Published online Dec 15, 1998. doi: 10.3748/wjg.v4.i6.506
Revised: November 22, 1998
Accepted: December 5, 1998
Published online: December 15, 1998
AIM: To set up cell lines of human hepatocellular carcinoma in nude mice for the research of cell biology and gene therapy.
METHODS: Xenotransplantation of human hepatoma into nude mice was carried out and the growth rate, histopathology and immunology of the nude mice were studied. The DNA from xenografts were analyzed by HBV gene and PCR amplification of a fragment of p53 gene exon 7, which were identified by dot blot hybridization, restriction fragments length polymorphism and DNA sequencing.
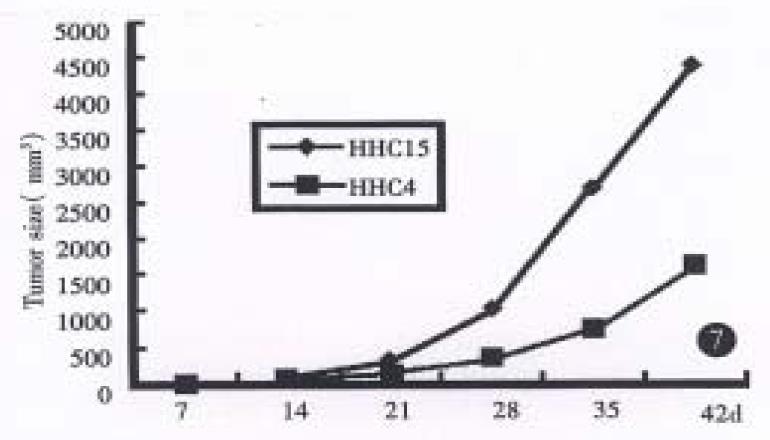
RESULTS: HHC4 and hHCC415 cell lines could be successively transplanted in nude mice and the population doubling time was 7 and 5 d respectively. These strains retained the original characteristics of histopathology, secreting AFP and heteroploid karyotypes in human hepatocellular carcinoma. The fragment of HBV gene was detected in the genomic DNA of both hHCC4 and hHCC15, however only hHCC4 secreted HBsAg. The mutation at 250 code (C→A) and 249 code (G→T) were detected respectively in the genomic DNA of HHC4 and HHC15.
CONCLUSION: The two cell lines are useful material for the studying of cell biology and gene therapy in human hepatocellular carcinoma and provide molecular biological trace of the relationship between high mortality of hepatoma and AFB1 severe pollution of the daily common foods in this district.
- Citation: Yang SM, Zhou H, Chen RC, Wang YF, Chen F, Zhang CG, Zhen Y, Yan JH, Su JH. Sequencing of p53 mutation in established human hepatocellular carcinoma cell line of HHC4 and HHC15 in nude mice. World J Gastroenterol 1998; 4(6): 506-510
- URL: https://www.wjgnet.com/1007-9327/full/v4/i6/506.htm
- DOI: https://dx.doi.org/10.3748/wjg.v4.i6.506
Although the etiopathology of human hepatocellular carcinoma (hHCC) is still unknown, a lot of evidence strongly suggested that infection of HBV and contamination of aflatoxin B1 (AFB1) were the inducing fuctos of the carcinogenesis of hHCC. Xenografts of human tumorous tissue in nude mice usually retain their original morphology, antigen, karyotype and function. These models can be used for several purposes, including assessment of the etiopathology, cell biology, sensitivity of chemotherapy, genetherapy, and so on. hHCC4 and hHCC15 cell lines were established from the patients of HHC in Tong’an district of Xiamen where there was high adjust death rates of hHCC (44.75 per one hundred thousand population from 1987 to 1989) which was the secondary mortality of hHCC in China. In this district, a lot of evidence has been shown that there was severe contamination of AFB1 in daily common foods, such as the oil of peanut (92.9%) and fermented soy beans (44.4%)[1]. Also high epidemic infection of HBV was presented in the population in this country (17.6%)[2]. Because the relationship between molecular biological changes and etiopathology of HHC in this district is still absent hHCC4 and hHCC15 cell lines in nude mice were studied by cell biology and molecular biology.
