Published online Jun 28, 2023. doi: 10.3748/wjg.v29.i24.3793
Peer-review started: April 18, 2023
First decision: April 26, 2023
Revised: May 4, 2023
Accepted: May 22, 2023
Article in press: May 22, 2023
Published online: June 28, 2023
Processing time: 71 Days and 1 Hours
Formyl peptide receptor 2 (Fpr2) is an important receptor in host resistance to bacterial infections. In previous studies, we found that the liver of Fpr2-/- mice is the most severely damaged target organ in bloodstream infections, although the reason for this is unclear.
To investigate the role of Fpr2 in liver homeostasis and host resistance to bacterial infections.
Transcriptome sequencing was performed on the livers of Fpr2-/- and wild-type (WT) mice. Differentially expressed genes (DEGs) were identified in the Fpr2-/- and WT mice, and the biological functions of DEGs were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) en-richment analysis. Quantitative real time-polymerase chain reaction (qRT-PCR) and western blot (WB) analyses were used to further validate the expression levels of differential genes. Cell counting kit-8 assay was employed to investigate cell survival. The cell cycle detection kit was used to measure the distribution of cell cycles. The Luminex assay was used to analyze cytokine levels in the liver. The serum biochemical indices and the number of neutrophils in the liver were measured, and hepatic histopathological analysis was performed.
Compared with the WT group, 445 DEGs, including 325 upregulated genes and 120 downregulated genes, were identified in the liver of Fpr2-/- mice. The enrichment analysis using GO and KEGG showed that these DEGs were mainly related to cell cycle. The qRT-PCR analysis confirmed that several key genes (CycA, CycB1, Cdc20, Cdc25c, and Cdk1) involved in the cell cycle had significant changes. The WB analysis confirmed a decrease in the expression of CDK1 protein. WRW4 (an antagonist of Fpr2) could inhibit the proliferation of HepG2 cells in a concentration dependent manner, with an increase in the number of cells in the G0/G1 phase, and a decrease in the number of cells in the S phase. Serum alanine aminotransferase levels increased in Fpr2-/- mice. The Luminex assay measurements showed that interleukin (IL)-10 and chemokine (C-X-C motif) ligand (CXCL)-1 levels were significantly reduced in the liver of Fpr2-/- mice. There was no difference in the number of neutrophils, serum C-reactive protein levels, and liver pathology between WT and Fpr2-/- mice.
Fpr2 participates in the regulation of cell cycle and cell proliferation, and affects the expression of IL-10 and CXCL-1, thus playing an important protective role in maintaining liver homeostasis.
Core Tip: Formyl peptide receptor 2 (Fpr2) is an important receptor in host resistance to bacterial infection. After Fpr2 deletion, the pathways involved in the cell cycle are affected, the expression of Cdk1 is downregulated, the proliferation activity of HepG2 cells is reduced, and the distribution of cell cycle is abnormal. After Fpr2 deletion, the permeability of the hepatocyte membrane increases, and the expression of interleukin-10 and Chemokine (C-X-C motif) ligand-1 decreases. These changes reflect the important protective effect of Fpr2 in maintaining liver homeostasis.
- Citation: Liu H, Sun ZY, Jiang H, Li XD, Jiang YQ, Liu P, Huang WH, Lv QY, Zhang XL, Li RK. Transcriptome sequencing and experiments reveal the effect of formyl peptide receptor 2 on liver homeostasis. World J Gastroenterol 2023; 29(24): 3793-3806
- URL: https://www.wjgnet.com/1007-9327/full/v29/i24/3793.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i24.3793
The liver plays an important role in human metabolism and immune homeostasis. Liver dysfunction and liver failure can directly lead to the progression of sepsis and death of patients. Pre-existing liver dysfunction is a risk factor for the progression of infection to sepsis[1]. The liver is an important organ that collects blood from the intestine via the portal vein. Once the intestinal barrier becomes dysfunctional, the liver is the first organ to encounter bacteria. The liver collects blood through the hepatic artery, so it is also an important organ involved in the removal of bacteria from the bloodstream in systemic infection[2,3].
As an organ mainly responsible for blood detoxification, the liver plays a key role in removing bacteria and toxins during sepsis. After intravenous injection, the liver captures 65% of bacteria in 10 min and over 80% in 6 h[4]. Two methods of liver clearance have been identified: Rapid and slow. The rapid clearance mainly relies on Kupffer cells to remove bacteria and does not depend on complement-mediated opsonophagocytosis or platelet combinations. The slow clearance ensures that the host not only removes a sufficient number of bacteria to prevent infection and damage but also maintains sufficient bacteria to induce host adaptive immunity. In addition to mesenteric lymph nodes, the liver is another important firewall against bacterial infections in the body[2,5,6].
Formyl peptide receptors (FPRs) are G-protein-coupled receptors that play an important role in host defense and inflammation. The most important biological activity of FPRs is the induction of leukocyte chemotaxis in infected or injured areas. In addition to chemotaxis, FPRs also mediate phagocytosis, production of reactive oxygen species, and production of various cytokines/chemokines. According to sequence similarity, FPRs are divided into three family members: FPR1, ALX/FPR2, and FPR3. Based on the ability of FPRs to recognize bacterial formylated peptides, they are considered to play an important role in host defense against microbial invasion.
Initially, research on FPRs mainly focused on regulating inflammation, but increasing evidence demonstrated that FPRs can also regulate the host’s defense process, including regulating the activation of neutrophils and dendritic cells and participating in host resistance to bacterial infection, tissue damage, and wound healing[7]. Liu et al[8] reported that during Listeria monocytogenes (Listeria) infection, FPRs can quickly recruit the first wave of neutrophils into the infected liver, followed by the second wave of neutrophil accumulation mediated by CXCR2. This discovery challenges the previously reported pattern of host defense against Listeria, that is, the receptor toll-like receptor 2 on the host cell is activated by bacterial lipoprotein, and then CXCR2 ligand is produced to induce neutrophil accumulation[9]. In sepsis, the biased expression of chemokine receptors CC chemokine receptor 5 (CCR5) and FPR2 in human and mouse neutrophils leads to the movement of neutrophils guided by CCR5 to aseptic tissue sites, whereas polymorphonuclear neutrophils guided by FPR2 to bacterial infection sites are significantly reduced[10]. Thus, inflammatory but non-functional neutrophils may increase the host’s morbidity and mortality, which may be related to infectious injury[11]. The above findings emphasize the importance of FPRs in the host defense against microorganisms.
Our previous research revealed that in group B streptococcus infection, Fpr2 activated the downstream key signal molecules and produced chemokine (C-X-C motif) ligand (CXCL)1/2 to attract neutrophils, indicating that Fpr2, as a chemotactic receptor, could not only attract neutrophils directly, but also indirectly control neutrophil chemotaxis by regulating the production of chemokines to resist infection[12]. The liver is the most important organ in the removal of bacteria, and Fpr2 is an important receptor involved in resisting bacterial infection. However, the effect of Fpr2 on the liver homeostasis remains unclear. In the present study, we evaluated the liver of Fpr2 knockout and wild-type (WT) mice in terms of transcriptome sequencing, histology, immunity, and biochemistry to understand the reason for the antibacterial activity of Fpr2.
Fpr2-/- (Fpr2 knockout) mice were purchased from Cyagen Biosciences (Guangzhou, China), who created this model according to previously reported methods[12]. The WT C57BL/6 mice (6-8 wk, 18-20 g body weight) were purchased from Charles River (Beijing, China). These mice were bred in the animal center of the Academy of Military Medical Sciences under specific pathogen free conditions. The animals were acclimatized to laboratory conditions (23 °C, 12 h/12 h light/dark, 50% humidity, and ad libitum access to food and water) for 2 wk, prior to experimentation. All animals were euthanized by isoflurane inhalation for tissue collection. The animal protocol was designed to minimize pain or discomfort to the animals. This research was conducted in compliance with the guidelines for laboratory animal care approved in China. All experimental procedures were approved by the Animal Ethics Committee of the Academy of Military Medical Sciences.
The HepG2 cell line was cultured in a high-glucose DMEM medium (Gibco, Rockville, MD, United States) supplemented with 10% fetal bovine serum (Gibco, Rockville, MD, United States). Cell counting kit-8 (CCK8; APExBIO, United States) assay was employed to investigate cell survival following the manufacturer’s instructions. In brief, HepG2 cells were plated in 96-well plates at a density of 3 × 103 cells/well. On the second day, cells were treated with different concentrations of WRW4 [Sangon Biotech (Shanghai) Co., Ltd. China] for 24 h. Then, 10 μL CCK-8 was added to each well and cells were incubated at 37 °C for 2 h. Absorbance was measured at 450 nm using a microplate spectrophotometer (Molecular Devices LLC, Sunnyvale, CA, United States).
The cell cycle detection kit (KeyGEN BioTECH, Nanjing, China) was used to measure the distribution of cell cycles. HepG2 cells were plated in 6-well plates and treated with WRW4 (1 μM) for 24 h. Dimethyl sulfoxide used as a control to dissolve WRW4. Next, cells were collected, washed, and fixed overnight at 4 °C in 70% ethanol. The samples were then incubated with RNase A and propidium in the dark for 30 min and cell cycle distribution was analyzed using flow cytometry.
The mice were sacrificed after anesthesia, and the liver was taken for subsequent RNA sequencing. RNA extraction, library preparation, DNA sequencing, and result analysis were conducted by Nevogene Co., Ltd (Beijing, China). Briefly, total RNA was extracted using TRIzol reagent (Invitrogen, Shanghai, China). RNA integrity was assessed using the RNA Nano 6000 Assay Kit on a Bioanalyzer 2100 system (Agilent Technologies, CA, United States).
To select cDNA fragments of 370-420 bp, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, United States). Polymerase chain reaction (PCR) was performed using Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. Finally, PCR products were purified (AMPure XP system), and library quality was assessed on the Agilent Bioanalyzer 2100 system. Clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina). The library preparations were sequenced on an Illumina Novaseq platform, and 150 bp paired-end reads were generated.
After data cleaning (to obtain clean reads with high quality), reads were mapped to the reference genome. Reference genome and gene model annotation files were downloaded from the genome website directly. The index of the reference genome was built using Hisat2 v2.0.5, and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5. FeatureCounts vl.5.0-p3 was used to count the number of reads mapped to each gene. Fragments per kilobase millions of each gene were calculated based on the length of the gene and the read counts were mapped to the respective genes.
Differential expression analysis of the two groups (four biological replicates per group) was performed using the DESeq2 R package (1.20.0). Genes with an adjusted P < 0.05 found by DESeq2 were assigned as differentially expressed. Quantitative real time-PCR (qRT-PCR) analysis further validated gene expression patterns. The primers used for qRT-PCR analysis were listed in Table 1.
| Primer | Sequences (5’ to 3’) |
| CycA-F | GCCTTCACCATTCATGTGGAT |
| CycA-R | TTGCTCCGGGTAAAGAGACAG |
| CycB1-F | AGAGCTATCCTCATTGACTGGC |
| CycB1-R | AACATGGCCGTTACACCGAC |
| Cdk1-F | AGAAGGTACTTACGGTGTGGT |
| Cdk1-R | GAGAGATTTCCCGAATTGCAGT |
| Cdc25c-F | GTTTCAGCACCCAGTTTTAGGT |
| Cdc25c-R | AGAATGCTTAGGTTTGCCGAG |
| Cdc20-F | TTCGTGTTCGAGAGCGATTTG |
| Cdc20-R | ACCTTGGAACTAGATTTGCCAG |
| Mps1-F | GCAGTGTGACGATTGATTCCA |
| Mps1-R | TCGGCACAGATTTTAGACAAGC |
| GAPDH-F | CATGGCCTTCCGTGTTCCTA |
| GAPDH-R | GCGGCACGTCAGATCCA |
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes (DEGs) were implemented using the ClusterProfiler R package, in which gene length bias was corrected. GO terms with a corrected P < 0.05 were considered significantly enriched by DEGs. The ClusterProfiler R package was used to test the statistical significance of DEGs in KEGG enrichment pathways.
The liver tissues were taken and washed three times with phosphate buffer solution, then stored at -80 °C. Total proteins were extracted from the liver using RIPA buffer (Beyotime, Nantong, China), and the concentrations were estimated using the bicinchoninic acid protein assay kit (Pierce, Rockford, IL, United States). Next, 30 μg of protein was separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis on a 10% gel and transferred to PVDF membranes. The membranes were blocked in 3% nonfat dry milk and then probed with anti-CDK1 antibody (1:2000 dilution; ab32094; Abcam, Cambridge, MA, United States), and anti-beta actin antibody (1:3000; ab8227; Abcam, Cambridge, MA, United States) overnight at 4 °C. On the next day, membranes were incubated with appropriate secondary antibody for 1 h at room temperature. The protein bands were visualized using Amersham Hyperfilm™ ECL reagent (Thermo Fisher Scientific, MA, United States).
Fresh liver tissues from the mice were weighed and homogenized. Concentrations of cytokines in the supernatants of the liver were measured by multiplexed Luminex xMAP assay (Cytokine & Chemokine 36-Plex Mouse ProcartaPlex™ Panel 1A; Thermo Fisher Scientific, MA, United States), according to the manufacturer’s instructions.
The liver tissue was digested into single-cell suspension using type IV collagenase (Sigma-Aldrich, United States). The number of leukocytes in the liver were determined using a BD TruCount system (BD Biosciences, United States). The cells were stained with FITC-anti-mouse Ly6G (Biolegend, San Diego, CA, United States), PerCP-anti-mouse CD45 (Biolegend, San Diego, CA, United States), and PE-anti-mouse CD11b (Biolegend, San Diego, CA, United States) antibiodies. All of the above-stained cells were assayed using a BD FACSVerse flow cytometer and data were analyzed using FlowJo software.
The blood from mice was collected, and serum was separated by centrifugation (4 °C, 3000 rpm, 10 min). A total of 200 μL serum was obtained and sent to the experimental animal center of the Academy of Military Medical Sciences for the determination of liver biochemical indexes.
The liver tissue of mice was fixed with 4% formaldehyde, paraffin sections were prepared, stained with hematoxylin and eosin, and observed and photographed with an Olympus camera system. The image magnifications were 200 × and 400 ×.
All data were analyzed using GraphPad Prism (version 8.0; GraphPad Software, La Jolla, CA, United States). Differences between the groups were calculated using an unpaired t-test and a log-rank test. Data were shown as mean ± SD. Differences were considered statistically significant at P < 0.05.
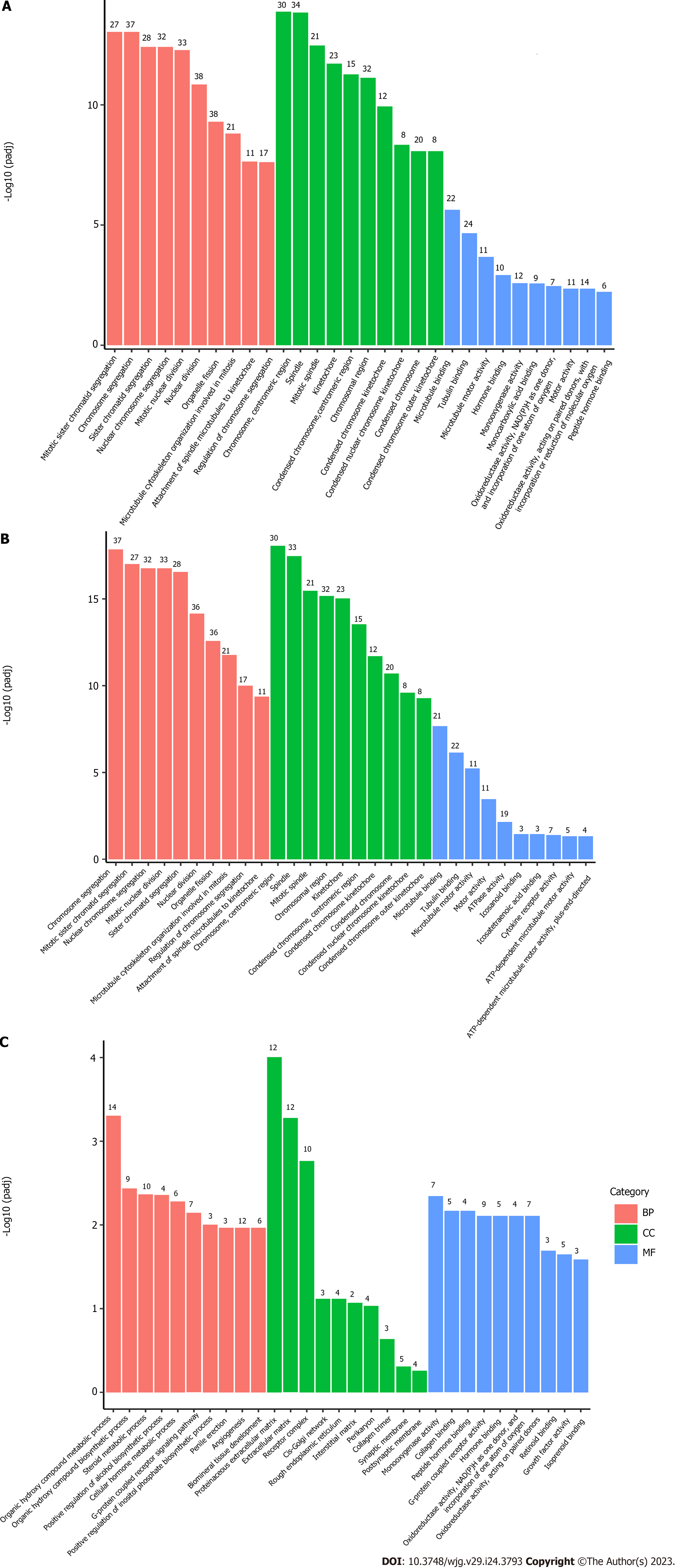
We used RNA-sequencing to analyze the liver transcriptome of Fpr2-/- mice and compared it with that of WT mice to determine DEGs. Taking |log2(fold change)| ≥ 1 and padj ≤ 0.05 as criteria, compared with the WT group, 445 DEGs, including 325 upregulated genes and 120 downregulated genes, were identified in Fpr2-/- mice (Figure 1A). Venn diagram analysis showed that 9578 genes showed identical expression between Fpr2-/- and WT mice. On the other hand, 541 and 371 specific genes were expressed in Fpr2-/- and WT mice, respectively (Figure 1B).
According to the results of the enrichment analysis, 179 GO terms were obtained, including 136 biological processes (BPs), 25 cell component (CC), and 18 molecular function (MF) terms. The results showed that the DEGs were mainly annotated to the cell cycle (Figure 2A). Further GO enrichment analysis of upregulated DEGs showed that a total of 156 GO terms were obtained, including 121 BPs, 25 CC, and 10 MF terms. The BP terms were significantly enriched in mitotic sister chromatid segregation, chromosome segregation, sister chromatid segregation, and nuclear chromosome segregation. The CC terms were significantly enriched in chromosomes, centromeric regions, spindles, mitotic spindles, kinetochores, condensed chromosomes, centromeric regions, and chromosomal regions. The MF terms were significantly enriched in microtubule binding, tubulin binding, and microtubule motor activity (Figure 2B). The results of the GO analysis suggest that the upregulated DEGs are significantly enriched in many phases of cell division. In addition, GO analysis of downregulated DEGs revealed that BP terms were significantly enriched in many BPs related to liver function, such as organic hydroxy compound metabolic process, organic hydroxy compound biosynthetic process, steroid metabolic process, positive regulation of alcohol biosynthetic process, cellular hormone metabolic process, G-protein coupled receptor signaling pathway, and coupled to cyclic nucleotide second messenger (Figure 2C).
Four pathways with significant differences obtained based on KEGG analysis are listed in Table 2. These pathways included cell cycle, neuroactive ligand-receptor interaction, oocyte meiosis, and progesterone-mediated oocyte maturation (Figure 3). Both GO function analysis and KEGG pathway enrichment analysis showed that core DEGs mainly regulated the cell cycle of the liver tissue in Fpr2-deficient mice, which revealed that Fpr2 deficiency may cause cell division cycle disorder to some extent.
| Term | Description | Sample number | P value | Gene_name |
| mmu04110 | Cell cycle | 16/182 | 0.00009 | Cdc20 Ccnb1 Ccnb2 Plk1 Ccna2 Ttk Mad2l1 Wee1 Bub1b Cdk1 Mcm5 Cdkn1a Cdc25c Mcm2 Espl1 Bub1 |
| mmu04080 | Neuroactive ligand-receptor interaction | 16/182 | 0.00079 | Fpr2 Mtnr1a Gzma P2ry1 Lpar2 Chrna4 Grid1 Gabra3 Adcyap1r1 Vipr1 Calcrl Pth1r Ednra Hcrtr2 Gpr35 P2ry13 |
| mmu04114 | Oocyte meiosis | 13/182 | 0.00079 | Cdc20 Ccnb1 Ccnb2 Plk1 Mad2l1 Camk2b Adcy1 Ar Cdk1 Cdc25c Adcy7 Espl1 Bub1 |
| mmu04914 | Progesterone-mediated oocyte maturation | 11/182 | 0.00132 | Ccnb1 Ccnb2 Plk1 Ccna2 Kif22 Mad2l1 Adcy1 Cdk1 Cdc25c Adcy7 Bub1 |
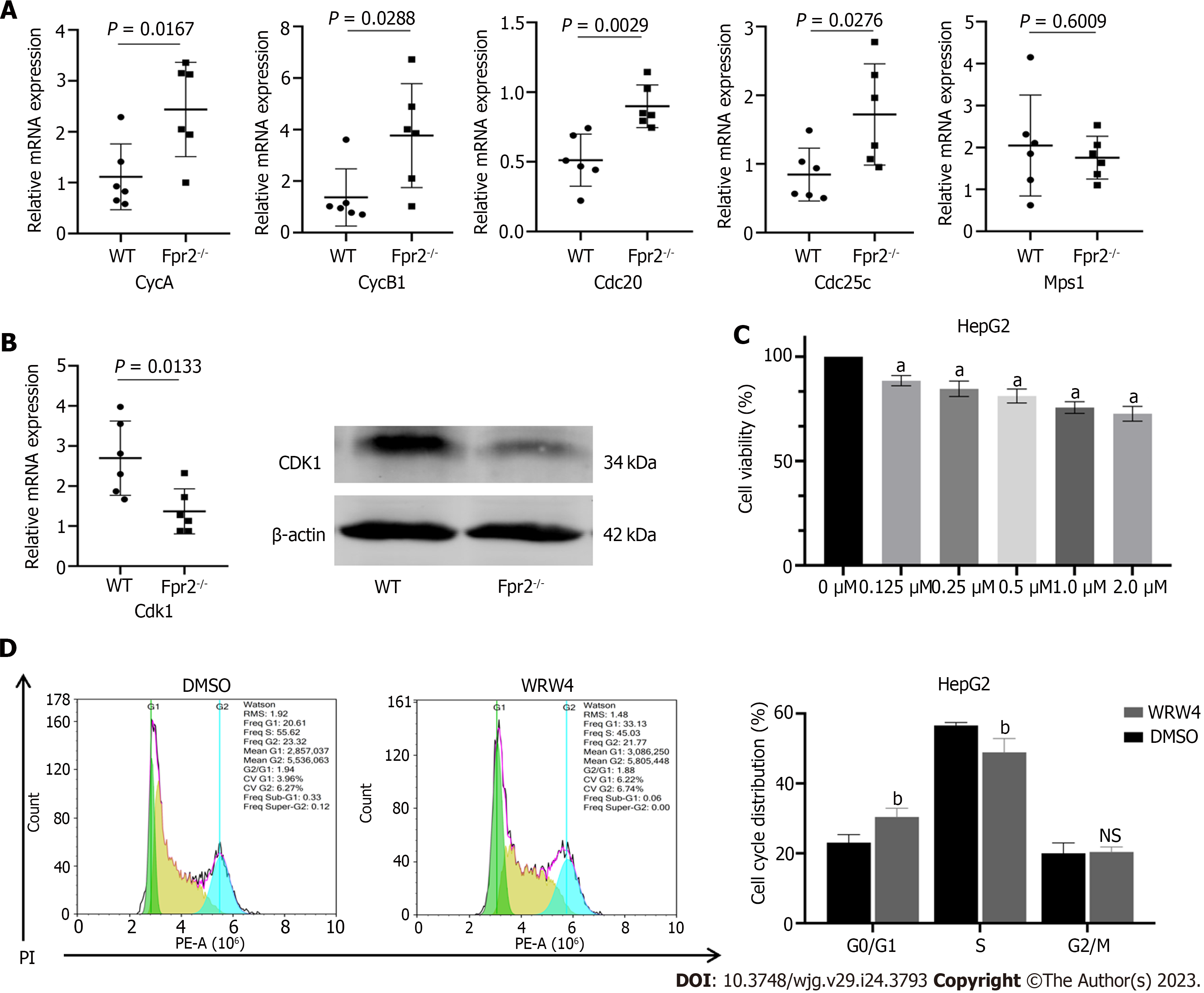
To further validate the results of RNA-sequencing, we selected several key genes CycA, CycB1, Cdc20, Cdc25c, Mps1, and Cdk1, which are closely related to the cell cycle, for qRT-PCR analysis. The results showed that the expression of CycA, CycB1, Cdc20, and Cdc25c was upregulated (Figure 4A), while the expression of Cdk1 was downregulated in the Fpr2-/- mice (Figure 4B). There was no significant difference in the expression of Mps1 between Fpr2-/- and WT mice (Figure 4A). The WB analysis further confirmed a decrease in CDK1 expression (Figure 4B). To clarify the impact of Fpr2 deficiency on liver cell proliferation, WRW4, an antagonist of Fpr2, was used to inhibit Fpr2 in the liver cells. CCK-8 further clarified the effect of different concentrations of WRW4 on the proliferation of HepG2 cells. Figure 4C indicates that WRW4 at all concentrations (0.125 μM, 0.25 μM, 0.5 μM, 1.0 μM, and 2.0 μM) inhibited the proliferation of HepG2 cells in a concentration dependent manner. WRW4 at 1 μM was used to analyze the distribution of cell cycles. Flow cytometry analysis showed that the number of cells in G0/G1 phase increased, and the number of cells in S phase decreased (Figure 4D). From the above analysis, it can be surmised that when Fpr2 is absent, some genes involved in cell cycle regulation are affected and these may inhibit cell proliferation by blocking the cell cycle.
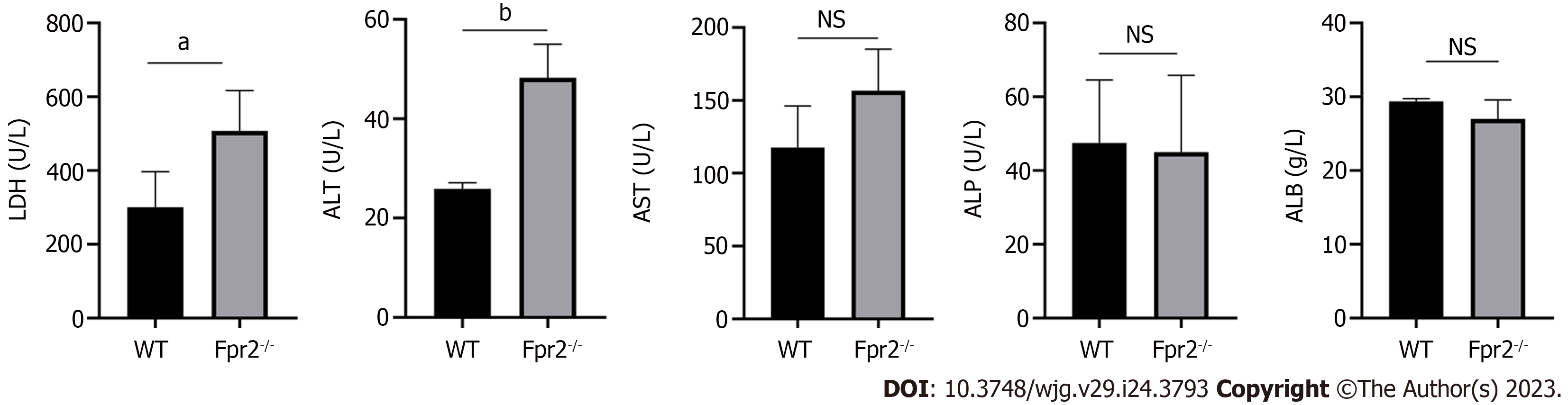
Considering that the lack of Fpr2 can lead to disruption of the liver cell cycle, we tested biochemical indicators that reflect liver function. Biochemical indicators related to liver function include determination of the levels of alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), lactate dehydrogenase (LDH) and albumin (ALB). In Fpr2-/- mice, we detected higher levels of serum ALT and LDH compared to those in the WT mice, with no significant changes in AST, ALP, and ALB in both mice (Figure 5). When the permeability of the liver cell membrane changes, ALT overflows from the cells into the bloodstream. Therefore, the increase of serum ALT in Fpr2-/- mice suggest that the stability of liver cell membrane is affected.
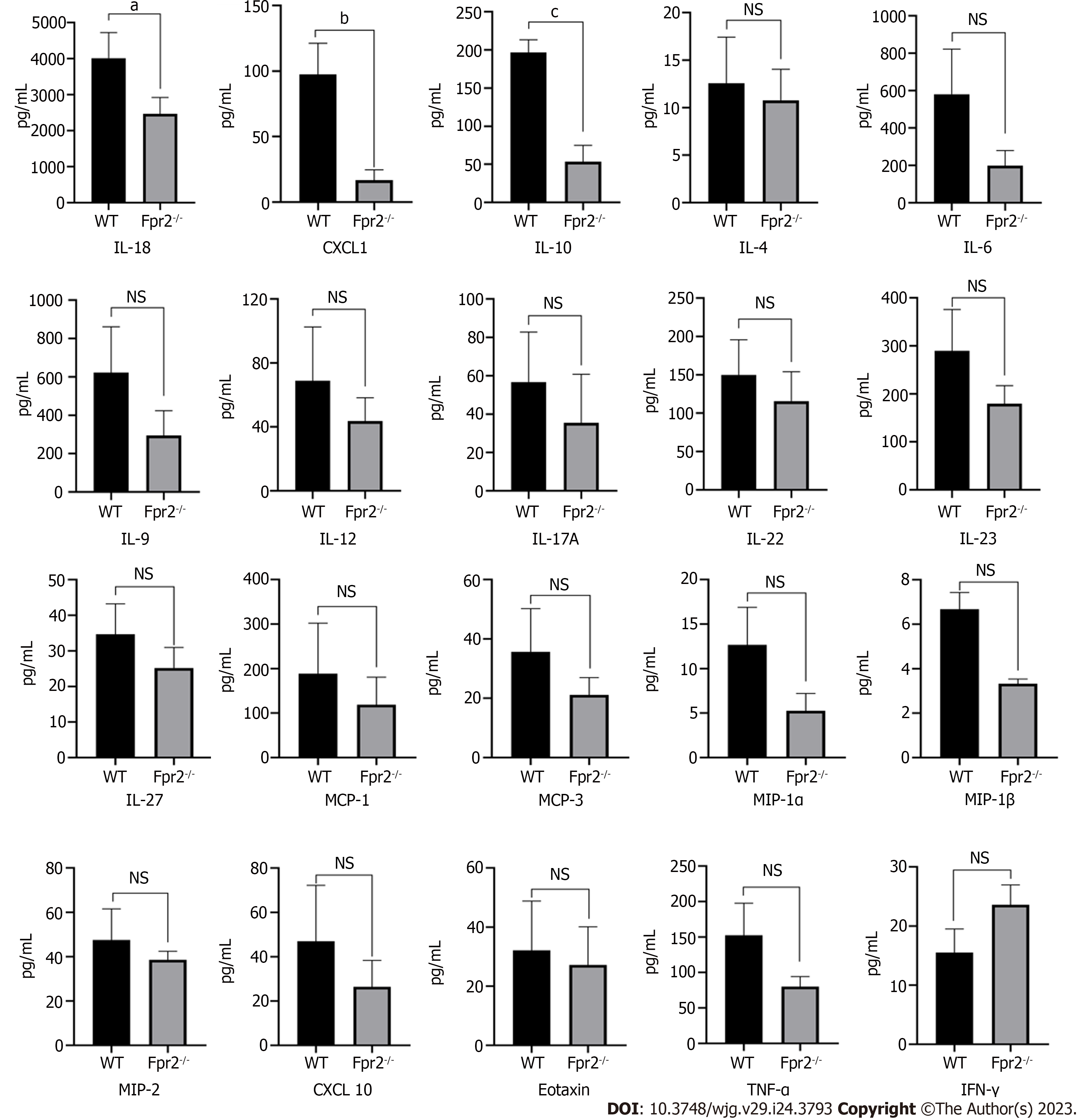
In the GO analysis, in addition to cell cycle, ERK1 and ERK2 cascade, myeloid leukocyte activation, and positive regulation of myeloid leukocyte differentiation were also significantly enriched in BP terms, and G-protein coupled receptor activity was significantly enriched in MF terms. These results prompted us to consider whether disruptions in the cell cycle and cell membrane stability may cause inflammation. The Luminex assay was performed to measure the cytokine levels in the liver. The results indicated that in addition to the increased levels of interferon gamma, the levels of interleukin (IL)-4, IL-6, IL-9, IL-12, IL-17A, IL-22, IL-23, IL-27, monocyte chemoattractant protein (MCP)-1, MCP-3, macrophage inflammatory protein (MIP)-1α, MIP-1β, MIP-2, CXCL10, Eotaxin, and tumor necrosis factor-α were decreased at different degrees in the Fpr2-/- mice (Figure 6). The decreases in IL-10, IL-18, and CXCL-1 levels were statistically significant.
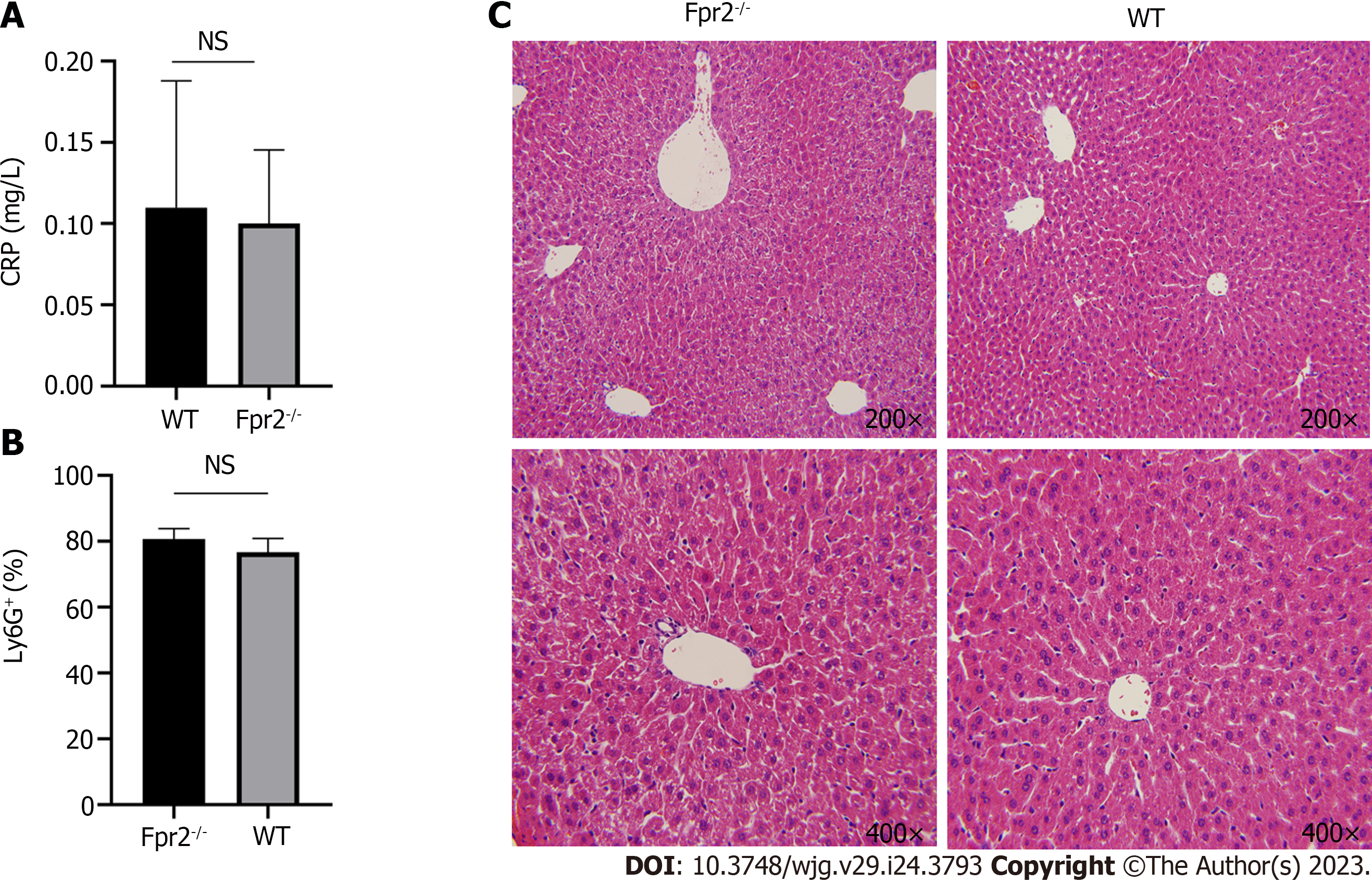
Based on the above results, the inflammatory status of the liver was further evaluated. We measured the C-reactive protein (CRP) levels in the peripheral blood and the number of neutrophils in the liver and performed histopathological analysis of the liver. The results showed that there was no significant difference in the CRP levels and the number of neutrophils in the liver between WT and Fpr2 -/- mice (Figures 7A and B), and there was no inflammatory cell infiltration in the histopathology of the liver (Figure 7C). The above results indicate that although the absence of Fpr2 can affect the cell cycle and membrane stability, it does not cause significant inflammation in the liver.
In this study, we focused on the effects of Fpr2 on liver homeostasis in mice. According to the results of RNA-sequencing, the deletion of Fpr2 mainly affects the cell cycle of the liver, which is clearly manifested by the significant changes in the expression of several key genes during cell division and enrichment of several related pathways of the cell cycle. The influence of Fpr2 deletion on the cell cycle may be multifaceted. From the perspective of function, DEGs contain many key genes in each stage of the cell cycle (CycA, CycB, Cdk1) and important cell cycle regulatory genes (Cdc25, Cdc20). In terms of distribution, DEGs are distributed in processes related to spindles, chromatin, and kinetochores. Cdk1 is essential for G1/S and G2/M phase transitions of eukaryotic cell cycle[13]. In Fpr2-/- mice, the expression level of Cdk1 is decreased. When FPR2 of HepG2 cells is blocked by WRW4, the cell proliferation activity decreased, the number of cells in G0/G1 phase increased, and the number of cells in S phase decreased, which indicates that FPR2 can affect cell proliferation by blocking cell cycle.
Most research on FPR2 has focused on its role in the regulation of immune cells in inflammation. Recently, it has also been reported that FPR2 is involved in regulating the migration and proliferation of many types of stem cells. Han et al[14] found that the proliferation of hair follicle stem cells and dermal papilla cells in Fpr2-/- mice was significantly reduced, which induced hair loss. Chen et al[15] found that Fpr2-/- mice displayed shortened colonic crypts and reduced acute inflammatory responses. This is because intestinal commensal bacteria mediate the proliferation and renewal of epithelial cells through Fpr2. In the present study, we found that the absence of this receptor in liver cells caused abnormal cell cycle. These findings provide evidence for the new function of Fpr2.
Although we found that Fpr2 deficiency can lead to abnormal liver cell cycle and membrane permeability, the infiltration of inflammatory cells in the liver was not observed when probed further. This is consistent with the fact that mice can still grow and reproduce normally with Fpr2 deficiency and do not die due to infection. Therefore, we speculate that these abnormal manifestations after Fpr2 deficiency may be offset by certain protective factors in the body. However, based on our previous research[12], the ability of Fpr2-/- mice to control infection was significantly lower than that of WT mice in the event of bacterial infections, with the liver becoming a severely damaged target organ. Therefore, our current findings may provide an explanation that the imbalance of cellular homeostasis caused by Fpr2 deficiency leads to insufficient defense function of the liver when encountering bacterial infections, making it a target organ for infection damage.
The levels of IL-10 and CXCL-1 were significantly reduced in the liver of Fpr2-/- mice. IL-10 is an important anti-inflammatory factor that can resist the pro-inflammatory effects of other cytokines, thus controlling inflammation. The anti-inflammatory reaction mediated by IL-10 is a fundamental homeostatic mechanism that controls the degree and duration of inflammation. IL-10 is an important factor in maintaining immunological tolerance in the liver[16]. IL-10 also plays an important role in liver regeneration and repair[17]. In addition, various cells, such as hepatocytes, endothelial cells, Kupffer cells, and hepatic stellate cells, are involved in the production of IL-10[18]. CXCL-1 is an important chemokine in neutrophils. Our previous research found that the loss of Fpr2 leads to a decrease in the recruitment of neutrophils and weakening of the host’s early immune response when resisting infection with Streptococcus lactis[12]. Therefore, the decline in repair ability and the decrease in chemotaxis to neutrophils may be another reason why Fpr2-/- mice are seriously injured when infected by bacteria. IL-18 was first discovered in Kupffer cells as a pro-inflammatory factor[19,20]. In Fpr2-/- mice, we observed a decrease in the levels of anti-inflammatory factor IL-10 and pro-inflammatory factor IL-18, and also observed varying degrees of decreases in the levels of other interleukin members, tumor necrosis factors, and chemokines. Considering the cell cycle disruption caused by Fpr2 deficiency, we postulate that the impact of Fpr2 deficiency on cell cycle may interfere with cytokine synthesis, although further validation is needed to test this hypothesis. In short, Fpr2 plays an important role in maintaining liver homeostasis, which may help the body resist bacterial infections.
Some limitations of this study should be noted. First, we used an immortal liver cell line for our study. Recent studies have reported that mature liver cells located in the middle region of liver lobules are the main contributors to the production of new cells in the entire liver[21,22]. Therefore, it is necessary to further evaluate the regulatory effect of Fpr2 on cell cycle using primary liver cells. Second, further research is required to establish whether Fpr2 regulates cell cycle directly or indirectly.
Fpr2 plays an important role in maintaining liver homeostasis by participating in cell cycle regulation. Fpr2 may play a positive role in liver tissue repair and neutrophil chemotaxis by regulating the expression of IL-10 and CXCL-1. Our results provide a basis and novel perspective for further understanding the functional diversity of Fpr2.
Formyl peptide receptor 2 (Fpr2) plays an important role in host’s defense and inflammatory response, as it can help the body control bacterial infections. Studies have found that in Fpr2-/- mice, the liver is the most severely damaged target organ in bloodstream infections. However, the reason for this is unclear.
To verify the role of Fpr2 in liver homeostasis, to provide new clues for the decreased ability of the liver to clear bacteria after Fpr2 deficiency, and to provide new ideas for future drug research and development.
To investigate the role of Fpr2 in liver homeostasis.
The differentially expressed genes (DEGs) in Fpr2-/- and wild-type (WT) mice were determined by transcriptome sequencing, and the biological functions of DEGs were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Quantitative real-time polymerase chain reaction (qRT PCR) and western blot (WB) were used to further validate the expression levels of differential genes. The Cell Counting Kit-8 assay was used to study cell proliferation. A cell cycle detection kit was used to measure the distribution of cell cycles. Luminex assay was used to analyze cytokine levels in the liver. A fully automated biochemical analyzer was used to detect the levels of serum biochemical indicators and C-reactive protein in WT and Fpr2-/- mice. The number of neutrophils in the liver was detected by flow cytometry. The histopathology of the liver was analyzed by hematoxylin-eosin staining.
Transcriptome sequencing showed that, compared with WT group, 445 genes in the liver of Fpr2-/- mice had significant changes in the expression level, including 325 upregulated genes and 120 downregulated genes. The functional enrichment analysis of GO and KEGG indicated that these DEGs were mainly related to cell cycle. qRT-PCR validated that after Fpr2 deletion, the expression levels of CycA, CycB1, Cdc20, and Cdc25c genes were upregulated. Both qRT-PCR and WB confirmed a decrease in the expression of CDK1. WRW4 (an antagonist of Fpr2) could inhibit the proliferation of HepG2 cells in a concentration dependent manner. The levels of serum alanine aminotransferase levels increased in Fpr2-/- mice with statistical significance. Luminex assay measurements showed that the expression levels of interleukin (IL)-10, IL-18, and CXCL-1 were significantly reduced in the liver of Fpr2-/- mice.
Fpr2 may exert a protective effect on the liver by participating in the regulation of cell cycle and proliferation, as well as affecting the expression of IL-10 and CXCL-1.
Fpr2 plays an important role in maintaining liver homeostasis, and its agonist may be a potential drug against bacterial infection.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Heij LR, Germany; Kuboki S, Japan S-Editor: Wang JJ L-Editor: A P-Editor: Cai YX
| 1. | Yan J, Li S. The role of the liver in sepsis. Int Rev Immunol. 2014;33:498-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 396] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 2. | Llorente C, Schnabl B. Fast-Track Clearance of Bacteria from the Liver. Cell Host Microbe. 2016;20:1-2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | Seki E, Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590:447-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 340] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 4. | Gonnert FA, Kunisch E, Gajda M, Lambeck S, Weber M, Claus RA, Bauer M, Kinne RW. Hepatic Fibrosis in a Long-term Murine Model of Sepsis. Shock. 2012;37:399-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Broadley SP, Plaumann A, Coletti R, Lehmann C, Wanisch A, Seidlmeier A, Esser K, Luo S, Rämer PC, Massberg S, Busch DH, van Lookeren Campagne M, Verschoor A. Dual-Track Clearance of Circulating Bacteria Balances Rapid Restoration of Blood Sterility with Induction of Adaptive Immunity. Cell Host Microbe. 2016;20:36-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 6. | Wood NJ. Liver: the liver as a firewall--clearance of commensal bacteria that have escaped from the gut. Nat Rev Gastroenterol Hepatol. 2014;11:391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Zhang M, Gao JL, Chen K, Yoshimura T, Liang W, Gong W, Li X, Huang J, McDermott DH, Murphy PM, Wang X, Wang JM. A Critical Role of Formyl Peptide Receptors in Host Defense against Escherichia coli. J Immunol. 2020;204:2464-2473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 8. | Liu M, Chen K, Yoshimura T, Liu Y, Gong W, Wang A, Gao JL, Murphy PM, Wang JM. Formylpeptide receptors are critical for rapid neutrophil mobilization in host defense against Listeria monocytogenes. Sci Rep. 2012;2:786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Chen K, Bao Z, Gong W, Tang P, Yoshimura T, Wang JM. Regulation of inflammation by members of the formyl-peptide receptor family. J Autoimmun. 2017;85:64-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 10. | Gobbetti T, Coldewey SM, Chen J, McArthur S, le Faouder P, Cenac N, Flower RJ, Thiemermann C, Perretti M. Nonredundant protective properties of FPR2/ALX in polymicrobial murine sepsis. Proc Natl Acad Sci U S A. 2014;111:18685-18690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 104] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 11. | Diao N, Zhang Y, Chen K, Yuan R, Lee C, Geng S, Kowalski E, Guo W, Xiong H, Li M, Li L. Deficiency in Toll-interacting protein (Tollip) skews inflamed yet incompetent innate leukocytes in vivo during DSS-induced septic colitis. Sci Rep. 2016;6:34672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Sun Z, Huang W, Zheng Y, Liu P, Yang W, Guo Z, Kong D, Lv Q, Zhou X, Du Z, Jiang H, Jiang Y. Fpr2/CXCL1/2 Controls Rapid Neutrophil Infiltration to Inhibit Streptococcus agalactiae Infection. Front Immunol. 2021;12:786602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Krek W, Nigg EA. Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: identification of major phosphorylation sites. EMBO J. 1991;10:305-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 243] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 14. | Han J, Lee C, Jung Y. Deficiency of Formyl Peptide Receptor 2 Retards Hair Regeneration by Modulating the Activation of Hair Follicle Stem Cells and Dermal Papilla Cells in Mice. Dev Reprod. 2021;25:279-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 15. | Chen K, Liu M, Liu Y, Yoshimura T, Shen W, Le Y, Durum S, Gong W, Wang C, Gao JL, Murphy PM, Wang JM. Formylpeptide receptor-2 contributes to colonic epithelial homeostasis, inflammation, and tumorigenesis. J Clin Invest. 2013;123:1694-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 16. | Nguyen NT, Umbaugh DS, Sanchez-Guerrero G, Ramachandran A, Jaeschke H. Kupffer cells regulate liver recovery through induction of chemokine receptor CXCR2 on hepatocytes after acetaminophen overdose in mice. Arch Toxicol. 2022;96:305-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 17. | Yin S, Wang H, Park O, Wei W, Shen J, Gao B. Enhanced liver regeneration in IL-10-deficient mice after partial hepatectomy via stimulating inflammatory response and activating hepatocyte STAT3. Am J Pathol. 2011;178:1614-1621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Wan S, LeClerc JL, Schmartz D, Barvais L, Huynh CH, Devière J, DeSmet JM, Vincent JL. Hepatic release of interleukin-10 during cardiopulmonary bypass in steroid-pretreated patients. Am Heart J. 1997;133:335-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 57] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Abbate A, Toldo S, Marchetti C, Kron J, Van Tassell BW, Dinarello CA. Interleukin-1 and the Inflammasome as Therapeutic Targets in Cardiovascular Disease. Circ Res. 2020;126:1260-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 503] [Article Influence: 100.6] [Reference Citation Analysis (0)] |
| 20. | Dinarello CA. The IL-1 family of cytokines and receptors in rheumatic diseases. Nat Rev Rheumatol. 2019;15:612-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 255] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 21. | He L, Pu W, Liu X, Zhang Z, Han M, Li Y, Huang X, Han X, Liu K, Shi M, Lai L, Sun R, Wang QD, Ji Y, Tchorz JS, Zhou B. Proliferation tracing reveals regional hepatocyte generation in liver homeostasis and repair. Science. 2021;371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 148] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 22. | Wei Y, Wang YG, Jia Y, Li L, Yoon J, Zhang S, Wang Z, Zhang Y, Zhu M, Sharma T, Lin YH, Hsieh MH, Albrecht JH, Le PT, Rosen CJ, Wang T, Zhu H. Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science. 2021;371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 160] [Article Influence: 40.0] [Reference Citation Analysis (0)] |