Copyright
©The Author(s) 2025.
World J Stem Cells. Feb 26, 2025; 17(2): 96893
Published online Feb 26, 2025. doi: 10.4252/wjsc.v17.i2.96893
Published online Feb 26, 2025. doi: 10.4252/wjsc.v17.i2.96893
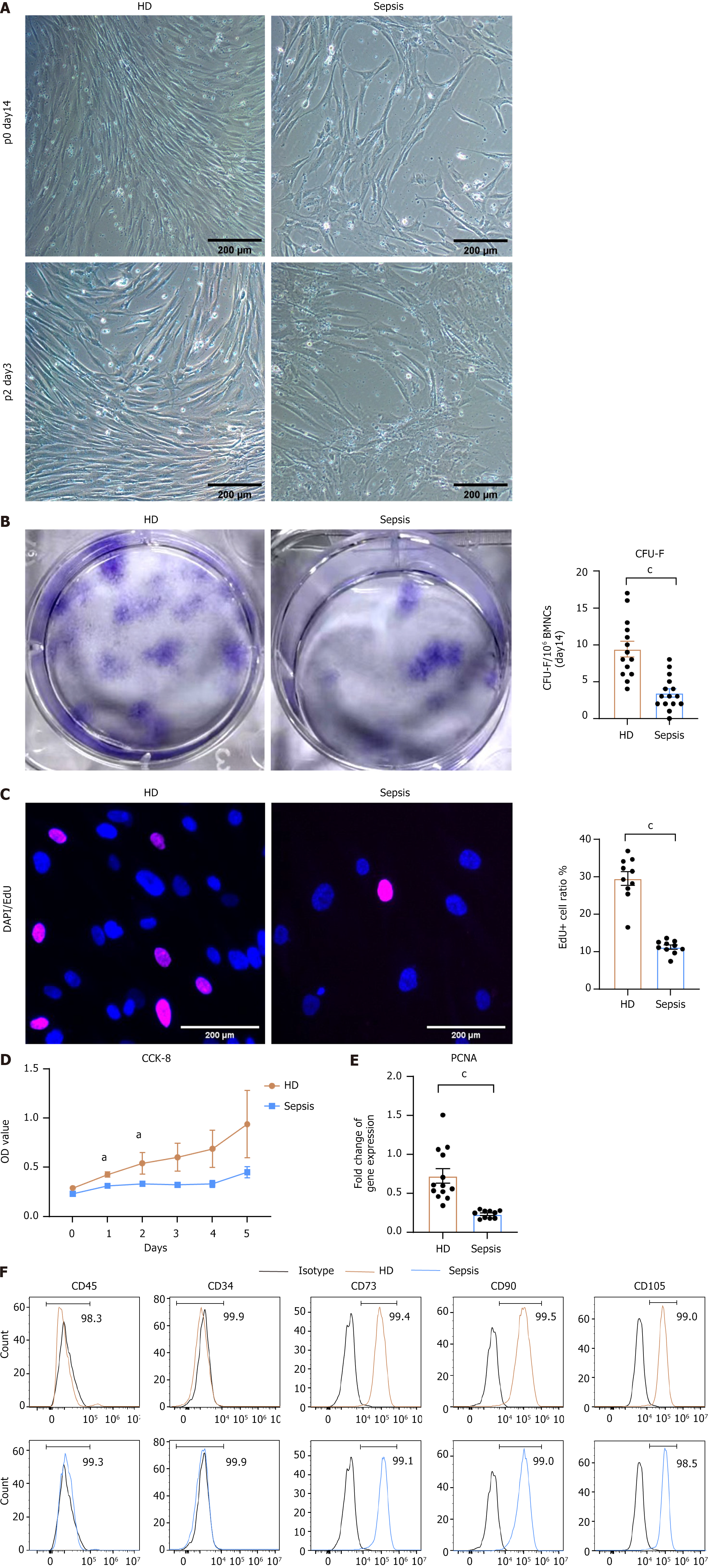
Figure 1 Morphology, clonogenic capacity, proliferation and characterization of bone marrow mesenchymal stem cells.
A: Representative images depicting the morphology of in vitro-expanded bone marrow mesenchymal stem cells (BMSCs) from healthy donors (HD) and patients with sepsis of various generations; B: Representative images of colony-forming units-fibroblast assays, with results expressed as the number of colony-forming units-fibroblast per one million bone marrow mononuclear cells for HD samples (n = 14) and septic samples (n = 15); C-E: Proliferation of HD and septic BMSCs, C: Merged channel image showing EdU (magenta) and DAPI (blue) staining and the percentage of EdU-positive cells relative to the total number of cells in HD BMSCs (n = 10) and septic BMSCs (n = 10) (C). Growth curves were assessed using the CCK-8 assay for HD BMSCs (n = 10) and septic BMSCs (n = 10) (D). Expression levels of proliferating cell nuclear antigen detected by quantitative real-time polymerase chain reaction (HD: n = 13; sepsis: n = 10) (E); F: Representative flow cytometry images illustrating typical BMSC markers. Positive markers: CD73, CD90, CD105; negative markers: CD34 and CD45. Each error bar represents the mean ± SEM. P values were determined using a t-test (aP < 0.05, cP < 0.001). HD: Healthy donors; CFU-F: Colony-forming units-fibroblast; PCNA: Proliferating cell nuclear antigen.
Figure 2 Osteogenic and adipogenic differentiation of bone marrow mesenchymal stem cells.
A: Alizarin Red S staining (original magnification, × 4); B: Oil Red O staining (original magnification, × 4); C: Quantification was carried out using cetylpyridinium chloride, and absorbance was measured using a microplate reader at 562 nm; D: Quantification was conducted using isopropanol, and the absorbance was measured using a microplate reader at 500 nm; E: Expression of related genes (runt-related transcription factor 2, alkaline phosphatase, and Sp7 transcription factor 7); F: Expression of related genes (peroxisome proliferator-activated receptor gamma, fatty acid binding protein 4, and perilipin-1). HD: n = 8; sepsis: n = 10. The expression of all genes was assessed using quantitative real-time polymerase chain reaction. Results were expressed as fold change relative to undifferentiated bone marrow mesenchymal stem cells. Each error bar shows the mean ± SEM. P values were determined using a t-test (aP < 0.05, cP < 0.001). HD: Healthy donors; RUNX2: Runt-related transcription factor 2; ALP: Alkaline phosphatase; SP7: Sp7 transcription factor 7; PPARG: Peroxisome proliferator-activated receptor gamma; FABP4: Fatty acid binding protein 4; PLN1: Perilipin-1.
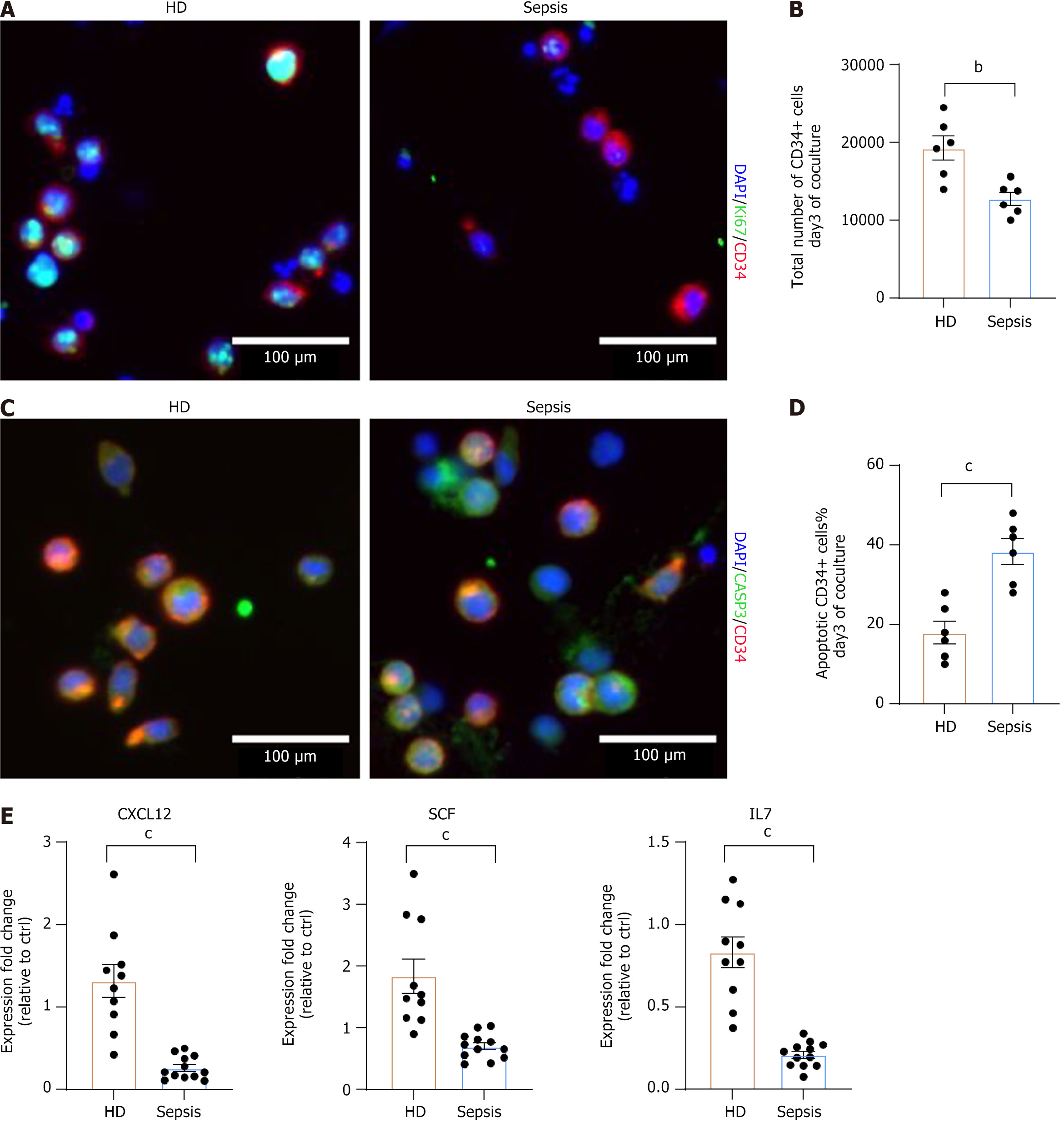
Figure 3 Supportive capacity of bone marrow mesenchymal stem cells.
A: Representative fluorescence image showing proliferating hematopoietic stem/progenitor cells (HSPCs) co-cultured with healthy donors (HD) and septic bone marrow mesenchymal stem cells (BMSCs) after 3 days in vitro. Ki67 (green), CD34 (red), DAPI (blue); B: Total number of CD34-positive HSPCs after co-culture with HD BMSCs (n = 6) or septic BMSCs (n = 6), starting with an initial number of 10000 cells; C: Representative fluorescence image of apoptotic HSPCs after co-culture; D: Percentages of apoptotic HSPCs (CD34+ and caspase-positive) relative to the total number of cells (HD: n = 6; sepsis: n = 6); E: Expression levels of supportive cytokines (C-X-C chemokine ligand 12, stem cell factor, and interleukin 7) were evaluated by fold change relative to one of the HD samples (HD: n = 10; sepsis: n = 12). Data were mean ± SEM. P values were determined using a t-test (bP < 0.01, cP < 0.001). HD: Healthy donors; CXCL12: C-X-C chemokine ligand 12; SCF: Stem cell factor; IL7: Interleukin 7.
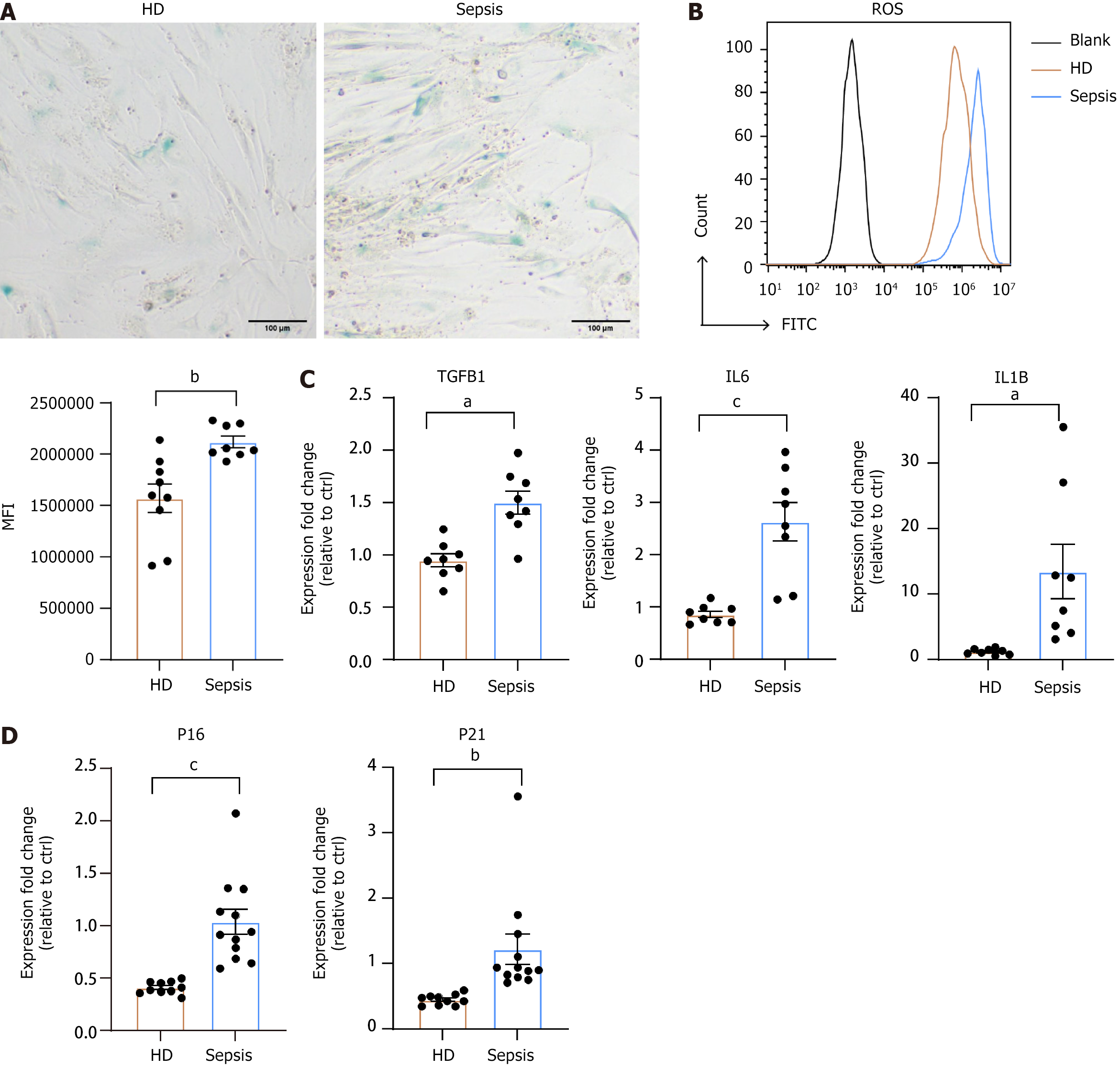
Figure 4 Senescence behavior in healthy donors and septic bone marrow mesenchymal stem cells.
A: Representative β-gal staining images of healthy donors (HD) and septic bone marrow mesenchymal stem cells (BMSCs); B: In vitro reactive oxygen species levels assessed by flow cytometry, expressed as mean fluorescence intensity of forward scatter for HD BMSCs (n = 9) and septic BMSCs (n = 8); C: Relative senescence-associated secretory phenotype content (transforming growth factor β1, interleukin 6, and interleukin 1β) compared to HD samples in both groups (n = 8); D: Detection of senescence markers (P16, P21) in HD BMSCs (n = 10) and septic BMSCs (n = 12). Data were presented as mean ± SEM. P values were determined by t-test (aP < 0.05, bP < 0.01, cP < 0.005). HD: Healthy donors; ROS: Reactive oxygen species; MFI: Mean fluorescence intensity; TGF: Transforming growth factor; IL: Interleukin.
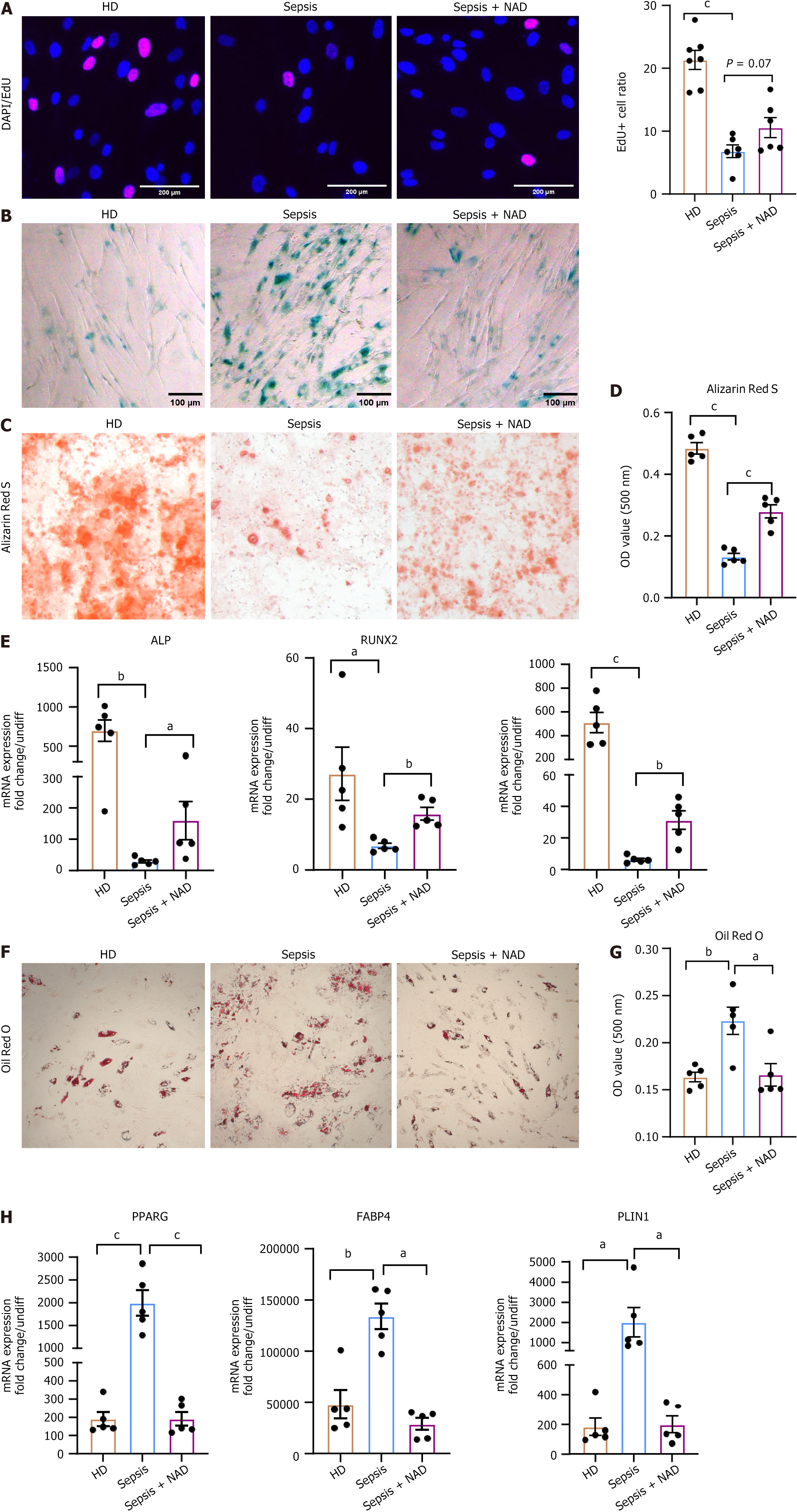
Figure 5 Nicotinamide adenine dinucleotide treatment rescue function of septic bone marrow mesenchymal stem cells.
A: Representative images of EdU incorporation, EdU (magenta), and DAPI (blue) staining. The percentage of EdU-positive cells increased after treatment with nicotinamide adenine dinucleotide (NAD) [healthy donors (HD): n = 7; sepsis: n = 6; sepsis with NAD: n = 6]; B: Cellular senescence assessed by β-gal staining in bone marrow mesenchymal stem cells (BMSCs) from three groups; C and D: Alizarin Red S staining (original magnification, × 4) and quantification of calcium precipitation; E: Quantitative real-time polymerase chain reaction analysis of osteogenic genes (runt-related transcription factor 2, alkaline phosphatase, and Sp7 transcription factor 7) in HD BMSCs (n = 5), septic BMSCs (n = 5), and NAD-treated septic BMSCs (n = 5); F and G: Oil Red O staining (original magnification, × 10) and lipid droplet quantification; H: Quantitative real-time polymerase chain reaction analysis of adipogenic genes (peroxisome proliferator-activated receptor gamma, fatty acid binding protein 4, and perilipin-1) in HD BMSCs (n = 5), septic BMSCs (n = 5), and NAD-treated septic BMSCs (n = 5). Results were presented as fold change relative to undifferentiated BMSCs. Error bars represent the mean ± SEM. P values were determined using One-Way ANOVA (aP < 0.05, bP < 0.01, cP < 0.001). HD: Healthy donors; NAD: Nicotinamide adenine dinucleotide; RUNX2: Runt-related transcription factor 2; ALP: Alkaline phosphatase; SP7: Sp7 transcription factor 7; PPARG: Peroxisome proliferator-activated receptor gamma; FABP4: Fatty acid binding protein 4; PLN1: Perilipin-1.
Figure 6 Nicotinamide adenine dinucleotide treatment improves the antioxidant capacity of mitochondria via the nicotinamide adenine dinucleotide/sirtuin 3/superoxide dismutase pathway.
A: Expression level of sirtuin 3 in healthy donors, septic and nicotinamide adenine dinucleotide (NAD)-treated septic bone marrow mesenchymal stem cells (BMSCs) (n = 6); B: In vitro reactive oxygen species levels were expressed as mean fluorescence intensity of forward scatter for healthy donor-, septic-, and NAD-treated septic BMSCs (n = 5); C: Superoxide dismutase activity of every 20 μg of protein from different BMSCs (n = 5); D: Measure mitochondrial membrane potential with TMRE staining to show mitochondrial integrity. TMRE staining (red) and Hoechst 33342 staining (blue). The mean intensity of TMRE staining was calculated with Image J; E: Graph of NAD/sirtuin 3/superoxide dismutase pathway. Error bars represent the mean ± SEM. P values were determined using One-Way ANOVA (aP < 0.05, bP < 0.01, cP < 0.001). HD: Healthy donors; NAD: Nicotinamide adenine dinucleotide; MFI: Mean fluorescence intensity; SIRT3: Sirtuin 3; ROS: Reactive oxygen species; SOD: Superoxide dismutase.
- Citation: Xia X, Zhou K, An LY, Zhao M, Tian BL, Zhao JY, Zhou ZG, Tong Y. Nicotinamide adenine dinucleotide rejuvenates septic bone marrow mesenchymal stem cells. World J Stem Cells 2025; 17(2): 96893
- URL: https://www.wjgnet.com/1948-0210/full/v17/i2/96893.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v17.i2.96893