Male and female nude mice, about 4 to 6 weeks old, with BALB/C genetic background were provided by Medical Experimental Animal Laboratory, Cancer Research Center, Xiamen University, where the mice were bred and maintained in vinyl box isolated under specific pathogene free condition. The sterilized food pellet and tap water were given ad libitum.
Tumor tissues of patients, who underwent partly hepatotomy in the Min-Hai Hospital, Tong’an, were dissected from the primary site in the liver and aseptically minced and placed in cooled culture medium. Several tumor tissue fragments about 2 mm in diameter were transplanted, with trocar, into the subcutaneous tissue of the back of 6 mice within 3 h after surgical removal of the tumor. Afterwards, a piece of tumor tissue from hHCC15 was orthotopically transplanted into the liver of 21 nude mice for studying the ability of secreting AFP from tumor.
Tumors in the subcutaneous tissue or liver were measured in 3 dimensions with calipers for every 7 d. Tumor size was plotted on a graph.
For light microscopy and transition electron microscopy (TEM) the primary tumors of liver and xenografts were fixed and stained by the standard methods.
The Sera from patients and tumor bearing mice were analyzed radioimmunologically with AFP Diagnosis Kit (The Institute of Biochemical Assay and Product, Shanghai). In brief, 100 μL of blood was obtained weekly for 6 weeks, by orbital venipuncturing from mice. Two cell lines were inoculated subcutaneously or into liver (for the later only with hHCC15) respectively. HBsAg and HBcAg in the sear of tumor-bearing mice were analyzed with HBsAg and HBcAg Kit (New and Advanced Co. Ltd Xiamen) by ELISA method.
According to the standard method of preparing metaphase cell treated with colchicine, chromosome in hHCC4 and hHCC15 cell line was carried out. The number of chromosomes in each spread cell was counted and all of spread cells were done at least for 100.
The DNA from xenografts were extracted with the standard method of Sambrook and assayed by the HBV Diagnosis Kit (a kind gift from professor Wu Bing-Qun, Dept. of Pathology Beijing Medical University, China). Forward and reverse primer for HBV fragment were designed as follow: F 5’-GGGTGGAGCCCTCAGGCTCAGGGCA-3’, R 5’-GAAGATGAGGCATAGCAGCAGGAT-3’. The positive and negative samples were amplified simultaneously as control. The products of PCR were run in 12 g/L agarose gel stained with ethidimum bromine and photographed.
DNA of two strains was a mplified by PCR to produce target of a 110 bp at seventh exon of p53 gene using primers of P1 5’-GTTGGCTCTGACTGTACCAC-3’ and P2 5’-CTGGAGTCTTCCAGTGTGAT-3’ on DNA Thermal Cycler 480 (Perkin-Elmer/Cetus). The product of 110 bp DNA fragment was identified by 20 g/L agarose gel electrophoresis and DNA dot blot hybridization which was performed with hDIG-labeled p53 cDNA probe (2.0 kb, cut from reconstructed plasmid ph p53β, a kind gift from Professor Liu Si-Li, Tianjin Medical College). The probe was labeled as the described method of DIG DNA Labeling and Detection Kit (Boehringer Mannhemi).
Five to 10¦Ìl of above PCR amplified products were digested with 7 to 10 unit Hae III restriction enzyme at 37 °C for 6 h, then precipitated with cooled ethanol. The sediments were analyzed with 150 g/L non-denatural polyacrylamide gel electrophoresis, and then stained with ethidium bromide and visualized under UV light.
The 110 bp of PCR amplification fragments were purified by standard low-melting point agarose gel electrophoresis method and labeled with fluorescence according to the description of Tag Dye DeoxyTM Terminator Cycle Sequencing Kit. The DNA sequence of PCR fragments were analyzed and edited by Applied Biosystems 373A DNA Sequencer.
The neoplasams of hHCC4 and hHCC15 were presented in the back of 1 in 6 and all 6 mice respectively and showed rapid growth after several generations. A 58% and 100% rate of tumor transplantation were presented in hHCC4 and in hHCC15 respectively. Xenografts were shown a short latency (18.7 d ± 4.9 d and 17.5 d ± 1.6 d respectively) and almost stable after successive generations. The population doubling time in hHCC4 and hHCC15 cell line was about 7 and 5 days respectively, the growth curves are shown in (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6) Figure 7.
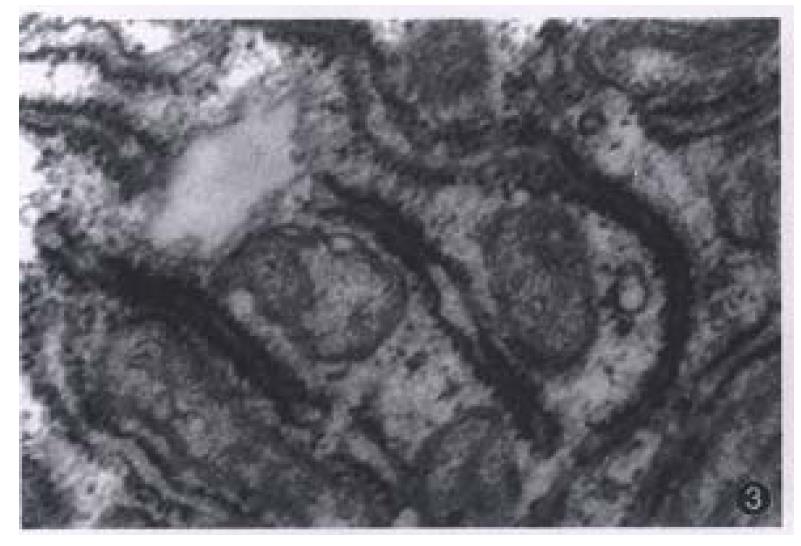
Most of the transplanted tumors retained approximately the original morphological characteristics (Figure 1). No metastasis foci was presented in the liver and lung of each tumor-bearing mice within 6 weeks. Ultrastructurally, sinusoid like structure and bile canaliculi were scattered between two cells (Figure 2). Under TEM, fibrillary structure of HBsAg could be seen in the rough endoplasmic reticulum (RER) of carcinoma cells in hHCC4 (Figure 3).
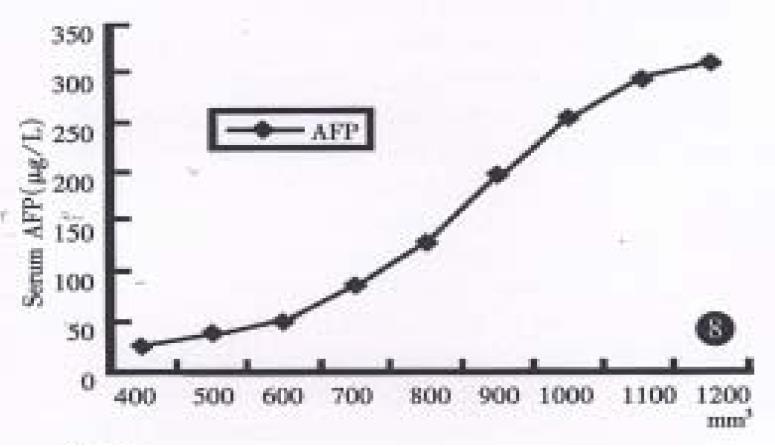
Radioimmunoassay disclosed human AFP in sera of both groups mice bearing xenografts of both strains, (until as large as 100 mm3), but not for the control group, and their values increased progressively in relation to orthotopic growth of the tumor in hHCC15 (Chart 2). HBsAg could be detected in hHCC4, but not for HBcAg. HBcAg and HBsAg could not be undetected in hHCC15 by ELISA immunoassay.
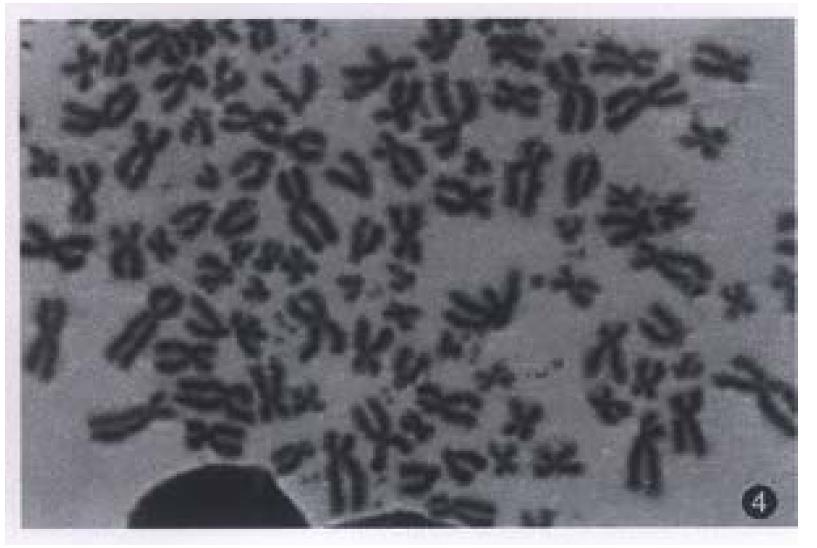
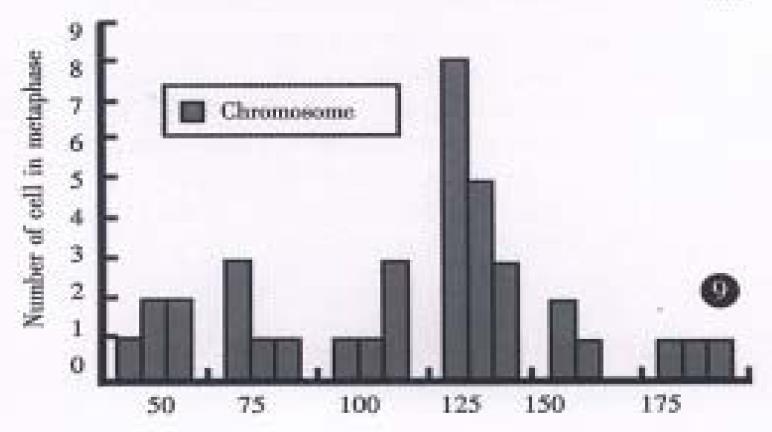
All evaluable chromosomes of metaphase in hHCC4 and hHCC15 were human chromosomes, and no any mouse chromosomes were seen (Figure 4). A histogram of chromosome counts of 100 cells disclosed the number of chromosomes ranging between 50 to 175, the median number of 106 to 126 in hHCC15 (Figure 8, Figure 9) and 110 to 134 in hHCC4 (data not shown) cell line respectively.
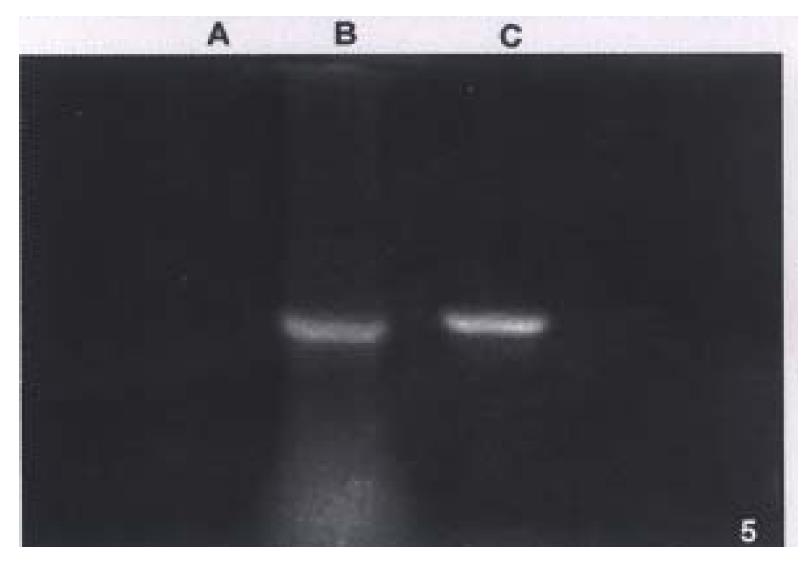
PCR products amplified from both hHCC4 (data not shown) and hHCC15 or positive control had similar band running in the same distance in agarose gel, but not any product in negative control sample (Figure 5).
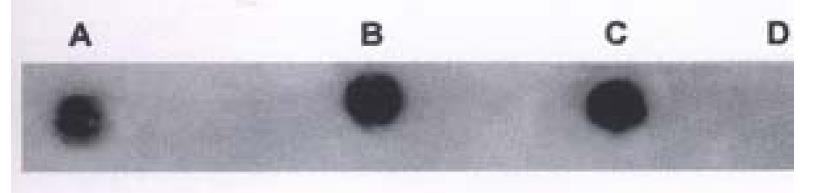
DNA extracted from hHCC4 and hHCC15 xencgrafts and plasmid ph p53β containing wild p53 cDNA were amplified respectively, then the PCR products were analyzed by DNA dot blot. Figure 6 showed positive hybridization of the PCR products from hHCC4, hHCC15 and ph p53β plasmid with the control negative from plasmid pBR322, which meant the good specificity of the PCR system. With the method of RFLP, it was shown that the product of PCR from ph p53β plasmid, as a contrast of wild p53 cDNA, was digested into two bands of 75 bp and 35 bp (Line A) and undigested two of 110 bp from hHCC4 and hHCC15 (Figure 10). It was suggested that the mutation at seventh exon of p53 gene could be presented in the xenografts of hHCC4 and hHCC15.
The results showed in Figure 11 and Figure 12 were the partial DNA sequence of fragments amplified from hHCC4 and hHCC15 genomic DNA respectively. Figure 11 revealed a CCC→ACC point mutation in 250 code of p53 gene from xenograft of hHCC4, while Figure 12 showed an AGG→AGT mutation of p53 gene from xenograft of hHCC15.
Xenografts of hHCC in nude mice usually retain their original morphology, antigen, karyotype and function, such as secreting AFP. It has been reported that both AFP and HBsAg can not be detected simultaneously in nude mouse transplanted with hHCC, but are presented in the cell lines of hHCC in vitro[3]. The presenting fibrae of HBsAg in the RER of carcinoma cell support the specific function of cells from hHCC4.
A variety of p53 mutations have been found in a wide spectrum of sporadic tumors, in which the cause of carcinogenesis was still unknown and direct evidence is absent. It has been reported that mutation of p53 gene was the most common inducing factor in primary advanced hHCC. AFB1 was the most inducing factor of carcinogenesis in hHCC and specially associated with the mutation of code 249 in p53 gene, being notable as “hot spots” in hHCC, spreading over South Africa or southeast coast of Asia on the earth[4,5].
Multiple evidence support a closely relationship between AFB1 and HHC. Rats fed with AFB1 developed hepatoma in a dose-depend fashion[6]. AFB1 intake might lead the liver to acute necrosis and proliferation of hepatoid-oval cells[7]. The more AFB1 intake daily was, the more necrosis appeared and the oval cells possessing the ability of division continuously divided.
According to the telomere hypothesis of cellular senescence theory, somatic cells, which continuously divided, lead to cell cycle exit and significant telomere erosion and shortage at which the crisis state (M1) of cell arrived, which was induced by activating p53 and pRb cascade. It is conceivable that p53 is proposed to signal a growth cheekpoint allowing cell to arrest in G0 or G1 state (replicative senescence) by inducing p21 expression which in turn inactivate cdk/cyclin complex leading to underphosphorylation of the Rb proteins[8]. It is reasonable that p53 gene mutation caused by AFB1 have the ability to allow cells to overcome M1, leading to an extended lifespan until a second growth checkpoint, M2 is reached. This rare event, M2, is most often associated with the reactivation of telomerase. During the past few years, there has been mounting evidence that the activation of telomerase, a ribonucleoprotein enzyme, is important in maintaining telomere length stability and necessary for the sustained growth of the most cancer[9]. It has been reported recently that due to reactivating telomerase, mammary epithelial cells, which were transfected with mutation p53 gene, could be survive and become immortal in vitro[10]. The two cell lines provided molecular biological trace of relationship between high mortality of hepatoma and AFB1 in this district. It shown that carcinoma cells of hHCC4 and hHCC15 possessing telomerase activity (unpublished data) supported the events of p53 mutation discovered by us.
The carcinogenesis of hHCC is closely related to chronic hepatitis B in which the molecular machanism of hHCC is still a mystery. Futhermore, AFB1 treatment of trangenic mice integrated with hepatitis B DNA greatly enhanced the development of hepatoma as compared with the mice not treated with AFB1[11]. It is wise to use two cell lines for the disclosing the contribution of carcinogenesis of human hepatoma in coordination of HBV integration and p53 mutation which were presented in both of the cell lines. Also they are the useful material for cell biology, sensitivity of chemotherapy and gene therapy.
Project supported by the Science Foundation of Department of Publish Health of Fujian, No. 85003-01-11
| 1. | Lin L, Sun CS, Kang TS, Xu WQ, Zhen ZY, Shi MD. Investigation on four common foods polluted by AFTB1 at Tong'an country. J Fujian Med Coll. 1997;26:216-218. |
| 2. | Shun CS, Ren JX, Xie CG, Zhen JY, Hong ZN, Zhong WL. Logistic regression analysis of dangerous factors contributed to the carcinogenesis of hepatoma in Tong'an country Fujian province. J Med Fujian. 1994;16:63-65. |
| 3. | Alexander JJ. Human hepatoma cell lines. In: Okuda K, Ishak KG, eds. Neo-plasms of the liver. Springer Verlag. 1987;46-56. [DOI] [Full Text] |
| 4. | Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 969] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 5. | Holletein M, Sidrausky D, Vo yelstein B, Harris CC. p53 mutations in human cancer. Science. 1991;253:49-53. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5532] [Cited by in RCA: 5489] [Article Influence: 161.4] [Reference Citation Analysis (0)] |
| 6. | Groopman JD, Cain LG, Kensler TW. Aflatoxin exposure in human populations: measurements and relationship to cancer. Crit Rev Toxicol. 1988;19:113-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 139] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 7. | Busby WF, Wogan GN. Aflatoxins. In: Searle CE, ed. Chemical carcinogens, 2dn ed. Washington: American Chemical Society. 1984;945-1136. |
| 8. | Wright WE, Shay JW. Time, telomeres and tumours: is cellular senescence more than an anticancer mechanism. Trends Cell Biol. 1995;5:293-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Kim NW, Piatyszek MA, Prowse KR, Hartey CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011-2015. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5156] [Cited by in RCA: 5234] [Article Influence: 168.8] [Reference Citation Analysis (0)] |
| 10. | Gollahon LS, Shay JW. Immortalization of human mammary epithelial cells transfected with mutant p53 (273(his)). Oncogen. 1996;12:715-725. |
| 11. | Sell S, Hunt JM, Dunsford HA, Chisari FV. Synergy between hepatitis B virus expression and chemical hepatocarcinogens in transgenic mice. Cancer Res. 1991;51:1278-1285. [PubMed] |