Published online Jun 26, 2020. doi: 10.4252/wjsc.v12.i6.406
Peer-review started: January 16, 2020
First decision: April 7, 2020
Revised: April 29, 2020
Accepted: May 27, 2020
Article in press: May 27, 2020
Published online: June 26, 2020
Processing time: 160 Days and 18.3 Hours
Since the first publication regarding the existence of stem cells in cancer [cancer stem cells (CSCs)] in 1994, many studies have been published providing in-depth information about their biology and function. This research has paved the way in terms of appreciating the role of CSCs in tumour aggressiveness, progression, recurrence and resistance to cancer therapy. Targeting CSCs for cancer therapy has still not progressed to a sufficient degree, particularly in terms of exploring the mechanism of dynamic interconversion between CSCs and non-CSCs. Besides the CSC scenario, the problem of cancer dissemination has been analyzed in-depth with the identification and isolation of microRNAs (miRs), which are now considered to be compelling molecular markers in the diagnosis and prognosis of tumours in general and specifically in patients with non-small cell lung cancer. Paracrine release of miRs via “exosomes” (small membrane vesicles (30-100 nm), the derivation of which lies in the luminal membranes of multi-vesicular bodies) released by fusion with the cell membrane is gaining popularity. Whether exosomes play a significant role in maintaining a dynamic equilibrium state between CSCs and non-CSCs and their mechanism of activity is as yet unknown. Future studies on CSC-related exosomes will provide new perspectives for precision-targeted treatment strategies.
Core tip: The role of cancer stem cells (CSCs) in tumour aggressiveness, progression, recurrence and resistance to cancer therapy is well appreciated. However, therapeutic strategies to target CSCs for cancer therapy has still not progressed sufficiently, particularly in terms of exploring the mechanism of dynamic interconversion between CSCs and non-CSCs. Similar to other cells, CSCs also release exosomes loaded with microRNAs (miRs) as part of their paracrine activity. Our review focusses on the exosomal payload of miRs released by cancer cells and their role in the diagnosis as well as prognosis of lung cancer patients.
- Citation: Aramini B, Masciale V, Haider KH. Defining lung cancer stem cells exosomal payload of miRNAs in clinical perspective. World J Stem Cells 2020; 12(6): 406-421
- URL: https://www.wjgnet.com/1948-0210/full/v12/i6/406.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i6.406
Stem cells related to cancer [cancer stem cells (CSCs)] were first reported in 1994 during a study on human acute myeloid leukaemia (AML)[1]. The authors of the study identified a rare population of AML-initiating cells isolated from AML patients post-engraftment in severe immunocompromised SCID mice. These cells expressed CD34+/CD38- surface markers and were less mature than the colony-forming cells besides possessing a higher proliferative capacity[1]. Subsequently, in 2003, human CSCs were identified in solid tumours in various organs, including the breast and brain[2-4]. Hence, a small CSC population (approximately 100 cells) developed into a tumour in an immunodeficient mice model[2]. A consensus regarding the criteria for classifying CSCs has not been established as yet. However, they are defined as a sub-population in a given tumour with the ability to self-renew and produce cells that can differentiate[4]. There is growing evidence that CSCs are highly resistant to different types of stresses, including anti-cancer therapy[5], and hence, they are generally associated with an increased risk of cancer relapse, metastasis and an overall low survival rate[5]. CSCs have also been linked with tumours identified in various cell lines but not so frequently in all human tissues (e.g., in human lung cancer).
According to recently published reports, in 2018, there were 121680 new lung cancer cases for men and 112680 for women in the United States, totaling 234030 cases[6], which is equivalent to an average of 641 lung cancer cases diagnosed per day. These epidemiologic data rank lung carcinoma as the 2nd most prevalent in men behind prostate cancer and for women after breast cancer[6]. On a positive note, Siegel et al[7] stated that there was an overall 27% decline in cancer-related death cases between 1991 and 2016, translating into more than 2 million fewer deaths than expected if the rate had stayed at the range of the standard values[7-9]. Conventional therapeutic strategies including surgery, radiotherapy, and chemotherapy are used for lung cancer treatment either singly or in combination at different cancer pathological stages. However, issues associated with chemotherapy and radiotherapy resistance are well known, and recurrence is still a challenge in advanced lung cancer patients. This inability to be 100% curative has been attributed to the sub-populations of stem cells that are capable of self-renewal, undergoing differentiation and producing multi-lineage progenies that may be tumourigenic or non-tumourigenic. This sub-population of cells contributes to the establishment and maintenance of tumours. Although the underlying molecular mechanisms behind these CSCs’ properties are less well-defined, the intrinsic resistance of CSCs to therapy is now generally ascribed to a lack of the capacity to induce the apoptotic signalling, increasing telomere length as well as interfering with the cell membrane’s ability to act as a transporter, favoring cell migration, and metastasis[10-12]. Owing to the multifactorial mechanistic nature of resistance, the current treatment methods are inadequate for cancer treatment, and hence, warrant in-depth molecular studies in the future to improve the contemporary therapeutic regimens[13].
Given their significant role in poor prognosis, relapse and drug resistance in cancer patients[14,15], the current treatment modalities are being focussed to target CSCs. Some of these emerging treatment modalities include immunotherapy directed against the CSCs’ specific surface antigens, interference with signalling pathways (i.e., Notch signalling) and epigenetic approaches[16,17]. For example, a recent pre-clinical study targeted the self-renewal regulator BMI1 to attenuate CSCs’ self-renewal and tumour-initiating potential in oral cavity squamous cell carcinoma[18]. Based on the promising results during pre-clinical studies, these therapeutic strategies are now entering into the clinical phase of assessment. Another interesting area of research targeting CSCs involves analyzing the underlying mechanisms regulating the dynamic interconversion between CSCs and non-CSCs (Figure 1). An understanding of the molecular mechanism underlying such a bidirectional interconversion of cells would have significant implications on the future development of therapeutic strategies[19,20].
Besides the CSCs paradigm, the issue of cancer dissemination has been analyzed in-depth with the identification and isolation of microRNAs (miRs). miRs are short (20-24 nucleotides), non-coding RNAs, which are now suggested to be the most promising molecular markers for the diagnosis and prognosis of tumours[21]. Their regulatory role in various cellular processes is now well-established, although some of the known miRs are dispensable for the normal functioning of cells but have instrumental participation in the initiation as well as the progression of diseases. The main function of miRs is to play an epigenetic role in regulating gene expression at the post-transcription stage[22]. In the context of their role in cancers, they are grouped as oncomirs that either function as oncogenes or as tumour suppressors. Given the pivotal role of miRs in the physiological functioning of a cell, their dysregulation has been associated with various pathologies, including the initiation and progression of cancer[23]. Their prognostic value as qualitative and quantitative biomarkers in plasma, either in free form or encapsulated in the microvesicles, has been reported in various cancers[24]. There have been attempts to classify human cancers based on miR expression profiles[25].
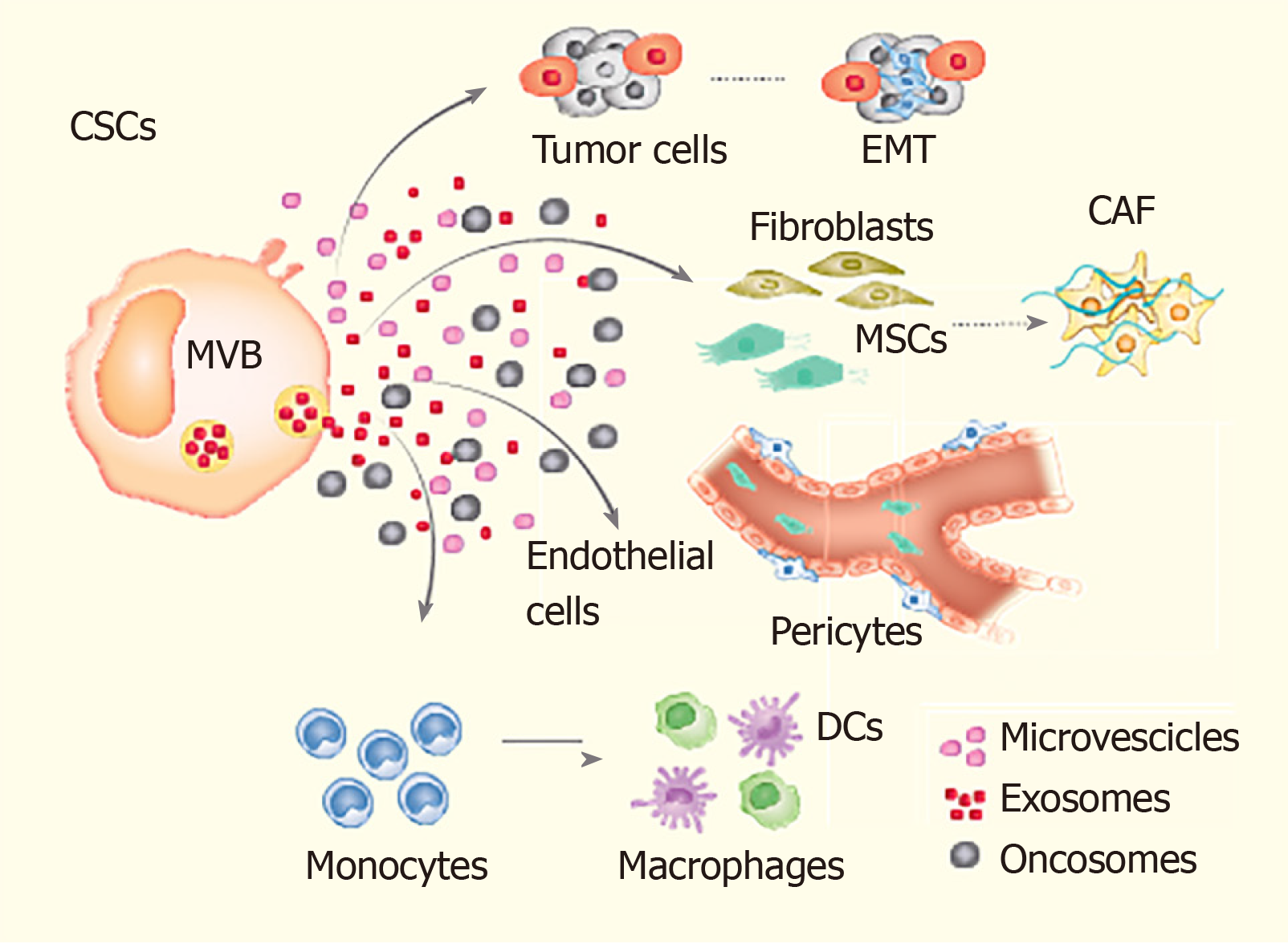
Exosomes are extracellular vesicles with a small size of approximately 30 to 100 nm. They are formed by the fusion of intracellular components surrounded by the plasma membrane and released from cells[26]. It is now becoming well-established that exosomes, besides other components, also transfer their nucleic acid payload including miRs, from the cell of their origin to the recipient cells. The importance of exosomes is mainly due to their capacity to transport miRs into the body, and this process forms an important focus for providing a deep understanding of the possible genetic implications between cancer and non-cancer cells[27]. The miRs thus transferred significantly affect the gene expression and cellular signalling pathways in the recipient cells, including maintenance of a dynamic equilibrium between CSCs and non-CSCs by delivering their miR-payload[28,29].
Mature miRs that are about 70 nucleotides long are derived from pre-miRs composed of 100 nucleotides and then transcribed[30]. Exosomes can transport small intracellular components, such as proteins and lipids, which are included by an endocytosis process from pre- to late-mature exosomes[31]. Hence, exosomes are currently the smallest cellular components carrying miRs from the cell to the human organs. They are detected in many fluids, such as urine[32,33], blood and saliva, and this peculiarity makes them a unique mediator against tumour development and progression. Each miR has various targets, although different miRs may have a single target[34], and this characteristic highlights their significant involvement in many genetic and cellular processes, such as in particular the preservation of cellular differentiation. The most interesting characteristic is that many human genes depend on miRs; this aspect reflects the roles of these small molecules in the genome[35,36].
In recent studies, it has been found that operable non-small cell lung cancer (NSCLC) patients showed a significant association between recurrence and survival[37-40]. In adenocarcinoma patients, a miR score has been defined for distinguishing between patients in stage I developing recurrence within two years from surgery, and those patients that were disease-free after three years[41,42]. We believe that determining the roles of exosomes and their miR-payload in the prevention and cure of cancer is the future of personalized cancer medicine. However, targeting exosomes and monitoring miRs in biological fluids will be the pillar for the setting of new approaches in terms of follow-up for cancer patients, with the focus being to determine more details in the history of each person affected from this disease[43-45].
The amount of miRs in exosomes is influenced by the intracellular components as well as by the physiological context[46-50]. In particular, exosomes can maintain the stability of miRs by preventing their degradation[48]. The consequence of this exosome-driven role is to drive them in cancer progression as miR-142-3p, miR-150, and miR-451, all of which have been found in exosomes in gastric cancer cells[46]. Similarly, microsomal miR-21, let-7f, miR-20b, and miR-30e-3p are differentially expressed in patients with solid tumours as compared with healthy people[46,49]. The role of exosomal miRs is undoubted in terms of cancer growth, development and recurrence. It is apparent that miRs also important players in angiogenesis and metastasis as they can influence host immunity-inducing chemoresistance and tumour microenvironment (TME) re-adaptation[51].
We know that the metastasis of cancer cells is an intricate process that implicates the colonization of cancer cells from their primary site to a secondary location[52]. It encompasses a cascade of molecular events that facilitates cell migration, invasion, angiogenesis and epithelial to mesenchymal transition (EMT)[53]. From amongst the oncomirs, specific miRs are also grouped as “metastamirs” due to their association with the molecular processes and signalling pathways that underlie cancer cell metastasis[54]. For example, Coebergh et al[55] have recently identified signature miRs, including let-71 and miR-10, which can serve as biomarkers to predict colon cancer patients who are at a risk of metastatic cancer spread. Both miRs’ signatures were successfully used for prediction of hepatic recurrence of cancer in stage-I and -II patients. Similarly, miR-126 suppresses EMT to influence lung cancer cell metastasis[56]. Molecular studies have revealed the inhibition of the PI3K/Akt Snail pathway with the involvement of miR-126 that could be a potential therapeutic target for lung cancer treatment. Similar observations have also been reported with a possible role of miR-30a via targeted regulation of Snail[57]. Further, the role of Wnt/β-catenin in EMT has been reported in human colorectal carcinoma metastasis that involved GNA13 and PTP4A genes’ regulation via β-catenin signalling and which are targeted by the miR-126 pathway via ERK/GSK3β/ β-catenin and Akt/GSK3/β-catenin signalling pathways[58]. The role of β-catenin in EMT has also been reported in a recently published study that involved miR-1246 as a regulator of EMT in A549 cells by inhibiting E-cadherin expression via regulation of the Wnt/β-catenin pathway through GSK3b/β-catenin targeting[59]. These data provide vivid evidence for the significant participation of miRs in supporting the metastatic spread of cancers from their primary origin.
There has been a recent interest in miR dissemination through exosomes. In this regard, an important role is played by the cancer-associated fibroblasts into the TME, a process that seems to release exosomes, inducing tumour development or control depending on the presence of some nutrients[60]. Besides EMT, angiogenesis is important for tumour maintenance and recurrence. In this context, exosomes released by cancer contribute to increased angiogenesis and tumour growth through the transforming growth factor β1-dependent pathway, which induces the fibroblast evolution process[61,62].
In lung cancer, exosomal miR-23a from hypoxic lung cancer cells and hypoxamir-210 from exosomes derived from such cells can improve permeability of the vessel membranes and increase vascularization through the STAT3 mechanism, which can transform normal bronchial cells into malignant ones[63]. One of the mechanisms that may induce tumour progression involves tumour-derived exosomal interactions with TME. For example, it has been shown that tumour-derived exosomes in lung cancer may induce bone marrow-derived mesenchymal stem cells to change themselves into a phenotype stimulating inflammation[64]. Hence, the immune system inside TME may be affected by the tumour-derived exosomes with the final result being tumour progression, most likely due to the reprogramming of the immune cells influenced by tumour exosomes[64-66]. Akin to other cells, the exchange of exosomal miRs from cancer cells to endothelial cells (ECs) significantly influences their angiogenic activity. Tumour cell-released miR-221-3p facilitates lymphangiogenesis in cervical squamous cell carcinoma by its transfer to lymphatic ECs[67]. Similarly, cancer cell-derived exosomes transfer miR-25-3p to the ECs and regulate VEGF expression by targeting KLF2 and KLF4, thus promoting angiogenesis[68].
As discussed before that the exosomes carrying miRs drive angiogenesis and cancer progression[69]. For example, it has been shown that miR-103 enhanced angiogenesis and induces tumour metastasis in hepatocarcinoma patients. This process involves several endothelial target proteins, such as VE-cadherin, p120-catenin and zonula occludens 1 in ECs[70]. In other blood diseases, such as leukaemia, exosomal miR210 secreted by hypoxic leukaemia cells have an important impact on angiogenesis through the receptor tyrosine kinase ligand Ephrin-A3 of ECs[71]. In contrast, exosomes may include miRs that can harm leukaemia cells, influencing motility and their capacity to adhere. This process is induced by the loss of C-X-C motif chemokine ligand 12 and vascular cell adhesion molecule-1 proteins in ECs[72].
Several exosomal miRs are essential in the process of recurrence. In particular, in metastatic breast cancer, exosomal miR-210 is involved in EC transport as well as improving angiogenesis[73]; in nasopharyngeal carcinoma (NPC) cells, miR-23a exosome enhances tumour growth and recurrence[74], although exosomal miR-9 suppresses NPC cell migration and the consequent vascular formation by targeting midkine and modulating the phosphoinositide-dependent protein kinase/protein kinase B (Akt)-signalling pathway[75].
Due to their already demonstrated crucial participation in metastatic processes and their presence into human fluids, exosomal miRs are the future of personalized medicine as biological biomarkers[76]. Exosomal miRs are already in practice as reliable biomarkers for the diagnosis of lung cancer patients[77-79]. Cazzoli et al[77] performed a thorough exosomal miR-analysis of 30 plasma samples (including n = 10 each from lung-adenocarcinoma, lung-granuloma and healthy-smoker subjects) and all the donors were matched for age and sex. The expression level of four miRs distinguished between tumour and healthy-smoker subjects[77]. These findings were subsequently used on a larger group of patients with 96% sensitivity and nearly 70% specificity. Several upregulated miRs-derived exosomes (such as miR-21 and miR-155) have been found in patients who developed lung cancer recurrence[78]. In particular, Li et al[63] used a qPCR-based array to analyze plasma from 10 patients affected by lung adenocarcinoma, and he found an increased level of 3 exosomal miRs (miR-23b-3p, miR-10b-5p, and miR-21-5p) which seemed to be correlated with decreased survival[63]. The expression profile of specific exosomal miRs in lung adenocarcinoma patients vindicated the previously published results that circulating exosomal miRNAs were a useful and possible marker for further diagnostic and therapeutic purposes in lung cancer[79]. Dejima et al[80] profiled plasma exosomal miRs derived from lung cancer patients to demonstrate that miR-21 and miR-4257 were significantly upregulated as compared with those patients without recurrence and healthy individuals as controls. The microarray data also showed that exosomal miR-21 and miR-4257 exosomes were significantly associated with tumour growth and metastatic invasion in lung cancer. These data were also supported by the low percentage of disease-free survival in patients with high expression of both exosomes levels[80,81].
Besides the recent reports regarding the pivotal role of exosomal miRs in driving recurrence and invasion of cancer in patients, their use as biomarkers seems to represent an effective method for diagnostic and prognostic purposes. That is justified by the fact that there are currently no markers that can satisfactorily diagnose presence of tumour in the early stage as well as predict long-term survival with respect to many solid cancers[82].
Although quantification of a panel of miRs in the plasma or serum of patients at high-risk to develop lung cancer has been proposed, the difficulty in discriminating between miRs for normal and cancerous tissue remains an obstacle in the development of an effective screening method[83]. Nigita et al[84] analyzed data from 87 NSCLC patients, attempting for the first time to identify and differentiate miRNAs for normal and tumour tissues; in particular, miR-441-5p was the most consistently detected among the NSCLC exosomes.
One of the mechanisms by which exosomal miRs affect the functioning of CSCs is via regulation of the interaction between CSCs and their microenvironment[85]. The exosomes serve as carriers of the genetic information (i.e., miRs) required for regulation of the signalling involved in the transformation of cancer cells into CSCs to achieve a dynamic equilibrium between the two cell types.
Reflecting their role in metastatic processes, it is apparent that exosomal miRs have an important impact on TME[86]. It has been demonstrated that exosomes driven by specific miRs, such as miR-223 derived from macrophages, may induce drug resistance in the hypoxic TME[87].
The interactions between cancer cells, exosomal miRs, and the TME rely on a complex network that has yet to be satisfactorily clarified. In particular, exosomes can mediate immune regulation, and several studies are attempting to understand the direct effects of exosomes in T-cell activation[88]. However, there is currently a lack of understanding regarding the identification of the connections between the immune system, miRs, exosomes, and CSCs. Recently, a correlation between tumour-infiltrating lymphocytes (TILs) and CSCs in NSCLC patients has been demonstrated[89]. This is important to highlight the existence of interactions and relations of these cellular components to further connect exosomes carrying miRs, CSCs and TME. Further clarification of the associations between TILs and CSCs will be helpful for the development of targeted therapies that may focus on miR exosomes, CSCs, and TME cells.
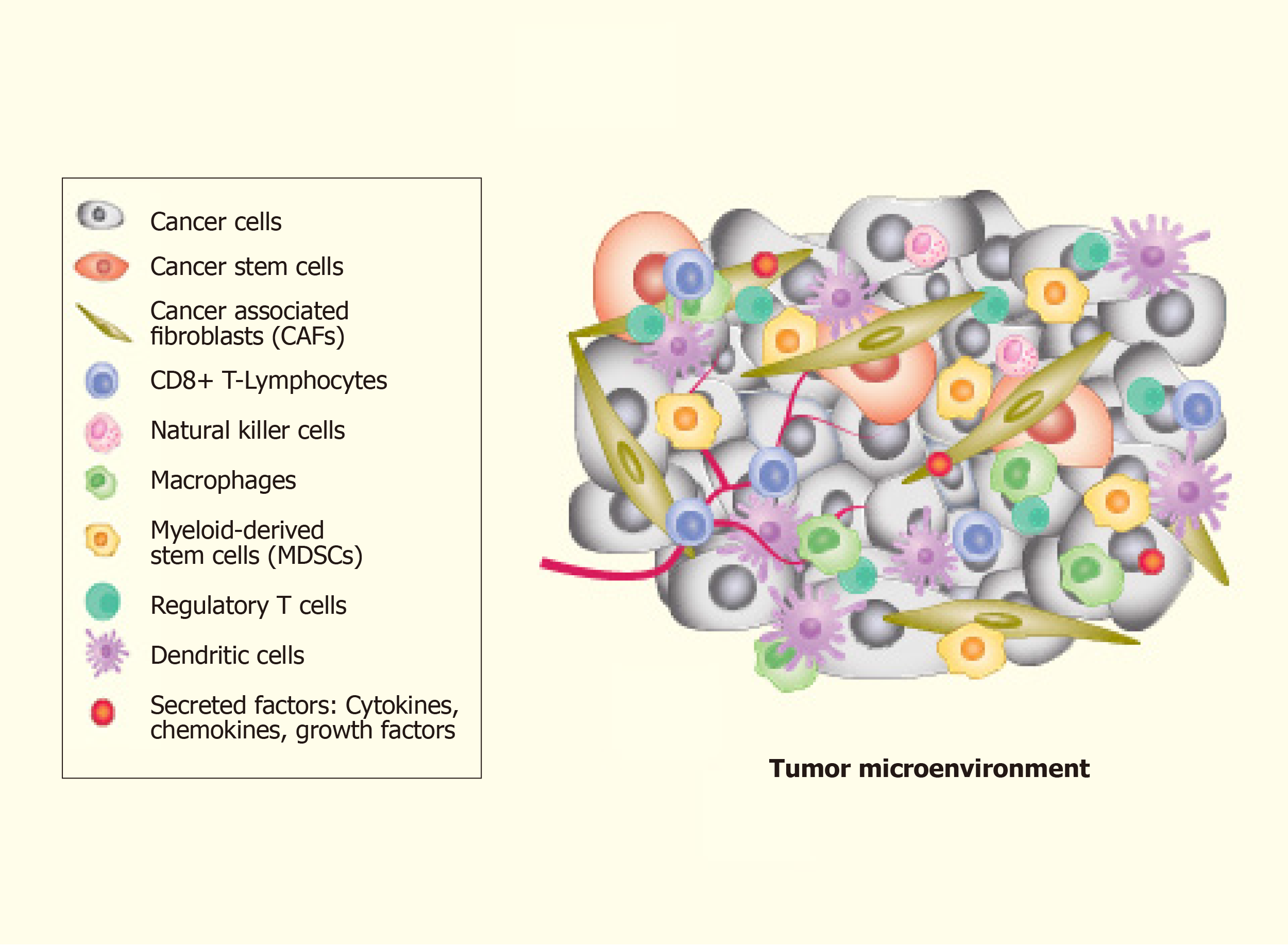
The complexity of TME leads to difficulty in understanding the mechanisms involved. TME is an amalgam of both cellular and non-cellular components that encompasses the surrounding microvascular structures, immune cells, fibroblasts, ECs, cancer cells, signalling molecules and the extracellular matrix (ECM) (Figure 2). Put together, these components offer a growth factor and cytokine-rich TME that is conducive for phenotypic plasticity, immune surveillance, survival, angiogenesis and cancer cell metastasis[90,91]. More importantly, TME contributes significantly to the spatiotemporal dynamics of pattern formation and is one of the primary factors that substantially influence tumour heterogeneity[92]. The link between TME and CSCs generation is represented by the EMT[93].
The discovery of the TME implies the possibility of a novel treatment strategy that goes beyond the paradigm of cancer genetics that restricts its focus only on cancer cells[94]. The presence of CSCs in the TME is crucial for ‘tumour nutrition’ due to their capacity to reproduce themselves, inducing tumour growth in NOD-SCID mice and facilitating the spread and chemoresistance of the tumour. There is mounting evidence that conservative therapeutic strategies (i.e., radiotherapy and chemotherapy) largely fail to eliminate CSCs, which are now associated with minimal residual disease and cancer relapse[95,96].
The physiological functioning of cells is affected by a multitude of physical factors that alter both genetic and epigenetic states. These molecular changes influence the intracellular regulatory circuitry that enables the body to achieve as well as sustain an appropriate response to the changing environment[97-99]. In this regard, ECM is an elastic barrier able to change the mechanical properties of its proteins through genes’ profile mutations. Studies of the TME have led to immunotherapy and other new-generation immune therapies, such as CAR-T cells[100-102] and anti-cancer vaccines[103-105]. The mechanism underlying the role of TME-associated transformation of normal cells into tumour cells and then converting to malignancy upon cancer initiation is less well defined[94]. However, this mechanism is associated with the mechanical/scaffold (elasticity) effect of ECM[106]. Li et al[94] suggested that the elasticity is an important aspect in ECM development, playing a key-role in miR exosome expression, regulating tumour gene expression and tumour growth. Further understanding of the role of tumour–TME interaction will significantly help in understanding the acquired resistance of cancer cells towards contemporary cancer therapies including surgery, radiotherapy, chemotherapy and anti-angiogenic therapy[107]. This understanding will open up a new avenue to target the CSC niche within the TME. In this context, in particular, the exosomes derived from the tumour (TEX) seem to have a strong influence on the TME for their capacity to expand themselves in other tissues and contributing to cancer dissemination to the organs[108]. Additionally, besides TME, Javeed et al[109] hypothesized the presence of a tumour “macroenvironment” (TMaE) that results from the pathological interaction between tumour cells with other organs and systems of the body to mediate immunosuppression and promote genetic and metabolic reprogramming of the cells through exosomal-miR payloads[109]. Further understanding of the TMaE would be helpful for two reasons: first, it would help in identifying those patients who would benefit from systemic therapy and second, it would help in the future development of novel systemic therapies[110].
One of the most interesting aspects in this context is that TEX is associated with some antigens, such as programmed death-ligand 1(PD-L1), which induces the pre-metastatic pathway for tumour dissemination[111]. PD-L1 expression has been reported to be enhanced by exosomes from melanoma, and this justifies the roles of exosomes in tumour growth, metastasis and immune modulation[112,113]. There is interest in establishing an “exosome screening” approach due to their presence in many fluids of the human body, and this aspect is highly important in terms of providing potential biomarkers as well as drug carriers for targeted drug delivery[114-116].
“Tumour aggressiveness” is typically used to define a highly incurable end-stage cancer that is also able to resist the standard treatments[117-119]. Dysregulated miRs can be oncogenes (oncomirs) if upregulated, or tumour suppressors (anti-oncomirs) if downregulated, depending on their target transcripts[120,121]. Tumour aggressiveness is generally associated with altered miRs profile that has already been reported in various solid tumours[121-125]. Specifically, miRs have been implicated in a substantial number of intracellular mechanisms that include tumour growth, proliferation, recurrence, tumour metabolic alterations and chemo- and radio-resistance. However, they are also influenced by many extracellular factors, such as hypoxia, which seems to contribute to poor clinical outcomes by increasing the recurrence risk[126-128].
The mechanism of oxygen sensing at the cellular level has remained an area of intense investigation, as evidenced by the two Nobel Prizes won by Otto Warburg in 1931 and Corneille Heymans in 1938 for their findings on the role of enzymes and the nervous system in respiratory cellular mechanisms[126]. Moreover, the genetic component of adaptation to oxygen flux remained oblivious for the major part of this century. More recent studies have successfully addressed this issue to elucidate the molecular biology of hypoxia-inducible factor (HIF) signalling as part of the body’s hypoxic response in health and disease[127,128]. HIF-1 signalling and HIF-1-dependent miRs, hypoxamirs, are being extensively studied for their role in cell survival and angiogenesis in the context of cancer cell biology[129-132] as well as regenerative medicine[133]. The data thus generated has been exploited to promote stem cell survival and angiogenesis either as part of physical or genetic manipulation of cells for hypoxamir expression[134-138]. In this context, the homeostasis of reactive oxygen species (ROS) is the key for maintaining normal biological processes[139]. Higher-level oxidative stress produces irreversible damage to intracellular and cellular components, thus altering the stability of the genome with the induction of malignant cell transformation[140]. For example, altered production of ROS is also associated with EMT, tumourigenesis and tumour progression[141]. In particular, the mechanism of ROS on CSC mechanism regulation is not yet fully clarified.
The precise mechanism by which ROS regulates CSCs and EMT characteristics with the HIF-mediated pathways is unclear[140-142]. It has been documented that normal stem cells, as well as CSCs, have a low level of ROS, potentially due to their strong defense system against DNA damage[140-143]. Low-level ROS in CSC-like cells is associated with higher free radical scavenger production. The inhibition of ROS scavengers by drug treatment in mouse breast Thy-1 + CD24 + Lin- CSC-enriched cells markedly decreased their clonogenicity with increased radio-sensitization[144]. Hence, low levels of ROS and enhanced ROS defense may contribute to tumour radio-resistance as compared to non-tumourigenic counterparts[145]. These findings strongly suggest that ROS may play an important role in the pathogenesis of CSCs.
Surprisingly, there is a link between ROS and miRs. It has been demonstrated that H2O2 treatment can dysregulate the expression of certain miRs in vascular smooth muscle cells and macrophages. Moreover, it has also been demonstrated that miR-30e contributes to regulating oxidative stress and ROS levels through SNAI1 mRNA in human umbilical endothelial vein cells[146]. Given the significant regulatory role of miRs in metabolic processes in the cells, many of the oncomirs, such as miR-21, are directly involved in the formation of ROS and hence, promote tumourgenesis[147]. Both ROS and miRs control each other’s expression in cancer cells to maintain a balance that is supportive of cancer cells in terms of their ability to produce the hallmarks of cancer[148,149]. More investigations are mandatory to clarify the possible importance of ROS in CSC regulation.
Although recent studies have identified the presence of CSCs in ADENO and SQUAMO cell carcinomas of the human lung, with this aspect being important in terms of understanding that CSCs are present in all NSCLC tumours[150-152], even in neuroendocrine tumours of the lung[153], the possibility to target CSCs is still debated. In particular, the scientific community is focusing attention on the characterization of miR exosomes for their intrinsic characteristics as complex paracrine factors[153]. In fact, it has been found that stem cell-derived exosomal miRs can be used to modulate the therapeutic response to stroke and may increase their therapeutic potential[77]. This aspect may be important in future if we consider the possibility of targeting CSC-derived exosomal miRs, in consideration of CSCs’ characteristics related to chemo- and radioresistance – this approach would have a key role in new cancer treatments.
Lung cancer cell-derived exosomal miRs are at the centre of interest in the present-day research for future roles as predictors of recurrence as well as biomarkers in the early stage or for their prognostic role in advanced-treated patients[13,154-157]. The prime objective is to explore the possibility of their use as biomarkers for early diagnosis of lung cancer combined with target therapy[158,159]. In one of the studies, immunostaining of exosomes from lung cancer tissue and chronic lung disease showed that 80% of these specimen-derived exosomes were positive for EGFR, although 2% of the inflammatory tissue was positive. This point suggested a possible role of EGFR exosomes as potential biomarkers in lung cancer[61,160,161]. A similar result has also been identified in ALK-EML4 translocated exosomes, which are markers for first-generation treatment ALK-TKIs[162,163].
On the same note, exosomes from the plasma of NSCLC patients (n = 276) indicated that exosomes may have a prognostic role as biomarkers in NSCLC[156]. Similar results were found in plasma exosomal miRs analyzed from lung adenocarcinoma patients (n = 84) versus healthy controls. These results showed increased levels of exosomal miR-10b-5p, miR-23b-3p, and miR-21-5p were significantly associated with poor prognosis, thus suggesting their significance as biomarkers of NSCLC prognosis[157]. The use of exosomal miRs as prognostic biomarkers is the basis for new-generation target therapy against NSCLC, particularly in lung cancer for highly important proteins such as EGFR, KRAS and RAB family.
The most relevant aspect related to targetable exosomes is connected to the precision treatment of NSCLC[164]. In this regard, exosomes can be produced as new cellular molecules of delivery for medical treatment as well as against tumours to reduce the negative collateral effects in the human body[165,166]. Lai et al[167] have established a method for loading exosomes with a drug or genetically manipulating cells with genes of interest to alter the exosomal payload derived from these cells. Mendt et al[168] have reported a standard operating procedure to engineer exosomes that could target KRAS (iExosomes), with a particularly good response in terms of improved survival. The possibility to target genes such as KRAS is highly innovative because it is responsible for the highest mortality rate in NSCLC patients.
The diversity of exosomal-payload and their functions relevant to NSCLC may provide future pioneering target treatments. Yang et al[169] determined that induced expression of miR-let-7 in exosomes for NSCLC treatment was specific and effective in tumour suppression[169,170]. Similarly, exosomes isolated from H460 cells and that had been transduced for the restoration of LKB1 (liver kinase B1) had an increased ability to restore lung cancer cell migration as compared to exosomes isolated from H460 cells lacking in LKB1 activity[171]. While elucidating the process, it was observed that H460 cells with restored LKB1 supported the emigrational activity of lung cancer cells by the suppression of exosomal secretion of migration-suppressing miRs, including miR-125a, miR-126a, and let7b. These data highlight the significance of LKB1 as a new target for future cancer therapy. In an interesting new development, antibody therapy with anti-CD9 or anti-CD63 to target tumour-derived exosomes effectively inhibited the progression of breast cancer in mice, suggesting a successful further treatment in lung cancer[172].
Characterization of CSC-derived exosomes in terms of their payload and the use of exosomes for CSC targeting have emerged as potential strategies for cancer theranostics. For example, the safety and feasibility of exosome targeting were first assessed in phase I clinical trial for metastatic melanoma[173]. The patients were treated with autologous dendritic cell-derived exosomes (DEX) loaded with the melanoma antigen gene (MAGE) by intradermal and subcutaneous[173,174]. Although no significant outcome was reported, the data vividly demonstrated that exosome-based treatment may represent a new approach for curing cancer patients. The results from phase I trial evidenced that the immune system of treated patients was active and that exosome-treated patients showed limited disease progression[174]. Based on these encouraging data, DEX has progressed to Phase II trials as maintenance immunotherapy after first-line chemotherapy in NSCLC patients[175]. Besides the value and importance of exosomes as mediators against anticancer effects, it is necessary to study their clinical effects in the human body to guarantee a better standardization of the methods of processing and ensure optimal and reproducible anti-tumour immune responses after exosome-based therapy in the clinical perspective[176,177].
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: Saudi Arabia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gazouli M, Tanabe S S-Editor: Dou Y L-Editor: A E-Editor: Xing YX
| 1. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3316] [Cited by in RCA: 3387] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 2. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7727] [Article Influence: 351.2] [Reference Citation Analysis (0)] |
| 3. | Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821-5828. [PubMed] |
| 4. | Yu Z, Pestell TG, Lisanti MP, Pestell RG. Cancer stem cells. Int J Biochem Cell Biol. 2012;44:2144-2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 498] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 5. | Prabavathy D, Swarnalatha Y, Ramadoss N. Lung cancer stem cells-origin, characteristics and therapy. Stem Cell Investig. 2018;5:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 6. | de Groot PM, Wu CC, Carter BW, Munden RF. The epidemiology of lung cancer. Transl Lung Cancer Res. 2018;7:220-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 448] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 7. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15470] [Article Influence: 2578.3] [Reference Citation Analysis (2)] |
| 8. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20512] [Article Influence: 2051.2] [Reference Citation Analysis (20)] |
| 9. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13161] [Article Influence: 1880.1] [Reference Citation Analysis (4)] |
| 10. | Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3291] [Cited by in RCA: 3382] [Article Influence: 281.8] [Reference Citation Analysis (0)] |
| 11. | Peters GJ. Cancer drug resistance: a new perspective. Cancer Drug Resist. 2018;1:1-5. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Ji X, Lu Y, Tian H, Meng X, Wei M, Cho WC. Chemoresistance mechanisms of breast cancer and their countermeasures. Biomed Pharmacother. 2019;114:108800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 216] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 13. | Masciale V, Grisendi G, Morandi U, Dominici M, Aramini B. Stem cells and lung cancer: between advanced diagnostics and new therapeutics. In: Haider HKh. Stem Cells: From Myth to Reality and Evolving. Berlin, Boston: Walter de Gruyter GmbH & Co KG, 2019; Chapter-2: 14-33. |
| 14. | Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018;2018:5416923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 474] [Cited by in RCA: 631] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 15. | Zhao Y, Dong Q, Li J, Zhang K, Qin J, Zhao J, Sun Q, Wang Z, Wartmann T, Jauch KW, Nelson PJ, Qin L, Bruns C. Targeting cancer stem cells and their niche: perspectives for future therapeutic targets and strategies. Semin Cancer Biol. 2018;53:139-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 16. | Annett S, Robson T. Targeting cancer stem cells in the clinic: Current status and perspectives. Pharmacol Ther. 2018;187:13-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 17. | Desai A, Yan Y, Gerson SL. Concise Reviews: Cancer Stem Cell Targeted Therapies: Toward Clinical Success. Stem Cells Transl Med. 2019;8:75-81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 127] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 18. | Hu J, Mirshahidi S, Simental A, Lee SC, De Andrade Filho PA, Peterson NR, Duerksen-Hughes P, Yuan X. Cancer stem cell self-renewal as a therapeutic target in human oral cancer. Oncogene. 2019;38:5440-5456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | van Neerven SM, Tieken M, Vermeulen L, Bijlsma MF. Bidirectional interconversion of stem and non-stem cancer cell populations: A reassessment of theoretical models for tumor heterogeneity. Mol Cell Oncol. 2016;3:e1098791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Yang G, Quan Y, Wang W, Fu Q, Wu J, Mei T, Li J, Tang Y, Luo C, Ouyang Q, Chen S, Wu L, Hei TK, Wang Y. Dynamic equilibrium between cancer stem cells and non-stem cancer cells in human SW620 and MCF-7 cancer cell populations. Br J Cancer. 2012;106:1512-1519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 21. | Filipów S, Łaczmański Ł. Blood Circulating miRNAs as Cancer Biomarkers for Diagnosis and Surgical Treatment Response. Front Genet. 2019;10:169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 22. | Haider HKh, Khan M, Sen CK. MicroRNAs with Mega Functions in Cardiac Remodeling and Repair: The Micromanagement of Matters of the Heart. In: Sen CK. MicroRNA in Regenerative Medicine. Academic Press. 2015;569-600. |
| 23. | Leng Q, Lin Y, Jiang F, Lee CJ, Zhan M, Fang H, Wang Y, Jiang F. A plasma miRNA signature for lung cancer early detection. Oncotarget. 2017;8:111902-111911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 24. | Zheng H, Liu JY, Song FJ, Chen KX. Advances in circulating microRNAs as diagnostic and prognostic markers for ovarian cancer. Cancer Biol Med. 2013;10:123-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 25. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 26. | Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1505] [Cited by in RCA: 1407] [Article Influence: 234.5] [Reference Citation Analysis (0)] |
| 27. | Bhome R, Del Vecchio F, Lee GH, Bullock MD, Primrose JN, Sayan AE, Mirnezami AH. Exosomal microRNAs (exomiRs): Small molecules with a big role in cancer. Cancer Lett. 2018;420:228-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 28. | Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 708] [Article Influence: 70.8] [Reference Citation Analysis (0)] |
| 29. | Sun Z, Wang L, Dong L, Wang X. Emerging role of exosome signalling in maintaining cancer stem cell dynamic equilibrium. J Cell Mol Med. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 30. | Felekkis K, Touvana E, Stefanou Ch, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236-240. [PubMed] |
| 31. | Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193-208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1613] [Cited by in RCA: 1747] [Article Influence: 249.6] [Reference Citation Analysis (0)] |
| 32. | Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412-9420. [PubMed] |
| 33. | Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3378] [Cited by in RCA: 3537] [Article Influence: 321.5] [Reference Citation Analysis (0)] |
| 34. | Wu KL, Tsai YM, Lien CT, Kuo PL, Hung AJ. The Roles of MicroRNA in Lung Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 175] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 35. | Yang N, Ekanem NR, Sakyi CA, Ray SD. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev. 2015;81:62-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 185] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 36. | Nana-Sinkam SP, Geraci MW. MicroRNA in lung cancer. J Thorac Oncol. 2006;1:929–931. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 37. | Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res. 2014;3:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 272] [Reference Citation Analysis (0)] |
| 38. | Tanaka F, Yoneda K, Kondo N, Hashimoto M, Takuwa T, Matsumoto S, Okumura Y, Rahman S, Tsubota N, Tsujimura T, Kuribayashi K, Fukuoka K, Nakano T, Hasegawa S. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res. 2009;15:6980-6986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 250] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 39. | Rud AK, Borgen E, Mælandsmo GM, Flatmark K, Le H, Josefsen D, Solvoll I, Schirmer CB, Helland Å, Jørgensen L, Brustugun OT, Fodstad Ø, Boye K. Clinical significance of disseminated tumour cells in non-small cell lung cancer. Br J Cancer. 2013;109:1264-1270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 40. | Bessa X, Elizalde JI, Boix L, Piñol V, Lacy AM, Saló J, Piqué JM, Castells A. Lack of prognostic influence of circulating tumor cells in peripheral blood of patients with colorectal cancer. Gastroenterology. 2001;120:1084-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 41. | Nakagawa M, Uramoto H, Oka S, Chikaishi Y, Iwanami T, Shimokawa H, So T, Hanagiri T, Tanaka F. Clinical significance of IGF1R expression in non-small-cell lung cancer. Clin Lung Cancer. 2012;13:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 42. | Boyd JA, Hubbs JL, Kim DW, Hollis D, Marks LB, Kelsey CR. Timing of local and distant failure in resected lung cancer: implications for reported rates of local failure. J Thorac Oncol. 2010;5:211-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 72] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Ingenito F, Roscigno G, Affinito A, Nuzzo S, Scognamiglio I, Quintavalle C, Condorelli G. The Role of Exo-miRNAs in Cancer: A Focus on Therapeutic and Diagnostic Applications. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 44. | Zhang W, Yang J, Cao D, You Y, Shen K, Peng P. Regulation of exosomes released from normal ovarian epithelial cells and ovarian cancer cells. Tumour Biol. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 45. | King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 650] [Cited by in RCA: 802] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 46. | Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 311] [Cited by in RCA: 588] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 47. | Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015;25:198-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 559] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 48. | Challagundla KB, Fanini F, Vannini I, Wise P, Murtadha M, Malinconico L, Cimmino A, Fabbri M. microRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev Mol Diagn. 2014;14:565-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 49. | Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 912] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 50. | Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2218] [Cited by in RCA: 2382] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 51. | Liu Q, Peng F, Chen J. The Role of Exosomal MicroRNAs in the Tumor Microenvironment of Breast Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 52. | Hunter KW, Crawford NP, Alsarraj J. Mechanisms of metastasis. Breast Cancer Res. 2008;10 Suppl 1:S2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 53. | Massagué J, Batlle E, Gomis RR. Understanding the molecular mechanisms driving metastasis. Mol Oncol. 2017;11:3-4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 54. | Ma L. MicroRNA and Metastasis. Adv Cancer Res. 2016;132:165-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 55. | Coebergh van den Braak RRJ, Sieuwerts AM, Lalmahomed ZS, Smid M, Wilting SM, Bril SI, Xiang S, van der Vlugt-Daane M, de Weerd V, van Galen A, Biermann K, van Krieken JHJM, Kloosterman WP, Foekens JA; MATCH study group*, Martens JWM, IJzermans JNM. Confirmation of a metastasis-specific microRNA signature in primary colon cancer. Sci Rep. 2018;8:5242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Jia Z, Zhang Y, Xu Q, Guo W, Guo A. miR-126 suppresses epithelial-to-mesenchymal transition and metastasis by targeting PI3K/AKT/Snail signaling of lung cancer cells. Oncol Lett. 2018;15:7369-7375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 57. | Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130:2044-2053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 233] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 58. | Zhang JX, Mai SJ, Huang XX, Wang FW, Liao YJ, Lin MC, Kung HF, Zeng YX, Xie D. MiR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann Oncol. 2014;25:2196-2204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 59. | Yang F, Xiong H, Duan L, Li Q, Li X, Zhou Y. MiR-1246 Promotes Metastasis and Invasion of A549 cells by Targeting GSK-3β‒Mediated Wnt/β-Catenin Pathway. Cancer Res Treat. 2019;51:1420-1429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 60. | Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, Alvarez H, Gupta S, Maiti SN, Cooper L, Peehl D, Ram PT, Maitra A, Nagrath D. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 505] [Cited by in RCA: 724] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 61. | Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng J, Fan S. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J Exp Clin Cancer Res. 2018;37:226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 62. | Webber JP, Spary LK, Sanders AJ, Chowdhury R, Jiang WG, Steadman R, Wymant J, Jones AT, Kynaston H, Mason MD, Tabi Z, Clayton A. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2015;34:290-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 343] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 63. | Li J, Gong W, Zhu W, Shao X, Zhang C. The functional role of exosome microRNAs in lung cancer. Open Life Sci. 2017;12:223–227. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 64. | Tian W, Liu S, Li B. Potential Role of Exosomes in Cancer Metastasis. Biomed Res Int. 2019;2019:4649705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 65. | Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol. 2017;189:259-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 66. | Gun SY, Lee SWL, Sieow JL, Wong SC. Targeting immune cells for cancer therapy. Redox Biol. 2019;25:101174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 158] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 67. | Zhou CF, Ma J, Huang L, Yi HY, Zhang YM, Wu XG, Yan RM, Liang L, Zhong M, Yu YH, Wu S, Wang W. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene. 2019;38:1256-1268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 163] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 68. | Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, Hu J, Zhu X, Yang W, Liao W, Li G, Ding Y, Liang L. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 707] [Article Influence: 101.0] [Reference Citation Analysis (0)] |
| 69. | Vautrot V, Chanteloup G, Elmallah M, Cordonnier M, Aubin F, Garrido C, Gobbo J. Exosomal miRNA: Small Molecules, Big Impact in Colorectal Cancer. J Oncol. 2019;2019:8585276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 70. | Wang M, Yu F, Ding H, Wang Y, Li P, Wang K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:791-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 71. | Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem. 2013;288:34343-34351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 294] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 72. | Taverna S, Amodeo V, Saieva L, Russo A, Giallombardo M, De Leo G, Alessandro R. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol Cancer. 2014;13:169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 73. | Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849-10859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 497] [Cited by in RCA: 617] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 74. | Bao L, You B, Shi S, Shan Y, Zhang Q, Yue H, Zhang J, Zhang W, Shi Y, Liu Y, Wang X, Liu D, You Y. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene. 2018;37:2873-2889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 75. | Lu J, Liu QH, Wang F, Tan JJ, Deng YQ, Peng XH, Liu X, Zhang B, Xu X, Li XP. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2018;37:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 76. | Iliescu FS, Vrtačnik D, Neuzil P, Iliescu C. Microfluidic Technology for Clinical Applications of Exosomes. Micromachines (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 77. | Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, Pass HI. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8:1156-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 365] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 78. | Munagala R, Aqil F, Gupta RC. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biol. 2016;37:10703-10714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 96] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 79. | Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 901] [Cited by in RCA: 1010] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 80. | Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett. 2017;13:1256-1263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 81. | Zhou J, Guo H, Yang Y, Zhang Y, Liu H. A meta-analysis on the prognosis of exosomal miRNAs in all solid tumor patients. Medicine (Baltimore). 2019;98:e15335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 82. | Han Y, Li H. miRNAs as biomarkers and for the early detection of non-small cell lung cancer (NSCLC). J Thorac Dis. 2018;10:3119-3131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 83. | Moretti F, D'Antona P, Finardi E, Barbetta M, Dominioni L, Poli A, Gini E, Noonan DM, Imperatori A, Rotolo N, Cattoni M, Campomenosi P. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget. 2017;8:94980-94996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 84. | Nigita G, Distefano R, Veneziano D, Romano G, Rahman M, Wang K, Pass H, Croce CM, Acunzo M, Nana-Sinkam P. Tissue and exosomal miRNA editing in Non-Small Cell Lung Cancer. Sci Rep. 2018;8:10222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 85. | Xu J, Liao K, Zhou W. Exosomes Regulate the Transformation of Cancer Cells in Cancer Stem Cell Homeostasis. Stem Cells Int. 2018;2018:4837370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 86. | Spugnini EP, Logozzi M, Di Raimo R, Mizzoni D, Fais S. A Role of Tumor-Released Exosomes in Paracrine Dissemination and Metastasis. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 87. | Zhu X, Shen H, Yin X, Yang M, Wei H, Chen Q, Feng F, Liu Y, Xu W, Li Y. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J Exp Clin Cancer Res. 2019;38:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 295] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 88. | Xie F, Zhou X, Fang M, Li H, Su P, Tu Y, Zhang L, Zhou F. Extracellular Vesicles in Cancer Immune Microenvironment and Cancer Immunotherapy. Adv Sci (Weinh). 2019;6:1901779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 209] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 89. | Masciale V, Grisendi G, Banchelli F, D'Amico R, Maiorana A, Sighinolfi P, Pinelli M, Lovati E, Stefani A, Morandi U, Dominici M, Aramini B. Correlating tumor-infiltrating lymphocytes and lung cancer stem cells: a cross-sectional study. Ann Transl Med. 2019;7:619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 90. | Kise K, Kinugasa-Katayama Y, Takakura N. Tumor microenvironment for cancer stem cells. Adv Drug Deliv Rev. 2016;99:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 91. | Masciale V, Grisendi G, Banchelli F, D'Amico R, Morandi U, Dominici M, Aramini B, Haider HKh. In: Haider HKh. Stem Cells: From Myth to Reality and Evolving. Haider HKh. Berlin, Boston: Walter de Gruyter GmbH & Co KG, 2019; Chapter-6. |
| 92. | Dai Z, Locasale JW. Metabolic pattern formation in the tumor microenvironment. Mol Syst Biol. 2017;13:915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 93. | Bocci F, Gearhart-Serna L, Boareto M, Ribeiro M, Ben-Jacob E, Devi GR, Levine H, Onuchic JN, Jolly MK. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc Natl Acad Sci USA. 2019;116:148-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 218] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 94. | Li SC, Vu LT, Luo JJ, Zhong JF, Li Z, Dethlefs BA, Loudon WG, Kabeer MH. Tissue Elasticity Bridges Cancer Stem Cells to the Tumor Microenvironment Through microRNAs: Implications for a "Watch-and-Wait" Approach to Cancer. Curr Stem Cell Res Ther. 2017;12:455-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 95. | Turdo A, Veschi V, Gaggianesi M, Chinnici A, Bianca P, Todaro M, Stassi G. Meeting the Challenge of Targeting Cancer Stem Cells. Front Cell Dev Biol. 2019;7:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 96. | Valent P, Bonnet D, De Maria R, Lapidot T, Copland M, Melo JV, Chomienne C, Ishikawa F, Schuringa JJ, Stassi G, Huntly B, Herrmann H, Soulier J, Roesch A, Schuurhuis GJ, Wöhrer S, Arock M, Zuber J, Cerny-Reiterer S, Johnsen HE, Andreeff M, Eaves C. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer. 2012;12:767-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 526] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 97. | Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9969] [Cited by in RCA: 9689] [Article Influence: 509.9] [Reference Citation Analysis (0)] |
| 98. | Mazzio EA, Soliman KF. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics. 2012;7:119-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 99. | Yosef N, Regev A. Writ large: Genomic dissection of the effect of cellular environment on immune response. Science. 2016;354:64-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 100. | Ozen M, Gündüz M, Haider HKh. Chimeric Antigen Receptor (CAR) T-cells as a therapeutic modality. In: Haider HKh. Stem cells: From Hype to Hope. Singapore: World Scientific, 2020: 211-236. |
| 101. | Romero D. Immunotherapy: CAR T cells ready to go mainstream. Nat Rev Clin Oncol. 2016;13:396-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 102. | Onea AS, Jazirehi AR. CD19 chimeric antigen receptor (CD19 CAR)-redirected adoptive T-cell immunotherapy for the treatment of relapsed or refractory B-cell Non-Hodgkin's Lymphomas. Am J Cancer Res. 2016;6:403-424. [PubMed] |
| 103. | Riedmann EM. Agenus’ brain cancer vaccine doubles survival rate in GBM patients. Hum Vaccin Immunother. 2014;10:2139-2140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 104. | Carlson B. Therapeutic vaccine for brain cancer succeeds using a unique approach. Biotechnol Healthc. 2011;8:31-32. [PubMed] |
| 105. | Spira A, Disis ML, Schiller JT, Vilar E, Rebbeck TR, Bejar R, Ideker T, Arts J, Yurgelun MB, Mesirov JP, Rao A, Garber J, Jaffee EM, Lippman SM. Leveraging premalignant biology for immune-based cancer prevention. Proc Natl Acad Sci USA. 2016;113:10750-10758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 106. | Lekka M. Discrimination Between Normal and Cancerous Cells Using AFM. Bionanoscience. 2016;6:65-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 267] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 107. | Huang CY, Ju DT, Chang CF, Muralidhar Reddy P, Velmurugan BK. A review on the effects of current chemotherapy drugs and natural agents in treating non-small cell lung cancer. Biomedicine (Taipei). 2017;7:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 281] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 108. | Tung KH, Ernstoff MS, Allen C, Shu S. A Review of Exosomes and their Role in The Tumor Microenvironment and Host-Tumor "Macroenvironment". J Immunol Sci. 2019;3:4-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 109. | Javeed N, Mukhopadhyay D. Exosomes and their role in the micro-/macro-environment: a comprehensive review. J Biomed Res. 2017;31:386-394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 110. | Castaño Z, Tracy K, McAllister SS. The tumor macroenvironment and systemic regulation of breast cancer progression. Int J Dev Biol. 2011;55:889-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 111. | Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1418] [Article Influence: 157.6] [Reference Citation Analysis (0)] |
| 112. | Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1806] [Cited by in RCA: 2054] [Article Influence: 293.4] [Reference Citation Analysis (0)] |
| 113. | Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384-3391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 1140] [Article Influence: 114.0] [Reference Citation Analysis (0)] |
| 114. | Hu C, Chen M, Jiang R, Guo Y, Wu M, Zhang X. Exosome-related tumor microenvironment. J Cancer. 2018;9:3084-3092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 115. | Gurunathan S, Kang MH, Jeyaraj M, Qasim M, Kim JH. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 843] [Cited by in RCA: 784] [Article Influence: 130.7] [Reference Citation Analysis (0)] |
| 116. | Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 306] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 117. | Macharia LW, Wanjiru CM, Mureithi MW, Pereira CM, Ferrer VP, Moura-Neto V. MicroRNAs, Hypoxia and the Stem-Like State as Contributors to Cancer Aggressiveness. Front Genet. 2019;10:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 118. | Lima FR, Kahn SA, Soletti RC, Biasoli D, Alves T, da Fonseca AC, Garcia C, Romão L, Brito J, Holanda-Afonso R, Faria J, Borges H, Moura-Neto V. Glioblastoma: therapeutic challenges, what lies ahead. Biochim Biophys Acta. 2012;1826:338-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 119. | Arpino G, Milano M, De Placido S. Features of aggressive breast cancer. Breast. 2015;24:594-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 120. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 121. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1980] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 122. | Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 240] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 123. | Cui JG, Zhao Y, Sethi P, Li YY, Mahta A, Culicchia F, Lukiw WJ. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol. 2010;98:297-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 124. | Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, Zhong Y, You Y, Pu P, Kang C. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 72] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 125. | Bao S, Wang X, Wang Z, Yang J, Liu F, Yin C. MicroRNA-30 mediates cell invasion and metastasis in breast cancer. Biochem Cell Biol. 2018;96:825-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 126. | Nobel Foundation. Nobel Prize in Physiology or Medicine 2019: How cells sense and adapt to oxygen availability. Science Daily. 2019. Available from: https://www.sciencedaily.com/releases/2019/10/191007095520.htm. |
| 127. | Pugh CW, Ratcliffe PJ. New horizons in hypoxia signaling pathways. Exp Cell Res. 2017;356:116-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 128. | Kaelin WG, Ratcliffe PJ, Semenza GL. Pathways for Oxygen Regulation and Homeostasis: The 2016 Albert Lasker Basic Medical Research Award. JAMA. 2016;316:1252-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 129. | Saito S, Lin YC, Tsai MH, Lin CS, Murayama Y, Sato R, Yokoyama KK. Emerging roles of hypoxia-inducible factors and reactive oxygen species in cancer and pluripotent stem cells. Kaohsiung J Med Sci. 2015;31:279-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 130. | Triner D, Shah YM. Hypoxia-inducible factors: a central link between inflammation and cancer. J Clin Invest. 2016;126:3689-3698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 131. | Tong WW, Tong GH, Liu Y. Cancer stem cells and hypoxia-inducible factors (Review). Int J Oncol. 2018;53:469-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 132. | Hajizadeh F, Okoye I, Esmaily M, Ghasemi Chaleshtari M, Masjedi A, Azizi G, Irandoust M, Ghalamfarsa G, Jadidi-Niaragh F. Hypoxia inducible factors in the tumor microenvironment as therapeutic targets of cancer stem cells. Life Sci. 2019;237:116952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 133. | Gündüz D, Aslam M. MicroRNAs as modulators of endothelial differentiation of stem cells: role in vascular regenerative medicine. In: Haider HKh. Stem Cells - From Drug to Drug Discovery. Berlin Boston: De Gruyter, 2017: 63-84. |
| 134. | Kim HW, Haider HKh, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem. 2009;284:33161-33168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 270] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 135. | Kim HW, Jiang S, Ashraf M, Haider HKh. Stem cell-based delivery of Hypoxamir-210 to the infarcted heart: implications on stem cell survival and preservation of infarcted heart function. J Mol Med (Berl). 2012;90:997-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 136. | Haider HKh, Kim HW, Ashraf M. Hypoxia-inducible factor-1alpha in stem cell preconditioning: mechanistic role of hypoxia-related micro-RNAs. J Thorac Cardiovasc Surg. 2009;138:257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 137. | Kim HW, Mallick F, Durrani S, Ashraf M, Jiang S, Haider HKh. Concomitant activation of miR-107/PDCD10 and hypoxamir-210/Casp8ap2 and their role in cytoprotection during ischemic preconditioning of stem cells. Antioxid Redox Signal. 2012;17:1053-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 138. | Ye L, Zhang W, Su LP, Haider HKh, Poh KK, Galupo MJ, Songco G, Ge RW, Tan HC, Sim EK. Nanoparticle based delivery of hypoxia-regulated VEGF transgene system combined with myoblast engraftment for myocardial repair. Biomaterials. 2011;32:2424-2431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 139. | Bao B, Azmi AS, Li Y, Ahmad A, Ali S, Banerjee S, Kong D, Sarkar FH. Targeting CSCs in tumor microenvironment: the potential role of ROS-associated miRNAs in tumor aggressiveness. Curr Stem Cell Res Ther. 2014;9:22-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 140. | Sun HR, Wang S, Yan SC, Zhang Y, Nelson PJ, Jia HL, Qin L-X and Dong Q-Z. Therapeutic Strategies Targeting Cancer Stem Cells and Their Microenvironment. Front Oncol. 2019;9:1104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 141. | Yeo CD, Kang N, Choi SY, Kim BN, Park CK, Kim JW, Kim YK, Kim SJ. The role of hypoxia on the acquisition of epithelial-mesenchymal transition and cancer stemness: a possible link to epigenetic regulation. Korean J Intern Med. 2017;32:589-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 82] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 142. | Bao B, Azmi AS, Ali S, Ahmad A, Li Y, Banerjee S, Kong D, Sarkar FH. The biological kinship of hypoxia with CSC and EMT and their relationship with deregulated expression of miRNAs and tumor aggressiveness. Biochim Biophys Acta. 2012;1826:272-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 143. | Zhou D, Shao L, Spitz DR. Reactive oxygen species in normal and tumor stem cells. Adv Cancer Res. 2014;122:1-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 269] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 144. | Chang L, Graham P, Hao J, Ni J, Deng J, Bucci J, Malouf D, Gillatt D, Li Y. Cancer stem cells and signaling pathways in radioresistance. Oncotarget. 2016;7:11002-11017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 145. | Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780-783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2102] [Cited by in RCA: 1934] [Article Influence: 120.9] [Reference Citation Analysis (0)] |
| 146. | Magenta A, Greco S, Gaetano C, Martelli F. Oxidative stress and microRNAs in vascular diseases. Int J Mol Sci. 2013;14:17319-17346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 143] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 147. | Zhang X, Ng WL, Wang P, Tian L, Werner E, Wang H, Doetsch P, Wang Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Cancer Res. 2012;72:4707-4713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 148. | He J, Jiang BH. Interplay between Reactive oxygen Species and MicroRNAs in Cancer. Curr Pharmacol Rep. 2016;2:82-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 149. | R Babu K, Tay Y. The Yin-Yang Regulation of Reactive Oxygen Species and MicroRNAs in Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 150. | Masciale V, Grisendi G, Banchelli F, D'Amico R, Maiorana A, Sighinolfi P, Stefani A, Morandi U, Dominici M, Aramini B. Isolation and Identification of Cancer Stem-Like Cells in Adenocarcinoma and Squamous Cell Carcinoma of the Lung: A Pilot Study. Front Oncol. 2019;9:1394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 151. | Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, Schuster K, Shao C, Larsen JE, Sullivan LA, Honorio S, Xie Y, Scaglioni PP, DiMaio JM, Gazdar AF, Shay JW, Wistuba II, Minna JD. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937-9948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 327] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 152. | Masciale V, Grisendi G, Banchelli F, D'Amico R, Maiorana A, Morandi U, Dominici M, Aramini B. Cancer stem-neuroendocrine cells in an atypical carcinoid case report. Transl Lung Cancer Res. 2019;8:1157-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 153. | Haider HKh, Aramini B. Mircrining the injured heart with stem cell-derived exosomes: an emerging strategy of cell-free therapy. Stem Cell Res Ther. 2020;11:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 154. | Tai YL, Chen KC, Hsieh JT, Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. 2018;109:2364-2374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 309] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 155. | Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525-1541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 236] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 156. | Sandfeld-Paulsen B, Aggerholm-Pedersen N, Bæk R, Jakobsen KR, Meldgaard P, Folkersen BH, Rasmussen TR, Varming K, Jørgensen MM, Sorensen BS. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol. 2016;10:1595-1602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 202] [Article Influence: 22.4] [Reference Citation Analysis (1)] |
| 157. | Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, Xiang Y, Wu N, Wu L, Bai L, Li Y. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017;8:13048-13058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 205] [Article Influence: 25.6] [Reference Citation Analysis (1)] |
| 158. | Reclusa P, Taverna S, Pucci M, Durendez E, Calabuig S, Manca P, Serrano MJ, Sober L, Pauwels P, Russo A, Rolfo C. Exosomes as diagnostic and predictive biomarkers in lung cancer. J Thorac Dis. 2017;9:S1373-S1382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 159. | Taverna S, Giallombardo M, Gil-Bazo I, Carreca AP, Castiglia M, Chacártegui J, Araujo A, Alessandro R, Pauwels P, Peeters M, Rolfo C. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget. 2016;7:28748-28760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 160. | Huang SH, Li Y, Zhang J, Rong J, Ye S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. 2013;31:330-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 161. | Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. Oncoimmunology. 2015;4:e1027472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 182] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 162. | Brinkmann K, Enderle D, Koestler T, Bentink S, Emenegger J, Spiel A, Mueller R, O'Neill V, Skog J, Noerholm M. Abstract 545: Plasma-based diagnostics for detection of EML4-ALK fusion transcripts in NSCLC patients. Cancer Res. 2015;75:545. [DOI] [Full Text] |
| 163. | Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, Iyer S, Reisman A, Wilner KD, Tursi J, Blackhall F; PROFILE 1014 Investigators. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167-2177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2160] [Cited by in RCA: 2469] [Article Influence: 224.5] [Reference Citation Analysis (0)] |
| 164. | Srivastava A, Amreddy N, Razaq M, Towner R, Zhao YD, Ahmed RA, Munshi A, Ramesh R. Exosomes as Theranostics for Lung Cancer. Adv Cancer Res. 2018;139:1-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 165. | Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 788] [Article Influence: 78.8] [Reference Citation Analysis (1)] |
| 166. | Harding CV, Heuser JE, Stahl PD. Exosomes: looking back three decades and into the future. J Cell Biol. 2013;200:367-371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 357] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 167. | Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31:543-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 403] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 168. | Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu CC, Gagea M, Yang S, Blanko EVR, Peng Q, Ma X, Marszalek JR, Maitra A, Yee C, Rezvani K, Shpall E, LeBleu VS, Kalluri R. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 577] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 169. | Yang G, Zhang W, Yu C, Ren J, An Z. MicroRNA let-7: Regulation, single nucleotide polymorphism, and therapy in lung cancer. J Cancer Res Ther. 2015;11 Suppl 1:C1-C6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 170. | Zhang C, Xiao X, Chen M, Aldharee H, Chen Y, Long W. Liver kinase B1 restoration promotes exosome secretion and motility of lung cancer cells. Oncol Rep. 2018;39:376-382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 171. | Nishida-Aoki N, Tominaga N, Takeshita F, Sonoda H, Yoshioka Y, Ochiya T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol Ther. 2017;25:181-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 168] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 172. | Motokawa Y, Kokubo M, Kuwabara N, Tatematsu KI, Sezutsu H, Takahashi H, Sakakura K, Chikamatsu K, Takeda S. Melanoma antigen family A4 protein produced by transgenic silkworms induces antitumor immune responses. Exp Ther Med. 2018;15:2512-2518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 173. | Escudier B, Dorval T, Chaput N, André F, Caby MP, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, Boccaccio C, Bonnerot C, Dhellin O, Movassagh M, Piperno S, Robert C, Serra V, Valente N, Le Pecq JB, Spatz A, Lantz O, Tursz T, Angevin E, Zitvogel L. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med. 2005;3:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 763] [Cited by in RCA: 1006] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 174. | Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, Valente N, Shreeniwas R, Sutton MA, Delcayre A, Hsu DH, Le Pecq JB, Lyerly HK. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 678] [Cited by in RCA: 882] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 175. | Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, Le Chevalier T, Livartoski A, Barlesi F, Laplanche A, Ploix S, Vimond N, Peguillet I, Théry C, Lacroix L, Zoernig I, Dhodapkar K, Dhodapkar M, Viaud S, Soria JC, Reiners KS, Pogge von Strandmann E, Vély F, Rusakiewicz S, Eggermont A, Pitt JM, Zitvogel L, Chaput N. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. 2016;5:e1071008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 604] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 176. | Wang J, Zheng Y, Zhao M. Exosome-Based Cancer Therapy: Implication for Targeting Cancer Stem Cells. Front Pharmacol. 2016;7:533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 177. | Markov O, Oshchepkova A, Mironova N. Immunotherapy Based on Dendritic Cell-Targeted/-Derived Extracellular Vesicles-A Novel Strategy for Enhancement of the Anti-tumor Immune Response. Front Pharmacol. 2019;10:1152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |