Published online Oct 26, 2021. doi: 10.4252/wjsc.v13.i10.1394
Peer-review started: March 18, 2021
First decision: June 16, 2021
Revised: June 21, 2021
Accepted: September 14, 2021
Article in press: September 14, 2021
Published online: October 26, 2021
Processing time: 221 Days and 18 Hours
Alternative ribonucleic acid (RNA) splicing can lead to the assembly of different protein isoforms with distinctive functions. The outcome of alternative splicing (AS) can result in a complete loss of function or the acquisition of new functions. There is a gap in knowledge of abnormal RNA splice variants promoting cancer stem cells (CSCs), and their prospective contribution in cancer progression. AS directly regulates the self-renewal features of stem cells (SCs) and stem-like cancer cells. Notably, octamer-binding transcription factor 4A spliced variant of octamer-binding transcription factor 4 contributes to maintaining stemness properties in both SCs and CSCs. The epithelial to mesenchymal transition pathway regulates the AS events in CSCs to maintain stemness. The alternative spliced variants of CSCs markers, including cluster of differentiation 44, aldehyde dehydrogenase, and doublecortin-like kinase, α6β1 integrin, have pivotal roles in increasing self-renewal properties and maintaining the pluripotency of CSCs. Various splicing analysis tools are considered in this study. LeafCutter software can be considered as the best tool for differential splicing analysis and identification of the type of splicing events. Additionally, LeafCutter can be used for efficient mapping splicing quantitative trait loci. Altogether, the accumulating evidence re-enforces the fact that gene and protein expression need to be investigated in parallel with alternative splice variants.
Core Tip: The alternative splicing machinery can produce various variants, associated with stemness characteristics of both stem cells (SCs) and cancer SCs. In this study, the role of spliced variants in SCs and stem-like cancer cells is reviewed. We highlight the importance of transcript-based expression concurrent with the gene and protein expression that leads to better understanding of self-renewal features of tumor cells.
- Citation: Ebrahimie E, Rahimirad S, Tahsili M, Mohammadi-Dehcheshmeh M. Alternative RNA splicing in stem cells and cancer stem cells: Importance of transcript-based expression analysis. World J Stem Cells 2021; 13(10): 1394-1416
- URL: https://www.wjgnet.com/1948-0210/full/v13/i10/1394.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i10.1394
Alternative ribonucleic acid (RNA) splicing is an emerging topic in molecular and clinical studies[1]. Alternative splicing (AS) is the key mechanism to generate a large number of messenger RNA (mRNA) transcripts from the relatively low number of human genes, which can lead to the assembly of different protein isoforms with distinct functions. This structural modification of gene transcripts and their encoded proteins is considered a vital process that increases diversity of protein functions in order to generate the complex cellular proteome[2,3].
Stem cells (SCs) are undifferentiated cells that are able to self-renewal, or differentiate into any types of differentiated cells. SCs can be found in both embryos and adult cells[4]. The self-renewal characteristics also can be found in cancer SCs (CSCs) within the tumor environment[5].
In this review, we discuss the significance of AS in determining the final fate of SCs. Also, the impact of AS events in promoting stemness features in CSCs is highlighted. To address all aspects related to AS, this review provides comprehensive detail regarding various types of AS tools.
The outcome of AS can result in a complete loss of function or the acquisition of new functions[2,3]. AS can also change the gene expression pattern in cancer cells. Exon-skipping (a form of alternative RNA splicing) in tumour suppressor genes can lead to truncated proteins, similar to classical nonsense mutations, resulting in cancer-specific AS in the absence of genomic mutations[6]. It has been demonstrated that nearly half of all active AS events are altered in ovarian and breast tumour cells compared to normal tissue[3].
There is ample evidence that AS coordinates significant changes in protein isoform expression and is the main cause of the functional diversity in proteins and proteome[7-10]. In humans, it is estimated that up to 94% of genes undergo AS, resulting in more than 100000 transcripts[7-9].
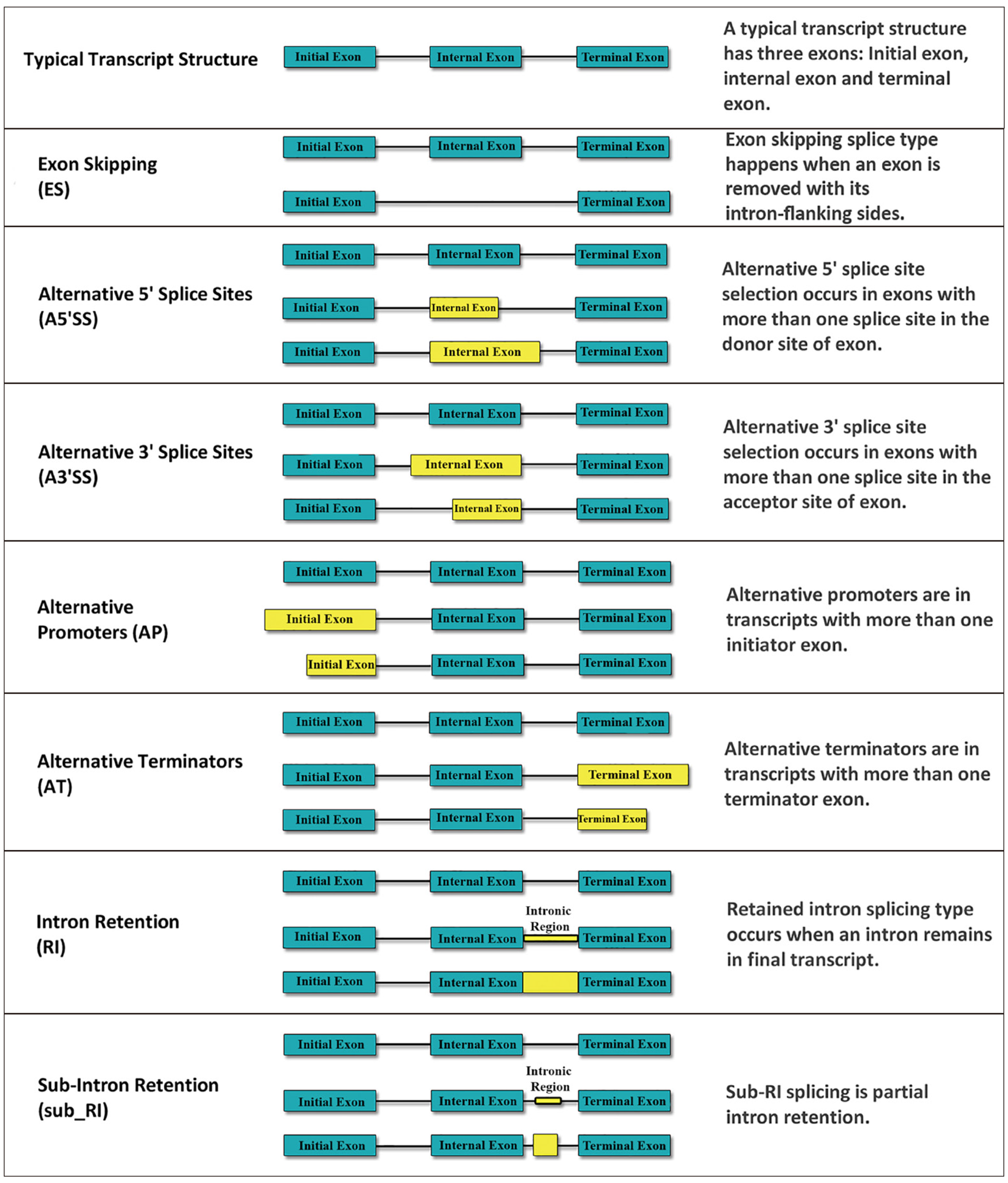
As presented in Figure 1, transcripts are products of precursor mRNAs (pre-mRNAs) splicing processes, where novel transcripts are discovered with increasing regularity and added to public databases, providing a valuable resource for analysis of AS[11]. During the transcription process, pre-mRNAs are produced. Then, through the RNA splicing process, the non-coding regions of pre-mRNAs (introns) are removed, and coding regions (exons) are joined, and they produce mature mRNAs[12]. Also, it is well-established that the more complex process (AS) may increase the diversity of the mRNAs which are expressed from a single gene. During AS, different exons can be extended or skipped, or introns can be retained within spliced transcripts and produce different mRNAs[13]. The majority (approximately ninety percent) of human genes are alternatively spliced[14]. AS has been considered as a gene expression regulator and miss-splicing can contribute to risk for various human diseases, like cancer[14]. With the advent of high-throughput sequencing methods, analysis of human genomic and transcriptomic has been efficiently developed. Using bioinformatics tools, the sequenced transcripts can be aligned to the genomic reference sequences to find AS events[15,16]. Five standard forms of AS events have been identified including skipped exon (SE, also known as cassette exons), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), retained intron (RI) or intron retention (IR), mutually exclusive exons (MXE). Also, alternative first exons (AFE), and alternative last exons (ALE) are considered as less common AS events[17].
Exon skipping is the most common AS event, in which the exon as a whole is skipped from the mature mRNA transcript[18]. During A3SS and A5SS events, exons are flanked on one side by a constitutive splice site (fixated) and on the opposite side are flanked by two (or more) competing for alternative splice sites, leading to an alternate region (extension) that either is included within the transcript or is excluded[17]. IR and MXE are the least abundant subtypes among all five major AS events. Normally, during pre-mRNA splicing, introns are fully spliced and excluded from the mature mRNA. However, during IR splicing, introns are retained in mature mRNAs. IR-spliced isoforms have different destinations, some of them may degrade by the nonsense-mediated decay (NMD) pathway, due to the existence of premature termination codons within introns. In some cases, further splicing is applied to the IR isoforms and helps them to escape NMD. NMD escaped-IR isoforms are translated into truncated proteins which have fewer or extra domains in their structure and increase the risk of diseases[19]. In the splicing of MXEs, exons are spliced in a coordinated manner. MXEs can leave the size of the spliced isoform products unchanged. In these isoforms, 1 out of 2 exons (or 1 group out of 2 exon groups) is retained, while the other one is spliced out[20].
There are more events that are not categorised among five standard forms including alternative AFE (alternative promoters) and ALE (alternative 3′ terminal exons). An AFE is the first exon of one splice variant of a gene, which is located downstream of some isoforms of this gene, or this exon is excluded from another isoform because it is located in the intronic region[21]. The AFE definition is also referred to as ALE in which the last exon (3′ terminal exon) may be retained or be excluded in different variants of a single gene[22].
There is a gap in knowledge of abnormal estrogen-associated RNA splice variants in breast cancer, and their prospective contribution to breast cancer progression while breast cancer is mainly a disease in which the sex hormone estrogen stimulates uncontrolled growth. Furthermore, their relative abundance and their potential to use AS in cancer diagnosis and treatment has been neglected.
In addition to the novelty of the subject, the major reasons for this shortcoming are: (1) Heavy computation and high computer skills are required for AS detection and analysis; and (2) A high depth of sequencing is required for comprehensive profiling of splicing events. Recently published in Scientific Reports (2018)[11], we developed a Windows-based, user-friendly tool for identifying AS events without the need for advanced computer skills. Additionally, this tool operates as an online module, and employs the SpliceGraphs module without the need for additional resource updates. First, SpliceGraph generates data based on the frequency of active splice sites in the pre-mRNA. Then, the presented approach compares the query transcript exons to SpliceGraph exons in the online genome browser ENSEMBL.
Our team in the Dame Roma Mitchell Cancer Research Laboratories is exceptionally well positioned to unravel the complexity of AS in breast cancer, and thereby, utilise the aberrant alternative splice variants unique to breast cancer as an accurate diagnosis tool. Moreover, this proposed project has the capacity to employ alternative splice variants as a determinant of better outcomes of disease—a completely novel paradigm of disease prevention and diagnosis.
There are various types of SCs. The highest differentiation potential which allows SCs to differentiate into any cell type of a whole organism is found in totipotent SCs, like a zygote. totipotent SCs can generate embryo and extra-embryonic structures in cells[23]. Pluripotent SCs (PSCs) are the other types of SCs that are not able to form extra-embryonic structures in cells, but they have the potential to differentiate into cells of three germ layers (endoderm, ectoderm, and mesoderm)[24]. Embryonic SCs (ESCs) and induced PSCs (iPSCs) are categorised in PSCs. ESCs are derived from the inner cell mass of a blastocyst (preimplantation embryos) and the indefinite self-renewal ability and plasticity are their vivid characteristics[25]. iPSCs are artificially derived from somatic cells, and their function and features are similar to PSCs. iPSCs have shown promising impacts on present and future regenerative medicine[26]. Another SC types are multipotent SCs which have limited differentiation abilities than PSCs and they only differentiate into a specific cell lineage. Haematopoietic SCs (HSCs) are multipotent SCs which can only differentiate into blood cells. Unipotent SCs have the least differentiation capabilities which they can only form one cell type, like dermatocytes[27].
Due to the similarities between cancer state and embryonic development, several studies have focused on the existence of CSCs within the tumor environment[5]. It has been well-established that tumor progression, anti-tumor drug resistance, and post-treatment tumor regeneration are driven by a special cancerous cell type, called CSCs. Generally, CSCs characterized by self-renewing, multipotent, and tumor-initiating properties. It is really difficult to detect CSCs within a tumor environment because some CSCs that lack specific markers have tumor regeneration abilities[28]. To illustrate, cluster of differentiation (CD) 133 marker firstly was utilized to isolate CSCs from colon carcinoma, but some CD133- metastatic colon carcinoma cells were found which had self-renewal properties as well as CD133+ CSCs[29]. Then, studies showed that CSCs could contain some specific subpopulation, like CD133- metastatic colon carcinoma cells, they are known as migrating SCs subpopulation[30]. So, in several solid tumors, the subpopulation of migrating and resident SCs can be found which they have been identified by various surface markers including CD24, CD29, CD44, CD90, CD133, aldehyde dehydrogenase 1 (ALDH1), and epithelial-specific antigen. These markers can be used as a potential target for developing anti-CSCs specific therapies[31].
The differentiation of SCs and their progenitors relies on various molecular controls associated with the gene expression regulation, including chromatin modification, transcription factors, post-transcriptional regulation by AS, microRNAs (miRNAs), and post-translational modifications. Advanced technologies, as an illustration of high-throughput RNA sequencing (RNA-seq), have demonstrated the contribution of AS patterns in SCs maintenance and differentiation[32].
ESCs are pluripotent cells which able to self-renew, and they have the ability to differentiate into the endoderm, the ectoderm, and the mesoderm cells[33]. Hence, they are the best model for monitoring early embryonic development. Also, ESCs are the potential source for developing differentiated cells for therapeutic approaches in regenerative medicine. One of the first genome-wide studies on splicing patterns in the ESCs reported more than thousands of AS events in ESCs[32]. The results of one study revealed that human and mouse ESC-related AS events are mostly found in genes associated with the cytoskeleton (dystonin, adducin 3), plasma membrane (dynamin 2, integrin subunit alpha 6), and kinase activity (calcium/calmodulin dependent serine protein kinase, microtubule affinity regulating kinase 2, and mitogen-activated protein kinase kinase 7)[34].
Both RNA-seq and splicing microarrays studies reported AS events, particularly those associated with changing protein sequence[15]. Also, the gene expression alteration of the proteins that regarded as splicing regulators was detected during developmental stages, suggesting various AS events during the process of differentiation[16]. For example, increasing expression of splicing regulators like muscle blind-like (MBNL1 and MBNL2) RNA binding proteins (RBPs) was shown during ESC differentiation which is highly associated with ESC-differential AS. The presence of the MBNL motif downstream and upstream of the flanking intronic sequences is associated with exon skipping and exon inclusion in ESC respectively[34]. It has been revealed that the AS events of genes that they contribute to ESC differentiation foster this process, like embryonic SC-specific event in forkhead box p1 (FOXP1), FOXP1-ES. The FOXP1-ES spliced isoform stimulates the expression of genes, including octamer-binding transcription factor 4 (OCT4), nanog homeobox (NANOG), nuclear receptor subfamily 5 group A member 2, and growth differentiation factor 3 which these transcription factors are required for pluripotency. Along with maintaining ESC pluripotency, FOXP1-ES showed effective involvement in the reprogramming of somatic cells to iPSCs[35,36]. The fibroblast growth factor 4 splice isoform (FGF4si) of FGF4 is another example of transcription factor that play major roles in ESC fate determination and promotes differentiation at later stages of development[37].
One of the most important regulators of self-renewal in ESCs is POU domain proteins (POU class 5 homeobox 1, also known as OCT4) and its gene expression is stimulated by FOXP1-ES. OCT4 has various isoforms including OCT4A, OCT4B, OCT4B1, OCT4B2, and OCT4B4. The expression of OCT4A only has been detected in ESCs and embryonal carcinoma cells (ECCs). The OCT4B detected in differentiated cells and OCT4B1 had higher expression levels in ESCs and ECCs rather than non-pluripotent cells[38]. Also, it has been revealed that expression patterns of OCT4B2 and OCT4B4 are similar to OCT4A (they highly expressed in undifferentiated cells), and like OCT4B1, their expression is lower in differentiated cells[39,40].
AS events also have found as critical factors to control the hematopoiesis process which during this process the HSCs produce mature blood cells in the bone marrow. The importance of AS in hematopoiesis has been identified through the analysis of AS related to RUNX family transcription factor 1 (RUNX1) which is a critical transcription factor for this process. RUNX1a isoforms are generated from RUNX1 by exon 6 AS and increase the capacity of the HSC pool[41]. Moreover, multiple isoforms of the other HSC-specific genes (homeobox A9, Meis homeobox 1, PR/SET domain 16, and HLF transcription factor-PAR bZIP family member) have been found using whole-transcriptome splicing of murine HSCs[42]. Also, a bioinformatics analysis revealed that the IR in HSC-specific transcripts led to a decrease in the expression of genes involved in DNA binding and RNA processing. This process consequently promoted the NMD pathway in HSCs[43,44].
Along with ESCs and HSCs, there are neural SCs (NSCs) that are responsible for generating neurons and glia during embryonic development. The nervous system applies AS for cell differentiation, morphogenesis, and the formation of complex neuronal networks. The three major alternatively spliced isoforms of Quaking (Qki) have been found in NSCs. Qki5 is one of these isoforms which regulates NSC functions[45]. Another example of AS involvement in the transition of SC to neuron cells is the existence of exon 10 in the mRNA of polypyrimidine tract binding protein 2 (PTBP2) protein[46]. Interestingly, it has been cleared that the downregulation levels of the other paralog of this protein, polypyrimidine tract binding protein 1 (PTBP1), inhibit the existence of exon 10 of PTBP2[47]. Hence, in neural cells, the neuron-specific microRNA miR-124 contributes to the downregulation of PTBP1[48]. On the other hand, there is another ubiquitous RNA-binding protein which suppresses PTB
SCs in normal tissues are capable of renewing themselves, also, stem-like cells within tumors, which have been called CSCs have the ability of self-renewing to seed new tumors. Hence, they have also been termed “tumor-initiating cells”[51].
Due to gene expression regulation at the transcriptional level, the contribution of AS events has been clearly reported in tumor-related biological processes like proliferation, cell death, migration, and angiogenesis. AS changes transcriptome and proteome profile in human cells, therefore, its deregulation may greatly contribute to tumor plasticity[9]. Effectively, AS plays a regulatory role in maintaining the balance between pluripotency and differentiation of human ESCs during embryogenesis and tissue differentiation. Hence, defective AS machinery could mimic the oncogenic effects in non-tumorigenic cells. Results of an RNA-seq-based study on mammary cells revealed that AS events regulated by serine and arginine rich splicing factor 1 altered in human tumors. These defective AS events led to an enhancement of the proliferation and decreased apoptosis in MCF-10A cell cultures[52]. The involvement of defective AS events in controlling tumor heterogeneity suggesting that they could also lead to re-programming of stem pathways, triggering metastasis, and tumor progression[53].
Phenotypic conversions of cells between epithelial and mesenchymal states, known as epithelial-mesenchymal transition (EMT), is activated during metastasis and enhances the re-activation of stem pathways. Hence, EMT is associated with tumor aggressiveness and resistance of cancer cells to anti-tumor drugs[54]. Various types of regulators including cytokines and growth factors are dysregulated in cancer cells which mainly involved in promoting EMT. EMT influences the mRNA maturation of some splicing factor-like epithelial splicing regulatory protein (ESRP). This splicing factor regulates the Wnt signaling pathway through exon 4 skipping of T-cell factor 4 (TCF4). The activation of TCF4 is prompted by nuclear localization of beta-catenin and they are major transcriptional mediators of the canonical Wnt signaling pathway[55]. It has been revealed that, during EMT, the transactivation of exon 4 carrying TCF4 isoforms is reduced. On the contrary, the lack of exon 4 Led to surge Wnt signaling during EMT[56].
Moreover, ESRP-splicing factors can alter splicing of the Fibroblast growth factor receptor 2 (FGFR2)[57]. There are two different isoforms of FGFR2 including IIIb and IIIc which have pivotal roles in ligand binding specificity. ESRPs mainly help to the production of FGFR2-IIIb by inhibiting exon IIIc. The FGFR2-IIIc levels increase in the absence of ESRPs[58]. Spliced transcripts of FGFR2 were detected in primary tumors, and significantly enriched in metastases and tumor plasticity[59].
Also, the association between EMT and the emergence of CSCs has been identified[60]. Studies showed that EMT induces AS events in genes involved in stem-like cancer cells. NUMB endocytic adaptor protein (NUMB) is an endocytic adaptor protein that has various functions in cell polarity maintenance, cell migration, and EMT. The importance of NUMB AS has been reported in various cancers including breast[61,62], lung cancer[63], and hepatocellular carcinoma cells[64]. Four NUMB isoforms based on the AS of exon 3 and exon 9 have been identified in vertebrates[65]. Exon 9 mainly founds in SCs rather than differentiated cells, also enhanced expression of NUMB exon 9 has been clearly shown in various cancer types. Interestingly, MEK/ERK signaling pathway regulated NUMB exon 9 splicing[66]. In cancer cells, EMT triggers NUMB exon skipping, which leads to enhance invasive properties, and abnormal AS events in NUMB may also affect the balance between stem-like and non-stem cancer cells[67].
One of the most important SC factors which contribute to embryogenesis and pluripotency of cancer stem-like cells is a member of POU family genes called OCT4. This SC factor plays a pivotal role in self-renewal and pluripotency properties in ESCs and ECCs[68]. AS leads to produce three main isoforms of the human OCT4, including OCT4A, OCT4B, and OCT4B1[38]. OCT4A contributes to maintaining stemness properties in ESCs and ECCs. On the contrary, OCT4B isoforms are not able to show these features[38,39].
Another gene that its spliced variants have been found in CSCs is Kirsten rat sarcoma viral oncogene homolog (KRAS). The occurrence of mutation and AS in this gene results in the production of transformed proteins that promote malignancies[69]. The spliced variants of KRAS are KRAS4A and KRAS4B which are known as minor and major isoforms respectively and in CSCs the splicing process of KRAS is regulated by the RNA binding motif protein 39 splicing complex. Hypoxic state in tumor environment activates the expression of KRAS4A in CSCs and there is a correlation between the expression of KRAS4A isoform and SC marker ALDH in cells with stem-like properties. KRAS4A spliced variant controls the metabolic requirements in SCs particularly the level of adenosine triphosphate and lactate. These metabolites increase in the absence of KRAS4A. Furthermore, Chen et al[70] reported that Indisulam, which is a novel sulfonamide compound with potential antineoplastic activity, is an inhibitor of KRAS4A splicing, but has no obvious effect on KRAS4B. Also, previously the invol
Erb-B2 receptor tyrosine kinase 2 (HER2) is a member of the epidermal growth factor receptor family of receptor tyrosine kinases and its overexpression is a common feature of invasive breast carcinomas. Cells that express HER2 receptors considered as HER2-positive breast cancer. The full-length HER2 is known as wild-type HER2 (wtHER2) which has about 1255 amino acids. While the altered form of HER2 has been identified with the absence of sixteen amino acids from the extracellular domain (deletion of exon 16). This HER2 splice variant is known as d16HER2 which highly enriched in the regulation of the breast CSCs (BCSCs) activity by its functional interaction with the notch receptor 1 family members[72]. Also, d16HER2 spliced variants are involved in initiation and aggressiveness of tumors, CSC properties, EMT, and the trastuzumab susceptibility of HER2 positive BC cells compared with wtHER2[73].
Another example of EMT-promoting splicing changes is occurred by two splicing factors including QKI and RNA-binding Fox proteins (RBFOX1/RBFOX2). QKI and RBFOX2 are responsible for exon skipping of cortactin transcript during EMT[74]. The roles of QKI and RBFOX1 in establishing the SC features in breast tumor cells and regulating the EMT have been declared by applying exon 30 splicing (exon skipping) in the actin-binding protein FLNB[75].
One of the most well-known cell surface adhesion receptors of CSCs is CD44 which also famous as a CSC marker. CD44 is a non-kinase transmembrane glycoprotein comprised of 20 exons which AS leads to produce two different isoforms including the standard (CD44s) and variant (CD44v) isoforms[76]. Exons 1–5 and 16–20 are involved in CD44s isoform and make it the smallest isoform which is expressed by the mesenchymal cells. On the other hand, the middle nine exons (exons 6-15 of the genomic DNA) can be alternatively spliced and located between exons 1–5 domain and exons 16–20 region which form CD44v isoforms[77]. CD44 activates by binding to its main ligand, hyaluronic acid. Activated CD44 Leads to cell proliferation and metastasis[78]. CD44s isoforms predominantly express on hematopoietic and mesenchymal cells and the CD44s isoforms abundance in cancer cells induces stem-like features[79]. This protein is highly expressed in many cancer cells and its alternative spliced variants play a critical role in tumor progression.
Previously, it has been reported that switching from CD44v to CD44s isoform led to promoting EMT in cells. Cancer cells that undergo an EMT acquire SC-like properties, and CD44 expression increases in these cells, while the expression levels of splicing factor ESRP reduced[54]. Splicing factor ESRP contributes to switching CD44v isoform into CD44s which is required for promoting EMT[80]. Various studies have been done to reveal the exact role of CD44 isoforms in cancer cells. It has been reported that increasing tumor cell survival, invasiveness, and migration is associated with CD44s[81]. Ouhtit et al[82] showed the increased expression of CD44 in metastatic breast tumors and they provided in vivo evidence for the role of the CD44s isoform in promoting breast tumor invasion and metastasis to the liver. Moreover, Hiraga et al[83] declared that CD44 can induce functional properties in CSCs and its CD44s variant contributes to the promotion of bone metastases. Besides CD44s, there is some evidence that proved the role of CD44v in enhancing CSC activities[84,85]. Also, protecting CSCs from reactive oxygen species-induced stress by interacting with a glutamate-cystine transporter that subsequently promotes tumor growth[86].
One of the driving factors that control EMT-associated splicing changes are ESRP-1 and -2 splicing factors. ESRP1 inhibits CSC traits by inhibiting the production of CD44s. Increased expression of CD44s upon EMT-induction leads to the invasive phenotype in cancer cells. Also, the results of bioinformatics analysis of the Cancer Genome Atlas program of breast cancer showed that there is a significant negative correlation between the ratio of the CD44 isoforms and the ESRP1 splicing factor[79]. Also, it has been proved that the expression levels of ESRP1 decreased in triple-negative breast cancer (TNBC) compared with non-TNBC samples. Along with direct regulatory effects of ESR1 on CD44s isoform, this splicing factor also controls the CSCs functions by splicing α6β1 integrin. α6 integrin subunit (CD49f) is a well-known biomarker for BCSC and other CSCs and there are two splice variants for this subunit including α6Aβ1 and α6Bβ1 isoforms[87,88]. ESRP1 impedes CSC function by inducing the expression of the α6Aβ1 splice variant and repressing α6Bβ1 integrin isoform[58,89].
Despite the above arguments, there are some controversies about the role of ESRP1 in promoting[90] or inhibiting[89] CSC properties. The results of a study which was conducted by Yae et al[90] seem contradictory. They reported that ESRP1 contributes to breast cancer metastasis through a mechanism in which these splicing factors stimulate the upregulation of the CD44v isoforms and results in lung metastasis of breast cancer cells accompanied by the expansion of stem-like cancer cells[90]. Also, Hu et al[91] revealed that CD44v is expressed on CD24-/CD44+ breast CSCs which enhances the risk of metastasis to the lung, rather than cells that expressing CD44s.
ALDH is a common CSC marker which catalyses the oxidation of aldehydes. There are 19 various types of ALDH in the human genome. Upregulation of ALDHs has found in SCs and CSCs[92]. However, ALDH1 mainly considered as SCs and CSCs markers and contributes in self-renewal activity which has three isoenzymes ALDH1A1, ALDH1A2, and ALDH1A3. Among these isotypes, ALDH1A1 is the most prominent SC marker in renal cell carcinoma (RCC) tumor, breast cancer[93], colon cancer[94], and is linked to tumorigenesis, mortality, and self-renewal activity[95].
Another marker of CSCs in the gastrointestinal tract is doublecortin-like kinase 1 (DCLK1) which mostly correlated with tumor initiation, EMT, and progression[96,97]. In RCC tumor, DCLK1 alternative spliced variants (DCLK1 ASVs) overexpressed compared to control samples. DCLK1-long isoforms (Isoforms 2 and 4) are associated with RCC recurrence and they mainly co-expressed with renal tumor SC markers including ALDH1A1, C-X-C motif chemokine receptor 4, and CD44. It has been proved that a high level of DCLK1 alternative transcript (Isoform 2) promotes the expression of RCC SC markers and increases self-renewal activity[98].
In summary, the AS machinery could regulate the self-renewal features of stem-like cancer cells. This process is performed by producing spliced variants of cancer cell markers. The most important spliced variants of CSCs markers are CD44s, CD44v, and α6Bβ1 integrin because they have found on the surface of various CSCs. Also, the splicing factor ESRP is responsible for the splicing changes of both CD44 isoforms and α6Bβ1 integrin. The Table 1 represents the association of the alternative RNA splice variants and their activities in both SCs and CSCs[99-105].
| Type | Gene | Ensembl ID | Alternative RNA splice variants | Activities | Ref. |
| Non-cancerous stem cell | IGF1 | ENSG00000017427 | IGF1Ec, IGF1 Ea | IGF-IEc enhances proliferation and represses muscle progenitors differentiation, IGF-IEa activates anabolic pathways | [99] |
| POU5F1 (OCT4) | ENSG00000204531 | OCT4A, OCT4B, OCT4B1 | Play roles in pluripotency and self-renewal of embryonic stem cells | [38] | |
| RBM4, RBM14, CoAA | ENSG00000173933 | CoAZ, ncCoAZ | Influence co-transcriptional splicing | [100] | |
| DNMT3B | ENSG00000088305 | DNMT3B3, DNMT3B3∆5 | DNMT3B3∆5 expressed in ESCs and functionally distinct from DNMT3B3 | [101] | |
| VEGFA | ENSG00000112715 | VEGF120, VEGF164, VEGF188 | All promote MSC proliferation; some enhance paracrine signaling, osteogenic, or endothelial differentiation | [102] | |
| FGF4 | ENSG00000075388 | FGF4, FGF4si | FGF4 is important to stem cell maintenance, while FGFsi antagonizes some of FGF4’s activity | [37] | |
| PKCd | ENSG00000163932 | PKCdI, PKCdII | PKCdI is caspasecleavable and PKCdII is caspase in-cleavable | [103] | |
| POU2F2 (OCT2) | ENSG00000028277 | OCT2.2, OCT2.4 | Oct2.2 is sufficient to induce neural differentiation in mouse ESCs, Oct2.4 is able to block neural differentiation | [104] | |
| RMB14, CoAA | ENSG00000239306 | CoAA, CoAM | CoAA is downregulated in favor of CoAM during early embryonic development | [105] | |
| RUNX1 | ENSG00000159216 | RUNX1a | Increases the capacity of the HSC pool | [41] | |
| Qki | ENSG00000112531 | Qki5 | Regulates neural stem cell function | [45] | |
| PTB | ENSG00000117569 | nPTB (PTBP2) with exon 10 | Transition of stem cell to neuron cells | [46] | |
| FOXP1 | ENSG00000114861 | FOXP1-ES | Promotes the maintenance of ESC pluripotency and contributes to efficient reprogramming of somatic cells into induced pluripotent stem cells | [35] | |
| Cancer stem cell | NUMB | ENSG00000133961 | NUMB exon skipping by EMT | Affect the balance between stem-like and non-stem cancer cells | [67] |
| KRAS | ENSG00000133703 | KRAS4A | Enriched in the cancer stem cell population and helps to modulate the metabolic requirements and stress responses associated with the cancer stem-progenitor cell transition | [70] | |
| HER2 | ENSG00000141736 | D16her2 | Role in the regulation of the BC stem cells (BCSCs) activity through its functional interaction with the NOTCH family members | [72] | |
| CD44 | ENSG00000026508 | CD44s | CD44s isoforms in cancer cells induces stem-like features | [79] | |
| CD44 | ENSG00000026508 | CD44v | Promote CSC activities and protecting CSCs from ROS-induced stress | [86] | |
| α6β1 integrin (ITGA5, VLA-6, CD49f/CD29) | ENSG00000091409 | α6Bβ1 integrin | Promotes the function of breast CSCs and tumor initiation | [89] | |
| FLNB | ENSG00000136068 | FLNB exon 30 skipping | Associated with EMT gene signatures in basal-like breast cancer/ establishing the mesenchymal and stem-like cell state in breast cancers | [75] | |
| DCLK1 | ENSG00000133083 | DCLK1-long isoforms (Isoforms 2 and 4) | Promotes expression of RCC stem cell markers and increases self-renewal activity | [98] | |
| POU5F1 (OCT4) | ENSG00000204531 | OCT4A | Promotes stemness properties in embryonal carcinoma cells | [38] | |
| ALDH1 | ENSG00000165092 | ALDH1A1 | Promotes tumorigenesis, mortality, and self-renewal activity in RCC | [98] |
Since the pivotal role of AS events has been cleared in increasing self-renewal properties and maintaining the pluripotency of CSCs, it is highly important to detect differential splicing using computational approaches. Sequencing methods have paved the way to survey AS. Initial studies by microarray profiling and EST-cDNA sequence data reported that about two-thirds of human multi-exon genes are alternatively spliced[106]. Then, high-throughput sequencing technologies provide a high depth of coverage and sensitivity to identify human AS. For the first time to survey the splicing complexity, the Genome Analyzer system of Illumina was used by Pan et al[7] Their results proved the effectiveness of the RNA-seq method to analyze AS events. However, it has been found that the RNA-seq method is not sufficient itself due to its short sequencing reads (approximately 100-150 bp), and needs a number of computational approaches to monitor differential splicing[107].
There are three approaches to perform differential splicing analysis, including exon-based, isoform-based, and event-based methods. Mainly, exon-based and event-based methods are categorized in one strategy called the count-based method (Table 2)[108-146].
| Tools | Human/ plants/ animals | Types of detecting splices | Type of tool | Year of publish | Ref. | Citation |
| Isoform-based method | ||||||
| Cuffdiff2 | All | - | Linux | 2012 | [109] | 9550 |
| HMMSplicer | All | - | Linux | 2010 | [110] | 79 |
| PennDiff | All | - | Linux | 2018 | [111] | 9 |
| rSeqDiff | All | - | R | 2013 | [112] | 29 |
| DiffSplice | All | - | Windows/Linux | 2013 | [113] | 141 |
| NSMAP | All | - | MatLab package | 2011 | [114] | 46 |
| MISO | All | SE / A3SS / A5SS / MXE / TandemUTR / RI / AF / AL | Linux | 2010 | [115] | 1166 |
| Exon-based method | ||||||
| TopHaT | All | - | Linux | 2009 | [116] | 11026 |
| DEXSeq | All | - | R | 2012 | [117] | 1091 |
| edgeR | All | - | R | 2010 | [118] | 19012 |
| JunctionSeq | All | - | R | 2016 | [119] | 81 |
| limma | All | - | R | 2015 | [120] | 11033 |
| rSeqNP | All | - | R | 2015 | [121] | 10 |
| Event-based method | ||||||
| MAJIQ | All | SE / A5SS / A3SS | Linux | 2016 | [122] | 171 |
| rMATS | All | SE / A5SS / A3SS / RI / MXE | Linux | 2014 | [123] | 746 |
| SUPPA | All | SE / A5SS / A3SS / RI / MXE / AF / AL | Linux | 2015 | [124] | 134 |
| SUPPA2 | All | SE / A5SS / A3SS / RI / MXE / AF / AL | Linux | 2018 | [125] | - |
| ASGAL | All | SE / A5SS / A3SS / RI | Linux | 2018 | [126] | 12 |
| Astalavista version 3.0 | All | SE / A5SS / A3SS / RI | Linux | 2012 | [127] | 62 |
| LeafCutter | All | SE / A5SS / A3SS | Linux | 2018 | [128] | 200 |
| SpliceGrapher | All | SE / A5SS / A3SS / RI | Linux | 2012 | [129] | 132 |
| KisSplice | All | SNPs, indels and AS events (SE / A5SS / A3SS / RI ) | Linux | 2012 | [130] | - |
| Alt Event Finder | All | SE | Linux | 2012 | [131] | 24 |
| Matt | All | SE / A5SS / A3SS / RI | Linux | 2018 | [132] | 14 |
| ALEXA-seq | All | SE / A5SS / A3SS / RI | Linux | 2010 | [133] | 340 |
| Outrigger | All | SE / A5SS / A3SS / RI | Linux | 2016 | [134] | 86 |
| ASDT | All | SE / A5SS / A3SS / RI | Perl | 2018 | [135] | 5 |
| SplicePie | All | SE / RI | R | 2015 | [136] | 17 |
| VAST-TOOLS | All | SE / A5SS / A3SS / RI | R | 2017 | [137] | 118 |
| spliceR | All | SE / A5SS / A3SS / RI / AF/AL | R | 2014 | [138] | 80 |
| Pro-Splicer | Human | SE / A5SS / A3SS | Website (http://prosplicer.mbc.nctu.edu.tw/) | 2003 | [139] | 50 |
| ASPicDB | Human | SE / A5SS / A3SS / RI / AF/AL | Website (http://srv00.recas.ba.infn.it/ASPicDB/) | 2006 | [140] | 33 |
| H-DBAS | Human | SE / A5SS / A3SS / RI / AF/AL | Website (http://www.h-invitational.jp/h-dbas/) | 2007 | [141] | 56 |
| ASIP | Plant | SE / A5SS / A3SS / RI | Website (http://www.plantgdb.org/ASIP/) | 2006 | [142] | 535 |
| ASG | Human | SE / A5SS / A3SS / RI | Website (https://brcwebportal.cos.ncsu.edu/asg/) | 2004 | [143] | 116 |
| VastDB | vertebrate | SE / A5SS / A3SS / RI | Website (https://vastdb.crg.eu/wiki/Main_Page) | 2017 | [137] | 118 |
| Astalavista | Human/ Animals | SE / A5SS / A3SS / RI | Website (http://astalavista.sammeth.net/) | 2007 | [144] | 205 |
| SpliceDitector | Plant/ Human | SE / A5SS / A3SS / RI | Windows | 2018 | [11] | 1 |
| dSpliceType | All | SE / A5SS / A3SS / RI / MXE | Windows/ Linux | 2015 | [145] | 4 |
| AltAnalyze | All | SE / A5SS / A3SS / RI / MXE | Windows/ Linux | 2010 | [146] | 253 |
Isoform-based methods are based on the reconstruction of full-length transcripts at the first step, then these methods estimate the relative isoform abundance across samples or conditions. Following that, statistical testing is used to identify the significant differences in the relative transcript abundances among various experimental conditions and samples. It is noticeable to say that the effectiveness of this method relies on accurate transcript quantification. Some of the isoform-based tools have been developed including cuffdiff2, HMMSplicer, PennDiff, rSeqDiff, DiffSplice, NSMAP, and MISO. The last two mentioned tools are also able to detect splicing events while it is not possible for others.
Cufflinks is an isoform-based pipeline which contains three programs including Cufflinks, Cuffmerge, and Cuffdiff. Cufflinks first applies transcript assembly by generating overlap graphs with fragments and quantifying the aligned reads. Transcript abundance of a transcript is then estimated in form of FPKM (fragments per kilobase per million mapped fragments). Then, using Cuffmerge, collected assemblies are merged to create a consensus reference. Cuffdiff2 finally performs different tests for detecting differentially expressed genes and differential isoform changes are calculated by applying a one-sided t-test[147].
HMMSplicer is a precise algorithm for analyzing canonical and non-canonical splice junctions in short-read datasets. HMMSplicer firstly divides each read in half, then seeds the halves to the reference genome and based on Hidden Markov Model, finds the exon boundary. At final points, a score is assigned to each junction, based on the alignment strength, number, and quality of bases supporting the splice junction. The true and false positives can be distinguished perfectly using this scoring algorithm and lead to find novel both canonical and non-canonical splice junctions[110]. PennDiff, as an accurate statistical method takes information regarding both gene structures and pre-estimated isoform relative abundances into consideration, then analyzes differential AS or transcription for RNA-seq data[111].
rSeqDiff is implemented as an R package for the detection of differential isoform expression from multiple RNA-Seq samples using the hierarchical likelihood ratio test. rSeqDiff can considering three cases for analysis, including genes with no differential expression, genes with differential expression without differential splicing, and genes with differential splicing[112].
DiffSplice is additionally another isoform-based method for the detection and visualization of differential transcription. The DiffSplice approach is not based on transcript or gene annotations, it overcomes the requirement for full transcript inference and quantification, which may be a challenge due to short read length. So, what makes DiffSplice distinct from other methods is that this tool uses a divide-and-conquer approach to seek out the difference between transcriptomes within the variety of AS modules (ASMs), where transcript isoforms separate. The abundance of various AS isoforms existing in each ASM is calculated for every sample and is compared across sample groups. A non-parametric statistical test is used for each ASM to demonstrate significant differential transcription with a controlled false discovery rate (FDR)[113].
NSMAP (non-negativity and sparsity constrained maximum a posteriori) model is provided to estimate the expression levels of isoforms using RNA-seq data. Like DiffSplice, NSMAP does not require annotation information. This tool drives the structures of isoforms and estimates the expression levels simultaneously[114].
MISO (mixture of isoforms) model with the help of bayesian inference estimates the expression level of alternatively spliced genes from RNA-seq data. Also, it is a probabilistic framework that takes RNA-seq data of single-end or paired-end to perform more accurate AS analysis at either the exon or isoform level. Interestingly, MISO provides confidence intervals for estimates of exon and isoform abundance, detects differential expression, and uses latent information to enhance accuracy. Also, this tool using GFF annotations can generate various types of AS events including SE, A3SS/A5SS, MXE, TandemUTR, RI, AFE, and ALE[115].
Count-based methods comprised of both exon-based and event-based models. In exon-based approaches read counts are assigned to different features, such as exons or junctions. The main difference between these two approaches is that exon-based methods are not able to provide the type of splicing event; they just estimate the differentially expressed exons/junctions between samples. Tools which have been developed based on the exon-based methods are TopHaT, DEXSeq, edgeR, Junc
TopHat is a software package that finds spliced junctions ab initio by large-scale mapping of RNA-seq reads. TopHat performs mapping the reads using Bowtie (http://bowtie-bio.sourceforge.net), an ultra-fast short-read mapping program to reference genome[116]. Also, TopHat is considered as a prior separately running tool for other software including Alt Event Finder, FineSplice, SplicingCompass, and NSMAP[148]. Firstly, the RNA-Seq data is processed by TopHat to find the splicing junctions. Then, NSMAP re-counts all the possible isoforms formed by the combinations of collected exons from TopHat, and identifies the expressed isoforms and their expression level estimations[114].
DEXSeq is another exon-based method which uses a linear model to detect differential splicing genes from RNA-seq data[117]. Identifying differential expression levels of genes, exons, or transcripts is also done by the edgeR package. Moreover, using a negative binomial generalized log-linear model, edgeR can be used to analyze the count data. Then, the expression levels are calculated by comparing the logFC of an exon to the logFC of the entire gene[118]. The JunctionSeq package works based on the statistical approach of DEXSeq to calculate the differential exon usage and exon junctions[119].
Limma package is a well-known R package for detecting differential gene expression also provides differential splicing using exon count data. Limma integrates various statistical principles to perform large-scale expression studies accurately. First, this package applies a linear model to calculate differential expression tests for the exon-level expression data. Then, the exon-level statistics are turned to gene-level statistics for detecting differential spliced genes[120].
rSeqNP like other exon-based methods is able to test differential expression and differential splicing of genes. This tool uses standard non-parametric tests based on ranks of expression values of genes and isoforms[121].
Event-based methods through calculating the percentage spliced in (PSI) values can generate splicing events including SE, A3SS/A5SS, MXE, RI, AFE, and ALE. There are various types of software in this category which some are available as python, R, Perl packages, or others are accessible through online websites. Some of the most important tools are MAJIQ, rMATS, LeafCutter, SUPPA/SUPPA2, ASGAL, spliceR, SpliceDitector, etc.
MAJIQ (modeling alternative junction inclusion quantification) is able to find splice graphs and local splice variations (LSV) using RNA-seq data and transcriptome annotation file. An LSV can be determined as a split in a splice graph or from a single exon (reference exon). Single source LSV is related to splits from a reference exon to multiple 3’ splice sites in downstream exons, while, single target LSV is related to multiple 5’ splice sites spliced to an upstream reference exon. Both simple splicing events and complex transcript variations can be included in the LSVs. The MAJIQ builder and MAJIQ quantifier make up MAJIQ. The MAJIQ builder tries to detect known and novel LSVs and construct a splice graph for genes collected from RNA-seq data and transcriptome annotation. Following that, the MAIJQ quantifier applies Bayesian PSI modeling and bootstrapping to estimates PSI values for each quantified LSV. MAJIQ is mainly used with the Voila package which is a visualization tool that using the output of MAJIQ (builder and quantifier), creates interactive summary files with gene splice graphs, LSVs, and their quantification[122].
MATS (multivariate analysis of transcript splicing) is a Bayesian statistical framework for the identification of differential spliced genes obtained from RNA-Seq data. MATS applies multivariate uniform before estimating the correlation of exon splicing between samples, also it uses a Markov Chain Monte Carlo (MCMC) method along with a simulation-based adaptive sampling method to report the P value and FDR of differential AS[149]. rMATS is a developed version of the MATS method. rMATS uses a hierarchical framework to account for sampling uncertainty in individual replicates and variability among both paired and unpaired replicates between sample groups, also it can estimate the PSI of each event[123].
LeafCutter is an event-based method developed to analyze samples and population variation in intron splicing using short-read RNA-seq data. LeafCutter can detect differential splicing between sample groups, and used for mapping splicing quantitative trait loci (sQTLs). So, analyzing sQTLs by LeafCutter can help to identification of disease-associated variants. To compare LeafCutter and MAJIQ tools, MAJIQ is used to predict local splicing variation using split-reads and identification of complex splicing events, but MAJIQ does not scale properly more than thirty samples and has not been adapted to map sQTLs. The LeafCutter output of AS events is mainly focused on SEs, 5′ and 3′ alternative splice site usage, and additional complex events that can be summarized by differences in intron excision. Comparison of LeafCutter to other methods for differential splicing analysis revealed that the majority of the introns that LeafCutter reported as the most significant differentially spliced, shared a splice site with rMATS and MAJIQ. It can be considered that LeafCutter is able to detect the same differentially spliced events[123].
SUPPA is another event-based method which uses RNA-seq data to calculate PSI for differential spliced events[124]. SUPPA and its different version, SUPPA2, are able to consider AFE and ALE events coupled with other standard types of events. SUPPA2 has more advantages, including working with various replicates per condition and with different conditions. Also, SUPPA2 is able to cluster the differentially spliced events among various conditions to determine common splicing patterns and regulatory mechanisms[125].
Alternative Splicing Graph ALigner (ASGAL) is an event-based tool that uses RNA-Seq samples and a gene annotation to detect AS events. Firstly, ASGAL constructs the splicing graph from the gene annotations. Then, the RNA-reads alignment is done against the constructed splicing graph of the input gene. Finally, AS events are determined. The most prominent feature of ASGAL is that this tool can have higher accuracy to predict events. Because as compared with other tools which perform spliced alignment against a reference genome, ASGAL tries to detect novel splice sites based on a splicing graph[126].
SpliceR is an R package to detect AS events uses the full-length transcript output from RNA-seq data. For each event, spliceR annotates the genomic coordinates of the differentially spliced elements, to enhance downstream sequence analysis. Moreover, the possibility of the coding potential and NMD sensitivity of each transcript are determined by spliceR[138].
SpliceDetector is a windows-based user-friendly software for the identification of AS events directly from transcripts without any computer skill requirement or database download. Furthermore, to construct splicing graphs, data updating is not necessary because SpliceDetector uses the updated information deposited in the Ensembl database. To use SpliceDetector software, first, it takes human, plant, and model organisms transcript IDs as input. Then, it constructs a SpliceGraph based on all of the exon coordinates of the related gene. Finally, AS events are provided as output[11].
The critical role of AS in producing different mRNA variants, and even non-coding RNAs (ncRNAs), form one gene is well-established. AS can directly enhance transcriptomic and proteomic diversity. AS is regulated by RBPs which monitor the splicing machinery by binding to the pre-RNA. AS can be subjected to epigenetic regulation, like DNA methylation and histone modifications, and regulation by ncRNAs[150]. Long ncRNA (lncRNAs) are ncRNAs mainly longer than 200 nucleo
MiRNA are small non-coding RNAs, about 23 nucleotides in length, play role in gene expression regulation by pairing with complementary sequences in protein-coding transcripts[154]. The microRNA-290 cluster maintains the pluripotency and stemness properties of ESCs by monitoring AS, also known as a master regulator of AS. The splicing factors MBNL1/2 are targeted by one of the members of this cluster, miR-294, and the downregulation levels of MBNL1/2 have been found in ESCs and during reprogramming of iPSCs. Also, it has been reported that the majority of AS events are regulated by miR-294 through MBNL1/2 repression and this repression leads to up-regulation of other splicing factors in ESCs, so miR-294-dependent AS events are enhanced[155].
Also, the indirect involvement of some specific spliced variants in the miRNA expression regulation process has been demonstrated in CSCs. CSCs of human head and neck squamous cell carcinoma (HNSCC) are mostly characterized by CD44v3 isoforms which generate through alternative mRNA splicing of CD44. Also, ALDH1 markers along with the high expression of transcription factors OCT4, SRY-box transcription factor 2 (SOX2), and NANOG are the other features of HNSCCs. In CSCs, Oct4-Sox2-Nanog TFs are activated by binding the CD44v3 to its ligand hyaluronan and these TFs have binding sites on miR-302 promoter. So, the interaction between hyaluronan-induced CD44v3-spliced variants and mentioned TFs plays a pivotal role in the downregulation of miR-302 target genes (like epigenetic regulators: Lysine-specific histone demethylase and DNA methyltransferase 1), and this process is critical for the acquisition of CSC features[156].
Measuring the expression of alternatively spliced mRNAs, instead of overall gene expression, is considered as a new and more accurate approach for marker discovery in cancer research[157]. For example, many apoptotic regulatory genes, such as BCL-x, encode for alternatively spliced protein variants with opposing functions: One apoptotic and the other anti-apoptotic[158]. In recent years, there has been a discernible shift in cancer diagnosis and therapy due to the perception about the role of genes and their proteins. Cancer can occur irrespective of changes in expression of a gene or protein, but rather as a result of aberrant splice variants that are linked to cancer progression and/or drug resistance and is compensated by the decreased expression of other splice variants originating from that same gene.
AS contributes to a range of phenotypic traits of tumours as they progress and metastasis, and is a potential target for gene therapy[6,9]. With the advent of next-generation sequencing technology (NGS) and bioinformatics approaches, studying AS patterns has been performed in both cancerous and non-cancerous cells with increasing details. However, assembled transcripts obtained from NGS technology are not complete because this technology could sequence short reads. Hence, NGS platform may not be suitable enough for AS analysis. To overcome this limitation, the Pacific Bioscience (PacBio) platform has been developed based on the single-molecule real-time sequencing technology. PacBio technology provides full-length transcript sequencing without assembly. The PacBio full-length transcriptome data is the most accurate source to investigate AS[159]. AS analysis has been performed in various plant and mammalian species using PacBio platform[160-162].
Some cancer-specific differential spliced isoforms have been identified which can be used in cancer diagnosis and as potential targets for selective anti-tumor treatments in the future[163]. Currently, single-cell transcriptomic studies have paved the way for analyzing the gene-level expression and identification of isoforms originating from the same gene, to determine the distinct gene expression signatures within cells, especially tumor cells. Also, single-cell techniques have provided a powerful method for identifying CSCs[164]. There are some controversies regarding the AS pattern in single-cells. Some evidence reported that several spliced transcripts exist in single-cells. Faigenbloom et al[165] measured pairs of included and skipped isoforms obtained from spliced exons in single-cells. While, other studies demonstrated that single-cells express only one transcript variant or the dominance of a single transcript variant[166,167]. Further work in single-cell AS analysis is required to unravel the potential future of using AS events to develop personalized medicine for various diseases including cancer.
Unravelling AS opens a new avenue towards the establishment of new diagnostic and prognostic markers of cancer progression and metastasis as well as the development of a new generation of anticancer therapeutics: Treatments that inhibit specific splice variants, rather than targeting genes. Although the significant roles of alternatively spliced transcripts in promoting self-renewal properties of CSCs have been identified, more studies are needed to identify the whole CSCs-related splicing events to strengthen the therapeutic benefits of AS in the future.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: Statistical Society of Australia, 48799844.
Specialty type: Oncology
Country/Territory of origin: Australia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tanabe S, Zhang Q S-Editor: Chang KL L-Editor: A P-Editor: Yu HG
| 1. | Pajares MJ, Ezponda T, Catena R, Calvo A, Pio R, Montuenga LM. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 192] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 2. | Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 620] [Cited by in RCA: 673] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 3. | Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, Lucier JF, Thibault P, Rancourt C, Tremblay K, Prinos P, Chabot B, Elela SA. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 281] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 4. | Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 248] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 5. | Yu Z, Pestell TG, Lisanti MP, Pestell RG. Cancer stem cells. Int J Biochem Cell Biol. 2012;44:2144-2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 498] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 6. | Wang L, Duke L, Zhang PS, Arlinghaus RB, Symmans WF, Sahin A, Mendez R, Dai JL. Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer. Cancer Res. 2003;63:4724-4730. [PubMed] |
| 7. | Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2521] [Cited by in RCA: 2820] [Article Influence: 165.9] [Reference Citation Analysis (0)] |
| 8. | Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4232] [Cited by in RCA: 3870] [Article Influence: 227.6] [Reference Citation Analysis (0)] |
| 9. | Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311-5318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 493] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 10. | Lara-Pezzi E, Desco M, Gatto A, Gómez-Gaviro MV. Neurogenesis: Regulation by Alternative Splicing and Related Posttranscriptional Processes. Neuroscientist. 2017;23:466-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Baharlou Houreh M, Ghorbani Kalkhajeh P, Niazi A, Ebrahimi F, Ebrahimie E. SpliceDetector: a software for detection of alternative splicing events in human and model organisms directly from transcript IDs. Sci Rep. 2018;8:5063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1307] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 13. | Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 586] [Cited by in RCA: 910] [Article Influence: 113.8] [Reference Citation Analysis (0)] |
| 14. | Martinez-Montiel N, Rosas-Murrieta NH, Anaya Ruiz M, Monjaraz-Guzman E, Martinez-Contreras R. Alternative Splicing as a Target for Cancer Treatment. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 15. | Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, Clark TA, Williams A, Blume JE, Samal E, Mercola M, Merrill BJ, Conklin BR. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010;107:10514-10519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 183] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 16. | Revil T, Gaffney D, Dias C, Majewski J, Jerome-Majewska LA. Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics. 2010;11:399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 17. | Hiller M, Platzer M. Widespread and subtle: alternative splicing at short-distance tandem sites. Trends Genet. 2008;24:246-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Sorek R, Shemesh R, Cohen Y, Basechess O, Ast G, Shamir R. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617-1623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 90] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Zheng JT, Lin CX, Fang ZY, Li HD. Intron Retention as a Mode for RNA-Seq Data Analysis. Front Genet. 2020;11:586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 20. | Pohl M, Bortfeldt RH, Grützmann K, Schuster S. Alternative splicing of mutually exclusive exons--a review. Biosystems. 2013;114:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, Ishii S, Sugiyama T, Saito K, Isono Y, Irie R, Kushida N, Yoneyama T, Otsuka R, Kanda K, Yokoi T, Kondo H, Wagatsuma M, Murakawa K, Ishida S, Ishibashi T, Takahashi-Fujii A, Tanase T, Nagai K, Kikuchi H, Nakai K, Isogai T, Sugano S. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 390] [Cited by in RCA: 392] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 22. | Dutertre M, Chakrama FZ, Combe E, Desmet FO, Mortada H, Polay Espinoza M, Gratadou L, Auboeuf D. A recently evolved class of alternative 3'-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nat Commun. 2014;5:3395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Biehl JK, Russell B. Introduction to stem cell therapy. J Cardiovasc Nurs. 2009;24:98-103; quiz 104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 165] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 24. | Donovan PJ, Gearhart J. The end of the beginning for pluripotent stem cells. Nature. 2001;414:92-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 203] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 25. | Vazin T, Freed WJ. Human embryonic stem cells: derivation, culture, and differentiation: a review. Restor Neurol Neurosci. 2010;28:589-603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 26. | Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 378] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 27. | Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 937] [Cited by in RCA: 952] [Article Influence: 158.7] [Reference Citation Analysis (35)] |
| 28. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47087] [Article Influence: 3363.4] [Reference Citation Analysis (5)] |
| 29. | Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, Chadburn A, Murphy AJ, Valenzuela DM, Gale NW, Thurston G, Yancopoulos GD, D'Angelica M, Kemeny N, Lyden D, Rafii S. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111-2120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 429] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 30. | Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1047] [Cited by in RCA: 1056] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 31. | Xia P. Surface markers of cancer stem cells in solid tumors. Curr Stem Cell Res Ther. 2014;9:102-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 32. | Pritsker M, Doniger TT, Kramer LC, Westcot SE, Lemischka IR. Diversification of stem cell molecular repertoire by alternative splicing. Proc Natl Acad Sci U S A. 2005;102:14290-14295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 33. | Ramirez JM, Gerbal-Chaloin S, Milhavet O, Qiang B, Becker F, Assou S, Lemaître JM, Hamamah S, De Vos J. Brief report: benchmarking human pluripotent stem cell markers during differentiation into the three germ layers unveils a striking heterogeneity: all markers are not equal. Stem Cells. 2011;29:1469-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, Wang E, Trcka D, Thompson T, O'Hanlon D, Slobodeniuc V, Barbosa-Morais NL, Burge CB, Moffat J, Frey BJ, Nagy A, Ellis J, Wrana JL, Blencowe BJ. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 255] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 35. | Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, Nedelec S, Wichterle H, Woltjen K, Hughes TR, Zandstra PW, Nagy A, Wrana JL, Blencowe BJ. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 295] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 36. | Chen K, Dai X, Wu J. Alternative splicing: An important mechanism in stem cell biology. World J Stem Cells. 2015;7:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 37. | Mayshar Y, Rom E, Chumakov I, Kronman A, Yayon A, Benvenisty N. Fibroblast growth factor 4 and its novel splice isoform have opposing effects on the maintenance of human embryonic stem cell self-renewal. Stem Cells. 2008;26:767-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 38. | Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068-3074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 222] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 39. | Poursani EM, Mehravar M, Soltani BM, Mowla SJ. OCT4B2, a novel alternative spliced variant of OCT4, is significantly upregulated under heat-stress condition and downregulated in differentiated cells. Tumour Biol. 2017;39:1010428317724280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 40. | Poursani EM, Mehravar M, Soltani BM, Mowla SJ. Novel variant of OCT4B4 is differentially expressed in human embryonic stem and embryonic carcinoma cells. Gene. 2017;627:369-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 41. | Komeno Y, Yan M, Matsuura S, Lam K, Lo MC, Huang YJ, Tenen DG, Downing JR, Zhang DE. Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice. Blood. 2014;123:3760-3769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 42. | Goldstein O, Meyer K, Greenshpan Y, Bujanover N, Feigin M, Ner-Gaon H, Shay T, Gazit R. Mapping Whole-Transcriptome Splicing in Mouse Hematopoietic Stem Cells. Stem Cell Reports. 2017;8:163-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 43. | Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, Thoeng A, Khoo TL, Bailey CG, Holst J, Rasko JE. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 351] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 44. | Gao Y, Vasic R, Halene S. Role of alternative splicing in hematopoietic stem cells during development. Stem Cell Investig. 2018;5:26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 45. | Hayakawa-Yano Y, Suyama S, Nogami M, Yugami M, Koya I, Furukawa T, Zhou L, Abe M, Sakimura K, Takebayashi H, Nakanishi A, Okano H, Yano M. An RNA-binding protein, Qki5, regulates embryonic neural stem cells through pre-mRNA processing in cell adhesion signaling. Genes Dev. 2017;31:1910-1925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 46. | Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17:265-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 257] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 47. | Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636-1652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 432] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 48. | Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1149] [Cited by in RCA: 1085] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 49. | Lin JC, Tarn WY. RBM4 down-regulates PTB and antagonizes its activity in muscle cell-specific alternative splicing. J Cell Biol. 2011;193:509-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 50. | Su CH, D D, Tarn WY. Alternative Splicing in Neurogenesis and Brain Development. Front Mol Biosci. 2018;5:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 136] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 51. | Peiris-Pagès M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 364] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 52. | Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, Allain FH, Krainer AR. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol Cell. 2015;60:105-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 257] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 53. | Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 524] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 54. | Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6972] [Cited by in RCA: 6804] [Article Influence: 400.2] [Reference Citation Analysis (0)] |
| 55. | Hrckulak D, Janeckova L, Lanikova L, Kriz V, Horazna M, Babosova O, Vojtechova M, Galuskova K, Sloncova E, Korinek V. Wnt Effector TCF4 Is Dispensable for Wnt Signaling in Human Cancer Cells. Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 56. | Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wöhrle S, Hecht A. Alternative splicing of Tcf7 L2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucleic Acids Res. 2010;38:1964-1981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 57. | Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591-601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 497] [Cited by in RCA: 461] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 58. | Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286-3300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 299] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 59. | Zhao Q, Caballero OL, Davis ID, Jonasch E, Tamboli P, Yung WK, Weinstein JN; Kenna Shaw for TCGA research network, Strausberg RL, Yao J. Tumor-specific isoform switch of the fibroblast growth factor receptor 2 underlies the mesenchymal and malignant phenotypes of clear cell renal cell carcinomas. Clin Cancer Res. 2013;19:2460-2472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 60. | Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741-4751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2076] [Cited by in RCA: 2072] [Article Influence: 138.1] [Reference Citation Analysis (0)] |
| 61. | Zhang Y, Dho SE, Simpson CD, Othman K, Bondoc A, McGlade CJ. Isoform-specific functions of Numb in breast cancer progression, metastasis and proteome remodeling. 2021 Preprint. Available from: bioRxiv: 429237. [DOI] [Full Text] |
| 62. | Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 374] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 63. | Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, Pece S, Di Fiore PP. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293-22298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 317] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 64. | Lu Y, Xu W, Ji J, Feng D, Sourbier C, Yang Y, Qu J, Zeng Z, Wang C, Chang X, Chen Y, Mishra A, Xu M, Lee MJ, Lee S, Trepel J, Linehan WM, Wang X, Neckers L. Alternative splicing of the cell fate determinant Numb in hepatocellular carcinoma. Hepatology. 2015;62:1122-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 65. | Dho SE, French MB, Woods SA, McGlade CJ. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem. 1999;274:33097-33104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 159] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 66. | Rajendran D, Zhang Y, Berry DM, McGlade CJ. Regulation of Numb isoform expression by activated ERK signaling. Oncogene. 2016;35:5202-5213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 67. | Wang Z, Sandiford S, Wu C, Li SS. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J. 2009;28:2360-2373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 68. | Mohiuddin IS, Wei SJ, Kang MH. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 69. | Jancík S, Drábek J, Radzioch D, Hajdúch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol. 2010;2010:150960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 241] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 70. | Chen WC, To MD, Westcott PM, Delrosario R, Kim I-J, Philips M, Trans Q, Bayani N, Balmain A. Regulation of KRAS4A/B splicing in cancer stem cells by the RBM39 splicing complex. 2019 Preprint. Available from: bioRxiv: 646125. [DOI] [Full Text] |
| 71. | Pells S, Divjak M, Romanowski P, Impey H, Hawkins NJ, Clarke AR, Hooper ML, Williamson DJ. Developmentally-regulated expression of murine K-ras isoforms. Oncogene. 1997;15:1781-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 72. | Castagnoli L, Ghedini GC, Koschorke A, Chiodoni C, Nanni P, Tagliabue E, Pupa SM. Role of d16HER2 splice variant in breast cancer stem cells. Eur J Cancer. 2016;61:S42. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 73. | Volpi CC, Pietrantonio F, Gloghini A, Fucà G, Giordano S, Corso S, Pruneri G, Antista M, Cremolini C, Fasano E, Saggio S, Faraci S, Di Bartolomeo M, de Braud F, Di Nicola M, Tagliabue E, Pupa SM, Castagnoli L. The landscape of d16HER2 splice variant expression across HER2-positive cancers. Sci Rep. 2019;9:3545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 74. | Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2014;33:1082-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 129] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 75. | Li J, Choi PS, Chaffer CL, Labella K, Hwang JH, Giacomelli AO, Kim JW, Ilic N, Doench JG, Ly SH, Dai C, Hagel K, Hong AL, Gjoerup O, Goel S, Ge JY, Root DE, Zhao JJ, Brooks AN, Weinberg RA, Hahn WC. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. Elife. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 76. | Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 503] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 77. | Orian-Rousseau V, Sleeman J. CD44 is a multidomain signaling platform that integrates extracellular matrix cues with growth factor and cytokine signals. Adv Cancer Res. 2014;123:231-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 89] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 78. | Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 865] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 79. | Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, Deng Y, Hu X, Zhang J, Gao XD, Kang Y, Mercurio AM, Goel HL, Cheng C. CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 2019;33:166-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 158] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 80. | Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 506] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 81. | Zhao P, Xu Y, Wei Y, Qiu Q, Chew TL, Kang Y, Cheng C. The CD44s splice isoform is a central mediator for invadopodia activity. J Cell Sci. 2016;129:1355-1365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 82. | Ouhtit A, Abd Elmageed ZY, Abdraboh ME, Lioe TF, Raj MH. In vivo evidence for the role of CD44s in promoting breast cancer metastasis to the liver. Am J Pathol. 2007;171:2033-2039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 94] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 83. | Hiraga T, Ito S, Nakamura H. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res. 2013;73:4112-4122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 183] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 84. | Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G, Gulotta G, Dieli F, De Maria R, Stassi G. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 578] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 85. | Lau WM, Teng E, Chong HS, Lopez KA, Tay AY, Salto-Tellez M, Shabbir A, So JB, Chan SL. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014;74:2630-2641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 171] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 86. | Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, Masuko T, Shimizu T, Ishikawa T, Kai K, Takahashi E, Imamura Y, Baba Y, Ohmura M, Suematsu M, Baba H, Saya H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 837] [Cited by in RCA: 937] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 87. | Vieira AF, Ricardo S, Ablett MP, Dionísio MR, Mendes N, Albergaria A, Farnie G, Gerhard R, Cameselle-Teijeiro JF, Seruca R, Schmitt F, Clarke RB, Paredes J. P-cadherin is coexpressed with CD44 and CD49f and mediates stem cell properties in basal-like breast cancer. Stem Cells. 2012;30:854-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 88. | Goel HL, Pursell B, Chang C, Shaw LM, Mao J, Simin K, Kumar P, Vander Kooi CW, Shultz LD, Greiner DL, Norum JH, Toftgard R, Kuperwasser C, Mercurio AM. GLI1 regulates a novel neuropilin-2/α6β1 integrin based autocrine pathway that contributes to breast cancer initiation. EMBO Mol Med. 2013;5:488-508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 89. | Goel HL, Gritsko T, Pursell B, Chang C, Shultz LD, Greiner DL, Norum JH, Toftgard R, Shaw LM, Mercurio AM. Regulated splicing of the α6 integrin cytoplasmic domain determines the fate of breast cancer stem cells. Cell Rep. 2014;7:747-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 90. | Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, Wada T, Masuko T, Mogushi K, Tanaka H, Osawa T, Kanki Y, Minami T, Aburatani H, Ohmura M, Kubo A, Suematsu M, Takahashi K, Saya H, Nagano O. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 304] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 91. | Hu J, Li G, Zhang P, Zhuang X, Hu G. A CD44v+ subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death Dis. 2017;8:e2679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 92. | Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev Rep. 2011;7:292-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 405] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 93. | Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3173] [Cited by in RCA: 3062] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 94. | Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382-3389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 831] [Cited by in RCA: 828] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 95. | Wang K, Chen X, Zhan Y, Jiang W, Liu X, Wang X, Wu B. Increased expression of ALDH1A1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Med Oncol. 2013;30:574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 96. | Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, Isomura A, Kawada K, Sakai Y, Yanagita M, Kageyama R, Kawaguchi Y, Taketo MM, Yonehara S, Chiba T. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 330] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 97. | Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N, Matsui W, Maitra A, Leach SD. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. 2014;146:245-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 98. | Ge Y, Weygant N, Qu D, May R, Berry WL, Yao J, Chandrakesan P, Zheng W, Zhao L, Zhao KL, Drake M, Vega KJ, Bronze MS, Tomasek JJ, An G, Houchen CW. Alternative splice variants of DCLK1 mark cancer stem cells, promote self-renewal and drug-resistance, and can be targeted to inhibit tumorigenesis in kidney cancer. Int J Cancer. 2018;143:1162-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 99. | Ates K, Yang SY, Orrell RW, Sinanan AC, Simons P, Solomon A, Beech S, Goldspink G, Lewis MP. The IGF-I splice variant MGF increases progenitor cells in ALS, dystrophic, and normal muscle. FEBS Lett. 2007;581:2727-2732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 100. | Brooks YS, Wang G, Yang Z, Smith KK, Bieberich E, Ko L. Functional pre-mRNA trans-splicing of coactivator CoAA and corepressor RBM4 during stem/progenitor cell differentiation. J Biol Chem. 2009;284:18033-18046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 101. | Gopalakrishnan S, Van Emburgh BO, Shan J, Su Z, Fields CR, Vieweg J, Hamazaki T, Schwartz PH, Terada N, Robertson KD. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic DNA methylation patterns and displays altered DNA binding. Mol Cancer Res. 2009;7:1622-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 102. | Lin H, Shabbir A, Molnar M, Yang J, Marion S, Canty JM Jr, Lee T. Adenoviral expression of vascular endothelial growth factor splice variants differentially regulate bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2008;216:458-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 103. | Patel NA, Song SS, Cooper DR. PKCdelta alternatively spliced isoforms modulate cellular apoptosis in retinoic acid-induced differentiation of human NT2 cells and mouse embryonic stem cells. Gene Expr. 2006;13:73-84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 104. | Theodorou E, Dalembert G, Heffelfinger C, White E, Weissman S, Corcoran L, Snyder M. A high throughput embryonic stem cell screen identifies Oct-2 as a bifunctional regulator of neuronal differentiation. Genes Dev. 2009;23:575-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 105. | Yang Z, Sui Y, Xiong S, Liour SS, Phillips AC, Ko L. Switched alternative splicing of oncogene CoAA during embryonal carcinoma stem cell differentiation. Nucleic Acids Res. 2007;35:1919-1932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 106. | Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141-2144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1028] [Cited by in RCA: 1055] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 107. | Hooper JE. A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Hum Genomics. 2014;8:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 108. | Liu R, Loraine AE, Dickerson JA. Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinformatics. 2014;15:364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 109. | Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8810] [Cited by in RCA: 9310] [Article Influence: 716.2] [Reference Citation Analysis (0)] |
| 110. | Dimon MT, Sorber K, DeRisi JL. HMMSplicer: a tool for efficient and sensitive discovery of known and novel splice junctions in RNA-Seq data. PLoS One. 2010;5:e13875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 111. | Hu Y, Lin J, Hu J, Hu G, Wang K, Zhang H, Reilly MP, Li M. PennDiff: detecting differential alternative splicing and transcription by RNA sequencing. Bioinformatics. 2018;34:2384-2391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 112. | Shi Y, Jiang H. rSeqDiff: detecting differential isoform expression from RNA-Seq data using hierarchical likelihood ratio test. PLoS One. 2013;8:e79448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 113. | Hu Y, Huang Y, Du Y, Orellana CF, Singh D, Johnson AR, Monroy A, Kuan PF, Hammond SM, Makowski L, Randell SH, Chiang DY, Hayes DN, Jones C, Liu Y, Prins JF, Liu J. DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res. 2013;41:e39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 114. | Xia Z, Wen J, Chang CC, Zhou X. NSMAP: a method for spliced isoforms identification and quantification from RNA-Seq. BMC Bioinformatics. 2011;12:162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 115. | Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009-1015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1194] [Cited by in RCA: 1005] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 116. | Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105-1111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10271] [Cited by in RCA: 9376] [Article Influence: 586.0] [Reference Citation Analysis (0)] |
| 117. | Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008-2017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1033] [Cited by in RCA: 1117] [Article Influence: 85.9] [Reference Citation Analysis (0)] |
| 118. | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22632] [Cited by in RCA: 29079] [Article Influence: 1817.4] [Reference Citation Analysis (0)] |
| 119. | Hartley SW, Mullikin JC. Detection and visualization of differential splicing in RNA-Seq data with JunctionSeq. Nucleic Acids Res. 2016;44:e127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 120. | Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16184] [Cited by in RCA: 25645] [Article Influence: 2564.5] [Reference Citation Analysis (0)] |
| 121. | Shi Y, Chinnaiyan AM, Jiang H. rSeqNP: a non-parametric approach for detecting differential expression and splicing from RNA-Seq data. Bioinformatics. 2015;31:2222-2224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 122. | Vaquero-Garcia J, Barrera A, Gazzara MR, González-Vallinas J, Lahens NF, Hogenesch JB, Lynch KW, Barash Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5:e11752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 298] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 123. | Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A. 2014;111:E5593-E5601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1073] [Cited by in RCA: 1687] [Article Influence: 153.4] [Reference Citation Analysis (0)] |
| 124. | Alamancos GP, Pagès A, Trincado JL, Bellora N, Eyras E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA. 2015;21:1521-1531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 167] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 125. | Trincado JL, Entizne JC, Hysenaj G, Singh B, Skalic M, Elliott DJ, Eyras E. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018;19:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 221] [Cited by in RCA: 381] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 126. | Denti L, Rizzi R, Beretta S, Vedova GD, Previtali M, Bonizzoni P. ASGAL: aligning RNA-Seq data to a splicing graph to detect novel alternative splicing events. BMC Bioinformatics. 2018;19:444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 127. | Gonzàlez-Porta M, Calvo M, Sammeth M, Guigó R. Estimation of alternative splicing variability in human populations. Genome Res. 2012;22:528-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 128. | Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, Pritchard JK. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 2018;50:151-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 521] [Cited by in RCA: 458] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 129. | Rogers MF, Thomas J, Reddy AS, Ben-Hur A. SpliceGrapher: detecting patterns of alternative splicing from RNA-Seq data in the context of gene models and EST data. Genome Biol. 2012;13:R4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 101] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 130. | Sacomoto GA, Kielbassa J, Chikhi R, Uricaru R, Antoniou P, Sagot MF, Peterlongo P, Lacroix V. KISSPLICE: de-novo calling alternative splicing events from RNA-seq data. BMC Bioinformatics. 2012;13 Suppl 6:S5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 131. | Zhou A, Breese MR, Hao Y, Edenberg HJ, Li L, Skaar TC, Liu Y. Alt Event Finder: a tool for extracting alternative splicing events from RNA-seq data. BMC Genomics. 2012;13 Suppl 8:S10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 132. | Gohr A, Irimia M. Matt: Unix tools for alternative splicing analysis. Bioinformatics. 2019;35:130-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 133. | Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, Corbett R, Tang MJ, Hou YC, Pugh TJ, Robertson G, Chittaranjan S, Ally A, Asano JK, Chan SY, Li HI, McDonald H, Teague K, Zhao Y, Zeng T, Delaney A, Hirst M, Morin GB, Jones SJ, Tai IT, Marra MA. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7:843-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 206] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 134. | Song Y, Botvinnik OB, Lovci MT, Kakaradov B, Liu P, Xu JL, Yeo GW. Single-Cell Alternative Splicing Analysis with Expedition Reveals Splicing Dynamics during Neuron Differentiation. Mol Cell. 2017;67:148-161.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 120] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 135. | Adamopoulos PG, Theodoropoulou MC, Scorilas A. Alternative Splicing Detection Tool-a novel PERL algorithm for sensitive detection of splicing events, based on next-generation sequencing data analysis. Ann Transl Med. 2018;6:244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 136. | Pulyakhina I, Gazzoli I, 't Hoen PA, Verwey N, den Dunnen JT, Aartsma-Rus A, Laros JF. SplicePie: a novel analytical approach for the detection of alternative, non-sequential and recursive splicing. Nucleic Acids Res. 2015;43:e80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 137. | Tapial J, Ha KCH, Sterne-Weiler T, Gohr A, Braunschweig U, Hermoso-Pulido A, Quesnel-Vallières M, Permanyer J, Sodaei R, Marquez Y, Cozzuto L, Wang X, Gómez-Velázquez M, Rayon T, Manzanares M, Ponomarenko J, Blencowe BJ, Irimia M. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017;27:1759-1768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 287] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 138. | Vitting-Seerup K, Porse BT, Sandelin A, Waage J. spliceR: an R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinformatics. 2014;15:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 139. | Huang HD, Horng JT, Lee CC, Liu BJ. ProSplicer: a database of putative alternative splicing information derived from protein, mRNA and expressed sequence tag sequence data. Genome Biol. 2003;4:R29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 140. | Castrignanò T, Rizzi R, Talamo IG, De Meo PD, Anselmo A, Bonizzoni P, Pesole G. ASPIC: a web resource for alternative splicing prediction and transcript isoforms characterization. Nucleic Acids Res. 2006;34:W440-W443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 141. | Takeda J, Suzuki Y, Nakao M, Kuroda T, Sugano S, Gojobori T, Imanishi T. H-DBAS: alternative splicing database of completely sequenced and manually annotated full-length cDNAs based on H-Invitational. Nucleic Acids Res. 2007;35:D104-D109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 142. | Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci U S A. 2006;103:7175-7180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 409] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 143. | Leipzig J, Pevzner P, Heber S. The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977-3983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 144. | Foissac S, Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007;35:W297-W299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 248] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 145. | Zhu D, Deng N, Bai C. A generalized dSpliceType framework to detect differential splicing and differential expression events using RNA-Seq. IEEE Trans Nanobioscience. 2015;14:192-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 146. | Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755-W762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 253] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 147. | Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511-515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13157] [Cited by in RCA: 11569] [Article Influence: 771.3] [Reference Citation Analysis (0)] |
| 148. | Ding L, Rath E, Bai Y. Comparison of Alternative Splicing Junction Detection Tools Using RNA-Seq Data. Curr Genomics. 2017;18:268-277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 149. | Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40:e61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 272] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 150. | Zhou Y, Lu Y, Tian W. Epigenetic features are significantly associated with alternative splicing. BMC Genomics. 2012;13:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 151. | Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 265] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 152. | Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, Salinas RD, Zarabi H, Kriegstein AR, Lim DA. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 274] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 153. | Hu S, Shan G. LncRNAs in Stem Cells. Stem Cells Int. 2016;2016:2681925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 154. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16076] [Article Influence: 1004.8] [Reference Citation Analysis (2)] |
| 155. | Wu DR, Gu KL, Yu JC, Fu X, Wang XW, Guo WT, Liao LQ, Zhu H, Zhang XS, Hui J, Wang Y. Opposing roles of miR-294 and MBNL1/2 in shaping the gene regulatory network of embryonic stem cells. EMBO Rep. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 156. | Bourguignon LY, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2012;287:32800-32824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 238] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 157. | Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, Gendron D, Brosseau JP, Thibault P, Lucier JF, Tremblay K, Prinos P, Wellinger RJ, Chabot B, Rancourt C, Elela SA. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525-9531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 158. | Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J Biol Chem. 2001;276:16411-16417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 125] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 159. | Ren YP, Zhang JQ, Sun Y, Wu ZF, Ruan JS, HE BJ, Liu GQ, Gao S, Bu WJ. Full-length transcriptome sequencing on PacBio platform. Chinese Sci Bull. 2016;61:1250-1254. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 160. | Teng K, Teng W, Wen H, Yue Y, Guo W, Wu J, Fan X. PacBio single-molecule long-read sequencing shed new light on the complexity of the Carex breviculmis transcriptome. BMC Genomics. 2019;20:789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 161. | Zhang G, Sun M, Wang J, Lei M, Li C, Zhao D, Huang J, Li W, Li S, Li J, Yang J, Luo Y, Hu S, Zhang B. PacBio full-length cDNA sequencing integrated with RNA-seq reads drastically improves the discovery of splicing transcripts in rice. Plant J. 2019;97:296-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 162. | Chen SY, Deng F, Jia X, Li C, Lai SJ. A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Sci Rep. 2017;7:7648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 163. | Sebestyén E, Singh B, Miñana B, Pagès A, Mateo F, Pujana MA, Valcárcel J, Eyras E. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016;26:732-744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 201] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 164. | Horning AM, Wang Y, Lin CK, Louie AD, Jadhav RR, Hung CN, Wang CM, Lin CL, Kirma NB, Liss MA, Kumar AP, Sun L, Liu Z, Chao WT, Wang Q, Jin VX, Chen CL, Huang TH. Single-Cell RNA-seq Reveals a Subpopulation of Prostate Cancer Cells with Enhanced Cell-Cycle-Related Transcription and Attenuated Androgen Response. Cancer Res. 2018;78:853-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 165. | Faigenbloom L, Rubinstein ND, Kloog Y, Mayrose I, Pupko T, Stein R. Regulation of alternative splicing at the single-cell level. Mol Syst Biol. 2015;11:845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 166. | Yap K, Makeyev EV. Functional impact of splice isoform diversity in individual cells. Biochem Soc Trans. 2016;44:1079-1085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 167. | Liu W, Zhang X. Single-cell alternative splicing analysis reveals dominance of single transcript variant. Genomics. 2020;112:2418-2425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |