Copyright
©The Author(s) 2025.
World J Stem Cells. Mar 26, 2025; 17(3): 100079
Published online Mar 26, 2025. doi: 10.4252/wjsc.v17.i3.100079
Published online Mar 26, 2025. doi: 10.4252/wjsc.v17.i3.100079
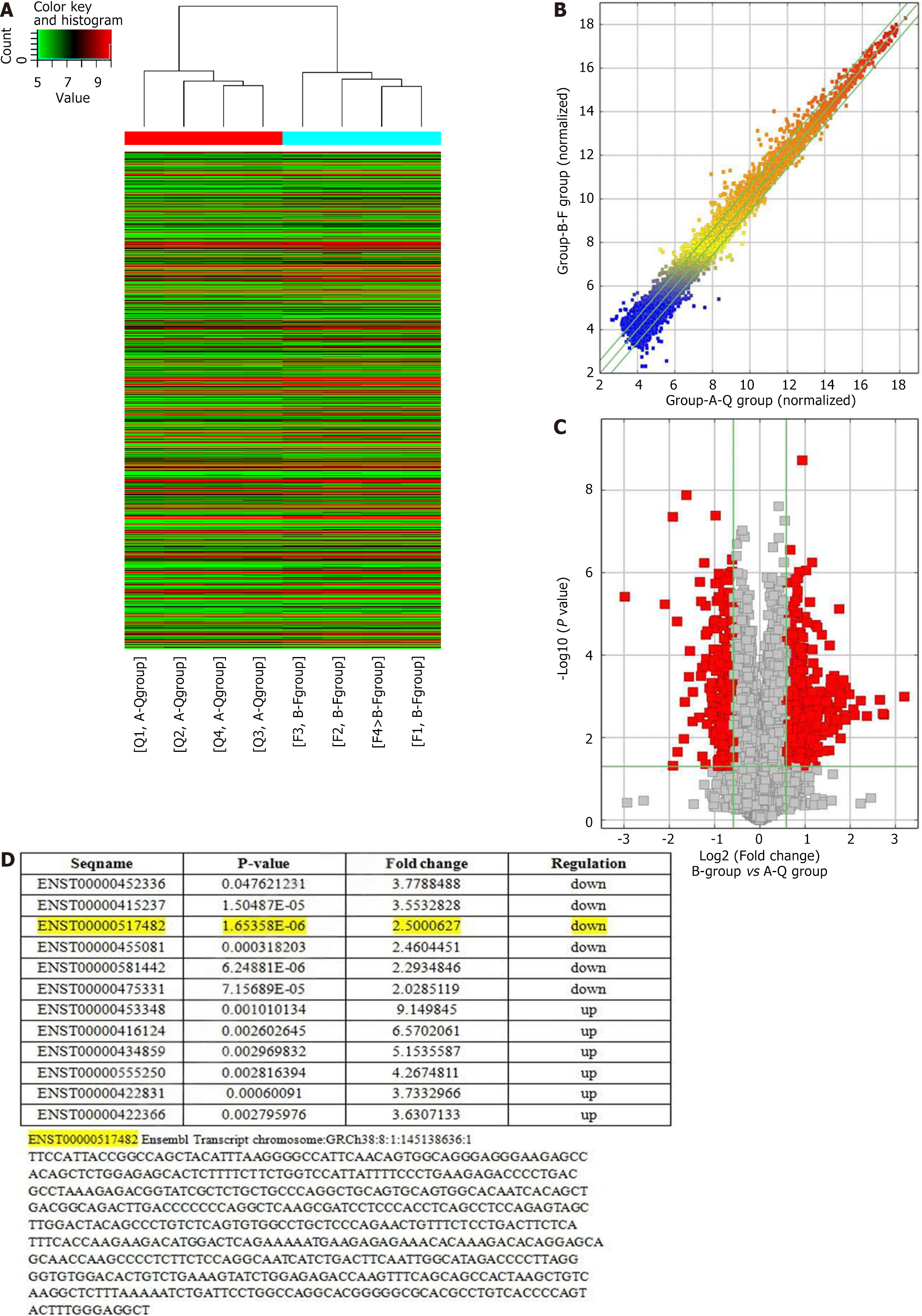
Figure 1 Screening and identification of ischemia, hypoxia, and oxidative stress-related long noncoding RNAs in lung injury.
A: Heatmap of differentially expressed long noncoding RNAs (lncRNAs) between lung injury groups and controls; B: Based on the normalized lncRNA expression levels in lung injury and control groups; C: Volcano plot of differentially expressed lncRNAs between lung injury and control groups; D: Sequences of lncRNAs and lncRNA-ENST screened by gene chip.
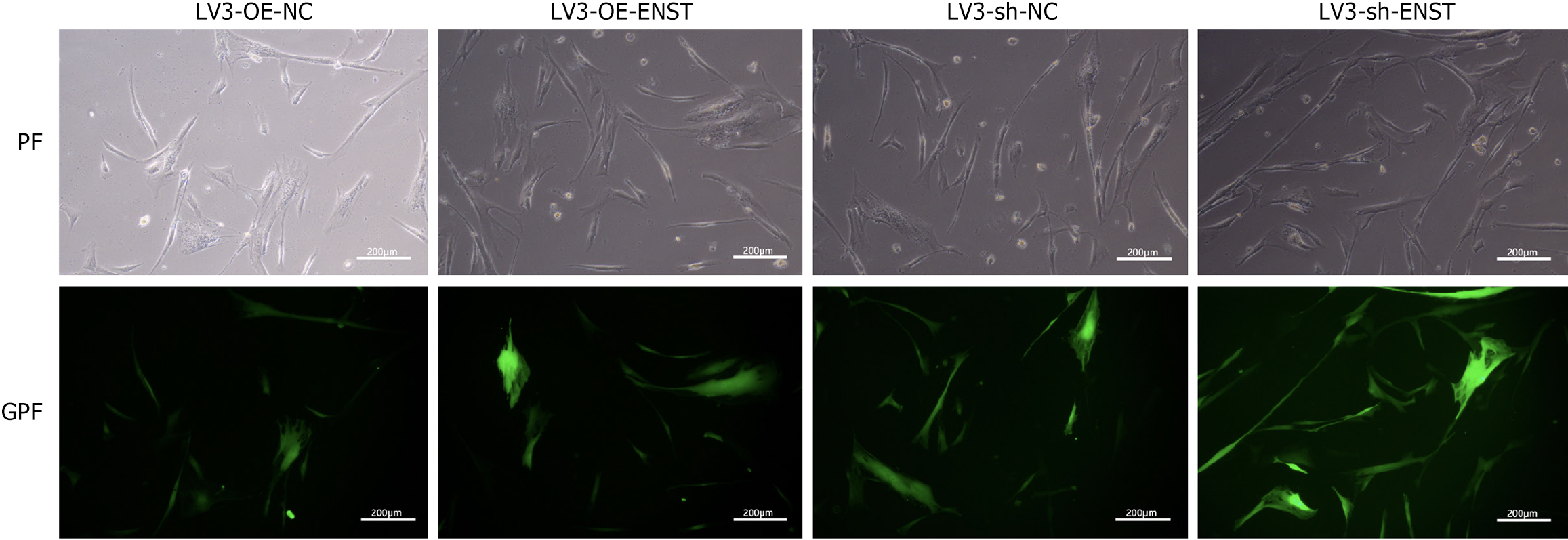
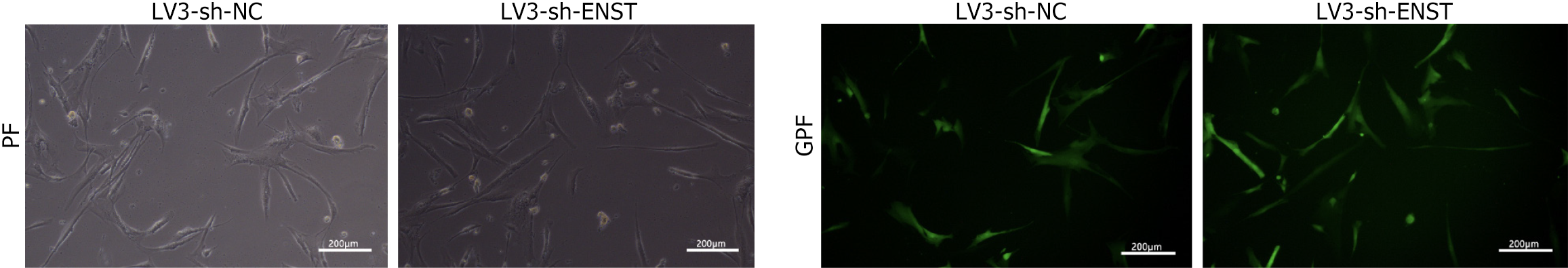
Figure 2 Inverted fluorescence microscopy was used to observe the cell lentiviral transfection efficiency.
“Transfection efficiency” was qualitatively evaluated based on green photo fluorescence, with cells exhibiting green photo fluorescence expression considered successfully transfected. “Quantitative analysis” was performed by flow cytometry, where the percentage of green photo fluorescence-positive cells was determined. The transfection efficiency was calculated to be “greater than 80%” based on flow cytometry results. NC: Negative control; PF: Photo fluorescence; GPF: Green photo fluorescence; OE: Overexpression.
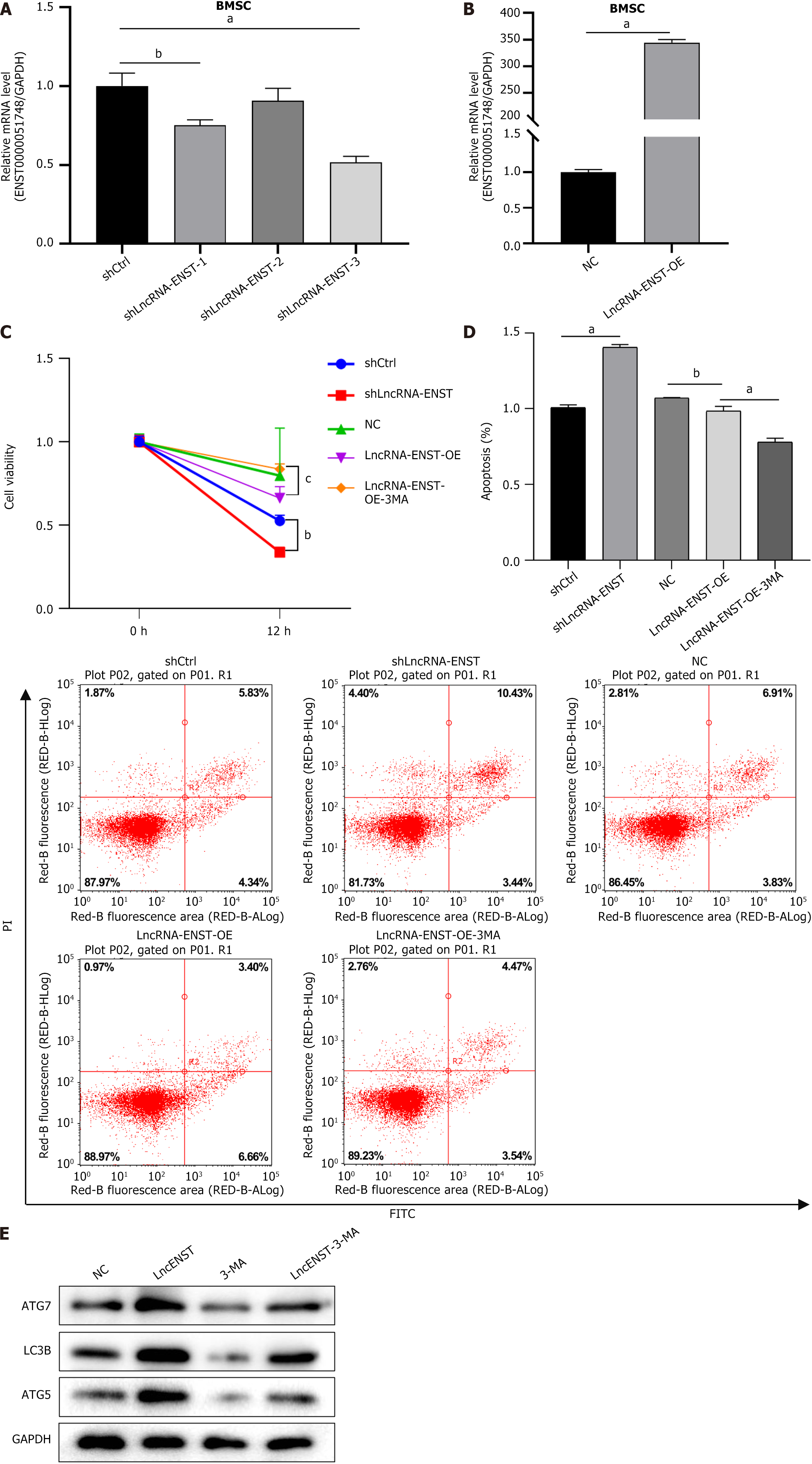
Figure 3 Long noncoding RNA-ENST affects the apoptosis and autophagy levels of bone marrow mesenchymal stem cells.
A: The quantitative polymerase chain reaction analysis revealed the knockdown efficiency of long noncoding RNA (lncRNA)-ENST (n = 3); B: The quantitative polymerase chain reaction analysis showed the overexpression efficiency of lncRNA-ENST (n = 3); C: The Cell Counting Kit-8 assay was used to evaluate cell viability and assess cell damage (n = 3); D: Flow cytometry dual staining was conducted to detect cell apoptosis (apoptosis level statistics chart and flow cytometry chart) (n = 3); E: Western blot analysis was performed to detect the expression of LC3B, ATG7, and ATG5 after knocking down lncRNA-ENST (n = 3). aP < 0.001, bP < 0.01, cP < 0.05. BMSC: Bone marrow mesenchymal stem cell; LncRNA: Long noncoding RNA; NC: Negative control; OE: Overexpression.
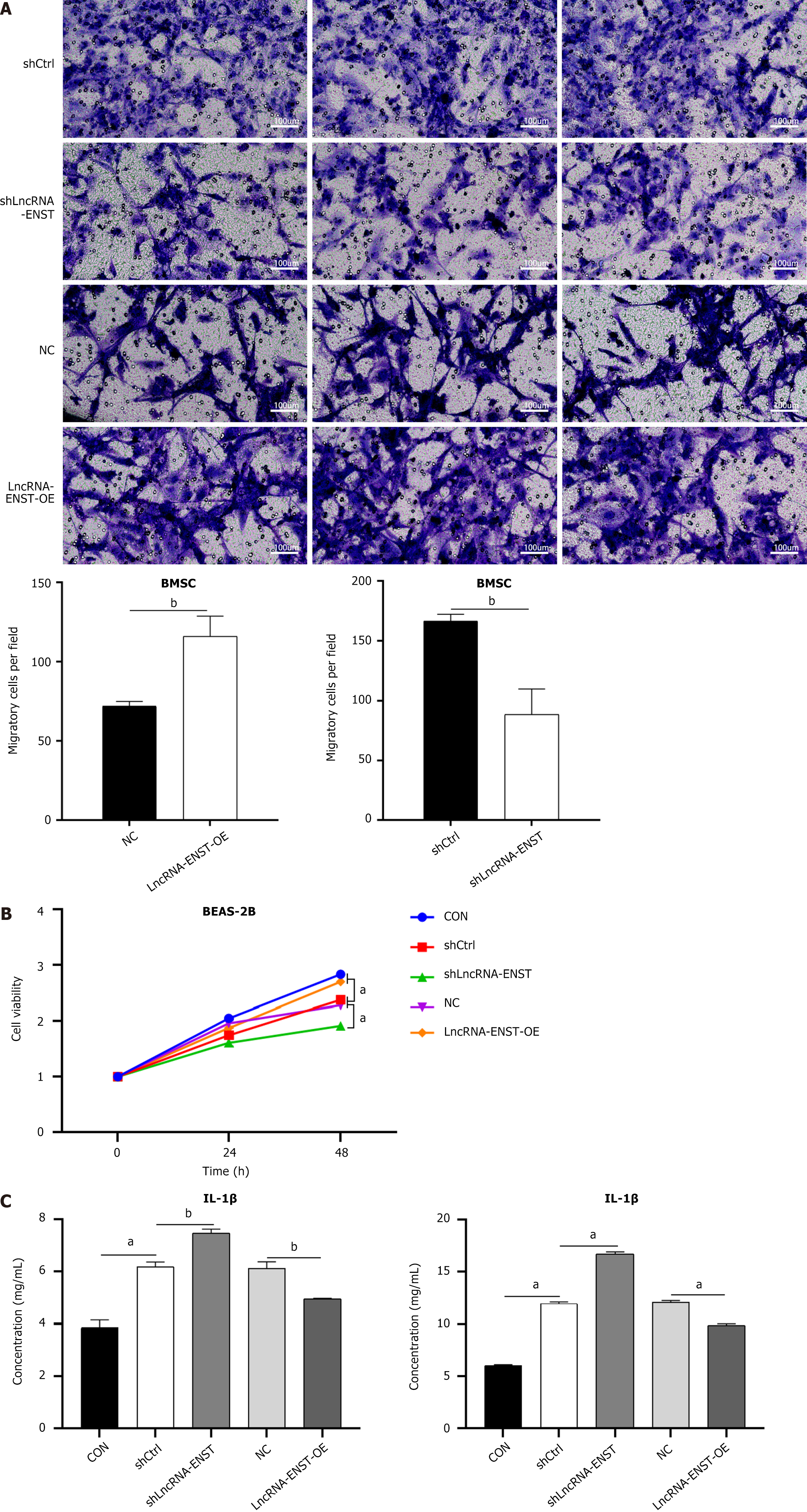
Figure 4 Bone marrow mesenchymal stem cells influence the migration of lung epithelial cells BEAS-2B in an indirect co-culture system.
A: Transwell assay was employed to evaluate the migration ability of bone marrow mesenchymal stem cells (n = 3); B: Cell Counting Kit-8 assay was performed to assess the viability of BEAS-2B cells (n = 3); C: Enzyme-linked immunosorbent assay was used to detect the expression of interleukin-1β and interleukin-6 (n = 3). aP < 0.001, bP < 0.01. BMSC: Bone marrow mesenchymal stem cell; LncRNA: Long noncoding RNA; NC: Negative control; OE: Overexpression; IL: Interleukin.
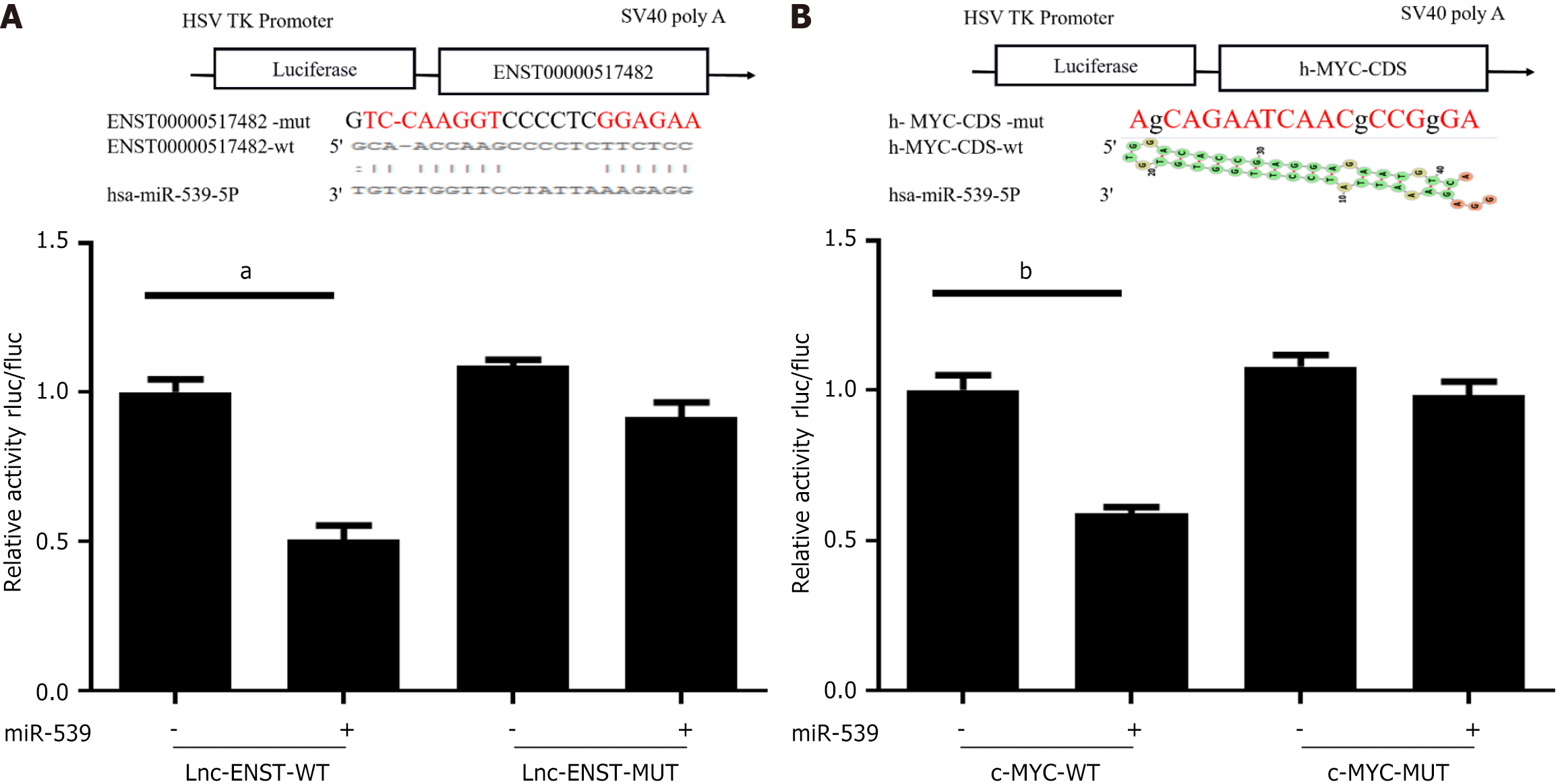
Figure 5 Dual-luciferase reporter assay was performed to investigate both the binding affinity and transcriptional regulation between lncRNA-ENST and miR-539.
A: The assay assessed the binding affinity between lncRNA-ENST and miR-539-5p; B: The same assay was performed to assess the interaction between c-MYC and miR-539-5p and its impact on luciferase activity. aP < 0.001, bP < 0.01.
Figure 6 Inverted fluorescence microscopy was used to assess lentiviral transfection efficiency.
The percentage of green photo fluorescence-positive cells was calculated using flow cytometry (n = 3). A transfection efficiency of “greater than 80%” was considered high, based on green photo fluorescence. Flow cytometry analysis confirmed the high transfection efficiency in both bone marrow mesenchymal stem cells and 293T cells. NC: Negative control; PF: Photo fluorescence; GPF: Green photo fluorescence.
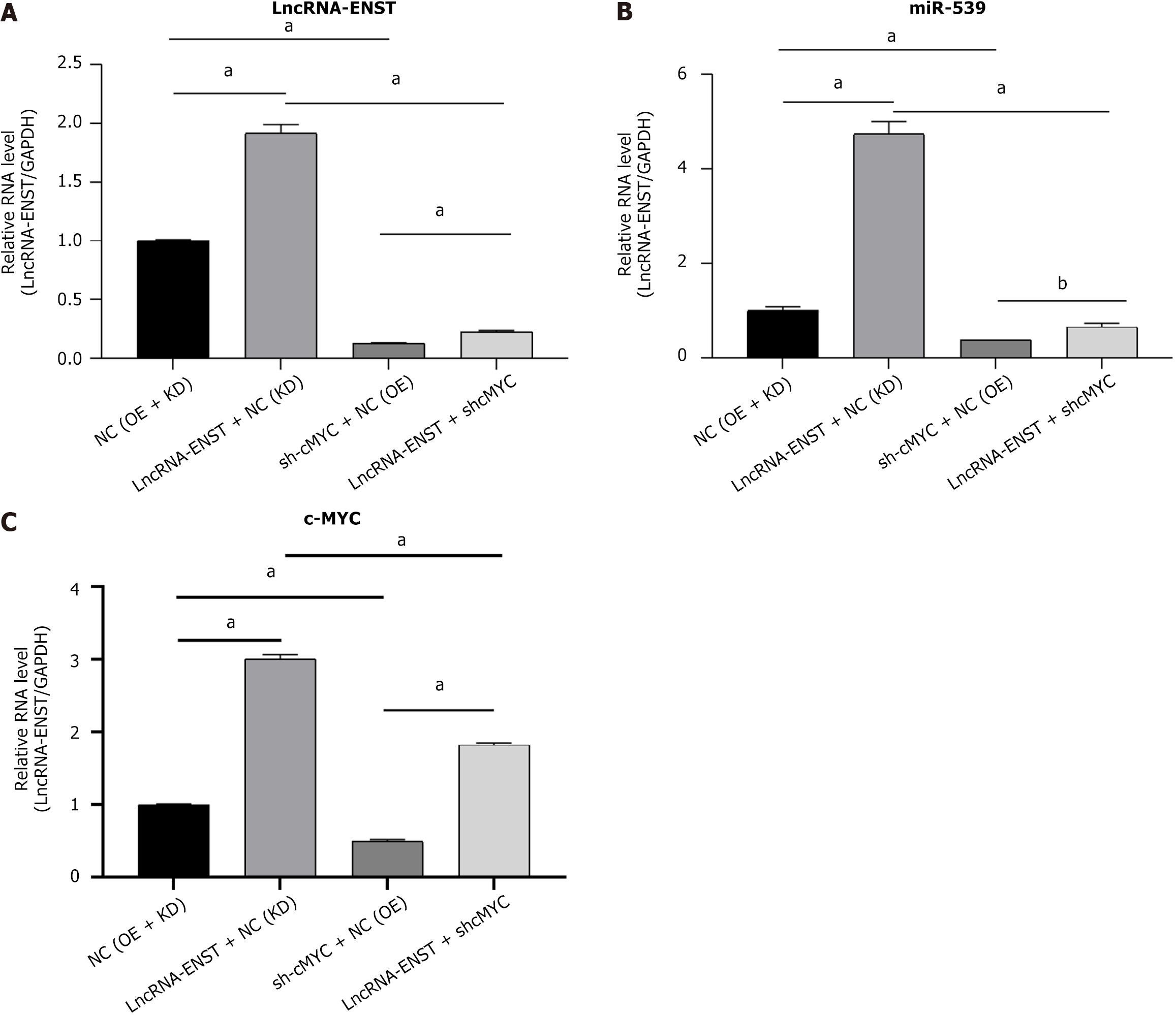
Figure 7 Long noncoding RNA ENST is involved in regulation through the miR-539-5p/c-MYC axis.
A: Quantitative polymerase chain reaction was conducted to measure the expression levels of long noncoding RNA-ENST and sh-cMYC (n = 3); B: Quantitative polymerase chain reaction was used to detect the expression levels of miR539 gene (n = 3); C: Quantitative polymerase chain reaction was conducted to measure the expression levels of c-MYC gene (n = 3). aP < 0.001, bP < 0.01. LncRNA: Long noncoding RNA; NC: Negative control; KD: Knockdown; OE: Overexpression.
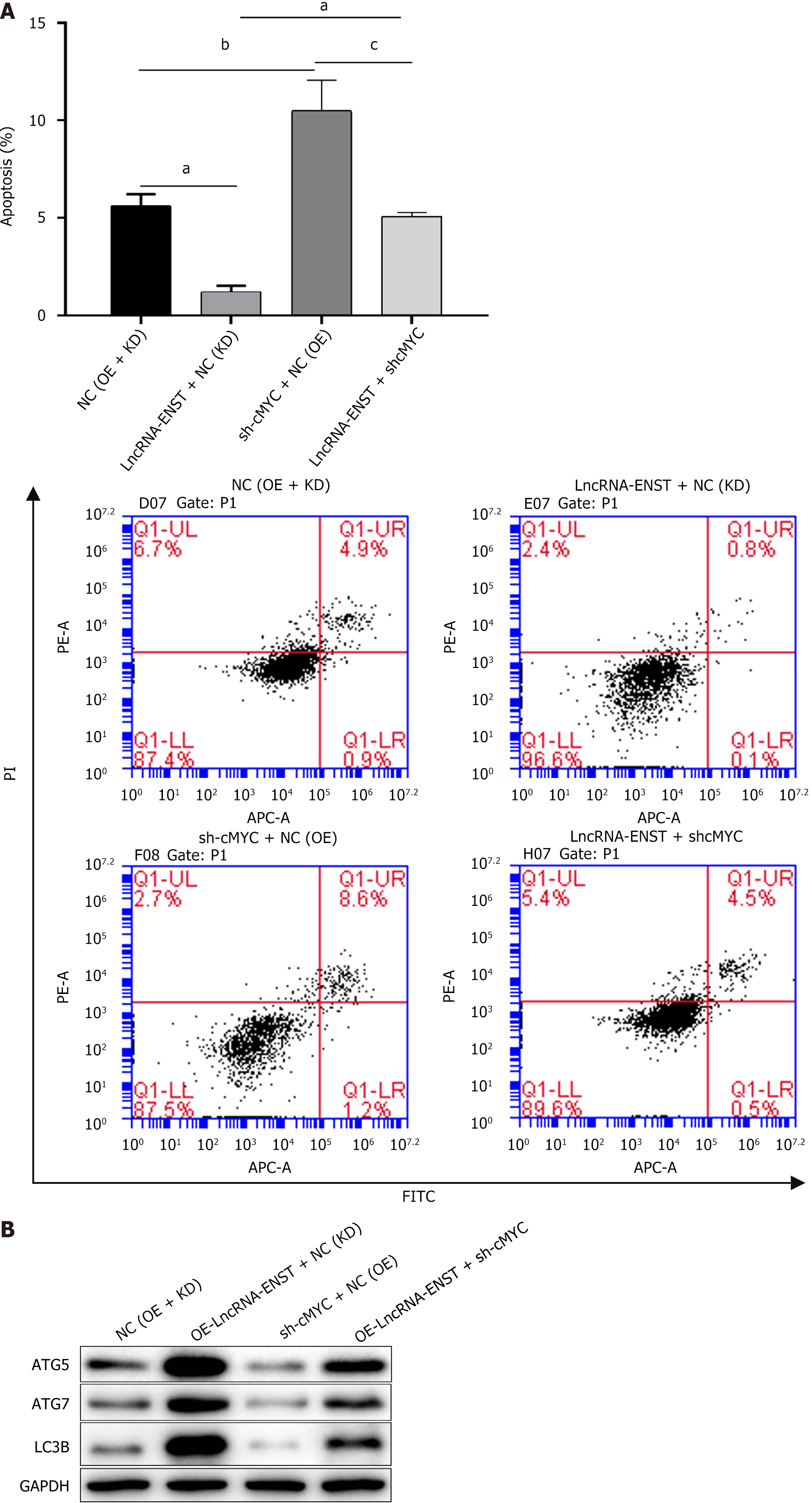
Figure 8 Long noncoding RNA ENST regulates autophagy and apoptosis of bone marrow mesenchymal stem cells through the miR-539-5p/c-MYC axis.
A: Flow cytometry was used to detect cell apoptosis (apoptosis level statistics chart and flow cytometry chart) (n = 3); B: Western blot was used to detect the protein expression of autophagy-related proteins LC3B, ATG7, and ATG5 (n = 3). aP < 0.001, bP < 0.01, cP < 0.05. LncRNA: Long noncoding RNA; NC: Negative control; KD: Knockdown; OE: Overexpression; PI: Propidium iodide.
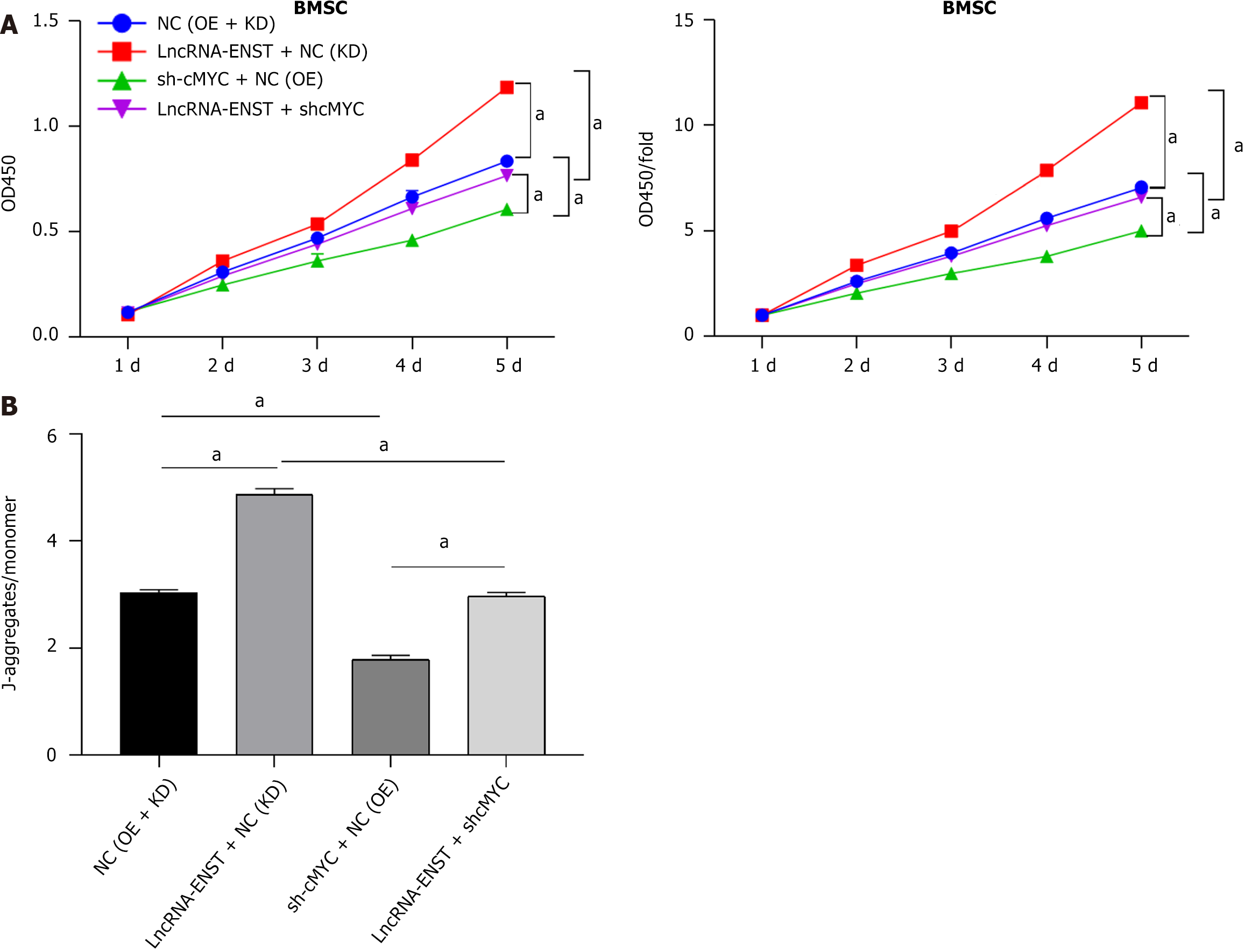
Figure 9 Long noncoding RNA ENST regulates the cell viability and mitochondrial membrane potential of bone marrow mesenchymal stem cells through the miR-539-5p/c-MYC axis.
A: Cell Counting Kit-8 assay was performed to measure the viability of bone marrow mesenchymal stem cells (n = 3); B: JC-1 staining was performed to detect mitochondrial membrane potential (n = 3). aP < 0.001. BMSC: Bone marrow mesenchymal stem cell; LncRNA: Long noncoding RNA; NC: Negative control; KD: Knockdown; OE: Overexpression.
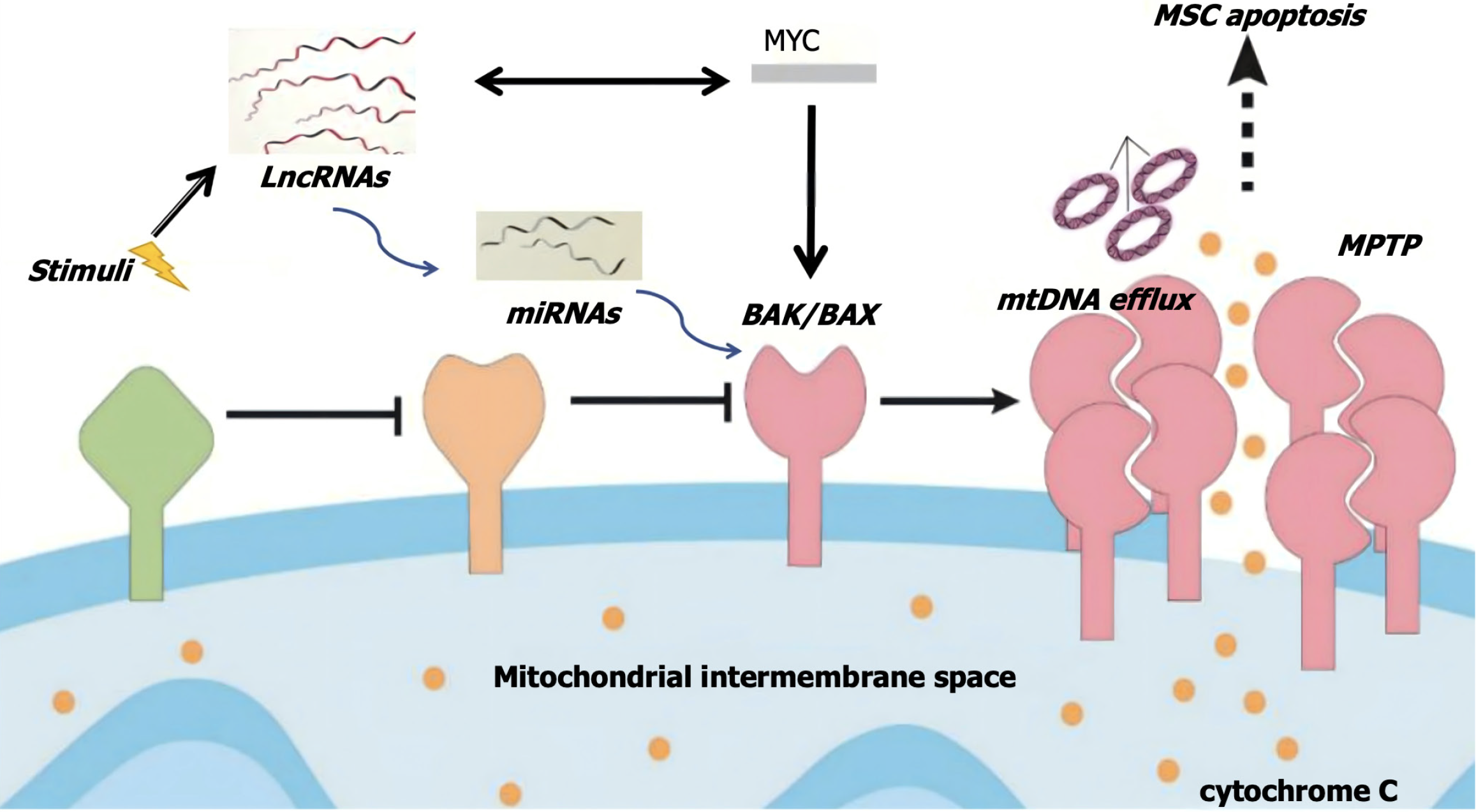
Figure 10 Schematic diagram of long noncoding RNA-microRNA-MYC-BAX/BAK-mitochondrial permeability transition pore apoptosis pathway.
LncRNAs: Long noncoding RNAs; miRNAs: MicroRNAs; MSC: Mesenchymal stem cell; MPTP: Mitochondrial permeability transition pore.
- Citation: Shen YZ, Yang GP, Ma QM, Wang YS, Wang X. Regulation of lncRNA-ENST on Myc-mediated mitochondrial apoptosis in mesenchymal stem cells: In vitro evidence implicated for acute lung injury therapeutic potential. World J Stem Cells 2025; 17(3): 100079
- URL: https://www.wjgnet.com/1948-0210/full/v17/i3/100079.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v17.i3.100079