation and projection (UMAP) method set by default, and subsequently, cell types were identified based on the expression of cellular markers. The data of SSC cell populations were extracted using the Subset function and re-clustered using UMAP. Monocle 3 (https://github.com/cole-trapnell-lab/monocle3) in R was used to perform pseudotime analysis of SSCs. The cell developmental trajectory begins in Subpopulation State 2. Dot, line, and violin plots were created and modified using ggplot2 (https://github.com/tidyverse/ggplot2) in R.
By transfecting Large T antigen into G protein-coupled receptor 125 (GPR125)-positive human undifferentiated spermatogonia, immortalized human SSC lines were established[18]. Immortalized human SSCs maintained many properties of their primary cells and expressed many markers of primary SSCs including GFRA1, RET, and promyelocytic leukemia zinc finger (PLZF). They did not express testicular endosomal cell markers such as SRY-box transcription factor 9[15]. The immortalized human SSCs were grown at 34 °C with 50 mL/L CO2 in an incubator, and the culture medium consisted of Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco, Grand Island, NY, United States) supplemented with 100 mL/L fetal bovine serum (FBS; Gibco). The cells were subcultured every 2 d or 3 d (0.5 g/L trypsin and 0.53 mmol/L Ethylenediaminetetraacetic acid (EDTA); Invitrogen, Carlsbad, CA, United States).
The total RNA of cells was isolated using RNAiso Plus reagent (Takara, Kusatsu, Japan) following the manufacturer’s instructions. Nanodrop (Thermo Fisher Scientific, Waltham, MA, United States) was employed to detect the quality and concentration of the extracted RNA. Then the reverse transcription of cDNA was conducted using commercial kits (Roche, Mannheim, Germany).
According to the manufacturer's instructions, quantitative PCR (qPCR) was performed using the ABI Prism 7700 system (Applied Biosystems, Foster City, CA, United States). The 2-△△(Ct) method was chosen to measure the relative levels of mRNAs, and actin beta was selected as an internal reference. Each sample was analyzed three times, and the results were averaged. All primer sequences were designed and listed in Supplementary Table 1.
Testis sections were deparaffinized with xylene and rehydrated with graded ethanol for immunohistochemistry. Then the heat-induced antigen retrieval method was conducted in 0.01 mol/L sodium citrate buffer at 98 °C for 18 min. After cooling and washing, the sections were incubated with 30 mL/L hydrogen peroxidase (Zsbio, Beijing, China) to block the endogenous peroxidase activity. After three washes with PBS, the tissue sections were permeated for 15 min with 2.5 mL/L Triton X-100 (Sigma, St. Louis, MO, United States), and 50 mL/L bovine serum albumin was applied to block nonspecific antigens for 1 h at room temperature (RT). Subsequently, sections were incubated with primary antibodies listed in Supplementary Table 2 at 4 °C overnight. After three washes with PBS, the sections were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody for 1 h at RT, and the 3,3’-diaminobenzidine chromogen kit (Dako, Glostrup, Denmark) was used for color development. Hematoxylin was used to stain the nucleus for 7 min at RT. For immunofluorescence, after incubation of the primary antibody for 16h at 4 °C, chromogenic development was performed using Alexa Fluor-conjugated secondary antibody, and 4,6-diamidino-2-phenylindole was used to counterstain the cell nuclei. The microscopic images of testicular sections were captured and analyzed using a Zeiss microscope (Zeiss, Jena, Germany).
For total protein extraction, testicular tissue and cells were lysed using RIPA (Thermo Fisher Scientific) or 15 min on ice, followed by centrifugation at 12000 g for 15 min and the collection of supernatants. According to the operating manual, the BCA Kit was utilized to detect total protein concentration. Twenty micrograms of total protein were taken from each sample for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis, as previously described[15]. Detailed antibody information is listed in Supplementary Table 2. Enhanced chemiluminescent chromogenic solution (Thermo Fisher Scientific) was used to visualize the protein band, and the chemiluminescent signal of bands was captured and analyzed with Fusion FX (Vilber Lourmat, Marne-la-Vallée, France). All samples were analyzed three times, and the results were averaged.
All small interfering RNAs (siRNAs) were designed and synthesized by Ribobio (Guangzhou, China), and the sequence of siRNAs was listed in Supplementary Table 3. Immortalized human SSCs were transfected with siRNAs (100 nmol/L) using Lipofectamine 3000 (Life Technologies, Carlsbad, CA, United States) according to the manufacturer’s instructions. After transfection for 48 h, cells were collected to extract protein and RNA for PCR and Western blot analysis.
The Cell Counting Kit-8 (CCK-8) Kit (Dojindo, Kumamoto, Japan) was used to detect SSC viability according to the manufacturer’s instructions. Cells were cultured for 3 h using the culture medium supplemented with 100 mL/L CCK-8 reagents. Then a microplate reader (Thermo Fisher Scientific) was used to detect the absorbance at 450 nm.
For the 5-ethynyl-2’-deoxyuridine (EdU) incorporation assay, DNA synthesis was detected with an EdU labeling kit (RiboBio). According to the manufacturer’s protocol, human SSCs were seeded into 96-well plates (5000 cells per well) in culture medium supplemented with 50 μmol/L EdU. After 12 h of incubation, cells were washed with DMEM and fixed in 40 g/L PFA. Next, cells were neutralized with glycine (2 mg/mL) and permeabilized with 5 mL/L Triton X-100 for 10 min at RT. Apollo staining reaction buffer was used for EdU visualization, and DAPI was employed for labeling cell nuclei. The microscopic images of EdU-positive cells were captured and analyzed using the Zeiss fluorescence microscope. A minimum of 500 cells per sample were assessed.
After transfection with SPOCD1-siRNA for 48 h, cells were digested using trypsin/EDTA and washed twice with ice-cold PBS. Next, according to the manufacturer’s instructions, at least 106 cells were resuspended in Annexin V binding buffer (BD Biosciences, Franklin Lakes, NJ, United States). The cells were incubated with 5 µL APC-labeled Annexin V for 15 min at RT. Before the assay, cells were incubated with 10 µL PI for 10 min. Cell apoptosis was evaluated on the C6 flow cytometer (BD Biosciences).
After transfection in human SSCs with SPOCD1-siRNA, an in situ cell death detection kit (Roche) was used to evaluate cell apoptosis according to the manufacturer’s instructions. Cells were fixed in PFA, and then incubated with proteinase K (20 mg/mL) for 15 min at RT. After washing, the cells were incubated with 50 µL terminal deoxynucleotidyl transferase (TdT) reaction buffer for 1 h away from light, and DAPI was used to label the cell nucleus. PBS free of TdT enzyme was utilized to treat the cells of the negative control group. At least 500 cells were counted per group using fluorescence microscopy (Zeiss).
The total RNA of cells was isolated using the Trizol Reagent Kit (Invitrogen). RNA quality was measured using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). To enrich eukaryotic mRNA, oligo (dT) beads were used, and ribosomal RNA (rRNA) was removed using a Magnetic Kit (Epicentre, Madison, WI, United States). Following this, the enriched mRNA was fragmented using the fragmentation buffer, and reverse transcription was performed using random hexamers. Subsequently, the cDNA was synthesized and purified using a commercial purification kit (Qiagen, Venlo, The Netherlands), followed by their end repair, poly (A) introduction, and ligation. Next, we utilized agarose electrophoresis to separate the ligation products, and after amplifying them using PCR, sequencing was performed on the Illumina HiSeq2500 system. Fastp (version 0.18.0) was used to filter the reads obtained from the sequencing machine. Bowtie2 (version 2.2.8) was applied to remove the rRNA-mapped reads. The remaining clean reads were used to assemble transcripts and determine gene abundance and mapped to the reference genome. Then the mapped reads were assembled using StringTie (version 1.3.1) in a reference-based strategy. DESeq2 software was used to assess differentially expressed genes (DEGs). ClusterProfiler in R was used to perform Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis on DEGs.
GraphPad Prism version 8.0 (GraphPad Software, La Jolla, CA, United States) was used for the data analyses. All assays were performed at least in triplicate. Data are shown as the mean ± SD. Differences between groups were evaluated using the t-test. P < 0.05 indicated statistical significance.
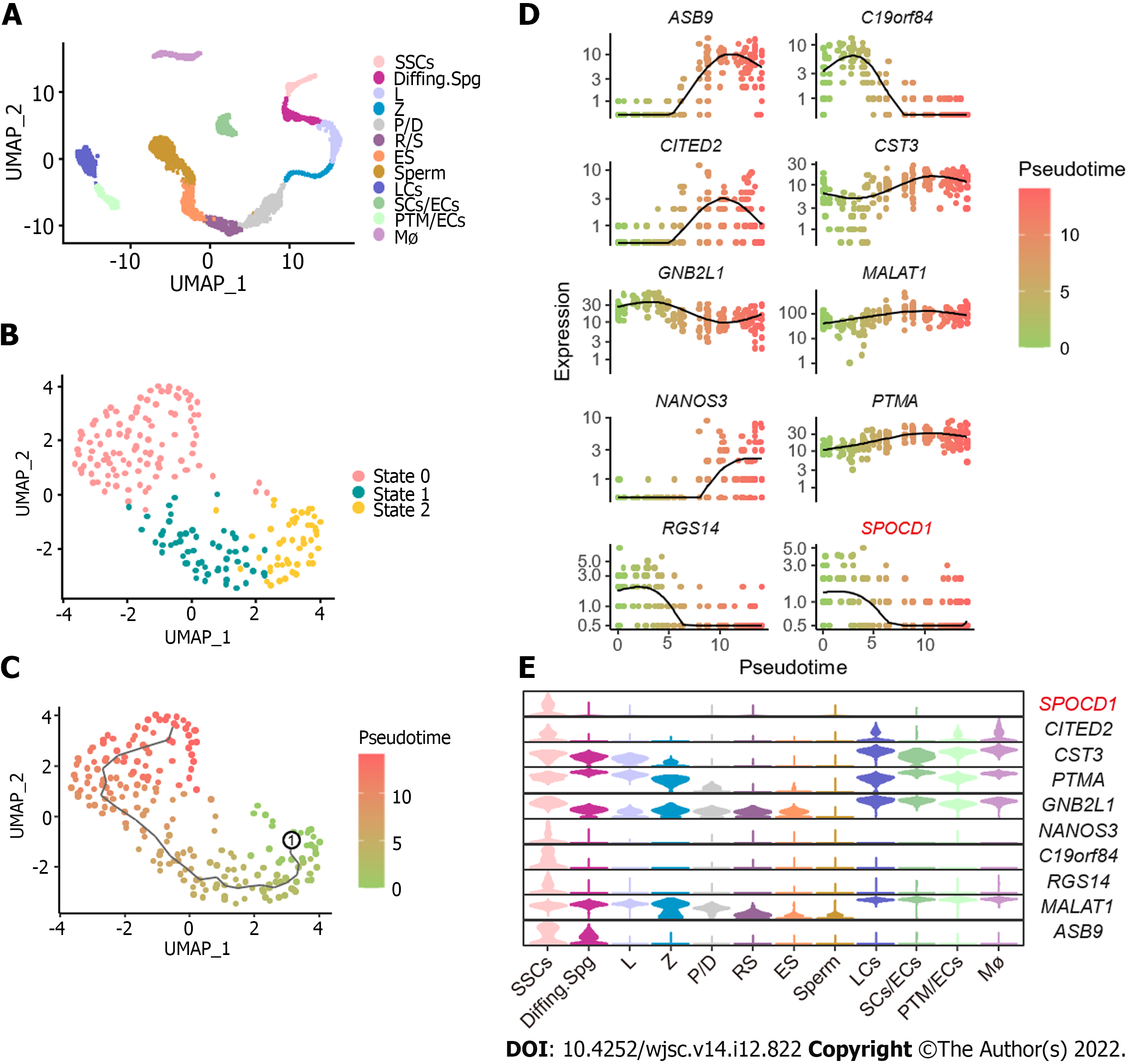
To explore the molecular mechanisms underlying the proliferation and self-renewal of human SSCs, we performed bioinformatics analysis on scRNA-seq datasets of normal adult testis from GSE149512 and GSE112013. By screening and integrating the data, 5176 testis cells and 23152 genes were identified. All cells were divided into 12 clusters using the Seurat package on R and identified according to the expression levels of a series of testicular cell marker genes including SSC markers (inhibitor of DNA binding 4, HLH protein), differentiating markers (KIT and stimulated by retinoic acid 8), meiosis markers (synaptonemal complex protein 3, SPO11, ovo-like zinc finger 2, and NME8), spermatid structure proteins (transition protein 2 and protamine 2), and some somatic markers and their respective cell clusters were identified. The 12 cell populations were SSCs, differentiating spermatogonia (Diffing. Spg), leptotene spermatocytes (L), zygotene spermatocytes (Z), pachytene/diplotene spermatocytes (P/D), round spermatids (RS), elongated spermatocytes (ES), sperm, Leydig cells (LCs), Sertoli cells/endothelial cells (SCs/ECs), peritubular myoid cells (PTM)/ECs, and macrophages (Mø) (Figure 2A). To further analyze the regulation of SSCs, reclustering of SSCs was performed using the Seurat package on R, and all SSCs were subdivided into three states, namely states 0, 1, and 2 (Figure 2B). Then, a monocle-based pseudotime analysis on SSCs was performed to create a developmental trajectory. According to the level of PIWI-like RNA-mediated gene silencing 4 and Nanos C2HC-type zinc finger 3, we assumed that State 2 was the developmental starting point, State 0 was late in development, and State 1 was the transitional period of development (Figure 2C). Differential gene expression analysis identified various genes including SPOCD1, ankyrin repeat and SOCS box containing 9, and chromosome 19 open reading frame 84 (Figure 2D). We also observed the distribution of these genes in all testicular cells using a Violin plot. Among these DEGs, SPOCD1 was specifically expressed in SSCs and progressively decreased with developmental trajectory (Figure 2E), indicating that SPOCD1 is associated with the SSC self-renewal and proliferation ability.
To validate the results of scRNA-seq analysis, we investigated the expression pattern of SPOCD1 in normal adult testicular tissue. Western blot analysis showed that SPOCD1 protein was moderately expressed in the testes of three OA patients with normal spermatogenesis (Figure 3A). Furthermore, we examined the localization of SPOCD1 in the normal testis using immunohistochemistry. The results demonstrated that the positive signal appeared in the nucleus of cells near the basal membrane of the seminal tubules, indicating that SPOCD1 is mainly expressed in spermatogonia (Figure 3B and C). Thus, we further analyzed the cell subtypes in which SPOCD1 was expressed using double immunofluorescence. The results showed that 91.11% ± 4.65% of SPOCD1-positive cells expressed glial cell derived neurotrophic factor family receptor alpha 1 (a marker of SSCs), and only 3.38% ± 1.54% of SPOCD1-positive cells weakly expressed KIT, a marker of differentiating spermatogonia. It should be noted that 84.60% ± 2.79% of SPOCD1-positive cells expressed proliferating cell nuclear antigen (PCNA), a feature of proliferating cells (Figure 3D and E). These data validated the results from bioinformatics analysis, showing that SPOCD1 was mainly localized to SSCs and may play roles in human SSC proliferation and self-renewal.
To examine the roles of SPOCD1 in SSC proliferation, an immortalized human SSC cell line was used. We used several siRNAs to repress SPOCD1 expression in cells and verified the knockdown efficiency of each siRNA by qPCR (Figure 4A) and Western blot analysis (Figure 4B and C). These results indicated that all three siRNAs inhibited the expression of SPOCD1, of which SPOCD1-siRNA2 had the best knockdown efficiency. Then, a CCK-8 assay was performed to investigate the proliferation of SPOCD1 siRNA2-transfected cells (Figure 4D). The results showed that SPOCD1 knockdown suppressed cell proliferation from Day 3 to Day 5 after transduction. We also examined the levels of various proteins associated with SSC proliferation, including promyelocytic leukemia zinc finger, cyclin D1, PCNA, and Thy-1 cell surface antigen, and found that all were significantly downregulated after the knockdown of SPOCD1 (Figure 4E and F). Likewise, 48 h after cell transfection, EdU incorporation assays were used to detect cell DNA synthesis. SPOCD1 inhibition induced a significant decrease in cellular DNA synthesis compared to the control group (34.73% ± 4.02% vs 21.56% ± 1.56%, P < 0.05) (Figure 4G and H).
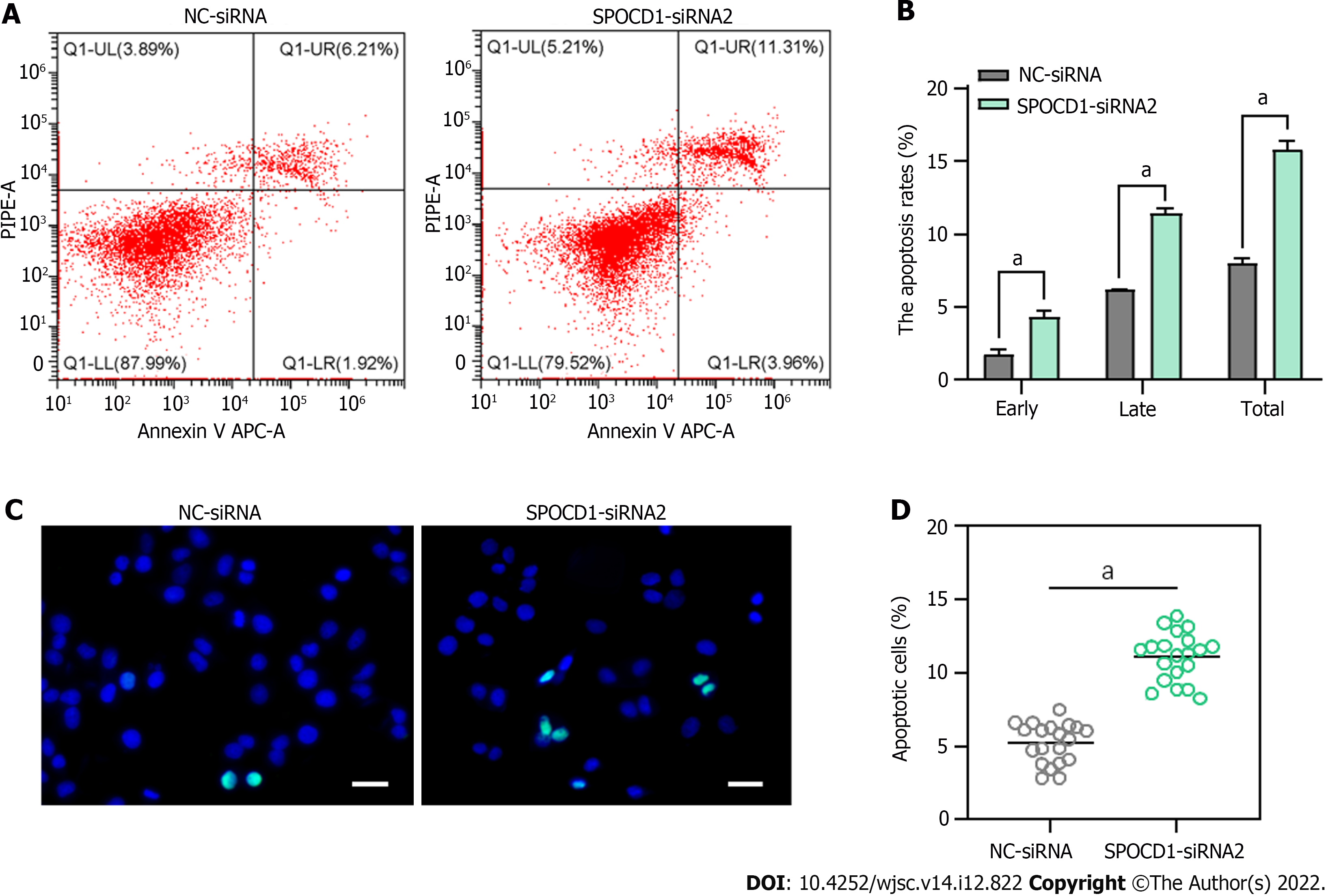
Following transfection with SPOCD1-siRNA2 for 48 h, we observed a significant increase in suspended cells and debris, so we examined cell apoptosis using Annexin V/propidium iodide staining and flow cytometry. The analysis showed that SPOCD1 knockdown led to a significant increase in early and late apoptosis compared to the control group (early apoptosis: 4.39% ± 0.40% vs 1.81% ± 0.29%, P < 0.05; late apoptosis: 11.43% ± 0.24% vs 6.24% ± 0.02%, P < 0.05, Figure 5A and B). Similar results were obtained with the TdT dUTP nick end labeling (TUNEL) assay, which showed a significant increase in the cellular DNA fragmentation rate (Figure 5C and D). These results suggest that inhibition of SPOCD1 expression triggers apoptosis in human SSC lines.
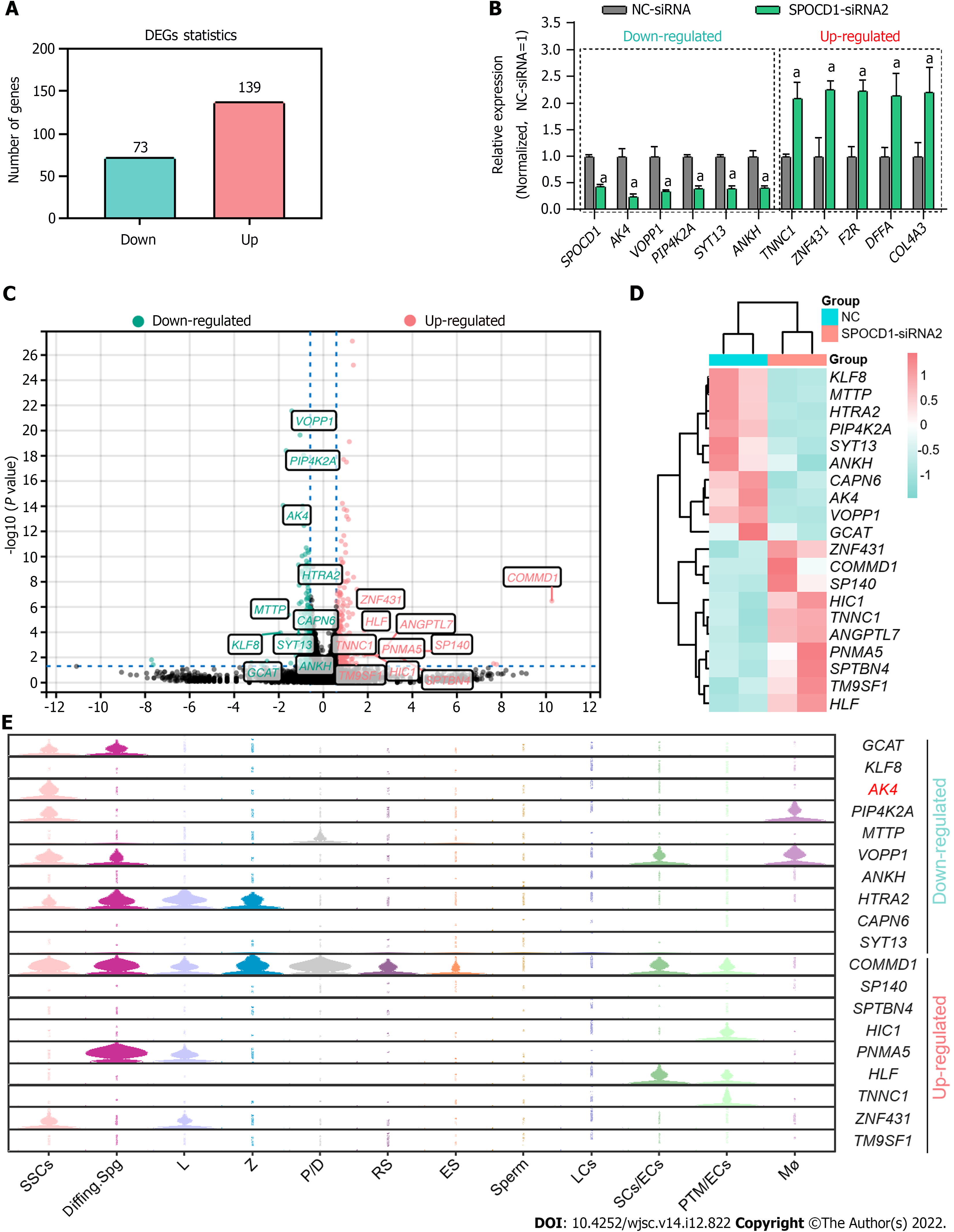
To explore the mechanisms of SPOCD1 in the proliferation and apoptosis of the SSC lines, we performed RNA-seq on cells after transfection with SPOCD1-siRNA. A total of about 20000 genes were detected. After excluding unrecognized reads and genes with fragments per kilobase of exon model per million reads mapped value < 0.001, 14556 genes were included for subsequent analysis (Supple
To verify the functions of AK4 in the SPOCD1-mediated proliferation of human SSCs, we re-expressed AK4 in SPOCD1-knockdown cells. Western blot analysis confirmed the transfection efficiency of SPOCD1-siRNA and AK4 expression plasmid (Figure 7A and B). CCK-8 (Figure 7C) and EdU (Figure 7D and E) results showed that the re-expression of AK4 significantly attenuated the growth inhibition conferred by SPOCD1 knockdown in human SSCs. Western blot analysis also showed that the re-expression of AK4 significantly restored the downregulation of PLZF and PCNA proteins caused by SPOCD1 knockdown (Figure 7F and G). We further examined the apoptosis level of the SSCs using fluorescence activated cell sorting. The results showed that the re-expression of AK4 significantly reversed the increased apoptosis resulting from SPOCD1 knockdown (Figure 7H and I), suggesting that AK4 is essential for SPOCD1-induced SSC proliferation.
NOA is one of the most serious male infertility disorders without effective treatment. According to the pathological examination of testicular tissue, NOA can be categorized as spermatogonia maturation arrest (Spg MA), spermatocyte maturation arrest (Spc MA), spermatid maturation arrest (Std MA), hypo-spermatogenesis (HS) and SC only syndrome (SCOS). SSCs are responsible for initiating adult spermatogenesis, and many studies have shown that the abnormal viability of SSCs impairs spermatogenesis. To explore whether SPOCD1 affected adult testicular function via SSCs, we examined the level and distribution of SPOCD1 in eight adult testes (Supplementary Figure 2) and the distribution changes of SPOCD1 in tissues by immunofluorescence staining with GFRA1 (Figure 8A). These findings revealed that the percentage of SPOCD1-positive cells was significantly decreased in testes diagnosed with Spc MA and Spg MA (Figure 8B). Additionally, there was no change in SPCOD1 intracellular localization; it remained in the nuclei. Western blot analysis showed that SPOCD1 levels were significantly downregulated in patients with Spg MA and Spc MA (Figure 8C and D). Our results indicate that SPOCD1 downregulation might be associated with spermatogenesis dysregulation in humans, but more evidence is needed to confirm these observations.
SSCs are responsible for long-term spermatogenesis by balancing self-renewal and differentiation[19]. Although many regulatory mechanisms were revealed in mouse SSCs and restored spermatogenesis in infertile mice by SSC transplantation[20], they were not conserved in humans and mice. Therefore, the regulatory mechanisms of human SSCs remain poorly understood. scRNA-seq has provided us with a transcriptional map of human SSCs, and various potential regulatory molecules of human SSCs have been discovered[21]. By analyzing the testis scRNA data from two studies and performing histological validation, SPOCD1 was found as a molecule specifically expressed in the early developmental stage of human SSCs.
SPOCD1 was first found to interact with testis protein phosphatase 1 in 2011[22]. It is a protein belonging to the transcription factor S-II family of transcription factors. SPOCD1 contains a SPOC domain that can regulate developmental progression and is considered a tumor-associated factor in various tumors[23]. It was shown to be significantly upregulated in many tumors including gastric cancer[24], glioblastoma[25], bladder cancer[26], and ovarian cancer[27]. Knockdown of SPOCD1 significantly inhibited the proliferation, migration, and invasion of gastric cancer cells in nude mice[24]. ADP ribosylation factor 5/Rab35 axis controlled the growth and invasiveness of glioblastoma by inhibiting the levels of SPOCD1[25]. SPOCD1 promotes ovarian cancer progression and inhibits apoptosis through the phosphoinositide 3-kinase/AKT pathway[28]. Recently, it was shown that the conditional knockout of SPOCD1 in the mice testis leads to spermatogenesis arrest in the pachytene stage[29]. Although the study demonstrated the importance of SPOCD1 in male fertility, it focused on the role of SPOCD1 in PIWI-interacting-directed de novo DNA methylation. The functions and mechanisms of SPOCD1 in spermatogenesis, especially in SSC fate determination, remain unknown. Our study found that SPOCD1 was mainly localized to human SSCs at an early stage. It was signi
AK4 is an adenylate kinase family member expressed in the mitochondrial matrix[30]. It is a phosphorylation enzyme that transfers phosphate from ATP or GTP to AMP, generating two molecules of ADP, which help to keep energy homeostasis by balancing the cellular adenine nucleotide composition[31]. AK4 plays important roles in energy metabolism and tumorigenesis. AK4 promotes lung adenocarcinoma metastasis by modulating oxidative stress and stabilizing hypoxia-inducible 1 alpha[30]. Increased expression of AK4 is involved in tamoxifen resistance through m6A-based epitranscriptomic mechanisms[32]. In addition, AK4 is also involved in energy metabolism, especially glycolysis[33-35]. Given that glycolysis is a major process for energy metabolism in SSCs to promote SSC self-renewal in mice[36], the association of AK4 with glycolysis in SSCs and its effects on SSC proliferation should be further confirmed in more studies.
Our study found many genes affected by SPOCD1 including AK4, KLF8, VOPP1, ZNF431, and COMMD1. We validated the functions of AK4 in SPOCD1-knockdown cells but did not clarify whether SPOCD1 influenced cell behavior through other pathways. SPOCD1 affects the proliferation of glioma cells via pentraxin 3[37]. Thus, whether SPOCD1 can mediate SSC fate determination through different ways remains further investigated. In addition, we tried to detect DNA fragments directly bound by SPOCD1 using the chromatin immunoprecipitation assay. Still, the results were not credible due to the lack of appropriate antibodies. In some NOA patients, we found the significant downregulation in SPOCD1, especially in Spg MA and Spc MA patients. However, it should be noted that the sample size included in our study was limited. According to recent reports[38,39], using computerized deep learning methods may help elucidate the relationship between SPOCD1 and NOA in large samples. A recent study showed that conditional silencing of the SPOCD1 gene in mouse testes resulted in blocked spermatogenesis at the pachytene spermatocyte stage[29]. However, we did not confirm whether SPOCD1 mutations or downregulation resulted in impaired spermatogenesis in humans. Further analysis of SPOCD1 mutations in NOA patients via whole-exome sequencing and validating the effects of SPOCD1 mutation would help to clarify the role of SPOCD1 in male reproduction.
sc-seq analysis showed that all testicular cells could be classified into 12 populations, with showed little difference from other reports[17,40]. During data quality control, we selectively retained cells where the percentage of mitochondrial genes was less than 15%. This may have contributed to fewer testicular cells being included. Furthermore, we divided SSCs into three states, which differ from other reports. According to Guo et al[17], there are five subtypes of SSCs and differentiating spermatogonia. Sohni et al[40] categorized all spermatogonium into five types. The difference could have been related to using different resolution parameters and reduction methods to reduce dimensionality, and only SSCs were included in our re-clustering analysis.
We demonstrated that SPOCD1 was predominantly localized to the human SSCs, and its downregulation suppressed cell proliferation and induced apoptosis. Re-expression of AK4 in SPOCD1 knockdown cells reversed the changes in cell proliferation and apoptosis. In addition, we also found that SPOCD1 was significantly downregulated in some patients with NOA. Thus, our study provides new insights into regulating human SSCs and new theories on the etiology of male infertility.
Spermatogonial stem cells (SSCs) are the origin of spermatogenesis, which continuously generates spermatozoa through self-renewal and differentiation. Although we have identified many molecules and pathways that regulate SSC function in mice, the mechanisms regulating human SSCs are not yet fully revealed.
To explore the regulatory mechanisms of human SSCs, we analyzed human testis single-cell RNA sequencing (scRNA-seq) data from the GSE149512 and GSE112013 datasets. We found that SPOC domain-containing protein 1 (SPOCD1) is differentially expressed in human SSCs. This study explored the role of SPOCD1 in human proliferation and apoptosis, which will help to expand the understanding of SSC regulation.
To investigate the functions and mechanisms of SPOCD1 in human proliferation and apoptosis, and to explore the potential effects on spermatogenesis.
In this study, scRNA-seq was used to detect differentially expressed genes in human SSCs, in which the SPOCD1 gene is highly expressed in human SSCs. Immunohistochemistry was used to investigate the expression pattern of SPOCD1 in human testicular tissue. Subsequently, we used small interfering RNA to knockdown SPOCD1 in human SSC lines and dissected the role of SPOCD1 in human SSCs by Cell Counting Kit-8, Western blot analysis, 5-ethynyl-2’-deoxyuridine, fluorescence-activated cell sorting, and terminal deoxynucleotidyl transferase dUTP nick end labeling. RNA-seq was used to explore gene expression alterations induced by SPOCD1 downregulation. Finally, we identified the functional target genes of SPOCD1 by rescue experiments.
The scRNA-seq and immunohistochemical results showed that SPOCD1 was predominantly localized to human SSCs. Knockdown of SPOCD1 in human SSC lines resulted in a significant decrease in cell proliferation and induced apoptosis. RNA-seq results showed that SPOCD1 knockdown caused the significant downregulation of genes such as adenylate kinase 4 (AK4) and affected pathways such as tumor necrosis factor and cyclic AMP. Overexpression of AK4 in SPOCD1 knockdown cells significantly responded to the changes in cell proliferation and apoptosis caused by SPOCD1 inhibition.
We demonstrated that SPOCD1 was predominantly localized to human SSCs and regulated its proliferation and apoptosis through AK4. Our study provides new insights into regulating human SSCs and potential novel targets for treating male infertility.
Future studies will explore the correlation between SPOCD1 and abnormal human spermatogenesis in large samples. These include screening for potentially curative mutations of SPOCD1 in azoospermia patients and exploring the association between abnormal SPOCD1 expression and azoospermia in large samples using deep learning.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Reproductive biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Abu Yousuf M, Bangladesh; Lomperta K, Poland S-Editor: Chen YL L-Editor: Filipodia P-Editor: Cai YX
| 1. | Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, Arafa M, Panner Selvam MK, Shah R. Male infertility. Lancet. 2021;397:319-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 629] [Article Influence: 157.3] [Reference Citation Analysis (0)] |
| 2. | Tharakan T, Luo R, Jayasena CN, Minhas S. Non-obstructive azoospermia: current and future perspectives. Fac Rev. 2021;10:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 3. | de Rooij DG. Stem cells in the testis. Int J Exp Pathol. 1998;79:67-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 237] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 4. | Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A. 1994;91:11303-11307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 811] [Cited by in RCA: 765] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 5. | Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 764] [Cited by in RCA: 758] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 6. | Medrano JV, Rombaut C, Simon C, Pellicer A, Goossens E. Human spermatogonial stem cells display limited proliferation in vitro under mouse spermatogonial stem cell culture conditions. Fertil Steril. 2016;106:1539-1549.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 7. | Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, Saarma M, Sariola H. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489-1493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1003] [Cited by in RCA: 985] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 8. | Yoshida S. Stem cells in mammalian spermatogenesis. Dev Growth Differ. 2010;52:311-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development. 2012;139:1734-1743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 10. | Chen W, Cui Y, Liu B, Li C, Du L, Tang R, Qin L, Jiang Y, Li J, Yu X, He Q, He Z. Hsa-miR-1908-3p Mediates the Self-Renewal and Apoptosis of Human Spermatogonial Stem Cells via Targeting KLF2. Mol Ther Nucleic Acids. 2020;20:788-800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Zhou F, Chen W, Cui Y, Liu B, Yuan Q, Li Z, He Z. miRNA-122-5p stimulates the proliferation and DNA synthesis and inhibits the early apoptosis of human spermatogonial stem cells by targeting CBL and competing with lncRNA CASC7. Aging (Albany NY). 2020;12:25528-25546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 12. | Zhou F, Yuan Q, Zhang W, Niu M, Fu H, Qiu Q, Mao G, Wang H, Wen L, Lu M, Li Z, He Z. MiR-663a Stimulates Proliferation and Suppresses Early Apoptosis of Human Spermatogonial Stem Cells by Targeting NFIX and Regulating Cell Cycle. Mol Ther Nucleic Acids. 2018;12:319-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Cui W, He X, Zhai X, Zhang H, Zhang Y, Jin F, Song X, Wu D, Shi Q, Li L. CARF promotes spermatogonial self-renewal and proliferation through Wnt signaling pathway. Cell Discov. 2020;6:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Du L, Chen W, Li C, Cui Y, He Z. RNF144B stimulates the proliferation and inhibits the apoptosis of human spermatogonial stem cells via the FCER2/NOTCH2/HES1 pathway and its abnormality is associated with azoospermia. J Cell Physiol. 2022;237:3565-3577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 15. | Zhou D, Fan J, Liu Z, Tang R, Wang X, Bo H, Zhu F, Zhao X, Huang Z, Xing L, Tao K, Zhang H, Nie H, Zhu W, He Z, Fan L. TCF3 Regulates the Proliferation and Apoptosis of Human Spermatogonial Stem Cells by Targeting PODXL. Front Cell Dev Biol. 2021;9:695545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen IC, Gildersleeve H, Lehle JD, Mayo M, Westernströer B, Law NC, Oatley MJ, Velte EK, Niedenberger BA, Fritze D, Silber S, Geyer CB, Oatley JM, McCarrey JR. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018;25:1650-1667.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 404] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 17. | Guo J, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie X, Guo Y, Takei Y, Yun J, Cai L, Kim R, Carrell DT, Goriely A, Hotaling JM, Cairns BR. The adult human testis transcriptional cell atlas. Cell Res. 2018;28:1141-1157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 443] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 18. | Hou J, Niu M, Liu L, Zhu Z, Wang X, Sun M, Yuan Q, Yang S, Zeng W, Liu Y, Li Z, He Z. Establishment and Characterization of Human Germline Stem Cell Line with Unlimited Proliferation Potentials and no Tumor Formation. Sci Rep. 2015;5:16922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 19. | de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21:776-798. [PubMed] |
| 20. | Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A. 1994;91:11298-11302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1102] [Cited by in RCA: 1080] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 21. | Chen Y, Zheng Y, Gao Y, Lin Z, Yang S, Wang T, Wang Q, Xie N, Hua R, Liu M, Sha J, Griswold MD, Li J, Tang F, Tong MH. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018;28:879-896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 305] [Cited by in RCA: 267] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 22. | Fardilha M, Esteves SL, Korrodi-Gregório L, Vintém AP, Domingues SC, Rebelo S, Morrice N, Cohen PT, da Cruz e Silva OA, da Cruz e Silva EF. Identification of the human testis protein phosphatase 1 interactome. Biochem Pharmacol. 2011;82:1403-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 23. | Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 390] [Cited by in RCA: 392] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 24. | Zhu M, Yan C, Ren C, Huang X, Zhu X, Gu H, Wang M, Wang S, Gao Y, Ji Y, Miao X, Yang M, Chen J, Du J, Huang T, Jiang Y, Dai J, Ma H, Zhou J, Wang Z, Hu Z, Ji G, Zhang Z, Shen H, Shi Y, Jin G. Exome Array Analysis Identifies Variants in SPOCD1 and BTN3A2 That Affect Risk for Gastric Cancer. Gastroenterology. 2017;152:2011-2021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 25. | Kulasekaran G, Chaineau M, Piscopo VEC, Verginelli F, Fotouhi M, Girard M, Tang Y, Dali R, Lo R, Stifani S, McPherson PS. An Arf/Rab cascade controls the growth and invasiveness of glioblastoma. J Cell Biol. 2021;220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | van der Heijden AG, Mengual L, Lozano JJ, Ingelmo-Torres M, Ribal MJ, Fernández PL, Oosterwijk E, Schalken JA, Alcaraz A, Witjes JA. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur J Cancer. 2016;64:127-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 27. | Wang C, Wang J, Shen X, Li M, Yue Y, Cheng X, Lu W, Wang X, Xie X. LncRNA SPOCD1-AS from ovarian cancer extracellular vesicles remodels mesothelial cells to promote peritoneal metastasis via interacting with G3BP1. J Exp Clin Cancer Res. 2021;40:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 28. | Liu D, Yang Y, Yan A. SPOCD1 accelerates ovarian cancer progression and inhibits cell apoptosis via the PI3K/AKT pathway. Onco Targets Ther. 2020;13:351-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 29. | Zoch A, Auchynnikava T, Berrens RV, Kabayama Y, Schöpp T, Heep M, Vasiliauskaitė L, Pérez-Rico YA, Cook AG, Shkumatava A, Rappsilber J, Allshire RC, O'Carroll D. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature. 2020;584:635-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 30. | Jan YH, Lai TC, Yang CJ, Lin YF, Huang MS, Hsiao M. Adenylate kinase 4 modulates oxidative stress and stabilizes HIF-1α to drive lung adenocarcinoma metastasis. J Hematol Oncol. 2019;12:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 31. | Jan YH, Tsai HY, Yang CJ, Huang MS, Yang YF, Lai TC, Lee CH, Jeng YM, Huang CY, Su JL, Chuang YJ, Hsiao M. Adenylate kinase-4 is a marker of poor clinical outcomes that promotes metastasis of lung cancer by downregulating the transcription factor ATF3. Cancer Res. 2012;72:5119-5129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Liu X, Gonzalez G, Dai X, Miao W, Yuan J, Huang M, Bade D, Li L, Sun Y, Wang Y. Adenylate kinase 4 modulates the resistance of breast cancer cells to tamoxifen through an m(6)a-based epitranscriptomic mechanism. Mol Ther. 2020;28:2593-2604. [RCA] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 33. | Wujak M, Veith C, Wu CY, Wilke T, Kanbagli ZI, Novoyatleva T, Guenther A, Seeger W, Grimminger F, Sommer N, Schermuly RT, Weissmann N. Adenylate Kinase 4-A Key Regulator of Proliferation and Metabolic Shift in Human Pulmonary Arterial Smooth Muscle Cells via Akt and HIF-1α Signaling Pathways. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 34. | Zhang S, Yamada S, Park S, Klepinin A, Kaambre T, Terzic A, Dzeja P. Adenylate kinase AK2 isoform integral in embryo and adult heart homeostasis. Biochem Biophys Res Commun. 2021;546:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 35. | Chin WY, He CY, Chow TW, Yu QY, Lai LC, Miaw SC. Adenylate Kinase 4 Promotes Inflammatory Gene Expression via Hif1α and AMPK in Macrophages. Front Immunol. 2021;12:630318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 36. | Kanatsu-Shinohara M, Tanaka T, Ogonuki N, Ogura A, Morimoto H, Cheng PF, Eisenman RN, Trumpp A, Shinohara T. Myc/Mycn-mediated glycolysis enhances mouse spermatogonial stem cell self-renewal. Genes Dev. 2016;30:2637-2648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 37. | Liu Q, Wang XY, Qin YY, Yan XL, Chen HM, Huang QD, Chen JK, Zheng JM. SPOCD1 promotes the proliferation and metastasis of glioma cells by up-regulating PTX3. Am J Cancer Res. 2018;8:624-635. [PubMed] |
| 38. | Aurna NF, Yousuf MA, Taher KA, Azad AKM, Moni MA. A classification of MRI brain tumor based on two stage feature level ensemble of deep CNN models. Comput Biol Med. 2022;146:105539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 39. | Ahamed KU, Islam M, Uddin A, Akhter A, Paul BK, Yousuf MA, Uddin S, Quinn JMW, Moni MA. A deep learning approach using effective preprocessing techniques to detect COVID-19 from chest CT-scan and X-ray images. Comput Biol Med. 2021;139:105014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 40. | Sohni A, Tan K, Song HW, Burow D, de Rooij DG, Laurent L, Hsieh TC, Rabah R, Hammoud SS, Vicini E, Wilkinson MF. The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep. 2019;26:1501-1517.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 229] [Article Influence: 45.8] [Reference Citation Analysis (0)] |