Published online Apr 7, 2023. doi: 10.3748/wjg.v29.i13.1994
Peer-review started: November 2, 2022
First decision: December 11, 2022
Revised: December 29, 2022
Accepted: March 20, 2023
Article in press: March 20, 2023
Published online: April 7, 2023
Processing time: 155 Days and 22.3 Hours
Celiac disease (CeD) is a multisystem immune-mediated multifactorial condition strongly associated with the intestinal microbiota.
To evaluate the predictive power of the gut microbiota in the diagnosis of CeD and to search for important taxa that may help to distinguish CeD patients from controls.
Microbial DNA from bacteria, viruses, and fungi, was isolated from mucosal and fecal samples of 40 children with CeD and 39 controls. All samples were sequenced using the HiSeq platform, the data were analyzed, and abundance and diversities were assessed. For this analysis, the predictive power of the microbiota was evaluated by calculating the area under the curve (AUC) using data for the entire microbiome. The Kruskal-Wallis test was used to evaluate the significance of the difference between AUCs. The Boruta logarithm, a wrapper built around the random forest classification algorithm, was used to identify important bacterial biomarkers for CeD.
In fecal samples, AUCs for bacterial, viral, and fungal microbiota were 52%, 58%, and 67.7% respectively, suggesting weak performance in predicting CeD. However, the combination of fecal bacteria and viruses showed a higher AUC of 81.8 %, indicating stronger predictive power in the diagnosis of CeD. In mucosal samples, AUCs for bacterial, viral, and fungal microbiota were 81.2%, 58.6%, and 35%, respectively, indicating that mucosal bacteria alone had the highest predictive power. Two bacteria, Bacteroides intestinalis and Burkholderiales bacterium 1-1-47, in fecal samples and one virus, Human_endogenous _retrovirus_K, in mucosal samples are predicted to be “important” biomarkers, differentiating celiac from nonceliac disease groups. Bacteroides intestinalis is known to degrade complex arabinoxylans and xylan which have a protective role in the intestinal mucosa. Similarly, several Burkholderiales species have been reported to produce peptidases that hydrolyze gluten peptides, with the potential to reduce the gluten content of food. Finally, a role for Human_endogenous _retrovirus_K in immune-mediated disease such as CeD has been reported.
The excellent predictive power of the combination of the fecal bacterial and viral microbiota with mucosal bacteria alone indicates a potential role in the diagnosis of difficult cases of CeD. Bacteroides intestinalis and Burkholderiales bacterium 1-1-47, which were found to be deficient in CeD, have a potential protective role in the development of prophylactic modalities. Further studies on the role of the microbiota in general and Human_endogenous _retrovirus_K in particular are needed.
Core Tip: Celiac disease (CeD) is known to be associated with the microbiota. In this study, the combination of bacterial and viral taxa in stools and mucosal bacterial taxa were the strongest predictors of celiac disease. In addition, we report important bacterial markers, namely, Bacteroides intestinalis and Burkholderiales bacterium 1-1-47, which were reduced in children with CeD, suggesting a protective role in children with CeD.
- Citation: El Mouzan M, Assiri A, Al Sarkhy A. Gut microbiota predicts the diagnosis of celiac disease in Saudi children. World J Gastroenterol 2023; 29(13): 1994-2000
- URL: https://www.wjgnet.com/1007-9327/full/v29/i13/1994.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i13.1994
Celiac disease (CeD) is an immune-mediated condition with multisystem clinical expression[1,2]. The disease is distributed worldwide, and the incidence is increasing. The global seroprevalence and biopsy-proven prevalence are estimated to be 1.4% and 0.7%, respectively[3]. In the Kingdom of Saudi Arabia (KSA), a seroprevalence between 1.5% and 3% and a biopsy-proven prevalence of 1% are some of the highest observed rates in the world[4,5]. The pathogenesis of CeD is multifactorial, requiring genetic susceptibility in the form of human leukocyte antigen DQ2 and DQ8 genotypes and exposure to gluten-containing food[6]. In the KSA, high prevalence of genetic susceptibility of 47% has been reported[7]. Although genetic susceptibility and exposure to gluten-containing food are necessary, not all genetically susceptible individuals develop CeD. Moreover, in some cases, CeD develops later in life after many years of gluten ingestion, indicating that other factors may be important in loss of tolerance to gluten and development of clinical disease[8,9]. Microbial dysbiosis associated with CeD is thought to be one of the important environmental factors contributing to loss of tolerance to gluten and thereby playing a role in pathogenesis of CeD[10,11]. However, identification of microbial markers and the predictive power of microbial dysbiosis in CeD have rarely been reported[12]. Therefore, the objectives of this study were to determine the predictive power of the gut microbial community in the diagnosis of CeD and to search for taxa that may be important in differentiating children with CeD from those without CeD.
Shotgun metagenomic analysis of fecal and duodenal mucosal samples from children with new onset CeD was performed for bacteria, viruses, and fungi. Briefly, there were 40 children with CeD. Patients were eligible if they had confirmed CeD by standard criteria and not received antibiotics for at least 6 mo. They were enrolled as they presented to clinics. There were 39 non-CeD controls, including 20 school children who were clinically healthy who provided stool samples and 19 from whom tissue samples from the second part of the duodenum were collected during diagnostic endoscopy performed for clinical indications. The children with CeD and controls were enrolled in the study after consent/assent. All children were recruited from King Khaled University Hospital and King Fahad Medical City, both institutions are in Riyadh (KSA). Microbial DNA was isolated and sequenced using the HiSeq platform. The results included abundance and diversity analyses for bacteria, viruses, and fungi that were recently reported[13-15].
For the purpose of this report, receiver operating characteristic (ROC) analysis with calculation of the area under the curve (AUC) was used to assess the predictive power of the gut microbiota in the diagnosis of CeD. ROC analysis and calculation of the AUC for discrimination using data regarding microbial communities, including bacteria, viruses, and fungi, in stool and mucosa were performed. Boruta analysis was used to identify important taxa that may differentiate children with CeD from non-CeD controls. In brief, the Boruta logarithm is a wrapper built around the random forest classification algorithm implemented in the R package random forest[16]. The Boruta process consists of assigning an ‘importance’ score to each variable and identifying a threshold above which the variables are deemed important and below which they are not. This process is repeated to establish reproducibility and robustness and therefore generates many ‘importance’ scores for each taxon. Species-level relative abundance data were used to generate shadow variables to predict taxa that may be important in distinguishing celiac from nonceliac groups[17]. Sensitivity and specificity were calculated based on the output from a random forest classifier[18]. Bioinformatics and statistical analyses were performed by specialists at Cosmos ID, United States (https://www.cosmosid.com/).
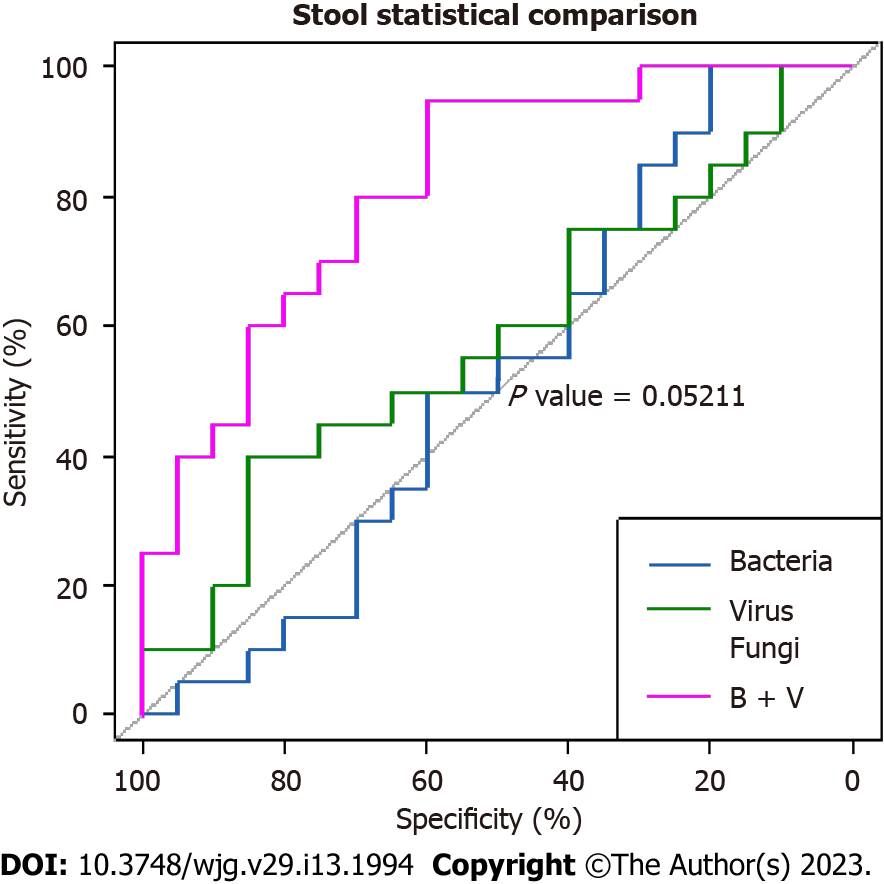
The areas under the curve for the bacterial, viral, and fungal microbiota in fecal samples were 52%, 58%, and 67.7%, respectively, suggesting poor performance for each, in predicting CeD. However, the combination of fecal bacteria and viruses revealed a higher AUC of 81.8 %, indicating a stronger predictive power for the diagnosis of CeD (Figure 1). Nevertheless, the difference between the AUC of the bacterial microbiome alone and the combined bacterial and viral microbiomes showed borderline significance (P = 0.05211).
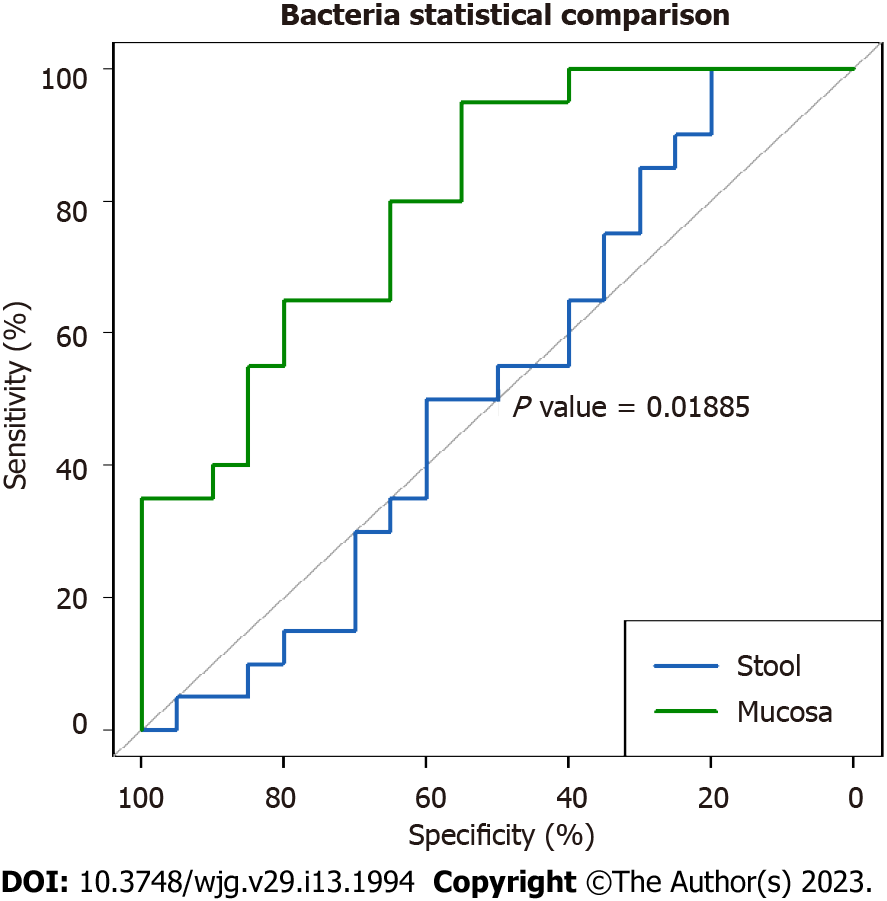
The areas under the curve for the bacterial, viral, and fungal microbiota in mucosal samples were 81.2%, 58.6%, and 35%, respectively, indicating the the highest predictive power for mucosal bacteria alone (Figure 2). The difference in AUC between mucosal and fecal bacteria was significant (P = 0.01885).
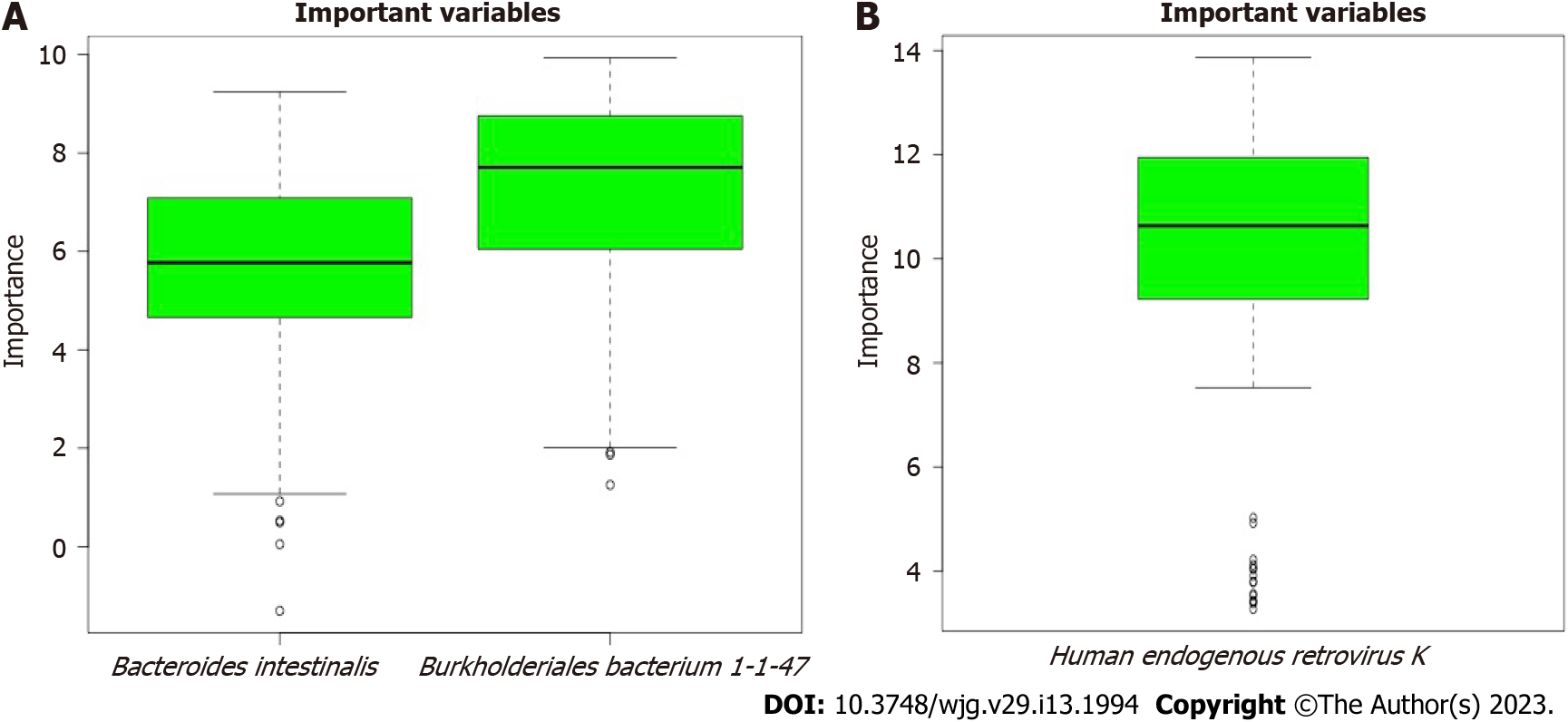
The scores for the confirmed important variables are summarized in Table 1, including the mean, median, minimum, and maximum importance values. The ‘Decision’ column indicates ‘Confirmed’ for the microbiota above the threshold set by Boruta analysis. The microbiota including two bacteria (Bacteroides intestinalis and Burkholderiales bacterium 1-1-47) and one virus (Human_endogenous _retrovirus_K), was confirmed to be important for distinguishing between CeD and non-CeD groups.
| Microbial species | Mean importance | Median importance | Minimum importance | Maximum importance | Decision |
| Bacteroides_intestinalis | 6.92517709 | 7.70811062 | 1.25328634 | 9.93239532 | Confirmed |
| Burkholderiales_bacterium_ 1_1_47 | 5.39952346 | 5.77233858 | -1.307345 | 9.24744767 | Confirmed |
| Human_endogenous _retrovirus_K | 9.95761324 | 0.6340023 | 3.26946721 | 3.8621105 | Confirmed |
The results of the Boruta random forest algorithm analysis are illustrated in Figure 3. Two bacteria (Bacteroides intestinalis and Burkholderiales bacterium 1-1-47) in fecal samples and one virus in mucosal samples (Human_endogenous _retrovirus_K) were predicted to be “important” for differentiating celiac from nonceliac disease groups. No fungal species were found to be important.
The role of the microbiota in predicting diseases in general has been largely reported. However, to our knowledge, the sensitivity and specificity of the gut microbiota in distinguishing CeD from non-CeD has not been reported thus far. In this study, the finding of a high AUC of the combination of bacteria and viruses in fecal samples (81.8%) indicates excellent predictive power with potential use in the diagnoses of difficult cases of CeD. Similarly, the significantly higher AUC for bacteria (81.2%) in the mucosal than in the fecal samples indicates stronger predictive power for mucosal bacteria (P = 0.01885). However, this was not surprising, as CeD is mainly a mucosal small bowel disease. These findings may have potential applications in the diagnosis of difficult cases of CeD.
Identification of the “important” specific microbiota in CeD in the form of Bacteroides intestinalis and Burkholderiales bacterium 1-1-47 has not been previously reported. Bacteroides intestinalis belongs to the Bacteroides genus, members of which are known to degrade complex arabinoxylans and xylan from dietary fibers, including wheat, rye, oat, and barley[19]. These degradation products, including butyrate and ferulic acid, have been shown to have a protective role in the intestinal mucosa[20-22]. Burkholderiales bacterium 1-1-47 is an unclassified bacterium belonging to the order Burkholderiales, class Betaproteobacteria and phylum Proteobacteria[23]. Several Burkholderiales species and Burkholderia gladioli in particular have been reported to produce peptidases that hydrolyze gluten peptides, with the potential to reduce the gluten content of food[24]. Accordingly, reports of decreased abundance of both Bacteroides intestinalis and Burkholderiales bacterium 1-1-47 in samples from children with CeD[13], indicate a potential protective role against the effects of gluten-containing grains.
Confirmation of Human_endogenous _retrovirus_K virus as important in differentiating CeD from non-CeD groups is interesting. This group of viruses has been suggested to have a role in immunity and autoimmune disorders. They can contribute to host protection or to damage, suggesting a subtle balance between the persistence of human endogenous retroviruses expression and maintenance of a basal immune alert[25]. Although a recent study found increased expression in children with CeD[26], identification of Human_endogenous _retrovirus_K as important and the significantly reduced abundance in children with CeD[13], should prompt further investigation of the role of these viruses in children with CeD.
This study had a relatively small sample size. In addition, the non-CeD controls were not completely healthy although they do not have CeD as TTG-A, endoscopy, and duodenal tissue histopathology were normal. However, the relatively small size might be partially compensated for by the use of the shotgun metagenomic analysis. Since this is the first report on microbiota accuracy and identification of important bacteria and viruses, but not fungi, further studies with larger sample sizes are needed.
The high AUCs of mucosal bacteria and the combination of fecal bacteria and viruses indicate a potential role in the diagnosis of difficult cases of CeD. In addition, identification of important bacteria as decreased abundances of Bacteroides intestinalis and Burkholderiales bacterium 1-1-47 in children with CeD, suggests a protective role with the potential for the development of preventive and adjuvant microbial therapy for CeD. The importance of Human_endogenous _retrovirus_K is interesting. However, further studies with larger sample sizes, are needed to improve our understanding of the role of the microbiota in CeD.
Dysbiosis associated with celiac disease (CeD) is well known and beneficial and harmful associations have been reported.
The role of the microbiota in predicting CeD has rarely been described.
To search for a microbial signature that may help in the diagnosis and prevention of CeD.
Metagenomic analysis of microbial DNA in mucosa and stool of children with newly diagnosed CeD calculation of the area under the curve to evaluate the predictive power of the whole microbiota and use of rendom forest analysis to identify important microbes in distinguishing CeD groups from controls.
Very high discriminatory power of combined bacteria and viruses (81.8%) in fecal samples and bacteria only in mucosal samples (81.2%). Bacteroides intestinalis and Burkholderiales bacterium 1-1-47 in fecal samples were demmed important.
The excellent predictive power of microbiota may help in the diagnosis of difficult cases of CeD. The identification of important specific bacterial species that are reduced in CeD may have a potential protective role.
Future research in this area with larger sample sizes is needed to clarify the role of microbiota in the diagnosis and prevention of CeD.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: Saudi Gastroenterology Association; Saudi Society for Pediatric Gastroenterology Hepatology and Nutrition.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Saudi Arabia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rashid R, Bangladesh; Sabelnikova EA, Russia S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Husby S, Koletzko S, Korponay-Szabó I, Kurppa K, Mearin ML, Ribes-Koninckx C, Shamir R, Troncone R, Auricchio R, Castillejo G, Christensen R, Dolinsek J, Gillett P, Hróbjartsson A, Koltai T, Maki M, Nielsen SM, Popp A, Størdal K, Werkstetter K, Wessels M. European Society Paediatric Gastroenterology, Hepatology and Nutrition Guidelines for Diagnosing Coeliac Disease 2020. J Pediatr Gastroenterol Nutr. 2020;70:141-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 677] [Article Influence: 135.4] [Reference Citation Analysis (0)] |
| 2. | Leffler DA, Green PH, Fasano A. Extraintestinal manifestations of coeliac disease. Nat Rev Gastroenterol Hepatol. 2015;12:561-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 186] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 3. | Singh P, Arora A, Strand TA, Leffler DA, Catassi C, Green PH, Kelly CP, Ahuja V, Makharia GK. Global Prevalence of Celiac Disease: Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol. 2018;16:823-836.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1076] [Cited by in RCA: 935] [Article Influence: 133.6] [Reference Citation Analysis (1)] |
| 4. | Al-Hussaini A, Troncone R, Khormi M, AlTuraiki M, Alkhamis W, Alrajhi M, Halal T, Fagih M, Alharbi S, Bashir MS, Chentoufi AA. Mass Screening for Celiac Disease Among School-aged Children: Toward Exploring Celiac Iceberg in Saudi Arabia. J Pediatr Gastroenterol Nutr. 2017;65:646-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 5. | Al Hatlani MM. Prevalence of celiac disease among symptom-free children from the Eastern Province of Saudi Arabia. Saudi J Gastroenterol. 2015;21:367-371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Tye-Din JA, Galipeau HJ, Agardh D. Celiac Disease: A Review of Current Concepts in Pathogenesis, Prevention, and Novel Therapies. Front Pediatr. 2018;6:350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 94] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 7. | Al-Hussaini A, Alharthi H, Osman A, Eltayeb-Elsheikh N, Chentoufi A. Genetic susceptibility for celiac disease is highly prevalent in the Saudi population. Saudi J Gastroenterol. 2018;24:268-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Saeed A, Assiri A, Assiri H, Ullah A, Rashid M. Celiac disease in Saudi children. Evaluation of clinical features and diagnosis. Saudi Med J. 2017;38:895-899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 9. | Alkhiari R, Aljameli SM, Almotairi DB, AlHarbi GA, ALmufadhi L, Almeathem FK, Alharbi AA, AlObailan Y. Clinical Presentation of Pediatric Celiac Disease Patients in the Qassim Region Over Recent Years. Cureus. 2022;14:e21001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 10. | Panelli S, Capelli E, Lupo GFD, Schiepatti A, Betti E, Sauta E, Marini S, Bellazzi R, Vanoli A, Pasi A, Cacciatore R, Bacchi S, Balestra B, Pastoris O, Frulloni L, Corazza GR, Biagi F, Ciccocioppo R. Comparative Study of Salivary, Duodenal, and Fecal Microbiota Composition Across Adult Celiac Disease. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 11. | Zafeiropoulou K, Nichols B, Mackinder M, Biskou O, Rizou E, Karanikolou A, Clark C, Buchanan E, Cardigan T, Duncan H, Wands D, Russell J, Hansen R, Russell RK, McGrogan P, Edwards CA, Ijaz UZ, Gerasimidis K. Alterations in Intestinal Microbiota of Children With Celiac Disease at the Time of Diagnosis and on a Gluten-free Diet. Gastroenterology. 2020;159:2039-2051.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 12. | Leonard MM, Valitutti F, Karathia H, Pujolassos M, Kenyon V, Fanelli B, Troisi J, Subramanian P, Camhi S, Colucci A, Serena G, Cucchiara S, Trovato CM, Malamisura B, Francavilla R, Elli L, Hasan NA, Zomorrodi AR, Colwell R, Fasano A; CD-GEMM Team. Microbiome signatures of progression toward celiac disease onset in at-risk children in a longitudinal prospective cohort study. Proc Natl Acad Sci U S A. 2021;118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 13. | El Mouzan M, Al-Hussaini A, Serena G, Assiri A, Al Sarkhy A, Al Mofarreh M, Alasmi M, Fasano A. Microbiota profile of new-onset celiac disease in children in Saudi Arabia. Gut Pathog. 2022;14:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 14. | El Mouzan M, Assiri A, Al Sarkhy A, Alasmi M, Saeed A, Al-Hussaini A, AlSaleem B, Al Mofarreh M. Viral dysbiosis in children with new-onset celiac disease. PLoS One. 2022;17:e0262108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (1)] |
| 15. | El Mouzan M, Al-Hussaini A, Fanelli B, Assiri A, AlSaleem B, Al Mofarreh M, Al Sarkhy A, Alasmi M. Fungal Dysbiosis in Children with Celiac Disease. Dig Dis Sci. 2022;67:216-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 16. | Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947-1958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1852] [Cited by in RCA: 1490] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 17. | Kursa MB, Rudnicki WR. Feature Selection with the Boruta Package. J Stat Soft. 2010;36:1-13. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1713] [Cited by in RCA: 1731] [Article Influence: 115.4] [Reference Citation Analysis (0)] |
| 18. | Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6456] [Cited by in RCA: 8291] [Article Influence: 592.2] [Reference Citation Analysis (0)] |
| 19. | Dhingra D, Michael M, Rajput H, Patil RT. Dietary fibre in foods: a review. J Food Sci Technol. 2012;49:255-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 660] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 20. | Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 515] [Cited by in RCA: 617] [Article Influence: 44.1] [Reference Citation Analysis (1)] |
| 21. | Hong PY, Iakiviak M, Dodd D, Zhang M, Mackie RI, Cann I. Two new xylanases with different substrate specificities from the human gut bacterium Bacteroides intestinalis DSM 17393. Appl Environ Microbiol. 2014;80:2084-2093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 22. | Yasuma T, Toda M, Abdel-Hamid AM, D'Alessandro-Gabazza C, Kobayashi T, Nishihama K, D'Alessandro VF, Pereira GV, Mackie RI, Gabazza EC, Cann I. Degradation Products of Complex Arabinoxylans by Bacteroides intestinalis Enhance the Host Immune Response. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 23. | Sánchez E, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal Bacteroides species associated with coeliac disease. J Clin Pathol. 2010;63:1105-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Liu YY, Lee CC, Hsu JH, Leu WM, Meng M. Efficient Hydrolysis of Gluten-Derived Celiac Disease-Triggering Immunogenic Peptides by a Bacterial Serine Protease from Burkholderia gladioli. Biomolecules. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Grandi N, Tramontano E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front Immunol. 2018;9:2039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 204] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 26. | Tovo PA, Opramolla A, Pizzol A, Calosso G, Daprà V, Galliano I, Calvi C, Pinon M, Cisarò F, Rigazio C, Calvo PL, Bergallo M. Overexpression of endogenous retroviruses in children with celiac disease. Eur J Pediatr. 2021;180:2429-2434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |