Published online May 15, 2003. doi: 10.3748/wjg.v9.i5.1077
Revised: November 23, 2002
Accepted: December 16, 2002
Published online: May 15, 2003
AIM: Allo-cell transplant rejection and autoimmune responses were associated with the presence of class II major histocompatibility complex (MHC II) molecules on cells. This paper studied the effect of Ribonuclease P (RNase P) against CIITA, which was a major regulator of MHCII molecules, on repressing the expression of MHCII molecules on hepatocyte.
METHODS: M1-RNA is the catalytic RNA subunit of RNase P from Escherichia coli. It were constructed that M1-RNA with guide sequences (GS) recognizing the 452, 3408 site of CIITA by PCR from pTK117 plasmid, then were cloned into the EcoR I/Bgl II or EcoR I/Sal I site of vector psNAV (psNAV-M1-452-GS, psNAV-M1-3408-GS) respectively. The target mould plate (3176-3560) of CIITA was obtained from Raji cell by RT-PCR, and then inserted into the Xho I/EcoR I of pGEM-7zf(+) plasmid (pGEM-3176). These recombinant plasmids were screened out by sequence analysis. psNAV-M1-452-GS, psNAV-M1-3408-GS and its target RNA pGEM-3176 were transcribed and then mixed up and incubated in vitro. It showed that M1-3408-GS could exclusively cleave target RNA that formed a base pair with the GS. Stable transfectants of hepatocyte cell line with psNAV-M1-3408-GS were tested for expression of class II MHC through FCM, for mRNA abundance of MHCII, Ii and CIITA by RT-PCR, for the level of IL-2 mRNA on T cell by mixed lymphocyte reaction.
RESULTS: When induced with recombinant human interferon-gamma (IFN-γ), the expression of HLA-DR, -DP, -DQ on psNAV-M1-3408-GS+ hepatocyte was reduced 83.27%, 88.93%, 58.82% respectively, the mRNA contents of CIITA, HLA-DR, -DP, -DQ and Ii decreased significantly. While T cell expressed less IL-2 mRNA in the case of psNAV-M1-3408-GS+ hepatocyte.
CONCLUSION: The Ribonuclease P against CIITA-M1-3408-GS could effectively induce antigen-specific tolerance through cleaving CIITA. These results provided insight into the future application of M1-3408-GS as a new nucleic acid drug against allo-transplantation rejection and autoimmune diseases.
- Citation: Guo R, Zou P, Fan HH, Gao F, Shang QX, Cao YL, Lu HZ. Repression of allo-cell transplant rejection through CIITA ribonuclease P+ hepatocyte. World J Gastroenterol 2003; 9(5): 1077-1081
- URL: https://www.wjgnet.com/1007-9327/full/v9/i5/1077.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i5.1077
The donor shortage has become the major restriction on liver transplantation with the increasing demands for it[1]. Now people are trying to produce artificial liver with hepatocytes and various biological materials[2,3], but are faced with the challenge of rejection in allo-hepatocyte transplantation[4]. Allo-transplant rejection, was associated with the presence of classII major histocompatibility complex (MHCII) on the tissues and organs[5,6]. MHCII played a critical role in the induction of immune responses by presenting fragments of alloantigenic peptides to CD4+ T lymphocytes, then resulting in the activation of CD8+ T lymphocytes. So it was more important for compatibility of MHCII in allo-transplantation. Moreover, the abnormal expression of MHCII molecules was associated with autoimmune disease too[7-9]. There are codominance and multiple allele for MHCII molecules which lead to their complicated polymorphism, so it is difficult to repress every MHCII molecules expression directly. MHC class II transactivator (CIITA) was the major rate-limiting factor for both constitutive and inducible MHCII expression., and with rare exceptions, its expression parallels to that of MHCII transcripts[10-12]. There was no rejection in allo-skin graft[5] or prolonged survival time in allo-cardiac graft[13] of CIITA(-/-) according to the latest investigation.
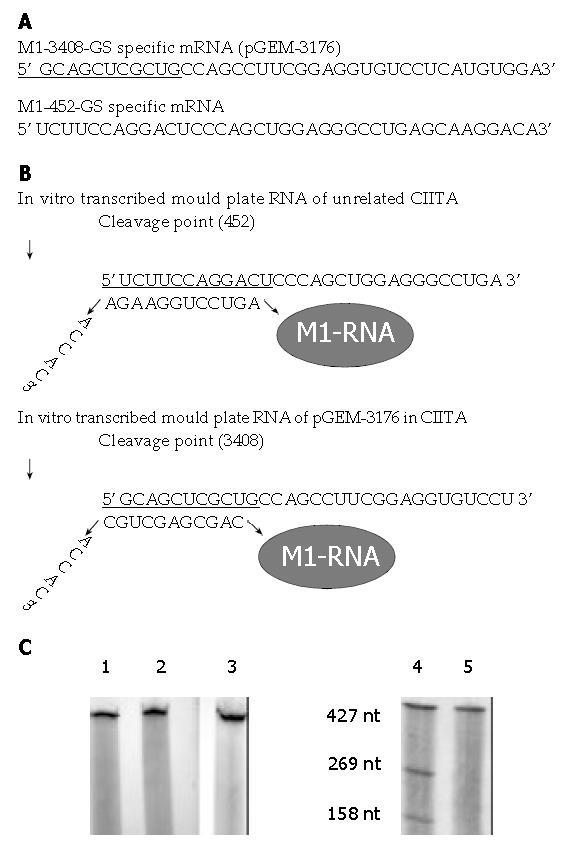
Ribozymes included hammerhead-, hairpin-, Ribonuclease P(RNase P), et al[14] M1-RNA was the catalytic RNA subunit of RNase P from Escherichia coli. RNase P was a ribonucleoprotein complex that catalyzed the hydrolysis reaction by removing a 5’ leader sequence from tRNA precursors. Hammerhead ribozymes required presence of specific nucleotide sequences in the target RNA to be cut[15,16], and these requirements could not always be fulfilled. M1-RNA could be used as a tool to cleave any specific mRNA sequence simply by the 3’terminal addition to the ribozyme sequence of a so-called guide sequence (GS) complementary to the target mRNA, that formed a base-pair with it and left a 5’-ACCAC-3’unpaired stretch needed for the M1-GS RNA to recognize and cleave this artificially created substrate (Figure 1B). Thus, M1-GS RNA, apart from some requirements to improve its cleavage efficiency, could be specifically directed to cut any mRNA sequences. This study represents, to our knowledge, the first gene therapy approach that makes use of the catalytic activity of M1-RNA for allo-cell transplant rejection in hepatic tissue engineering and the autoimmune diseases.
PTK117, a pUC19 derivative in which the DNA sequence coding for E. coli M1RNA is under the control of the T7 RNA polymerase promoter, was provided by Dr Chen BB[14]. pGEM-7zf(+) vector was purchased from Shanghai BioEngineering Company, adeno-associated virus vector (psNAV) was provided by Dr Lu HZ.
T4 DNA ligase and restriction endonucleases EcoR I, Sal I, Bgl II, Xho I were purchased from MBI; mouse anti-human isotype control (IgG2a)-FITC, HLA-DR (IgG2a, k), -DQ (IgG2a, k) monoclonal antibody and recombinant human IFN-γ were obtained from PharMingen, mouse anti-HLA-DP (IgG2b, BRA-FB6) from CYMBUS, GENETICIN (G418) from GIBCO. TRIZOLR from GIBCOBRL, TITANIUMR one-step RT-PCR kit from Clontech.
Cell line used included L-02 cell line (from human fetal liver) and Raji cell line (human B lymphoma), all purchased from Cell Bank of Shanghai Academy of Science. Cells were maintained in RPMI 1640 (GIBCOBRL) m ed iu m supplemented with 15% fetal calf serum (HyClone).
RNaseP construction The M1-RNA with the anti-CIITA-directed GS (M1-452-GS or M1-3408-GS) were constructed by the polymerase chain reaction (PCR) with the gene for M1-RNA as found in plasmid pTK117 as a template. The 5’ primer, OliT7: 5’-gcggaattcTAATACGACTCACTATAG-3’, annealing with the T7 promoter and providing a 5’EcoR I site for cloning. The 3’ primers contained the appropriate GS and were Olip3408: 5’-ACGCGTCGACGTGGTGCAGCTCGCTGATTACGCCAAGC-3’ and Olip452: 5’-GAAGATCT GTGGTTCTTCCAGGACTGCCAAGCTTGC-3’. The 3’ proximal sequences of 15 nucleotides in Olip3408 and 11 nucleotides in Olip452 served as primers for the PCR with the pTK117 sequence. The lowercase sequences, the underlined sequences, and the black ones corresponded, respectively, to the Sal I in Olip3408 or Bgl II in Olip452, the ACCAC-3’ sequence, and the GS. The two different PCR products were obtained according to the instructions of 2 × HiFi Master PCR kit (Shanghai Bioengineering Company), and cloned into psNAV (psNAV-M1-3408-GS, psNAV-M1-452-GS), then completely sequenced to exclude any mutation during the PCR reaction.
Construction of artificial substrate The CIITA (3176-3560) mould mRNA was obtained from Raji cell line by RT-PCR, according to instructions of TRIZOLR and TITANIUMR one-step RT-PCR kit. The 5’ primer: 5’-CCGCTCGAGAGCTGAAGTCCTTGGAA-3’, The 3’ primers: 5’-GCGGAATTCGAACATGCCTGTCCAGAGC-3’. The RT-PCR product, containing 384 nucleotides spanning at the point of 3408, was cloned into the Xho I/EcoR I site of pGEM-7zf(+) (pGM-3176).
Assays for cleavage by M1-GS RNA[16] Plasmid pGM-3176 was linearized with EcoR I, psNAV-M1-3408-GS and psNAV-M1-452-GS were linearized with Bgl II, then purified by EndoFreeR Plasmid Maxi kit(QIAGEN), and transcribed in vitro from the pGEM-7zf or M1-RNA T7 promoter in the presence of [32P]-GTP, with Ribomax large scale RNA production system-T7 (Promega), then transcribed ribozyme and substrate RNA were phenol chloroform extracted and precipitated with ethanol, and their integrity was checked by either acrylamide/urea or agarose electrophoresis. RNaseP RNA and uniformly labeled substrates were incubated for 4 h at 50 °C in 25 mM Tris HCl (pH7.5), 50 mM MgCl2, 50 mM NH4Cl buffer in a final reaction volume of 20 μl. Reactions were stopped by the addition of 6 μl of a solution containing 95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol FF boiled for 5 min and chilled on ice. The reaction products were analyzed on 10% polyacrylamide/8 mol/L urea gels (Figure 1C).
Hepatocytes were transfected with 0.4 μg psNAV-M1-3408-GS by nanometer vector. According to Effectene (QIAGEN) kit’s instructions, seeded 2.5 × 105 cells/well the day before transfection. The cell number seeded should produce 40%-80% confluence on the day of transfection. psNAV-M1-3408-GS were diluted in 100 μl EC buffer, mixed with Enhancer 3.2 μl, incubating 2-4 min at RT, then adding Effectene 10 μl, at RT 7-8 min, mixed with 600 μl medium containing serum and antibiotics, and immediately transferred the total volume to the above cells in the 6-well plate.
Hepatocytes were collected and washed with 1.5 g/L MPBS buffer (10 g/L BSA and 1 g/L NaN3) once at the density of 1 × 106/ml, adding IgG2a, HLA-DR, DP, DQ 10 μl respectively, incubating at 4 °C for 30 min, detecting the expression of MHCII molecules by Flow cytometry (COULTER, EPICSXL).
RT-PCR was done according to the instructions of TRIZOLR and TITANIUMR one-step RT-PCR kit. In a total 50 μl volume, 50 °C 1 h, 94 °C 5 min, 94 °C 30 s, 65 °C 30 s, 68 °C 1 min, 30 cycles, 72 °C extending 7 min. Primers sequences (synthesized by Shanghai Bioengineering Company) referred to Table 1.
| Primer | Sequence | Length |
| CIITA mould | L 5’-CCGCTCGAGAGCTGAAGTCCTTGGAA-3’ | 384 bp |
| R 5’-GCGGAATTCGAACATGCCTGTCCAGAGC-3’ | ||
| CIITA | L 5’-CCGCTCGAGGCTGCCTGGCTGGGATT-3’ | 410 bp |
| R 5’-GCGGAATTCCGATCACTTCATCTGGTCCTAT-3’ | ||
| M1-452-GS | L 5’-GCGGAATTCTAATACGACTCACTATAG-3’ | 446 bp |
| R 5’-GAAGATCTGTGGTTCTTCCAGGACTGCCAAGCTTGC-3’ | ||
| M1-3408-GS | L 5’-GCGGAATTCTAATACGACTCACTATAG-3’ | 454 bp |
| R 5’-ACGCGTCGACGTGGTGCAGCTCGCTGATTACGCCAAGC-3’ | ||
| HLA-DR | 5’-AATGGCCATAAGTGGAGTCC-3’ | 335 bp |
| 5’-GGAGGTACATTGGTGATCGG-3’ | ||
| HLA-DP | 5’-CAGAGCTGTGATCTTGAGAG-3’ | 197 bp |
| 5’-AGATGCCAGACGGTCTCCTT-3’ | ||
| HLA-DQ | 5’-CTCTGACCACCGTGATGAGC-3’ | 153 bp |
| 5’-CTCTCCAGGTCCACGTAGAA-3’ | ||
| Ii | 5’-CCAGATGCACAGGAGGAGAA-3’ | 714 bp |
| 5’-CCTCTGCTGCTCTCACATGG-3’ | ||
| Neo gene | L 5’-ACAATCGGCTGCTCTGAT-3’ | 349 bp |
| R 5’-CTCGCTCGATGCGATGTT-3’ | ||
| β-actin | L 5’-ATCATGTTTGAGACCTTCAA-3’ | 310 bp |
| R 5’-CATCTCTTGCTCGAAGTCCA-3’ |
It were incubated at 37 °C, 5% CO2 (keeping away light) after adding mitocin-C (25 μg/ml, Sigma) into IFN-γ induced psNAV-M1-3408-GS+ hepatocyte (1 × 107/ml), and washed with RPMI1640 twice, plated at a density of 1 × 106/well as stimulating cells. Then added peripheral blood mono-nucleated cells (PBMNC, 1 × 106) from healthy donors into above stimulating cells. The level of IL-2 mRNA from PBMNC after 48 h incubation was Detected through RT-PCR[17].
Hepatocyte without IFN-g induction The expression of HLA-DR, DP, DQ on hepatocyte was low, (0.14 ± 0.04)%, (26.76 ± 5.26)%, (2.12 ± 0.56)% respectively.
Hepatocyte after IFN-g induction The expression of HLA-DR, DP, DQ on hepatocyte with IFN-γ (40 ng/ml) induction for 3 d increased significantly, (18.68 ± 2.94)%, (41.78 ± 4.90)%, (4.78 ± 1.26)% respectively.
The expression of MHCIIon psNAV-M1-3408-GS+ hepatocyte after the induction of IFN-γ (40 ng/ml) for 3 d was repressed. Compared with void-vector+ hepatocyte, the expression of HLA-DR, DP, and DQ on psNAV-M1-3408-GS+ hepatocyte was inhibited 83.27%, 88.93%, and 58.82% respectively.
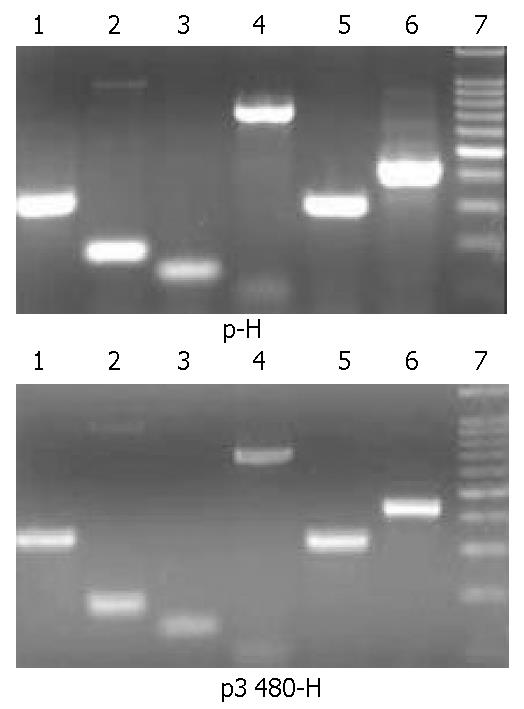
The induction of MHCII, Ii and CIITA mRNA on psNAV-M1-3408-GS+hepatocyte was detected through RT-PCR (Figure 2). Compared with void vector, the amount of MHCII and Ii mRNA of psNAV-M1-3408-GS+ hepatocyte was significantly down-modulated, the CIITA mRNA was down-modulated 71%.
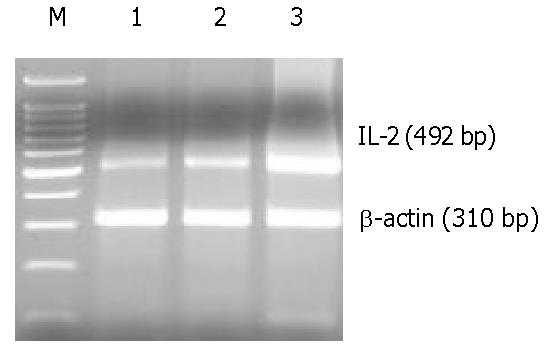
The excretion of IL-2 from PBMNC stimulated by psNAV-M1-3408-GS+ hepatocyte after the induction of IFN-γ for 3 d referred to Figure 3. Void-vector+ hepatocyte could induce PBMNC high amount of IL-2 mRNA, but negative control and psNAV-M1-3408-GS+ hepatocyte could hardly induce the excretion of IL-2 mRNA from PBMNC.
In allo-cell transplantation or autoimmune diseases, some cytokines such as IFN-γ, induced some low/non MHCII antigen expressing cells to express these molecules highly[18,19]. We selected IFN-γ to induce hepatocyte for 3 days, the expression of HLA-DR, DP, DQ antigens increased exactly, while the expression of HLA-DP was most apparent.
CIITA regulated the transcription of MHCII gene by interacting with the trans-acting factors such as RFX, X2BP and NFY. The expression of CIITA paralleled to that of MHCIImolecules and appeared only in the MHCII-positive cells[10,11]. In the hepatocyte detected by us, the expression of CIITA was consistent with that of MHCII molecules: without IFN-γ induction, all hepatocytes didn’t express MHCII molecules and CIITA gene; following IFN-γ induction, these cells expressed MHCII molecules and CIITA gene simultaneously; in the case of anti-CIITA psNAV-M1-3408-GS positive hepatocytes, the induced MHCII expression on their surface was nearly completely lost, and their CIITA mRNA detected by RT-PCR was also defect, perhaps the latter was the direct reason that MHCII expression didn’t react to IFN-γ induction. This view was coincidence with that of Luder et al[18,20]: TOXO plasma gondi parasite lowered the MHCII expression by inhibiting its induced CIITA expression. Moreover, HMG-CoA reductase inhibitors, cyclosporine and phosphatidylethanolamine-linked hyaluronic acid (HYPE) could completely repress MHCII expression of human microvascular endothelial cells by reducing its induced CIITA mRNA contents ex vitro[19,21-23].
Gene blocking techniques were mainly made up of anti-sense oligonucleatide, anti-sense RNA, ribozyme and RNA interference (RNAi), and so on. Anti-sense oligonucleatide referred to a small fragment of single-strand deoxyribonucleic acid (14-23 bases) synthesized artificially, which could hybridize with target DNA or mRNA. However, there were still some problems of stability and efficiency of entering cell in vivo about it. Anti-sense RNA, ribozyme and RNAi all took action on target mRNA, namely anti-sense complementation, cutting and interference respectively. The novel RNAi technique was the double-strand RNA connected by anti-sense RNA and sense RNA in essence, and was more efficient than single anti-sense RNA[24,25]. The mechanism of which was still not clear and might be related to activating ribonucleasep to degrade target mRNA. But when it is larger than 30 bp, the action of it was not specific[24]. Compared with above-mentioned gene blocking techniques, ribozyme not only sealed mRNA, but also cut mRNA with specificity. Moreover, ribozyme could be used repeatedly, so it had higher efficiency. Both hammerhead ribozyme and hairpin ribozyme require GUC sequence to identify in target sequence. However, RnaseP was not limited to this and could aim at any site in the target sequence, so it had wider selective range[14]. There was no report on RnaseP yet at home. According to human RNA sequences published in the NCBI Gene Bank, our experiment selected 452, 3408 site in the CIITA gene as target sites of M1-RNA after eliminating the possibility of their homology. PST and LRR regions initiated by 452, 3408 site were very essential to the transcription activation of CIITA. Moreover, the secondary structure around them was relatively simple and accessible. The GS of M1-3408-GS and M1-452-GS were programmed as 11 and 12 nucleotides respectively, to fit for the combination with their own substrate, the disconnection of cutting products with the ribozyme, and the specificity of the ribozyme. The 5’-terminal of M1-RNA had a TAATA box (T7 promoter), and M1-RNA was cloned into the psNAV vector (without T7 promoter), then the transcription was gone on owing to T7 promoter in the ribozyme itself. This could avoid supplementary sequence of the psNAV vector and objectively reflect the cutting activity of the Rnase P. The result of our experiment revealed the expected cutting stripes in the electrophoresis of cutting products of M1-3408-GS and CIITA mould plate ex vitro.
The reason why our experiment used nanometer-vector to mediate the transfection of M1-RNA into hepatocyte was that nanometer had the advantages of both virus vector and non-virus vector[26,27]. For instance, adenovirus vector[28,29] or retroviral vectors[30] could cause too strong immunological reaction of body to fit for the study of inhibiting the immunological rejection in our experiment; Especially, nanometer-vector could mediate exogenous gene to integrate into the chromosome DNA of host cell so that the long-term and stable expression of transgene could be obtained. In our experiment using the novel nanometer vector Effectene to transfect human hepatocyte, the rate was about 11%, and it could rise to 60%-80% after the screening with G418 for 1 wk. Nanometer vector, however, has just begun to be used in the field of gene therapy. So far, internationally, there has no report on it used in the gene therapy of clinical or pre-clinical study.
Moreover, hepatocytes induced by IFN-γ could stimulate the secretion of IL-2 mRNA from exogenous T cell, while psNAV-M1-3408-GS+ hepatocyte after IFN-γ induction lost this ability. Therefore, M1-3408-GS inhibited CIITA mRNA and thus the family of MHCII molecules regulated by CIITA, then down-regulated the ability of stimulating mixed lymphocyte reaction. In conclusion, our research will have important theoretical and practical meaning on the study of transplantation immune in the whole hepatocyte tissue engineering and the therapy of autoimmune diseases.
Edited by Yuan HT
| 1. | Zhu XF, Chen GH, He XS, Lu MQ, Wang GD, Cai CJ, Yang Y, Huang JF. Liver transplantation and artificial liver support in fulminant hepatic failure. World J Gastroenterol. 2001;7:566-568. [PubMed] |
| 2. | Wang YJ, Li MD, Wang YM, Nie QH, Chen GZ. Experimental study of bioartificial liver with cultured human liver cells. World J Gastroenterol. 1999;5:135-137. [PubMed] |
| 3. | Wang YJ, Wang XH, Li MD, Wang YM. Effect of cultured human hepatocytes on D-Gal induced acute hepatic failure in mice. Shijie Huaren Xiaohua Zazhi. 1998;6:383-385. |
| 4. | Bumgardner GL, Li J, Prologo JD, Heininger M, Orosz CG. Patterns of immune responses evoked by allogeneic hepatocytes: evidence for independent co-dominant roles for CD4+ and CD8+ T-cell responses in acute rejection. Transplantation. 1999;68:555-562. [PubMed] |
| 5. | Felix NJ, de Serres S, Meyer AA, Ting JP. Comparison of Abeta(b-/-), H2-DM(-), and CIITA(-/-) in second-set skin allograft rejection. J Surg Res. 2002;102:185-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Lei H, Madsen JC, Sachs DH. Strain variation of constitutive ex-pression of MHC class II on coronary vascular endothelium. Zhongguo Mianyixue Zazhi. 1996;12:350-352. |
| 7. | Tsark EC, Wang W, Teng YC, Arkfeld D, Dodge GR, Kovats S. Differential MHC class II-mediated presentation of rheumatoid arthritis autoantigens by human dendritic cells and macrophages. J Immunol. 2002;169:6625-6633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Czaja AJ. Understanding the pathogenesis of autoimmune hepatitis. Am J Gastroenterol. 2001;96:1224-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 89] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 9. | Stüve O, Youssef S, Slavin AJ, King CL, Patarroyo JC, Hirschberg DL, Brickey WJ, Soos JM, Piskurich JF, Chapman HA. The role of the MHC class II transactivator in class II expression and antigen presentation by astrocytes and in susceptibility to central nervous system autoimmune disease. J Immunol. 2002;169:6720-6732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 69] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Holling TM, van der Stoep N, Quinten E, van den Elsen PJ. Activated human T cells accomplish MHC class II expression through T cell-specific occupation of class II transactivator promoter III. J Immunol. 2002;168:763-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 120] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Waldburger JM, Suter T, Fontana A, Acha-Orbea H, Reith W. Selective abrogation of major histocompatibility complex class II expression on extrahematopoietic cells in mice lacking promoter IV of the class II transactivator gene. J Exp Med. 2001;194:393-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 107] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Nagarajan UM, Bushey A, Boss JM. Modulation of gene expression by the MHC class II transactivator. J Immunol. 2002;169:5078-5088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 13. | June Brickey W, Felix NJ, Griffiths R, Zhang J, Wang B, Piskurich JF, Itoh-Lindstrom Y, Coffman TM, Ting JP. Prolonged survival of class II transactivator-deficient cardiac allografts. Transplantation. 2002;74:1341-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Cobaleda C, Sánchez-García I. In vivo inhibition by a site-specific catalytic RNA subunit of RNase P designed against the BCR-ABL oncogenic products: a novel approach for cancer treatment. Blood. 2000;95:731-737. [PubMed] |
| 15. | Hao ZM, Luo JY, Cheng J, Wang QY, Yang GX. Design of a ribozyme targeting human telomerase reverse transcriptase and cloning of it's gene. World J Gastroenterol. 2003;9:104-107. [PubMed] |
| 16. | Sullivan JM, Pietras KM, Shin BJ, Misasi JN. Hammerhead ribozymes designed to cleave all human rod opsin mRNAs which cause autosomal dominant retinitis pigmentosa. Mol Vis. 2002;8:102-113. [PubMed] |
| 17. | Okimoto T, Yahata H, Fukuda Y, Dohi K. [The mechanism of donor specific unresponsiveness in renal transplant recipients]. Nihon Rinsho Meneki Gakkai Kaishi. 1998;21:118-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 18. | Lüder CG, Lang C, Giraldo-Velasquez M, Algner M, Gerdes J, Gross U. Toxoplasma gondii inhibits MHC class II expression in neural antigen-presenting cells by down-regulating the class II transactivator CIITA. J Neuroimmunol. 2003;134:12-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 64] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Yard BA, Yedgar S, Scheele M, van der Woude D, Beck G, Heidrich B, Krimsky M, van der Woude FJ, Post S. Modulation of IFN-gamma-induced immunogenicity by phosphatidylethanolamine-linked hyaluronic acid. Transplantation. 2002;73:984-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Lüder CG, Walter W, Beuerle B, Maeurer MJ, Gross U. Toxoplasma gondii down-regulates MHC class II gene expression and antigen presentation by murine macrophages via interference with nuclear translocation of STAT1alpha. Eur J Immunol. 2001;31:1475-1484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Youssef S, Stüve O, Patarroyo JC, Ruiz PJ, Radosevich JL, Hur EM, Bravo M, Mitchell DJ, Sobel RA, Steinman L. The HMG-CoA reductase inhibitor, atorvastatin, promotes a Th2 bias and reverses paralysis in central nervous system autoimmune disease. Nature. 2002;420:78-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 846] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 22. | Charreau B, Coupel S, Boulday G, Soulillou JP. Cyclosporine inhibits class II major histocompatibility antigen presentation by xenogeneic endothelial cells to human T lymphocytes by altering expression of the class II transcriptional activator gene. Transplantation. 2000;70:354-361. [PubMed] |
| 23. | Sadeghi MM, Tiglio A, Sadigh K, O'Donnell L, Collinge M, Pardi R, Bender JR. Inhibition of interferon-gamma-mediated microvascular endothelial cell major histocompatibility complex class II gene activation by HMG-CoA reductase inhibitors. Transplantation. 2001;71:1262-1268. [PubMed] |
| 24. | Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6971] [Cited by in RCA: 7022] [Article Influence: 292.6] [Reference Citation Analysis (0)] |
| 25. | Yamamoto T, Omoto S, Mizuguchi M, Mizukami H, Okuyama H, Okada N, Saksena NK, Brisibe EA, Otake K, Fuji YR. Double-stranded nef RNA interferes with human immunodeficiency virus type 1 replication. Microbiol Immunol. 2002;46:809-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Esfand R, Tomalia DA. Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications. Drug Discov Today. 2001;6:427-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1255] [Cited by in RCA: 1095] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 27. | Dennig J, Duncan E. Gene transfer into eukaryotic cells using activated polyamidoamine dendrimers. J Biotechnol. 2002;90:339-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 28. | Yang ZY, Wyatt LS, Kong WP, Moodie Z, Moss B, Nabel GJ. Overcoming immunity to a viral vaccine by DNA priming before vector boosting. J Virol. 2003;77:799-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 172] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 29. | Sailaja G, HogenEsch H, North A, Hays J, Mittal SK. Encapsulation of recombinant adenovirus into alginate microspheres circumvents vector-specific immune response. Gene Ther. 2002;9:1722-1729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 78] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Podevin G, Pichard V, Durand S, Aubert D, Heloury Y, Ferry N. In-vivo retroviral gene transfer to the liver is cancelled by an immune response against the corrected cells. Can it be avoided? Pediatr Surg Int. 2002;18:595-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |