Published online Jul 15, 2003. doi: 10.3748/wjg.v9.i7.1431
Revised: March 4, 2003
Accepted: March 28, 2003
Published online: July 15, 2003
AIM: To investigate the expression of NGF family and their receptors in gastric carcinoma and normal gastric mucosa, and to elucidate their effects on gastric carcinoma.
METHODS: RNA of gastric cancer tissues and normal gastric tissues was respectively isolated and mRNA was purified. Probes of both mRNA reverse transcription product cDNAs labled with α-33P dATP were respectively hybridized with Atlas Array membrane where NGF and their family genes were spotted on. Hybridized signal images were scanned on phosphor screen with ImageQuant 5.1 software after hybridization. Normalized values on spots were analyzed with ArrayVersion 5.0 software. Differential expression of NGF family and their receptors mRNA was confirmed between hybridized Atlas Array membranes of gastric cancer tissues and normal gastric mucosa, then their effects on gastric carcinoma were investigated.
RESULTS: Hybridization signal images on Atlas Array membrane appeared in a lower level of nonspecific hybridization. Both of NGF family and their receptors Trk family mRNA were expressed in gastric cancer and normal gastric mucosa. But adversely up-regulated expression in other tissues and organs. NGF, BDGF, NT-3, NT-4/5, NT-6 and TrkA, B and C were down-regulated simultaneously in gastric carcinoma in comparison with normal gastric mucosa. Degrees of down-regulation in NGF family were greater than those in their receptors Trk family. Down-regulation of NT-3 and BDGF was the most significant, and TrkC down-regulation level was the lowest in receptors Trk family.
CONCLUSION: Down-regulated expression of NGF family and their receptors Trk family mRNA in gastric cancer is confirmed. NGF family and their receptors Trk family probably play a unique role in gastric cancer cell apoptosis by a novel Ras or Raf signal transduction pathway. Their synchronous effects are closely associated with occurrence and development of gastric carcinoma induced by reduction of signal transduction of programmed cell death.
- Citation: Du JJ, Dou KF, Peng SY, Qian BZ, Xiao HS, Liu F, Wang WZ, Guan WX, Gao ZQ, Liu YB, Han ZG. Expression of NGF family and their receptors in gastric carcinoma: A cDNA microarray study. World J Gastroenterol 2003; 9(7): 1431-1434
- URL: https://www.wjgnet.com/1007-9327/full/v9/i7/1431.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i7.1431
Recently, NGF family and their receptors family have been found in other non-neural tissues of the body, and more and more attentions are being paid to their effects on these tissues, especially on tumor tissues. Gene expression of NGF and Trk families in gastric tissue and gastric carcinoma has been seldom documented. Simultaneous detection of NGF family and their receptors family expression has become possible since cDNA microarray was developed[1]. As abnormal expression of many genes is involved in gastric carcinogenesis[2-11], it contributes to a better understanding of their roles in the occurrence and development of gastric cancer and helps reveal mRNA expression of the family genes in gastric tissue and gastric carcinoma.
RNA of gastric cancer tissues and normal gastric mucosa was respectively isolated with Trizol (Gibco) in five cases of gastric cancer from Xijing Hospital, Fourth Military Medical University. To ensure good total RNA quality 28S/18S ≥ 1.5, samples were immediately placed into liquid nitrogen after being removed intraoperatively, and trituration of the samples was performed in liquid nitrogen. Then, mRNA was purified in Oligotex mRNA Kit (Qiagen). An equal mRNA mixture of gastric cancer tissues and normal gastric mucosa from five patients respectively constituted gastric cancer group and normal gastric mucosa group. At last, reverse transcription product (the first stranded cDNAs) of mRNA mixture was electrophoresed to evaluate its size and quality.
One μg of mRNA mixture of gastric cancer tissue and normal gastric mucosa from five patients was respectively transcripted into cDNAs as a probe labeled with 3.5 μL α-33P dATP (> 2, 500 kCi.mol-1, 10 Ci•l-1, Dupont) and 1 μL CDS primer 1 (Clontech) in a mixture containing 1 μL MMLV, 1 μL 10 × dNTP Mix (for dATP label), 0.5 μL DTT (100 mM), and 2 μL 5 × reaction buffer in a final volume of 10 μL was incubated for 0.25 h at 50 °C using an unregulated heat block (Eppendorf). Labeled reaction was stopped by adding 1 μL 10 × Termination Mix. To purify the labeled cDNA from unincorporated 33P-labeled nucleotides and small (< 0.1 kb) cDNA fragments, the above probe synthetic reactions were diluted to 200 μL total volume with Buffer NT2 included in Atlas cDNA Expression Arrays (Clontech), then transferred to a NucleoSpin Extraction Spin Column. 400 μL Buffer NT3 was added into the column after centrifugation at 14000 rpm for 1 min and the flowthrough was discarded. The procedure was repeated twice. To elute the labeled probe, 100NE was added into the column, centrifuged at 14000 rpm for 1 min. The flowthrough was obtained as the labeled probe.
Prehybridization of Atlas Array membrane was carried out in 0.5 mg heat-denatured sheared salmon DNA, and 5 mL prewarmed ExpressHyb solution (Clontech) in a hybridization bottle was incubated for 0.5 h at 68 °C. Then, heat-denatured cDNA probes were added into the above prehybridization bottle together with 5 μL heat-denatured C0t-1 DNA. The hybridization reactions were performed at 68 °C overnight. The next day, the Atlas Array membrane was washed three times in prewarmed wash solution 1 (2 × SSC, 1% SDS) with continuous agitation at 68 °C for 0.5 h, and in prewarmed wash solution 2 (0.1 × SSC, 0.5% SDS) at 68 °C for 0.5 h. The damp Atlas Array membrane was wrapped in a plastic wrap after the last washing in 2 × SSC at room temperature for 5 min.
Atlas Array hybridization membrane was exposed to phosphor screen at room temperature overnight. The hybridization signals were analyzed with ArrayVersion 5.0 software (MD) after the phosphor screen was scanned with ImageQuant 5.1 software (MD). Quantitative data of each hybridization signals were obtained.
Good total RNA quality was confirmed by 28S/18S ≥ 1.5. Size range of reverse transcription product cDNAs represented a smear from 0.2-4 kb both in gastric cancer and normal gastric mucosa (Figure 1).
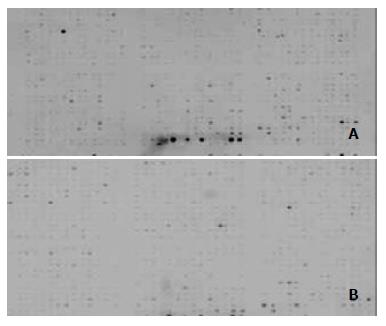
Hybridization signal images on Atlas Array membrane appeared in lower levels of nonspecific hybridization (Figure 2).
To quantify hybridization signals, signal intensity was detected after hybridized signal normalization of two hybridization Atlas Array membranes between gastric cancer and the normal mucosa. Signal intensity of NGF family and their receptors Trk family on Atlas Array membranes represented their mRNA expression level. NGF, BDGF, NT-3, NT-4/5, NT-6 and TrkA, B and C were down-regulated in gastric carcinoma in comparision with normal gastric mucosa. Degrees of down-regulation in NGF family were greater than those in their receptors Trk family. Down-regulation of NT-3 and BDGF was the most significant, and TrkC down-regulation level was the lowest in receptors Trk family (Table 1).
| Dot | Gene | CAVOL | NORnVOL | NORnVOL/CAnVOL |
| D07n | NT-3, BDGF | 0.031 | 0.311 | 10.128 |
| D11n | NGF | 0.045 | 0.272 | 6.036 |
| D08n | NT-4/5, NT-6 | 0.06 | 0.352 | 5.822 |
| D02i | TrkC | 0.029 | 0.119 | 4.172 |
| D03k | Trk | 0.038 | 0.157 | 4.093 |
| D14h | TrkA | 0.043 | 0.141 | 3.252 |
| D01i | TrkB | 0.026 | 0.051 | 1.926 |
Gene microarray has been rapidly and extensively used in detecting expression of genes, DNA sequence, novel genes and gene mutants, DNA polymorphism, and in screening drugs, diagnosing diseases and mapping gene library since Schena reported it in 1995[12-28]. Profiling of differentially expressed genes in human gastric carcinoma by cDNA expression array was also reported[29]. The study detected expression of NGF family and their receptors Trk family mRNA by using cDNA microarray. The Atlas Array membranes were provided by Clontech. A set of housekeeping genes was included on the Atlas Array membranes to normalize mRNA expression levels. Our good total RNA and mRNA quality, as well as successful synthesis and labeling of a cDNA probe with highly specific activity ensured the best possible results. ExpressHybTM hybridization solution was used in our hybridization experiments, a low-viscosity hybridization solution that significantly enhances the sensitivity of detection and reduces background.
NGF is composed of three subunit proteins (α, β and γ) among which β subunit represents an active form. NGF produced by targets of sympathetic neuron, sensory central neuron exerts an important effect on growth, development, differentiation of these neurons. Brain-derived nerve growth factor (BDNF), NT-3, NT-4/5 and NT-6 are members of the NGF family, and NGF has 50% of homology with BDNF. Difference between members of the family comes from the distribution of tissues, the early and/or later expression, and different receptors. NGF family receptors are subdivided into three types: Trk A, TrkB and TrkC. The structure of the three receptors consists of cellular external region, transcellular membrane region and cellular internal region. The receptors all are tyrosine kinase, and there is 66%-68% of homology between them. NGF binds TrkA, and BDNF, NT-3, NT-4/5 and NT-6 bind TrkB, in which binding of NT-3 is weaker and mainly with TrkC. As the functions of NGF family, TrkA, TrkB and TrkC can regulate growth, development and differentiation of corresponding neurons while receiving signals of NGF, BDNF, NT-3, NT-4/5 and NT-6. NT-3 and its receptor TrkC play a role in early growth, development and differentiation of neural systems.
Recently NGF family and their receptors family were found in other non-neural tissues of the body, and more and more attentions are being paid to their effects. It was reported that dermal pigment cells expressed NGF and Trk[30], hepatic cells expressed BDNF, NT-3, NT-4/5 as well as TrkA, which were related to liver pathophysiology. Hepatic stellate cells expressed BDNF, NT-3, NT-4/5, TrkB and TrkC that were involved in liver remodeling[31]. NT-3, NT-4/5, TrkB and TrkC expressed by microphages played a role in tissue inflammatory reaction and repair[32]. Cardiac myocytes expressed TrkC and NT-3, and early growth and development of the heart were retarded when blockage of TrkC was used[33]. Schneider et al[34] demonstrated TrkA and TrkC expression in pancreatic ducts and pancreatic islets, TrkB in apha-cells of islets, NGF in pancreatic ducts and pancreatic acinar cells, NT-3 and NT-4 respectively in capillary endothelia and ductule cells. Additionally, TrkB and TrkC were found in endocrine cells of gut epithelium and neural tissues in fish[35,36], and TrkA, TrkB and TrkC were all expressed in testis of rats. A current study also indicated NGF family and their receptors played an essential role in the development of tubulogenesis in embryonic kidney, spermatogenesis, hair follicle, heart and vascular differentiation and maintenance of blood and immune cells[37].
Similarly, more attentions are being paid to their effects on tumors. Antagonists of NGF family and their receptors are being applied to kill tumor cells experimentally. It was noted that NGF family and their receptors were not expressed in some normal tissues, but expressed in their corresponding tumors. For example, up-regulated expression of Trk was present in tumors originating from thyroid and ovary while absent in the normal tissues. When chromosomal translocation occurred, a fusion protein of Trk-T1 composed of carboxyl terminal tyrosine kinase domain of NTRK1 and amino terminal portion of TPR (translocated promoter region) was formed. Trk-T1 was oncogenic in vivo and contributed to the papillary neoplastic transformation of the thyroid[38]. These results suggest that NGF family and their receptors are involved in tumorigenicity. A recent study showed that pancreatic carcinoma cells could express NGF, TrkA and TrkC[34]. It is interesting that Trk was highly expressed in esophageal carcinoma, thyroid carcinoma and prostate carcinoma, and adversely Trk expression revealed significantly lower in gastric carcinoma and colon carcinoma[39,40]. Difference between NGF and Trk expression in various tumors suggests that NGF family and their receptors may play a different role or have directly reverse effects on various carcinoma originating from different tissues. Some studies verified the assumption that ocurrence of apoptosis could be inhibited and/or enhanced while cascade effect was brought out by intracellular signal transduction pathway blocking or inducing programmed cell death[41,42]. Its signal transduction pathways include Ras, the Cdc42/Rac/RhoG protein family, MAPK, PI3K and PLC-gamma. Adversely, other novel Ras and/or Raf pathways participate in signal transduction paths mediating programmed cell death. For example, prostate growth depends on autocrine NGF interaction with Trk expressed by itself, otherwise, apoptosis occurred in medulloblastoma when NGF was bound to Trk[43]. Therefore, inhibition or enhancement of apoptosis induced by Trk depended on cellular types and development periods of cells[44].
The study showed that expression of NGF family and their receptors were simultaneously down-regulated in gastric cancer tissues. NT-3 and BDGF down-regulation was the lowest in NGF family, Trk C down-regulation was the lowest in Trk family. The evidences support that NGF family and their receptors Trk family may play a unique apoptotic role in gastric cancer by a new Ras or Raf signal transduction pathway. These further substantiate that NGF family and Trk family have synchronous effects on the occurrence and development of gastric carcinoma induced by reduction in signal transduction of programmed cell death brought out by simultaneously down-regulated expression of NGF family and Trk family. It remains unclear that which cells in gastric mucosa secret NGF family, and whether NGF plays a role in the occurrence and development of gastric carcinoma in autocrine or paracrine way.
Edited by Zhang JZ and Wang XL
| 1. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5103] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 2. | Liu HF, Liu WW, Fang DC, Men RP. Expression and significance of proapoptotic gene Bax in gastric carcinoma. World J Gastroenterol. 1999;5:15-17. [PubMed] |
| 3. | To KF, Leung WK, Lee TL, Yu J, Tong JH, Chan MW, Ng EK, Chung SC, Sung JJ. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer. 2002;102:623-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Gao HJ, Yu LZ, Bai JF, Peng YS, Sun G, Zhao HL, Miu K, L XZ, Zhang XY, Zhao ZQ. Multiple genetic alterations and behavior of cellular biology in gastric cancer and other gastric mucosal lesions: H. pylori infection, histological types and staging. World J Gastroenterol. 2000;6:848-854. [PubMed] |
| 5. | Liu DH, Zhang XY, Fan DM, Huang YX, Zhang JS, Huang WQ, Zhang YQ, Huang QS, Ma WY, Chai YB. Expression of vascular endothelial growth factor and its role in oncogenesis of human gastric carcinoma. World J Gastroenterol. 2001;7:500-505. [PubMed] |
| 6. | He XS, Su Q, Chen ZC, He XT, Long ZF, Ling H, Zhang LR. Expression, deletion [was deleton] and mutation of p16 gene in human gastric cancer. World J Gastroenterol. 2001;7:515-521. [PubMed] |
| 7. | Kaneda A, Kaminishi M, Yanagihara K, Sugimura T, Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;62:6645-6650. [PubMed] |
| 8. | Ficorella C, Cannita K, Ricevuto E, Toniato E, Fusco C, Sinopoli NT, De Galitiis F, Di Rocco ZC, Porzio G, Frati L. P16 hypermethylation contributes to the characterization of gene inactivation profiles in primary gastric cancer. Oncol Rep. 2003;10:169-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 9. | Li HL, Chen DD, Li XH, Zhang HW, Lu YQ, Ye CL, Ren XD. Changes of NF-κB, p53, Bcl-2 and caspase in apoptosis induced by JTE-522 in human gastric adenocarcinoma cell line AGS cells: role of reactive oxygen species. World J Gastroenterol. 2002;8:431-435. [PubMed] |
| 10. | Xia L, Yuan YZ, Xu CD, Zhang YP, Qiao MM, Xu JX. Effects of epidermal growth factor on the growth of human gastric cancer cell and the implanted tumor of nude mice. World J Gastroenterol. 2002;8:455-458. [PubMed] |
| 11. | Fang DC, Luo YH, Yang SM, Li XA, Ling XL, Fang L. Mutation analysis of APC gene in gastric cancer with microsatellite instability. World J Gastroenterol. 2002;8:787-791. [PubMed] |
| 12. | Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 714] [Cited by in RCA: 593] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 13. | Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1643] [Cited by in RCA: 1327] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 14. | Shannon MF, Rao S. Transcription. Of chips and ChIPs. Science. 2002;296:666-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21:10-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1077] [Cited by in RCA: 887] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 16. | Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Boguski MS. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1467] [Cited by in RCA: 1313] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 17. | Favis R, Barany F. Mutation detection in K-ras, BRCA1, BRCA2, and p53 using PCR/LDR and a universal DNA microarray. Ann N Y Acad Sci. 2000;906:39-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Wang K, Gan L, Jeffery E, Gayle M, Gown AM, Skelly M, Nelson PS, Ng WV, Schummer M, Hood L. Monitoring gene expression profile changes in ovarian carcinomas using cDNA microarray. Gene. 1999;229:101-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 193] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 19. | Xu S, Mou H, Lü G, Zhu C, Yang Z, Gao Y, Lou H, Liu X, Cheng Y, Yang W. Gene expression profile differences in high and low metastatic human ovarian cancer cell lines by gene chip. Chin Med J (. Engl). 2002;115:36-41. [PubMed] |
| 20. | Wellmann A, Thieblemont C, Pittaluga S, Sakai A, Jaffe ES, Siebert P, Raffeld M. Detection of differentially expressed genes in lymphomas using cDNA arrays: identification of clusterin as a new diagnostic marker for anaplastic large-cell lymphomas. Blood. 2000;96:398-404. [PubMed] |
| 21. | Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7864] [Cited by in RCA: 5568] [Article Influence: 214.2] [Reference Citation Analysis (0)] |
| 22. | Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7015] [Cited by in RCA: 6307] [Article Influence: 252.3] [Reference Citation Analysis (0)] |
| 23. | Assersohn L, Gangi L, Zhao Y, Dowsett M, Simon R, Powles TJ, Liu ET. The feasibility of using fine needle aspiration from primary breast cancers for cDNA microarray analyses. Clin Cancer Res. 2002;8:794-801. [PubMed] |
| 24. | Hippo Y, Yashiro M, Ishii M, Taniguchi H, Tsutsumi S, Hirakawa K, Kodama T, Aburatani H. Differential gene expression profiles of scirrhous gastric cancer cells with high metastatic potential to peritoneum or lymph nodes. Cancer Res. 2001;61:889-895. [PubMed] |
| 25. | Tiwari G, Sakaue H, Pollack JR, Roth RA. Gene expression profiling in prostate cancer cells with Akt activation reveals Fra-1 as an Akt-inducible gene. Mol Cancer Res. 2003;1:475-484. [PubMed] |
| 26. | Mycko MP, Papoian R, Boschert U, Raine CS, Selmaj KW. cDNA microarray analysis in multiple sclerosis lesions: detection of genes associated with disease activity. Brain. 2003;126:1048-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 27. | Man K, Lo CM, Lee TK, Li XL, Ng IO, Fan ST. Intragraft gene expression profiles by cDNA microarray in small-for-size liver grafts. Liver Transpl. 2003;9:425-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 28. | Kawaguchi K, Kaneko S, Honda M, Kawai HF, Shirota Y, Kobayashi K. Detection of hepatitis B virus DNA in sera from patients with chronic hepatitis B virus infection by DNA microarray method. J Clin Microbiol. 2003;41:1701-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 29. | Liu LX, Liu ZH, Jiang HC, Qu X, Zhang WH, Wu LF, Zhu AL, Wang XQ, Wu M. Profiling of differentially expressed genes in human gastric carcinoma by cDNA expression array. World J Gastroenterol. 2002;8:580-585. [PubMed] |
| 30. | Innominato PF, Libbrecht L, van den Oord JJ. Expression of neurotrophins and their receptors in pigment cell lesions of the skin. J Pathol. 2001;194:95-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Cassiman D, Denef C, Desmet VJ, Roskams T. Human and rat hepatic stellate cells express neurotrophins and neurotrophin receptors. Hepatology. 2001;33:148-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 173] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 32. | Barouch R, Appel E, Kazimirsky G, Brodie C. Macrophages express neurotrophins and neurotrophin receptors. Regulation of nitric oxide production by NT-3. J Neuroimmunol. 2001;112:72-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Lin MI, Das I, Schwartz GM, Tsoulfas P, Mikawa T, Hempstead BL. Trk C receptor signaling regulates cardiac myocyte proliferation during early heart development in vivo. Dev Biol. 2000;226:180-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 34. | Schneider MB, Standop J, Ulrich A, Wittel U, Friess H, Andrén-Sandberg A, Pour PM. Expression of nerve growth factors in pancreatic neural tissue and pancreatic cancer. J Histochem Cytochem. 2001;49:1205-1210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Radaelli G, Domeneghini C, Arrighi S, Castaldo L, Lucini C, Mascarello F. Neurotransmitters, neuromodulators, and neurotrophin receptors in the gut of pantex, a hybrid sparid fish (Pagrus major x Dentex dentex). Localizations in the enteric nervous and endocrine systems. Histol Histopathol. 2001;16:845-853. [PubMed] |
| 36. | De Girolamo P, Lucini C, Vega JA, Andreozzi G, Coppola L, Castaldo L. Co-localization of Trk neurotrophin receptors and regulatory peptides in the endocrine cells of the teleostean stomach. Anat Rec. 1999;256:219-226. [PubMed] [DOI] [Full Text] |
| 37. | Sariola H. The neurotrophic factors in non-neuronal tissues. Cell Mol Life Sci. 2001;58:1061-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 96] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 38. | Russell JP, Powell DJ, Cunnane M, Greco A, Portella G, Santoro M, Fusco A, Rothstein JL. The TRK-T1 fusion protein induces neoplastic transformation of thyroid epithelium. Oncogene. 2000;19:5729-5735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 82] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Koizumi H, Morita M, Mikami S, Shibayama E, Uchikoshi T. Immunohistochemical analysis of TrkA neurotrophin receptor expression in human non-neuronal carcinomas. Pathol Int. 1998;48:93-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | Weeraratna AT, Arnold JT, George DJ, DeMarzo A, Isaacs JT. Rational basis for Trk inhibition therapy for prostate cancer. Prostate. 2000;45:140-148. [PubMed] [DOI] [Full Text] |
| 41. | Patapoutian A, Reichardt LF. Trk receptors: mediators of neurotrophin action. Curr Opin Neurobiol. 2001;11:272-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 825] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 42. | Chou TT, Trojanowski JQ, Lee VM. A novel apoptotic pathway induced by nerve growth factor-mediated TrkA activation in medulloblastoma. J Biol Chem. 2000;275:565-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Muragaki Y, Chou TT, Kaplan DR, Trojanowski JQ, Lee VM. Nerve growth factor induces apoptosis in human medulloblastoma cell lines that express TrkA receptors. J Neurosci. 1997;17:530-542. [PubMed] |
| 44. | Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169:107-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 361] [Article Influence: 15.0] [Reference Citation Analysis (0)] |